Abstract.
Vietnam has a high burden of tuberculosis (TB) and multidrug-resistant (MDR) TB, but drug resistance patterns and TB transmission dynamics among TB/human immunodeficiency virus (HIV) coinfected patients are not well described. We characterized 200 Mycobacterium tuberculosis isolates from TB/HIV coinfected patients diagnosed at the main TB referral hospital in Ho Chi Minh City, Vietnam. Phenotypic drug susceptibility testing (DST) for first-line drugs, spoligotyping, and 24-locus mycobacterial interspersed repetitive unit (MIRU-24) analysis was performed on all isolates. The 24-locus mycobacterial interspersed repetitive unit clusters and MDR isolates were subjected to whole genome sequencing (WGS). Most of the TB/HIV coinfected patients were young (162/174; 93.1% aged < 45 years) males (173; 86.5% male). Beijing (98; 49.0%) and Indo-Oceanic (70; 35.0%) lineage strains were most common. Phenotypic drug resistance was detected in 84 (42.0%) isolates, of which 17 (8.5%) were MDR; three additional MDR strains were identified on WGS. Strain clustering was reduced from 84.0% with spoligotyping to 20.0% with MIRU-24 typing and to 13.5% with WGS. Whole genome sequencing identified five additional clusters, or members of clusters, not recognized by MIRU-24. In total, 13 small (two to three member) WGS clusters were identified, with less clustering among drug susceptible (2/27; 7.4%) than among drug-resistant strains (25/27; 92.6%). On phylogenetic analysis, strains from TB/HIV coinfected patients were interspersed among strains from the general community; no major clusters indicating transmission among people living with HIV were detected. Tuberculosis/HIV coinfection in Vietnam was associated with high rates of drug resistance and limited genomic evidence of ongoing M. tuberculosis transmission among HIV-infected patients.
INTRODUCTION
Globally, Mycobacterium tuberculosis was the number one infectious disease killer in 2016, causing an estimated 1.3 million deaths.1 Tuberculosis (TB) is also the leading cause of death among people living with human immunodeficiency virus (HIV). Of the estimated 10.4 million new TB cases that occurred worldwide in 2016, more than one million (11%) were coinfected with HIV, with 370,000 TB-related deaths in this group.1 Large numbers of patients with multidrug-resistant (MDR; resistance to at least both isoniazid and rifampicin) TB, and potential epidemic spread of these drug-resistant strains, pose a particular challenge.2,3
In 2016, Vietnam was ranked 16th among high-burden TB countries and 13th among high-burden MDR-TB countries in the world.1 Multidrug-resistant TB was reported in 4.1% of new and 26% of retreatment TB cases.1 Multidrug-resistant TB can be acquired by patients initially infected with a drug-susceptible strain, usually as a result of inadequate treatment (defined as secondary resistance), or occur following infection with an MDR strain (primary or transmitted drug resistance).4 A common, but misplaced, assumption has been that MDR-TB among new cases provides an accurate reflection of transmitted drug resistance.5 However, it has been demonstrated that many retreatment cases with MDR-TB also represent transmitted drug resistance.6
The epidemic spread of MDR-TB is a particular concern in areas with poor control of ongoing TB transmission, especially among HIV-infected patients with severe immunodeficiency where health-care facilities can serve as epidemic amplifiers.7,8 Global TB control efforts have been complicated by increased drug resistance and the negative synergism of TB and HIV coinfection.8 Despite the rapid expansion MDR-TB services and national efforts to identify and treat MDR-TB cases in Vietnam, case numbers continue to rise.10 Furthermore, MDR-TB/HIV coinfection often leads to poor treatment outcomes,11 and the risk of MDR-TB transmission within settings that provide HIV care has been well documented.12
Of concern, TB/HIV coinfection in Vietnam has been associated with high rates of drug resistance,13 but the genetic determinants of drug resistance remain poorly described and a better understanding of TB transmission dynamics is required to guide prevention strategies. Traditional methods of M. tuberculosis genotyping, such as spoligotyping or 24-locus mycobacterial interspersed repetitive unit (MIRU-24) analysis, have been hampered by poor discriminatory power.14 Whole genome sequencing (WGS) offers unprecedented strain resolution, especially of lineage 2 (Beijing) strains which is the dominant lineage circulating in Vietnam.15 It also provides genome-wide view for assessment of drug resistance mutations. This study aimed to examine the transmission dynamics and drug resistance–conferring mutations in M. tuberculosis strains isolated from TB/HIV coinfected patients in Vietnam by using a variety of contemporary molecular methods, including WGS.
METHODS
We conducted a retrospective laboratory-based analysis of 200 M. tuberculosis isolates from TB/HIV coinfected patients in Ho Chi Minh City, Vietnam. Consecutive culture-positive isolates identified at the Pham Ngoc Thach Hospital Mycobacterial Reference Laboratory, during periods that no other studies were performed (2009–2010 and then again in 2014), were subjected to phenotypic drug susceptibility testing (DST), spoligotyping, MIRU-24, and WGS. Basic demographic data were acquired from the laboratory database. Whole genome sequencing data were compared with publicly accessible sequences from a recent study by Holt et al.17 that documented strain diversity in HIV-uninfected TB patients from Ho Chi Minh City in a similar time frame.
Study setting.
Ho Chi Minh City is the most populous metropolis in Vietnam, with an estimated population of 8.2 million.18 Pham Ngoc Thach Hospital is the largest (750-bed) lung disease treatment facility in Ho Chi Minh City and treated more than 7,600 TB patients in 2016. The hospital serves as a tertiary referral center for TB patients and people living with HIV throughout southern Vietnam, treating around 350 HIV-infected (including TB/HIV coinfected) patients per year.18 The hospital also hosts a World Health Organization–accredited regional mycobacterial reference laboratory.19
Drug susceptibility testing.
Primary mycobacterial cultures were grown on solid Lowenstein–Jensen (LJ) media (bioMérieux SA, Marcy-l’Étoile, France) or liquid culture (BACTEC™ or MGIT™ 960; Becton Dickinson & Co., Franklin Lakes, NJ). All isolates identified as M. tuberculosis underwent phenotypic DST against isoniazid, rifampicin, streptomycin, and ethambutol by the standard 1% proportion method.20 Cultures were grown on solid LJ media with the following critical drug concentrations: isoniazid (0.2 μg/mL), rifampin (40 μg/mL), streptomycin (4 μg/mL), and ethambutol (2 μg/mL).
Twenty four-locus mycobacterial interspersed repetitive unit and spoligotyping.
Spoligotyping was performed according to the standard protocol21 and phylogenetic lineages were assigned using the SIVITWEB (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/) and SPOTCLUST (http://cgi2.cs.rpi.edu/∼bennek/SPOTCLUST.html) databases. The 24-locus mycobacterial interspersed repetitive unit typing was also performed as described before22 and strain lineage assigned using the http://www.miru-vntrplus.org online database, which was also used to construct the minimum spanning tree. Two or more isolates sharing an identical MIRU-24 profile were considered a cluster.
Whole genome sequencing and bioinformatics analysis.
Genomic DNA from M. tuberculosis isolates were extracted as described before23 from all drug-resistant and MIRU-24–clustered isolates. Libraries were constructed using Nextera XT DNA preparation kit (Illumina, San Diego, CA) and genome sequencing was performed on the NextSeq500 (Illumina) at the Center for Infectious Diseases and Microbiology, New South Wales Health Pathology in Sydney, Australia. Sequencing reads were mapped to the reference genome H37Rv (Genbank NC_000962) using the RedDog pipeline (https://github.com/katholt/RedDog). An initial phylogenetic tree was inferred using FastTree v2.1.8.24 Variant calling was performed using Snippy version 3.1 (https://github.com/tseemann/snippy) and careful checks performed for low-frequency variants in the case of discrepant phenotypic and genotypic DST results. Insertions and deletions (indels) were assessed as possible markers of drug resistance, but were not included in the phylogenetic tree reconstruction. Genomes with ≤ 10 single nucleotide polymorphisms (SNP) differences (calculated as the number of SNP difference between individual members of the cluster compared with the ancestral node) were classified as a WGS cluster, and those with 11–20 SNPs as a “possible cluster,” indicative of recent transmission.15
Maximum likelihood phylogenetic trees were constructed using RAxML (version 8.2; Heidelberg, Germany) and FastTree (version 2.1.8) using the GTR model of nucleotide substitution and a Gamma model of rate heterogeneity to analyze a concatenated alignment of SNP alleles. Trees were visualized using Microreact (www.microreact.org) and Figtree v1.4.3. Ancestral sequence reconstruction was performed using FastML (version 3.1).
Statistical analysis and human research ethics approval.
Demographic and clinical characteristics of different M. tuberculosis lineages, as well as the odds ratio (OR) for clustering of drug susceptible and drug-resistant isolates, were examined using STATA version 13 (StataCorp, College Station, TX). Branch length comparisons were generated using R package phytools v0.5 (The Free Software Foundation, Boston, MA). The product of branch length and the number of substitutions (i.e., SNPs) per genome, mapped to each terminal branch, provided the branch length for each isolate. The boxplots comparing branch lengths of lineage-specific phylogenetic trees for M. tuberculosis strains isolated from HIV-infected and HIV-uninfected patients were plotted using the ggplot2 and metrics calculated using stats (R version 3.2.5) (The Free Software Foundation).25 The P value for branch length comparison between M. tuberculosis strains isolated from HIV-infected and HIV-uninfected patients was calculated with GraphPad prism version 7 (San Diego, CA) (using a hypothetical mean of 100).
The study was approved by the Pham Ngoc Thach Hospital Ethics Review Committee (approved on October 9, 2014) and the Human Research Ethics Committee of the University of Sydney (Project No 2015/522).
RESULTS
Table 1 provides an overview of the demographic characteristics and drug resistance profiles of TB/HIV coinfected patients included in the study. The majority (63.8%) were young adults (25–34 years of age), predominantly (86.5%) male. Beijing (98; 49.0%) and Indo-Oceanic (70; 35.0%) lineage strains were most common. Phenotypic resistance against any of the first-line drugs was identified in 84 (42.0%) isolates; 17 (8.5%) were MDR by phenotypic and 20 (10.0%) by genotypic assessment. Patients with MDR-TB were also predominantly young (17/20; 85.0% < 45 years of age) males (14/20; 70.0%), infected with Beijing (14/20; 70.0%) or Indo-Oceanic (3/20; 15.0%) lineage strains. Among phenotypically drug-resistant strains, resistance to streptomycin was near universal (80/84; 95.2%), with the only exception being two isolates with isoniazid mono-resistance and two with isoniazid as well as pyrazinamide resistance. Phenotypically, 10/84 (11.2%) drug-resistant isolates were pan resistant to all first-line drugs tested (isoniazid, rifampicin, ethambutol, and streptomycin), of which nine displayed concurrent genotypic resistance to pyrazinamide. No extensively drug-resistant (XDR; MDR with additional resistance to a flouroquinolone and second-line injectable drugs) or pre-XDR strains were identified by DST, but two streptomycin-resistant strains had gyrA mutations indicative of fluoroquinolone resistance.
Table 1.
Demographic characteristics and drug resistance profiles of TB/HIV coinfected patients in Ho Chi Minh City, Vietnam
| Characteristic (N = 200) | n (%) | |
|---|---|---|
| Gender | ||
| Male | 173 (86.5) | |
| Age distribution (N = 174) | ||
| Median age (range) (years) | 31 (17–57) | |
| Age category | ||
| 17–24 | 13 (7.5) | |
| 25–34 | 111 (63.8) | |
| 35–44 | 38 (21.8) | |
| ≥ 45 | 12 (6.9) | |
| Strain lineages* | ||
| Lineage 1 (Indo-Oceanic; East African-Indian, EAI) | 70 (35.0) | |
| Lineage 2 (East Asian; Beijing) | 98 (49.0) | |
| Lineage 3 (Indian/Central Asian; Delhi/CAS) | 0 (0) | |
| Lineage 4 (Euro-American, EA) | 22 (11.0) | |
| Other and unknown† | 10 (5.0) | |
| Drug resistance profiles | Phenotypic | Genotypic‡ |
| Any drug resistance | 84 (42.0) | 82 (41.0) |
| Resistance to a single drug (mono-resistance) | 37 (18.5) | 31 (15.5) |
| H | 4 (2.0) | 5 (2.5) |
| R | 0 | 0 |
| E | 0 | 0 |
| S | 34 (17.0) | 25 (12.5) |
| Z | NA | 0 |
| Resistance to multiple drugs (but not MDR) | 29 (14.5) | 32 (16.0) |
| HS | 28 (14.0) | 20 (10.0) |
| RS | 1 (0.5) | 0 |
| HZ | NA | 2 (1.0) |
| SZ | NA | 2 (1.0) |
| SQ | NA | 2 (1.0) |
| HSZ | NA | 4 (2.0) |
| HSEZ | NA | 2 (1.0) |
| Multidrug resistant (MDR) | 17 (8.5) | 20 (10.0) |
| HR | 0 | 0 |
| HRS | 7 (3.5) | 2 (1.0) |
| HRSE | 10 (5.0) | 8 (4.0) |
| HRSZ | NA | 1 (0.5) |
| HRSEZ | NA | 9 (4.5) |
E = ethambutol; H = isoniazid; HIV = human immunodeficiency virus; MDR = resistant to at least H and rifampicin; NA = not applicable, resistance to Z and Q not assessed; Q = quinolones; R = rifampicin; S = streptomycin; TB = tuberculosis; Z = pyrazinamide.
TB lineages classification as of Gagneux and Small.16
Spoligo and 24-locus mycobacterial interspersed repetitive unit typing unsuccessful.
All phenotypic drug-resistant isolates were sequenced—no resistance mutations were detected in two S mono-resistant strains.
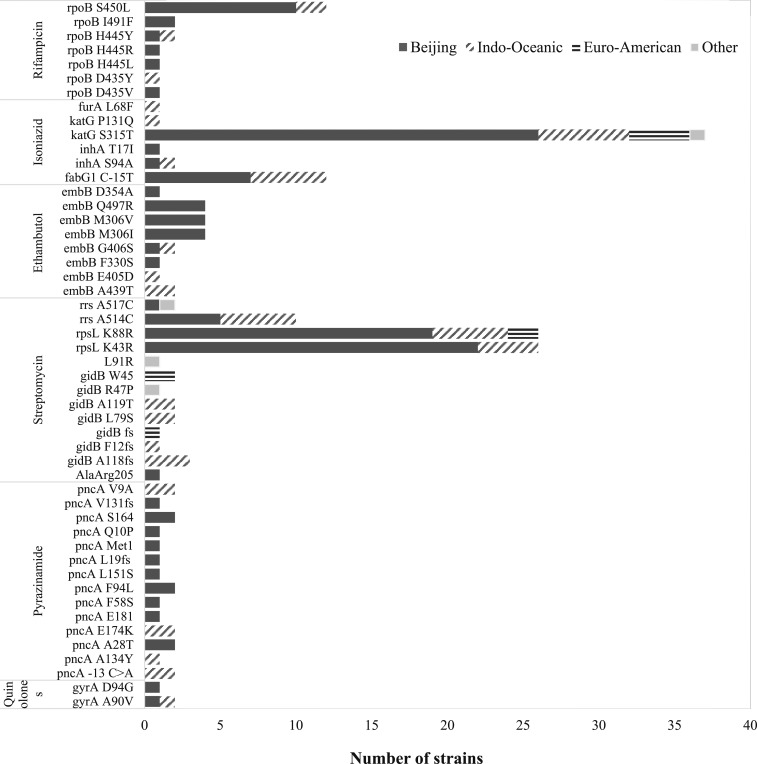
Drug resistance–conferring mutations identified in relevant M. tuberculosis strain lineages are presented in Figure 1. Among phenotypically isoniazid-resistant isolates, the katG S315T mutation was most common (38/54; 70.3%) and found in all phylogenetic lineages, followed by the C-15T mutation in fabG1-inhA regulatory region (12/54; 22.2%). The most common mutation responsible for resistance to rifampicin was rpoB S450L (11/20; 55%). Most rpoB mutations (S450L, H445Y/R/L, and D435Y/V) were identified inside the rifampicin resistance-determining region (RRDR) of rpoB gene targeted by Xpert MTB/RIF®. One mutation was found outside the RRDR (I491F) in two unrelated TB-HIV isolates. Three phenotypically drug-resistant but non-MDR isolates carried mutations associated with a high likelihood of resistance to isoniazid and rifampicin; two phenotypically rifampicin-susceptible isolates had non-synonymous rpoB mutations (D435Y and I491F) and one isoniazid susceptible isolate had a fabG1 C-15T inhA mutation (isolates 25, 31, and 56, Supplemental Table 1). Supplemental Table 1 summarizes results from phenotypic and genotypic resistance testing. Resistance-conferring mutations to ethambutol and pyrazinamide were highly variable with major discrepancies between phenotypic methods and susceptibility profiles inferred from WGS. Phenotypic resistance to pyrazinamide was not assessed.
Figure 1.
Drug resistance mutations identified in different Mycobacterium tuberculosis strain lineages.
Table 2 describes and compares the number and size of clusters identified by spoligotyping, MIRU-24, and WGS. Overall, 84% of isolates were clustered by spoligotyping, including all (98; 100.0%) Beijing lineage strains, which share a unique spoligotyping profile. The 24-locus mycobacterial interspersed repetitive unit clustering was identified in 20.0% of isolates (29.6% of Beijing lineage strains), with 16 clusters consisting of two to four isolates per cluster. Whole genome sequencing confirmed the presence of 13 (27 isolates) clusters (Table 3) and three (seven isolates) “possible clusters” (characterized by 11–20 SNP difference between genomes). The variable clustering identified by MIRU-24 and WGS is further explored in Table 3. Among 16 MIRU-24 clusters, four (25.0%) matched with WGS clusters (including “possible clusters”), six (37.5%) had a partial match, and six (37.5%) had no match. Given the increased resolution of WGS, it is expected that clustering will generally be reduced compared with MIRU-24; however, WGS identified five new clusters (or members of clusters) that were not recognized by MIRU-24 (Table 3).
Table 2.
Number and size of clusters identified by spoligotyping, MIRU-24, and WGS
| Cluster detection | Spoligotyping | MIRU-24* | WGS† |
|---|---|---|---|
| Clustered isolates (N = 200) | 168 (84.0%) | 40 (20.0%) | 27 (13.5%) |
| Number of clusters | 16 | 16 | 13 |
| Median cluster size (range) | 2.5 (2–93) | 3 (2–4) | 2 (2–3) |
| SNP range | NA† | 20–399 | 1–10 |
| Clusters with multidrug-resistant isolates | 6 | 6 | 3 |
| Lineage-specific clustering | |||
| Beijing (N = 98) | 98 (100.0%) | 29 (29.6%) | 12 (14.3%) |
| Indo-Oceanic (N = 70) | 53 (75.7%) | 7 (10.0%) | 9 (12.8%) |
| Euro-American (N = 22) | 14 (63.6%) | 4 (18.2%) | 6 (27.3%) |
| Other (N = 10) | 4 (40.0%) | 0 (0%) | 0 (0%) |
MIRU-24 = 24-locus mycobacterial interspersed repetitive unit typing; SNP = single nucleotide polymorphisms; WGS = whole genome sequencing.
Only identical MIRU-24 profiles were recognized as clusters.
Only MIRU-24 clusters and drug-resistant strains were sequenced—strains with ≤ 10 SNPs were identified as a cluster.
Table 3.
Variable clustering identified by MIRU-24 and WGS
| ID | MIRU-24 clusters ID | MIRU-24 cluster* size (SNP difference) | WGS cluster ID | WGS cluster† size (SNP difference) | SNP by WGS | MIRU-24 profile | Lineage |
|---|---|---|---|---|---|---|---|
| 10-V027 | Cluster 1 | 2 (0) | Cluster 1.WGS | 2 (0) | 0 | 244233342644425163353725 | Beijing |
| 10-V029 | 0 | 244233342644425163353725 | |||||
| 10-V109 | Cluster 2 | 2 (4) | Cluster 2.WGS | 2 (4) | 1 | 224225132134425112333732 | Euro-American |
| 14-V120 | 3 | 224225132134425112333732 | |||||
| 14-V045 | Cluster 3 | 2 (11) | Not clustered | Not clustered | 5 | 244233352644425173354823 | Beijing |
| 14-V057 | 6 | 244233352644425173354823 | |||||
| 10-V004 | Cluster 4 | 3 (16) | Not clustered | Not clustered | 7 | 244233382A44425173355823 | Beijing |
| 14-V052 | 2 | 244233382644425173355823 | |||||
| 14-V054 | 7 | 244233382644425173355823 | |||||
| 14-V078 | Cluster 5 | 3 (21) | Not clustered | Not clustered | 7 | 244233342644425163353723 | Beijing |
| 14-V189 | 11 | 244233342644425163353723 | |||||
| 14-V082 | Cluster 3.WGS | 2 (8) | 5 | 244233342644425163353723 | |||
| 14-V064 | Not clustered | Not clustered | 3 | 244235342674425163353723 | |||
| 10-V007 | Cluster 6 | 2 (52) | Not clustered | Not clustered | NA | 233213322534435163332632 | Euro-American |
| 14-V053 | Cluster 4.WGS | 2 (5) | 4 | 233213322534435163332632 | |||
| 14-V055 | Not clustered | Not clustered | 1 | 233213322534435463332632 | |||
| 14-V128 | Cluster 7 | 2 (67) | Not clustered | Not clustered | NA | 224624272273245422354613 | Indo-Oceanic |
| 14-V134 | Not clustered | Not clustered | NA | 224624272273245422354613 | |||
| 10-V026 | Cluster 8 | 2 (73) | Not clustered | Not clustered | NA | 324634252273245423354613 | Indo-Oceanic |
| 10-V025 | Cluster 5.WGS | 3 (3) | 1 | 324634252273245423354713 | |||
| 10-V028 | Not clustered | Not clustered | 1 | 244233342644425163353725 | |||
| 10-V010 | Cluster 9 | 3 (187) | 1 | 324634252273245223A54613 | |||
| 14-V079 | Not clustered | Not clustered | NA | 324634252273245223354613 | |||
| 14-V154 | Not clustered | Not clustered | NA | 324634252273245223354613 | |||
| 14-V034 | Cluster 10 | 2 (89) | Not clustered | Not clustered | NA | 244232352644225173353824 | Beijing |
| 09-V088 | Not clustered | Not clustered | NA | 244232352644225173353824 | |||
| 14-V118 | Cluster 11 | 2 (158) | Not clustered | Not clustered | NA | 244233352644425172353823 | Beijing |
| 10-V131 | Not clustered | Not clustered | NA | 244233352644425172353823 | |||
| 09-V085 | Cluster 12 | 4 (172) | Not clustered | Not clustered | NA | 244232352644425173353824 | Beijing |
| 14-V177 | Not clustered | Not clustered | NA | 244232352644425173353824 | |||
| 14-V178 | Not clustered | Not clustered | NA | 244232352644425173353824 | |||
| 10-V012 | Cluster 6.WGS | 2 (2) | 0 | 2442323526444251733538A4 | |||
| 14-V081 | Not clustered | Not clustered | 2 | 244232352642425173253824 | |||
| 14-V074 | Cluster 13 | 3 (194) | Cluster 7.WGS | 2 (7) | 2 | 244233352544425173353823 | Beijing |
| 14-V030 | 5 | 244233352544425173353823 | |||||
| 14-V083 | Cluster 8.WGS | 2 (1) | 0 | 244233352544425173353823 | |||
| 14-V060 | Cluster 14 | 2 (291) | 1 | 244233352544425173453823 | Beijing | ||
| 14-V037 | Not clustered | Not clustered | NA | 244233352544425163353723 | |||
| 14-V046 | Cluster 15 | 4 (238) | Not clustered | Not clustered | NA | 244233352644425173353823 | Beijing |
| 14-V051 | Not clustered | Not clustered | NA | 244233352644425173353723 | |||
| 14-V167 | Not clustered | Not clustered | NA | 244233352644425173353823 | |||
| 14-V041 | Not clustered | Not clustered | NA | 244233352644425173353823 | |||
| 10-V113 | Cluster 16 | 2 (399) | Not clustered | Not clustered | NA | 244233352544425172353823 | Beijing |
| 14-V146 | Not clustered | Not clustered | NA | 244233342544425172353223 | |||
| 14-V034 | Not clustered | No cluster by MIRU | Cluster 9.WGS | 2 (7) | 3 | 244232352644225173353824 | Beijing |
| 14-V039 | 4 | 244232352646425173353823 | |||||
| 14-V035 | Not clustered | No cluster by MIRU | Cluster 10.WGS | 2 (7) | 3 | 222624392393246324A65411 | Indo-Oceanic |
| 14-V038 | 4 | 2226243A2393246424365411 | |||||
| 10-V006 | Not clustered | No cluster by MIRU | Cluster 11.WGS | 2 (3) | 1 | 224633272273234223354813 | Indo-Oceanic |
| 14-V049 | 2 | 224633272273234423354813 | |||||
| 14-V044 | Not clustered | No cluster by MIRU | Cluster 12.WGS | 2 (5) | 2 | 224534272AA3245223346613 | Indo-Oceanic |
| 14-V056 | 3 | 224514272AA4225223346613 | |||||
| 14-V040 | Not clustered | No cluster by MIRU | Cluster 13.WGS | 2 (0) | 0 | 234232352274445223364624 | Indo-Oceanic |
| 14-V043 | 0 | 2442333826446251733548A3 |
MIRU-24 = 24-locus mycobacterial interspersed repetitive unit typing; SNP = single nucleotide polymorphisms; WGS = whole genome sequencing.
MIRU-24 cluster identified as identical profiles and tree-based identification MIRU-VNTRplus (http://www.miru-vntrplus.org/MIRU/index.faces).
WGS cluster identified as ≤ 10 SNPs difference, “possible cluster” as 10–20 SNPs difference, and “not clustered” > 20 SNPs difference.
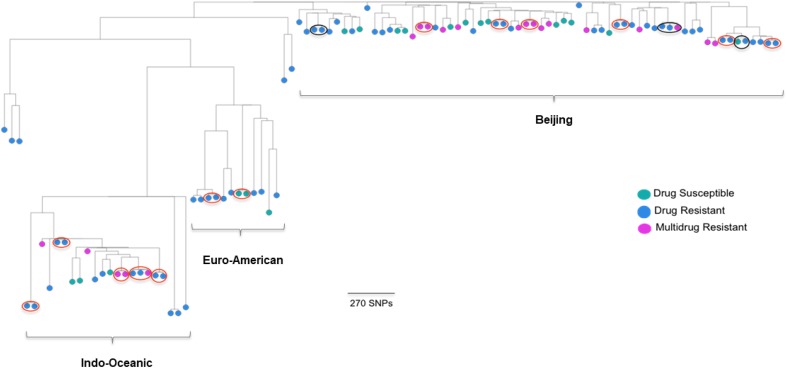
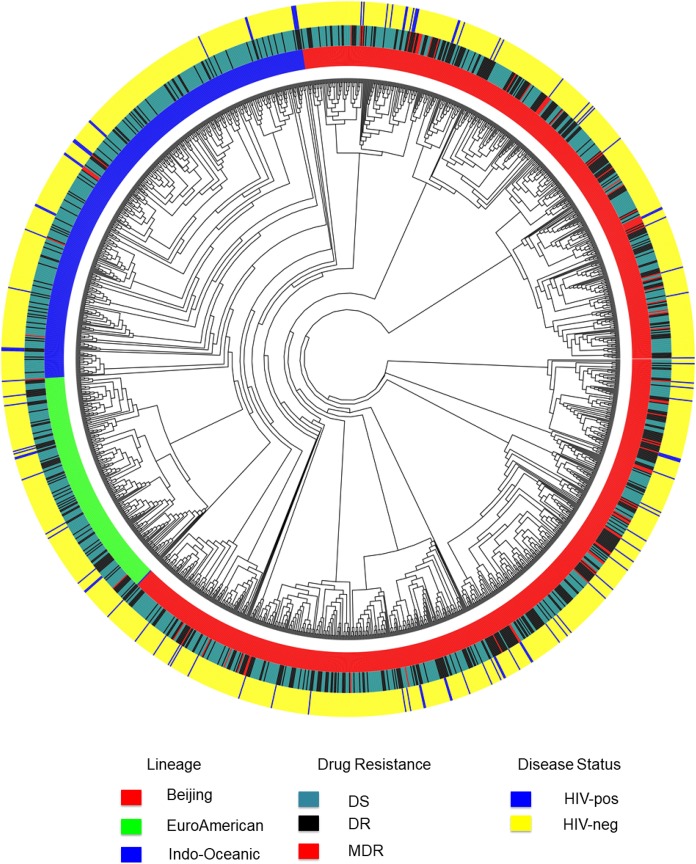
The comparison of M. tuberculosis genomes sequenced in this study identified 13 small (two to three strain) clusters dispersed among all the major strain lineages, with nearly all clusters (12/13; 92.3%) containing drug-resistant strains (Figure 2). We documented less clustering among drug-susceptible (2/27; 7.4%) than among drug-resistant strains (25/27; 92.6%) (OR 0.93, 95% confidence interval: 0.77–0.98). A combined analysis of our dataset and that of non-HIV–infected patients between 2008 and 2011 described by Holt et al.17 demonstrated that the strains from TB/HIV coinfected patients were intermingled with strains circulating in the general community (Figure 3). This comparison identified 21 clusters among HIV-infected and uninfected patients, including one big clonal outbreak with 17 isolates, four medium-sized clusters (5–10 isolates each), and 16 small clusters (two to four isolates each). Of the 102 sequenced strains from TB-HIV coinfected patients, probable recent transmission was identified in 36 (25.3%), with most of the transmitted strains (25/36; 69.4%) appearing to be acquired in the community. Supplemental Table 2 presents the M. tuberculosis strain lineage, drug resistance profile, and cluster characteristics of strains isolated from TB/HIV coinfected patients compared with those from the general community. All large clusters appeared to be independent of the patients’ HIV status. Multidrug-resistant TB transmission was noted in a number of small clusters, but unrelated to HIV infection status. Supplemental Table 3 which includes all sequenced strains reflects 3/13 (23.0%) potential “crossover” clusters between the two time periods 2010 and 2014.
Figure 2.
Phylogenetic tree of sequenced* Mycobacterium tuberculosis strains from patients with tuberculosis/human immunodeficiency virus coinfection in Ho Chi Minh City, Vietnam. Multidrug resistant (MDR): resistant to at least isoniazid and rifampicin; *All strains that were drug resistant (84) or clustered by 24-locus mycobacterial interspersed repetitive unit (40) were sequenced; Red circles indicate identified clusters; Black circles indicate possible cluster.
Figure 3.
Mycobacterium tuberculosis strains identified in tuberculosis/HIV coinfected patients in Ho Chi Minh City, Vietnam, compared with community strains identified among non-HIV–infected patients.* DR = drug resistant; DS = drug susceptible; HIV = human immunodeficiency virus; MDR = multidrug resistant. *Sequence data for community M. tuberculosis strains were derived from Holt et al.15
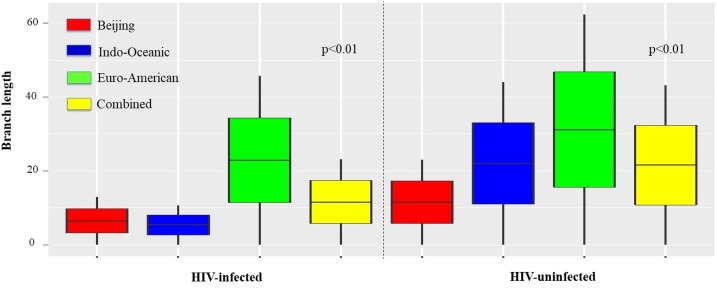
The average branch length of M. tuberculosis genomes derived from SNP-based phylogenetic trees in TB patients with and without HIV coinfection was used as a proxy measure for the recency of TB infection. The average branch length in strains from HIV-infected patients was significantly shorter than those from HIV-uninfected patients (P < 0.01). This observation was consistent across all phylogenetic lineages, but most pronounced in Indo-Oceanic strains (Figure 4). To assess the role of patients’ age in these differences, given the young age of TB/HIV coinfected patients, the same analysis was performed comparing HIV-infected patients in different age groups (Supplemental Figure 1). This demonstrated a pronounced age-related effect with shorter branch lengths noted in strains isolated from HIV-infected patients younger than 45 years compared with those ≥ 45 years of age (P < 0.01).
Figure 4.
Comparison of branch length by Mycobacterium tuberculosis strain lineage in HIV-infected and uninfected patients. HIV = human immunodeficiency virus. This figure appears in color at www.ajtmh.org.
DISCUSSION
This is the first study to assess the M. tuberculosis transmission dynamics among HIV-infected patients in Vietnam.8,16 We did not observe large strain clusters, indicative of recent transmission and potential poor infection control practices in health-care centers, as documented in KwaZulu-Natal, South Africa.26 Being able to compare strains from HIV-infected patients with those in the general community provided an unprecedented opportunity to map the dynamic exchange of M. tuberculosis strains between HIV-infected and uninfected individuals in the same population. The fact that strains from HIV-infected patients were interspersed with community strains indicates that most HIV-infected patients acquired M. tuberculosis infection in the general community, with limited person-to-person transmission among HIV-infected patients. Although the absence of big clonal outbreaks among TB/HIV coinfected patients is comforting from an infection control perspective, more effort is still justified to limit the TB exposure of HIV-infected patients within the community.
Given the limited resolution of MIRU-24 and spoligotyping to differentiate Beijing lineage clusters, WGS allowed more accurate cluster characterization.15,27 Whole genome sequencing revealed variation between isolates that were indistinguishable by MIRU-24 and illuminated new clusters that were not detected by MIRU-24. The observation that some strains with nonidentical MIRU-24 profiles, especially those with multiple loci differences, were recognized as part of a WGS cluster seems counterintuitive, given that MIRU-24 has reduced resolution compared with WGS. This might be explained by the fact that repetitive regions of the M. tuberculosis genome, including the variable number tandem repeats used for MIRU typing, are excluded from genomic comparisons and analysis (e.g., regions annotated as PE/PPE/PGRS and cysA genes, insertions sequences, transposases, and prophage components)15 and, therefore, MIRU-24 may capture some genome variability that is not detected by current WGS analysis methods. Compared with MIRU-24, WGS provided the greatest resolution improvement in Beijing lineage strains, which display the least MIRU-24 allelic diversity.15 In our study, WGS also demonstrated added value in the evaluation of Indo-Oceanic and Euro-American lineage strains, with the identification additional clusters not recognized by MIRU-24.
The significantly shorter phylogenetic tree branch lengths observed among TB/HIV coinfected patients, compared with strains from HIV-uninfected patients, suggest more recent transmission with a shorter time between infection and disease progression. As similar observation of Holt and others16 has previously been made comparing Beijing compared with other strain lineages. However, the use of branch lengths as a measure of recency of infection can be confounded by within host dynamics,28 as well as an age cohort effect among TB/HIV coinfected patients who were generally younger in our study. Most TB/HIV coinfected patients were less than 45 years of age, whereas HIV-uninfected TB patients in Vietnam are typically older.29
Although we found no evidence of a dominant drug-resistant clone, as reported recently in Papua New Guinea,30 the high rate of drug resistance within identified strain clusters is alarming because it confirms frequent transmission of drug-resistant strains within the community. The increased clustering of drug-resistant strains that we observed might have been biased by the fact that we did not sequence all drug-susceptible isolates. However, our findings are consistent with household contact surveys that suggested increased TB transmission within MDR-TB–affected households in Vietnam.31 Overall, the rate of drug-resistant disease among TB/HIV coinfected patients was high and the fact that streptomycin resistance was most common aligns with previous findings from Vietnam32,33 and Mongolia.34 In Vietnam, this can be partially explained by the perception that there is a preference for intramuscular injections and the fact that streptomycin was used during the intensive phase of standard first-line treatment in preference to ethambutol until 2016.32,35
Our findings also extended the catalog of significant genomic changes that can be used to infer drug resistance in strains circulating in Southeast Asia. As in other settings, mutations at codons 43 and 88 of the rpsL gene were most frequent and associated with high-level streptomycin resistance.36,37 Mutations in gidB, which has been associated with a low-level of streptomycin resistance,36,37 were variable and found in both phenotypically resistant and susceptible strains. Strains harboring gidB mutations associated with phenotypic streptomycin resistance, without concurrent rrs or rpsL mutations, were mostly found in non-Beijing isolates, similar to observations from Thailand.37 No rrs mutation associated with streptomycin resistance provided cross-resistance with second-line injectable drugs such as amikacin or kanamycin.38 The small number of discordant phenotypic and genotypic streptomycin resistance profiles may be due to imperfect phenotypic results or potential culture contamination.39,40
Whole genome sequencing identified three additional MDR-TB cases that were not recognized by routine phenotypic methods,12 which fuels debate about the most appropriate critical concentrations and reference standards for MDR-TB determination. It has been noted before that strains harboring “high-confidence” drug resistance mutations may sometimes be missed by phenotypic DST.41 In some discrepant cases, the critical concentrations of antibiotic used for phenotypic DST may be inadequate to detect lower levels of resistance, which is relevant in rpoB mutations associated with low-level rifampicin resistance.42 However, genotypic results may also be affected by incomplete catalogs of resistance associated mutations and/or potential misclassification of lineage-specific polymorphisms as resistance mutations.43 S450L has been the most common mutation associated with rifampicin resistance,44 and this was the case among HIV-infected and HIV-uninfected patients in Vietnam. Most rpoB mutations would have been detected by Xpert MTB/RIF® which is widely available and used in Vietnam (23), but the I491F mutation detected in two unrelated strains would have been missed, even by the new Ultra version.45 This mutation is outside the 81-bp RRDR and has been recognized as problematic by the CRyPTIC consortium,46 with potential for diagnostic selection observed in Swaziland.47
Culture-based DST for pyrazinamide and ethambutol remains challenging.48 We identified resistance-conferring mutations to ethambutol by WGS, but many of these strains appeared to be phenotypically susceptible. It has been suggested that the suboptimal sensitivity of phenotypic ethambutol resistance testing may be due to the presence of bacterial subpopulations49,50 or drug concentrations that are too high to detect all clinically relevant resistance.51 We also found three common gyrA mutations that have been associated with high-level resistance to fluoroquinolones52,53 in isolates without a MDR-TB phenotype. This may reflect a risk of using fluoroquinolone treatment of community-acquired pneumonia or other common infections, especially in HIV-infected patients at high risk of TB.54,55
Several study limitations have to be acknowledged. First, the fact that sample collection was noncontinuous, although consecutive samples were collected at times (2009–2010 and then again in 2014) when no other studies were in progress. However, we believe that our findings remain informative, given that consecutive sample collection was unbiased and the sampling periods were broadly comparable to the study of HIV-uninfected patients performed by Holt et al.17 We cannot exclude the possibility that some of the TB cases included in Holt’s study may have been included in our study as well, but, given good HIV and TB record keeping in Ho Chi Minh City, we believe the chances for this are small and unlikely to have had any significant influence on our results. Second, we have already commented on the predominance of young males in our study, which reflects the fact that the HIV epidemic in Vietnam primarily affects injecting drug users and men who have sex with men.56 This limits the generalizability of our findings to settings with different demographics, such as sub-Saharan Africa, where HIV transmission is mostly heterosexual and affects equal numbers of men and women.57
In conclusion, drug resistance in M. tuberculosis in TB-HIV coinfected patients was associated predominantly with Beijing and Indo-Oceanic lineage strains and the broad spectrum of resistance-conferring mutations. Mycobacterium tuberculosis transmission among people living with HIV in Ho Chi Minh City appears to be limited, but our findings suggest extensive M. tuberculosis transmission between HIV-infected and uninfected individuals within the community, with high rates of transmitted drug-resistant TB. These findings highlight the added value of genome sequencing–based analysis in tracking transmission pathways and encourage further efforts to reduce TB transmission within the general community, with specific emphasis on the optimal use of preventive therapy in HIV-infected individuals.
Supplementary Material
Supplemental figure and tables
Acknowledgments:
We thank the TB doctors and nurses from the Pham Ngoc Thach TB and Lung Disease Hospital for their help with data collection; NSW Mycobacterial Reference Laboratory, NSW Health Pathology, and the Centre for Infectious Diseases and Microbiology—Public Health, ICPMR, Westmead Hospital, Sydney, for technical assistance with MIRU-VNTR and genome sequencing.
Note: Supplemental figure and tables appear at www.ajtmh.org.
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The study is in partial fulfillment of a PhD degree.
REFERENCES
- 1.World Health Organization , 2017. Global Tuberculosis Report, 2017. Geneva, Switzerland: WHO. [Google Scholar]
- 2.Abubakar I, et al. 2013. Drug-resistant tuberculosis: time for visionary political leadership. Lancet Infect Dis 13: 529–539. [DOI] [PubMed] [Google Scholar]
- 3.McBryde ES, Meehan MT, Doan TN, Ragonnet R, Marais BJ, Guernier V, Trauer JM, 2017. The risk of global epidemic replacement with drug-resistant Mycobacterium tuberculosis strains. Int J Infect Dis 56: 14–20. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Yew W, 2009. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 13: 1320–1330. [PubMed] [Google Scholar]
- 5.Zhao Y, et al. 2012. National survey of drug-resistant tuberculosis in China. N Engl J Med 366: 2161–2170. [DOI] [PubMed] [Google Scholar]
- 6.Ragonnet R, Trauer JM, Denholm JT, Marais BJ, McBryde ES, 2017. High rates of multidrug-resistant and rifampicin-resistant tuberculosis among re-treatment cases: where do they come from? BMC Infect Dis 17: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marais BJ, Mlambo CK, Rastogi N, Zozio T, Duse AG, Victor TC, Marais E, Warren RM, 2013. Epidemic spread of multidrug-resistant tuberculosis in Johannesburg, South Africa. J Clin Microbiol 51: 1818–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, Zeller K, Andrews J, Friedland G, 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368: 1575–1580. [DOI] [PubMed] [Google Scholar]
- 9.Raviglione M, et al. 2012. Scaling up interventions to achieve global tuberculosis control: progress and new developments. Lancet 379: 1902–1913. [DOI] [PubMed] [Google Scholar]
- 10.Vietnam National Tuberculosis Control Program, Ministry of Health , 2015. Report of The National Tuberculosis Control Program in 2014. Hanoi, Vietnam: Vietnam National Tuberculosis Control Program, Ministry of Health.
- 11.Isaakidis P, Casas EC, Das M, Tseretopoulou X, Ntzani EE, Ford N, 2015. Treatment outcomes for HIV and MDR-TB co-infected adults and children: systematic review and meta-analysis. Int J Tuberc Lung Dis 19: 969–978. [DOI] [PubMed] [Google Scholar]
- 12.Basu S, Andrews JR, Poolman EM, Gandhi NR, Shah NS, Moll A, Moodley P, Galvani AP, Friedland GH, 2007. Prevention of nosocomial transmission of extensively drug-resistant tuberculosis in rural South African district hospitals: an epidemiological modelling study. Lancet 370: 1500–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai TQ, Van Anh NT, Hien NT, Lan NH, Giang DC, Hang PTT, Lan NTN, Marais BJ, Sintchenko V, 2017. Drug resistance and Mycobacterium tuberculosis strain diversity in TB/HIV co-infected patients in Ho Chi Minh city, Vietnam. J Glob Antimicrob Resist 10: 154–160. [DOI] [PubMed] [Google Scholar]
- 14.Gurjav U, Jelfs P, McCallum N, Marais BJ, Sintchenko V, 2014. Temporal dynamics of Mycobacterium tuberculosis genotypes in New South Wales, Australia. BMC Infect Dis 14: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurjav U, Outhred AC, Jelfs P, McCallum N, Wang Q, Hill-Cawthorne GA, Marais BJ, Sintchenko V, 2016. Whole genome sequencing demonstrates limited transmission within identified Mycobacterium tuberculosis clusters in New South Wales, Australia. PLoS One 11: e0163612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagneux S, Small PM, 2007. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 7: 328–337. [DOI] [PubMed] [Google Scholar]
- 17.Holt KE, et al. 2018. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat Genet 50: 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham Ngoc Thach TB and Lung Disease Hospital , 2016. Annual Report 2016—Pham Ngoc Thach TB and Lung Disease Hospital.
- 19.Caws M, Duy PM, Tho DQ, Lan NTN, Hoa DV, Farrar J, 2006. Mutations prevalent among rifampin- and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J Clin Microbiol 44: 2333–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel Aziz M, Laszlo A, Raviglione MC, Rieder HL, Espinal MA, Wright A, 2003. Guidelines for Surveillance of Drug Resistance in Tuberculosis. Geneva, Switzerland: World Health Organization. WHO/CDS/TB/2003.320.
- 21.Kamerbeek J, et al. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44: 4498–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankhurst LJ, et al. COMPASS-TB Study Group , 2016. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med 4: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price MN, Dehal PS, Arkin AP, 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickham H, 2009. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag New York.
- 26.Bantubani N, Kabera G, Connolly C, Rustomjee R, Reddy T, Cohen T, Pym AS, 2014. High rates of potentially infectious tuberculosis and multidrug-resistant tuberculosis (MDR-TB) among hospital inpatients in KwaZulu Natal, South Africa indicate risk of nosocomial transmission. PLoS One 9: e90868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamieson FB, Teatero S, Guthrie JL, Neemuchwala A, Fittipaldi N, Mehaffy C, 2014. Whole-genome sequencing of the Mycobacterium tuberculosis Manila sublineage results in less clustering and better resolution than mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) typing and spoligotyping. J Clin Microbiol 52: 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T, 2014. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol Biol Evol 31: 2472–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nhung NV, Hoa NB, Khanh PH, Hennig C, 2015. Tuberculosis case notification data in Viet Nam, 2007 to 2012. Western Pac Surveill Response J 6: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aia P, et al. 2016. The burden of drug-resistant tuberculosis in Papua New Guinea: results of a large population-based survey. PLoS One 11: e0149806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox GJ, Anh NT, Nhung NV, Loi NT, Hoa NB, Ngoc Anh LT, Cuong NK, Buu TN, Marks GB, Menzies D, 2017. Latent tuberculous infection in household contacts of multidrug-resistant and newly diagnosed tuberculosis. Int J Tuberc Lung Dis 21: 297–302. [DOI] [PubMed] [Google Scholar]
- 32.Nhu NT, Lan NT, Phuong NT, Chau Nv, Farrar J, Caws M, 2012. Association of streptomycin resistance mutations with level of drug resistance and Mycobacterium tuberculosis genotypes. Int J Tuberc Lung Dis 16: 527–531. [DOI] [PubMed] [Google Scholar]
- 33.Quy HT, Buu TN, Cobelens FGJ, Lan NTN, Lambregts CSB, Borgdorff MW, 2006. Drug resistance among smear-positive tuberculosis patients in Ho Chi Minh City, Vietnam. Int J Tuberc Lung Dis 10: 160–166. [PubMed] [Google Scholar]
- 34.Gurjav U, Erkhembayar B, Burneebaatar B, Narmandakh E, Tumenbayar O, Hill-Cawthorne GA, Marais BJ, Sintchenko V, 2016. Transmission of multi-drug resistant tuberculosis in Mongolia is driven by Beijing strains of Mycobacterium tuberculosis resistant to all first-line drugs. Tuberculosis (Edinb) 101: 49–53. [DOI] [PubMed] [Google Scholar]
- 35.Hang NTL, et al. 2013. Primary drug-resistant tuberculosis in Hanoi, Viet Nam: present status and risk factors. PLoS One 8: e71867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tudo G, et al. 2010. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis clinical isolates in the area of Barcelona. J Antimicrob Chemother 65: 2341–2346. [DOI] [PubMed] [Google Scholar]
- 37.Smittipat N, et al. 2016. Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J Glob Antimicrob Resist 4: 5–10. [DOI] [PubMed] [Google Scholar]
- 38.Coll F, et al. 2018. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat Genet 50: 307–316. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad S, Mokaddas E, Al-Mutairi N, Eldeen HS, Mohammadi S, 2016. Discordance across phenotypic and molecular methods for drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates in a low TB incidence country. PLoS One 11: e0153563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piersimoni C, Olivieri A, Benacchio L, Scarparo C, 2006. Current perspectives on drug susceptibility testing of Mycobacterium tuberculosis complex: the automated nonradiometric systems. J Clin Microbiol 44: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shea J, Halse TA, Lapierre P, Shudt M, Kohlerschmidt D, Van Roey P, Limberger R, Taylor J, Escuyer V, Musser KA, 2017. Comprehensive whole-genome sequencing and reporting of drug resistance profiles on clinical cases of Mycobacterium tuberculosis in New York state. J Clin Microbiol 55: 1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Deun A, Aung KJM, Bola V, Lebeke R, Hossain MA, de Rijk WB, Rigouts L, Gumusboga A, Torrea G, de Jong BC, 2013. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 51: 2633–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S, 2017. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep 7: 46327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Yew WW, 2015. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis 19: 1276–1289. [DOI] [PubMed] [Google Scholar]
- 45.Chakravorty S, et al. 2017. The new Xpert MTB/RIF Ultra: improving detection of Mycobacterium tuberculosis and resistance to rifampin in an assay suitable for point-of-care testing. MBio 8: e00812–e00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker TM, et al. Modernizing Medical Microbiology (MMM) Informatics Group , 2015. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 15: 1193–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Padilla E, Merker M, Beckert P, Jochims F, Dlamini T, Kahn P, Bonnet M, Niemann S, 2015. Detection of drug-resistant tuberculosis by Xpert MTB/RIF in Swaziland. N Engl J Med 372: 1181–1182. [DOI] [PubMed] [Google Scholar]
- 48.Van Deun A, Wright A, Zignol M, Weyer K, Rieder HL, 2011. Drug susceptibility testing proficiency in the network of supranational tuberculosis reference laboratories. Int J Tuberc Lung Dis 15: 116–124. [PubMed] [Google Scholar]
- 49.Kim SJ, 2005. Drug-susceptibility testing in tuberculosis: methods and reliability of results. Eur Respir J 25: 564–569. [DOI] [PubMed] [Google Scholar]
- 50.Jou R, Chiang CY, Yu CY, Wu MH, 2009. Proficiency of drug susceptibility testing for Mycobacterium tuberculosis in Taiwan. Int J Tuberc Lung Dis 13: 1142–1147. [PubMed] [Google Scholar]
- 51.Shi R, Zhang J, Otomo K, Zhang G, Sugawara I, 2007. Lack of correlation between embB mutation and ethambutol MIC in Mycobacterium tuberculosis clinical isolates from China. Antimicrob Agents Chemother 51: 4515–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Von Groll A, Martin A, Jureen P, Hoffner S, Vandamme P, Portaels F, Palomino JC, da Silva PA, 2009. Fluoroquinolone resistance in Mycobacterium tuberculosis and mutations in gyrA and gyrB. Antimicrob Agents Chemother 53: 4498–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Gao X, Luo T, Wu J, Sun G, Liu Q, Jiang Y, Zhang Y, Mei J, Gao Q, 2014. Association of gyrA/B mutations and resistance levels to fluoroquinolones in clinical isolates of Mycobacterium tuberculosis. Emerg Microbes Infect 3: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grossman RF, Hsueh P-R, Gillespie SH, Blasi F, 2014. Community-acquired pneumonia and tuberculosis: differential diagnosis and the use of fluoroquinolones. Int J Infect Dis 18: 14–21. [DOI] [PubMed] [Google Scholar]
- 55.Jeon CY, Calver AD, Victor TC, Warren RM, Shin SS, Murray MB, 2011. Use of fluoroquinolone antibiotics leads to tuberculosis treatment delay in a South African gold mining community. Int J Tuberc Lung Dis 15: 77–83. [PubMed] [Google Scholar]
- 56.Trinh QM, Nguyen HL, Do TN, Nguyen VN, Nguyen BH, Nguyen TVA, Sintchenko V, Marais BJ, 2016. Tuberculosis and HIV co-infection in Vietnam. Int J Infect Dis 46: 56–60. [DOI] [PubMed] [Google Scholar]
- 57.Dellar RC, Dlamini S, Karim QA, 2015. Adolescent girls and young women: key populations for HIV epidemic control. J Int AIDS Soc 18 (Suppl 1): 19408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables