Summary
Systems vaccinology has proven a fascinating development in the last decade. Where traditionally vaccine development has been dominated by trial and error, systems vaccinology is a tool that provides novel and comprehensive understanding if properly used. Data sets retrieved from systems‐based studies endorse rational design and effective development of safe and efficacious vaccines. In this review we first describe different omics‐techniques that form the pillars of systems vaccinology. In the second part, the application of systems vaccinology in the different stages of vaccine development is described. Overall, this review shows that systems vaccinology has become an important tool anywhere in the vaccine development chain.
Keywords: bioinformatics, proteomics, T‐cell, transcriptomics, vaccination
Introduction
There are many effective vaccines available to protect humans against infectious diseases. However, the development of vaccines against ‘difficult’ pathogens remains challenging. For example, vaccines against human immunodeficiency virus (HIV) and respiratory syncytial virus (RSV) are still under development. Furthermore, some existing vaccines require improved efficacy, e.g. vaccines against Bordetella pertussis, influenza viruses, Mycobacterium tuberculosis and Plasmodium species causing malaria. In addition, new infectious diseases such as severe acute respiratory syndrome and Ebola virus disease will continue to emerge, which require new vaccines to prevent epidemics. Also, therapeutic vaccines against non‐infectious diseases, such as cancer, are needed. The lack of in‐depth knowledge of the pathogen and requirements for protective immunity often hamper development.1 Application of systems biology during the development of vaccines, or systems vaccinology, can be an important tool to enhance insight into immune responses induced by (candidate) vaccines or identification of (early) correlates of protection.2, 3 Alan Aderem defined systems biology as a ‘comprehensive quantitative analysis of the manner in which all the components of a biological system interact functionally over time and space that is executed by an interdisciplinary team of investigators.’4 This definition can readily be applied to study the responses to vaccination. Systems‐based approaches are often labeled unbiased and broad (sometimes even the word ‘holistic’ is used). However, that is only true to a certain extent, as defining a research objective is already introducing bias. Often only a few analytical techniques are used (as described in the next section) to address the objectives (Fig. 1). Usually, there are also limitations in availability of materials. For example, in preclinical studies using mice the amount of blood that can be collected is very limited, whereas in clinical studies mainly body fluids can be used, although biopsies and mucosal lavages are sometimes available. Also, time is an essential parameter of systems vaccinology for studying kinetics and cause–effect relations, but repeated sampling is limited in humans. In this review, we describe current developments of the techniques that form the pillars of systems vaccinology and discuss the implementation of systems vaccinology in the vaccine research and development chain.
Figure 1.
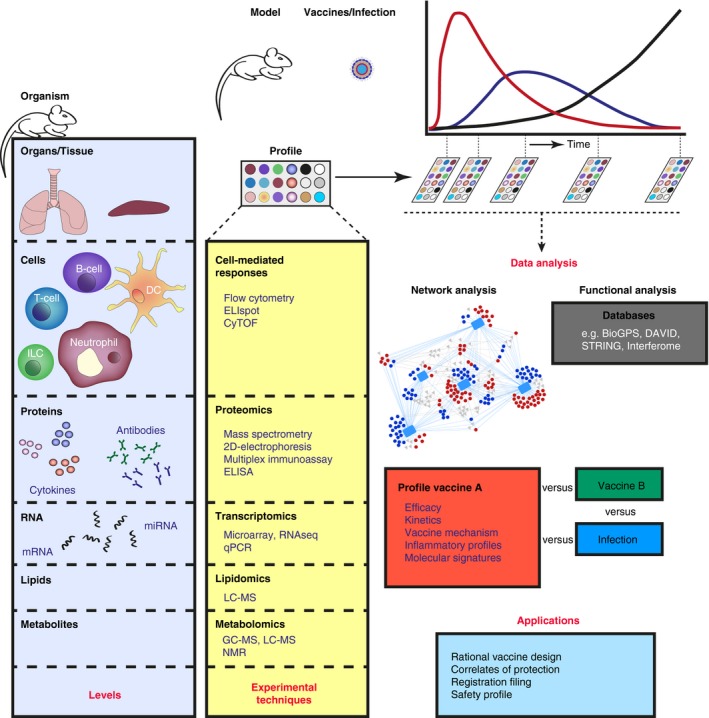
Systems vaccinology approach in a pre‐clinical setting. A biological system can range from a single cell to the complete human body consisting of different levels such as genes, proteins, cells, tissues and organs that interact with each other. The biological processes in these levels have distinct time‐ and space‐resolved kinetics. Information on the immune status can be acquired by analysis at the molecular level of the actors (i.e. gene expression, protein synthesis, lipid secretion and production of metabolites), or by determining the changes in cellular composition and morphology. To study the relationship and interaction between all distinct levels of a biological system, a comprehensive approach is required, using multiple analytical techniques. Data, preferably obtained during a time course of the same subject, are combined for further analysis. Network analysis (e.g. Cytoscape) is performed to determine co‐expression profiles, indicating interdependence. Functional analysis is executed in public databases, e.g. DAVID (http://www.david.ncifcrf.gov), STRING (http://www.string-db.org), BioGPS (http://www.biogps.org), and Interferome (http://www.interferome.org). Combined data form a response profile for a vaccine. Vaccine profiles can be compared with other vaccine or infection profiles and used for multiple applications as mentioned in Table 1.
Systems vaccinology techniques
Twenty years ago, Rino Rappuoli and co‐workers introduced reverse vaccinology, a genome‐based method to identify new leads with strong antibody responses.5 However, the role of immunogens is currently studied in conjunction with other factors, such as adjuvants and antigen delivery, as these influence immune responses and induction of immunological memory that are key for vaccine efficacy. Systems vaccinology addresses these aspects in order to understand why some vaccines function properly whereas other vaccines do not. In this section, the most important and widely used techniques applied in different levels of systems vaccinology are described. An overview of these techniques and applications thereof that are used for the systems vaccinology‐based approach, are listed in Table 1.
Table 1.
The omics in vaccinology
| Omics type | Application | Technology | Literature |
|---|---|---|---|
| Genomics | 142 | ||
| Single‐nucleotide polymorphism |
Restriction fragment length polymorphism (RFLP) Microarray analysis DNA sequencing |
143
144, 145 |
|
| Epigenomics | Age‐related immune responses, vaccine‐induced memory T‐cells | ChIP‐seq, DNA methylation | 146, 147, 148 |
| Personal genomics | Personalized vaccines | 149, 150, 151 | |
| Transcriptomics | 152, 153 | ||
| Single cell transcriptomics |
RNA‐seq CITE‐seq |
154
17 |
|
| Host–pathogen interaction | RNA‐seq, Microarray | 155, 156 | |
| Infection‐induced responses | RNA‐seq, Microarray, quantitative polymerase chain reaction (qPCR) | 12, 157 | |
| Transcriptional responses by vaccination | RNA‐seq, Microarray, qPCR | 13, 15, 116, 124, 126 | |
| In vivo analysis of transcriptome | Transcriptome in vivo analysis (TIVA) | 11 | |
| Proteomics | |||
| Immunoproteomics |
T‐cell epitope identification Antibody specificity (antigen identification) |
Liquid chromatography‐mass spectrometry (LC‐MS) ELISA, MIA, two‐dimensional Western blotting + LC‐MS |
20
59 |
| Interactomics and Ligandomics |
B‐cell epitope mapping Antibody epitope identification Antigen identification Cell‐to‐cell signaling |
X‐ray crystallography LC‐MS (epitope excision/extraction) Peptide arrays Immunoglobulin sequencing Chemical cross‐linking mass spectrometry (XL‐MS) |
158
159, 160 60 55 36 |
| Peptidomics | MHC epitope display | Mass spectrometry | 161 |
| Secretomics | Chemokine secretion | BONCAT | 40 |
| Systems serology | Antibody level, isotype, subtype, specificity, functionality, glycosylation | ELISA, MIA, LC‐MS, Western blotting | 72 |
| Metabolomics | 47 | ||
| Metabolomics |
Response to vaccination Predictive biomarker discovery |
Gas chromatography‐Mass spectrometry (GC‐MS) LC‐MS Nuclear magnetic resonance (NMR) |
46, 162, 163 |
| Lipidomics | Biomarker discovery | LC‐MS | 105, 164, 165 |
| Glycomics | Mapping antibody glycosylation |
Capillary electrophoresis LC‐MS |
166 |
| Cellomics |
Mechanism of action, Correlates of protection, Involved cell types B‐cell and T‐cell repertoire |
Flow cytometry, ELIspot Cytometry by time‐of‐flight MS High‐throughput single‐cell analysis |
12, 15
53, 54, 137 56, 57, 58 |
| Microbiomics |
Vaccine optimization Environmental effects Host‐interaction |
Next‐generation sequencing | 167, 168, 169, 170, 171, 172, 173, 174 |
Transcriptomics
For decades, RNA analysis has been applied to study vaccine‐induced responses6 and has evolved from (quantitative) polymerase chain reaction to high‐throughput methods like microarrays and next‐generation sequencing (RNAseq). The applications range from assessing transcriptional profiles in whole tissues and blood to purified cell populations7, 8 and, more recently, even single cells.9, 10 In contrast to conventional transcriptome analysis that results in loss of tissue or cells, transcriptome in vivo analysis allows us to monitor mRNA expression in situ in live cells or animals.11 Transcriptomics of the complete genome is unbiased, relatively straightforward and is offered as a service by many contract laboratories. Gene expression levels and kinetics provide information on involvement of genes following immunization.12, 13, 14, 15 However, mRNA is often not an effector molecule in itself, but needs translation into protein and mRNA levels often do not correlate with protein content. One study reported that in mice, under those conditions, only 27% of the mRNA transcript correlates with protein expression.16 To that end, transcriptomic profiling in vaccine research serves as an excellent tool to find potential markers of vaccine‐induced responses that subsequently need to be confirmed on protein or cellular level. Novel methods such as cellular indexing of transcriptomes and epitopes by sequencing (CITE‐seq) try to overcome this translational challenge by combining in parallel the targeting of single cells with protein markers with unbiased transcriptome profiling to obtain more specific information on gene expression in cellular phenotypes than RNA‐seq in the complete tissue.17 In addition to mRNA profiles, investigating the role of non‐coding microRNAs becomes relevant given their role in immunology.18
Proteomics
Contrary to the transcriptome, the proteome encompasses the effectors that exert and control the immune response. Receptors, ligands, enzymes, hormones and even structural proteins detect foreign substances, determine the identity of cells and communicate messages between cells and tissues. Profiling the subset of proteins and peptides involved in the immune response (termed immunoproteomics)19 is important to understand how vaccines work. During the last three decades, mass spectrometry (MS) ‐based proteomics has emerged as an essential tool to profile this immunoproteome in terms of protein identification, protein dynamics and protein–protein interactions. Hunt et al.20 pioneered the MS‐based analysis of immunogenic peptides presented by molecules of the major histocompatibility complex (MHC). Since then, many studies have profiled the human leucocyte antigen (HLA) ligand repertoire using immunoprecipitation techniques and subsequent MS‐based ligand identification to find new antigen leads and to understand the concept of immunogenicity. Apart from targeting infectious diseases,21, 22, 23, 24 MHC ligandome analysis also is applied in, for example, cancer immunotherapy.25 Recent studies, however, have shown that the plethora of MHC‐presented peptides is even more complex than expected, due to the formation of non‐linear peptide sequences (i.e. splicing variants) of these T‐cell epitopes as it is estimated that one‐third of the CD8+ T‐cell epitopes is comprised of proteasome‐generated spliced epitopes.26, 27, 28, 29
In addition to profiling the T‐cell epitope repertoire, assessing the antigenic determinants that interact with B‐cells and antibodies is of high significance in elucidating immune responses in vaccine development and vaccine design.30, 31, 32 Since B‐cell epitopes are often conformational or even discontinuous, their identification is complex and currently not possible using in silico tools. With instrumental improvement of MS and the supporting software tools, MS is now used as a versatile tool to determine B‐cell epitopes, that is, the areas where antibodies bind. Opuni et al.33 comprehensively reviewed MS‐based approaches to map B‐cell epitopes. Antibody–antigen binding is just one example of a protein–protein interaction. Large‐scale interaction analysis (interactomics) is rapidly developing to identify any type of protein–protein interaction.
A new class of MS‐cleavable cross‐linking reagents has a great potential in interactomics.34, 35, 36 Like all other cross‐linking reagents, these compounds stabilize transient protein–protein interactions by the formation of covalent bonds under physiological conditions. But they also allow for the unambiguous identification of the spatially distributed peptides of the interacting proteins, based on the presence of characteristic reporter ions in the mass spectra that are representative for intramolecular or intermolecular cross‐links. Liu et al. showed a good correlation between protein three‐dimensional‐structure data obtained from chemical cross‐linking MS (XL‐MS) using MS‐cleavable reagents and data from cryo‐electron microscopy (cryo‐EM) on purified protein assemblies. In addition, they also identified in HeLa cell lysates cross‐links from protein domains lacking X‐ray or cryo‐EM three‐dimensional‐structure data,37 illustrating the great potential of this strategy in identifying protein–protein interactions. To detect protein–protein interactions by XL‐MS, it is mandatory that the amino acids in the interaction area are reactive with the cross‐linker.
Secreted proteins play an essential role in communication between immune cells. The immune secretome consists of a wide range of cytokines such as tumor necrosis factors, interferons and interleukins. Multiplex immunoassays and ELISA methods target predefined proteins from the secretome, based on cytokine‐specific antibodies. In contrast, newly developed MS‐based proteomics methods like BioOrthogonal Non‐Canonical Amino acid Tagging (BONCAT) can target the full repertoire of secreted proteins, although limited to in vitro experiments only. In a BONCAT assay, an amino acid is replaced by an azido‐containing analogue (e.g. methionine replacement by azidohomoalanine). The azido moiety allows for purification of excreted, low abundance, newly synthesized proteins from cell culture medium by click‐chemistry using alkyne‐functionalized beads.38, 39 The amino acid modification does not affect the structure and function of the protein, nor does it alter the protein synthesis rate or cell viability.40, 41 In combination with multiplexing technologies based on Tandem Mass Tagging (TMT), N,N‐dimethyl leucine (DiLeu) tagging or neutron‐encoded reagents, up to 25 samples can be multiplexed in a single run, for unbiased identification and relative quantification of de novo‐synthesized proteins in the secretome.42, 43
Limitations of MS‐based proteomics lay in the fact that, like transcriptomics, it is dependent on the availability and quality of annotated protein databases, e.g. number of annotated genes or proteins. The sensitivity of the current generation of mass spectrometers is high, but could improve further to reach the levels achieved with, for example, quantitative polymerase chain reaction. However, as described here and by others,44 MS is currently very useful for the characterization of a wide range of immunological mechanisms in the context of infection and vaccination.
Miscellaneous techniques
The Systems Vaccinology approach is not limited to the common application of transcriptomics and proteomics and novel techniques and applications are more often applied (Table 1). For example, metabolomics is a high‐throughput technology to study metabolic pathways in biological systems. The metabolome covers all small molecules and intermediates (usually < 2000 dalton (Da) molecular weight) that are highly dynamic in the catabolic and anabolic processes of cells. In many immunological studies, metabolomics is now an integral part of system vaccinology, to predict biomarkers and to evaluate vaccine efficacy.45, 46 Untargeted metabolome analysis requires many different sample preparation procedures and analytical tools to identify all metabolites. Gas chromatography, liquid chromatography, capillary electrophoresis, MS, Fourier transform infrared spectroscopy and nuclear magnetic resonance are frequently used or combined to study the metabolome, although with limited precision and accuracy. Improved accuracy, precision and specificity of these assays can be obtained by using semi‐targeted or targeted metabolomics strategies if the chemical identities of the analytes are known beforehand.47
Humoral and cell‐mediated responses
In the context of systems vaccinology, conventional assays that are neither high throughput, nor unbiased, remain necessary to confirm hypotheses. Histology is still the common method to confirm the influx and the location of immune cells in tissues as a result of an infection or immunization. However, visualization of cells in tissue sections continually improves by better immunoimaging techniques48 such as confocal microscopy. Ex vivo, assays such as ELISA and flow cytometry are applied to identify and quantify specific cell subsets as well as their activation state and function (such as cytokine production or proliferation). These immunological techniques for studying these responses have also become more powerful. Flow cytometry equipment has improved, for example, by combining the measurement of the amount of proteins per cell (flow cytometry), with visualization of their localization within the cells (microscopy), so‐called imaging flow cytometry.49 Also, more fluorescent labels have become available, increasing the number of markers that can be investigated simultaneously. Multi‐parameter flow cytometry enables the identification of these specialized, often rare, subsets of cells.50 For example, innate cells that play a crucial role in pathogen recognition and in influencing the magnitude and direction of adaptive immunity, such as specific subtypes of antigen‐presenting cells and innate lymphoid cells. With respect to adaptive immune responses, the ability to identify rare subsets mainly focuses on antigen‐specific B‐cells51 and T‐cells,52 both circulating and tissue‐resident cells, and precursors as well as effector and memory cells.7, 8 Cytometry by Time‐of‐Flight mass spectrometry (CyTOF)53 uses rare‐metal isotopes instead of fluorescent labels for labeling, thereby almost abolishing the spectral overlap seen with fluorescent dyes and expanding the number markers that can be measured simultaneously even further. For example, Van Unen et al.54 identified 142 immune subsets in the human mucosal immune system.
In addition, the opportunity of sorting specific subsets of cells not only as a population, but also as single cells, in combination with high‐throughput analysis of the isolated cells, added yet another dimension. Lymphocyte epitope specificity necessarily is extremely diverse to fight the wide array of possible pathogens and is accomplished by randomness through several recombinatorial events and additional random changes during lymphocyte ontogeny. For example, the number of different antibodies human B‐cells can produce, outnumbers the B lymphocytes (~1011) in the human body.55 High‐throughput single‐cell analysis enabled detailed interrogation of these broad B‐cell and T‐cell repertoires.56 These techniques also revealed that priming might not always be predictive, as preferential expansion of B‐cells and T‐cells also plays an important role.57 This illustrates that enhanced understanding of lymphocyte responses not only leads to identification of novel early markers for efficacy of vaccines, but is also useful for their evaluation. Hypotheses with respect to B‐cell and T‐cell epitopes can be derived from MS‐based studies as described in the former paragraph. Moreover, current knowledge with respect to HLA specificity has also enabled the prediction of T‐cell epitopes in silico, decreasing the time needed for analysis.58 As B‐cell epitopes are often conformational, current knowledge does not yet enable in silico predictions to the same extent as for T‐cells. For B‐cell epitope analysis of complex multi‐protein vaccines, antibody–antigen binding can be investigated using two‐dimensional gel electrophoresis with Western blotting to pinpoint immunogenic antigens.59 Subsequently, peptide arrays can be applied to investigating the immunodominant epitopes of these discovered antigens.60
The ability to isolate single antibody‐producing B‐cells and subsequent DNA sequencing of immunoglobulin genes in high‐throughput settings (Ig‐seq) has greatly impacted the depth of understanding with respect to the antibody repertoire.55 Single‐cell sorting in combination with B‐cell cloning has enabled the isolation of specific antibodies able to neutralize pathogens, including rare antigens with characteristics that are of special interest, such as antibodies that are broadly cross‐reactive among highly variable viruses by binding to conserved regions. This was, for example, found for influenza hemagglutinin (HA) subtypes upon infection as well as vaccination.61, 62, 63, 64 In general, this has shifted the focus from the most immunogenic epitopes to the more conserved regions. However, it has proved challenging to design vaccines that induce strong responses against these regions. Challenges with respect to design of vaccines inducing such broadly neutralizing antibodies for HIV and influenza virus were extensively reviewed by Corti and Lanzavecchia.65 For example, other regions might need to be removed or de‐immunized and the conserved epitopes might not be easily accessible, preventing the vaccine‐induced antibodies from binding to the natural target. Another point of attention is that a focus on increased immunogenicity should not lead to a decrease in safety. For example, antibodies with long or charged CDR‐H3 regions enable binding to occluded sites on pathogens and mediate pathogen neutralization but are also more likely to be autoreactive.55
IgG antibody levels have served as correlates of protection for traditional vaccines,66 but in the context of systems vaccinology, not only antibody quantity, but also functionality is increasingly investigated. Antibodies may inhibit other functional antibodies or enhance disease by facilitating virus entry into cells. However, non‐neutralizing antibodies may still be functional and could have more impact on protection from disease than neutralizing antibodies, for instance by interaction of the antibody constant domain with Fc receptors. This interaction is determined by two characteristics, antibody subclass (irreversible) and antibody glycosylation (fast and reversible) that can for example impact the affinity for Fc receptors.67 The glycosylation profile of antigen‐specific antibodies is changed upon vaccination, whereas the glycosylation profile of total IgG is not affected. Interestingly, despite very different pre‐vaccination glycosylation levels in populations in different parts of the world,68 profiles induced by vaccination are similar, as was shown after influenza vaccination.69
However, different vaccine formulations can induce different glycosylation profiles.68 This is likely also influenced by the adjuvant present in the formulation, either directly or via cytokine induction.70 Though the biological implications of these differences are not yet clear, the recent developments in systems serology demonstrate that multiple parameters of the antibody response, such as IgG subtype and antibody glycosylation among others, influence antibody functionally and therefore vaccine efficacy.71, 72, 73
Data analysis and bioinformatics
One of the difficulties in systems approaches is the amount of data. Statistical evaluation, visualization and extracting meaningful conclusions are major challenges and require multiple tools and expertises.74, 75 New technologies in systems vaccinology such as metabolomics may require different statistical and bioinformatic analysis strategies compared with transcriptomics and proteomics data sets, as described by Ren et al.76 Visualization of data from full genomes or proteomes was initially performed with tools applying e.g. heatmaps, Venn diagrams and network analysis, e.g. with Cytoscape.77 These allowed visualization of numerous individual data points or groups, but also the clustering of related data points. Additionally, the generation of multiple systems‐scale data sets enables the comparison of immune responses induced by different vaccines in an unbiased manner.78, 79 Hierarchical Stochastic Neighborhood Embedding (HSNE) enables us to visualize data of cellular composition of millions of cells in detail up to the single‐cell level.80 Although these graphs provide an easier perspective on trends in the data sets, they are not designed to provide information on the biological function of individual data points. The development of data mining tools (Fig. 1) has given scientists a handle to translate their data sets into the description of relevant biological processes.81 This allows for investigation of the involvement and mutual interaction of pathways, processes and cell types. Tools that are focused on pathway analysis include DAVID,82 Pathway‐Express,83 Gene Onthology (GO)84 and the Kyoto Encyclopedia of Genes and Genomes (KEGG).85 With BioGPS, the involvement of cells or the different stages in cell activation can be investigated.86 This has also led to the development of specialized databases for specific research topics. For example, InnateDB contains data on innate immune responses87 while INTERFEROME focuses on interferon‐related processes88 and the CEMiTool can be used for co‐expression analyses and discovery of functionality of genes.89 For the discovery and analysis of the repertoire of T‐ and B‐cell receptor sequences, VDJviz can be applied for the routine analysis and quality control of sequencing of immune repertoires90 and VDJdb is designed for the annotation of T‐cell receptor repertoire data.91 These databases evolve and expand continuously based on novel data, scientific insights and technologies. Depending on the choice of data mining tools, the outcome of results might differ. However, within the Human Vaccines Project,92 researchers demonstrated that there was no difference in the outcome of their data when it was either analyzed using a tool based on prior knowledge, in this case the NetAnalyst platform, or with an unbiased tool, DIABLO.
In addition, to generate knowledge and understanding, large data sets are used in computational systems biology studies to predict host immune responses.75, 93 From the antigen point of view, machine‐learning tools are widely applied to predict B‐cell94 and T‐cell epitopes95 and novel tools such as the IL17eScan96 enable us to predict the capacity of epitopes to induce interleukin‐17. All these data can be gathered in silico prior to or simultaneous with laboratory studies to confirm the role of host biomarkers and antigen epitopes in vaccine responses.
Application of systems vaccinology in vaccine research
In each phase of vaccine development (Fig. 2), systems vaccinology techniques can be applied to improve the quality of the vaccine development process as well as decreasing the risk of late‐stage failure as a result of increased knowledge (Table 2). Apart from host–vaccine responses, a systems approach can also provide essential information on the vaccine itself, more specifically the antigen composition. Most vaccines belong to one of four groups: (i) live‐attenuated, (ii) whole inactivated, (iii) subunit and (iv) conjugate vaccines. Detailed vaccine characterization is mainly of interest for complex multi‐protein vaccines such as live‐attenuated and inactivated (bacterial) vaccines if for instance batch consistency must be proven.
Figure 2.
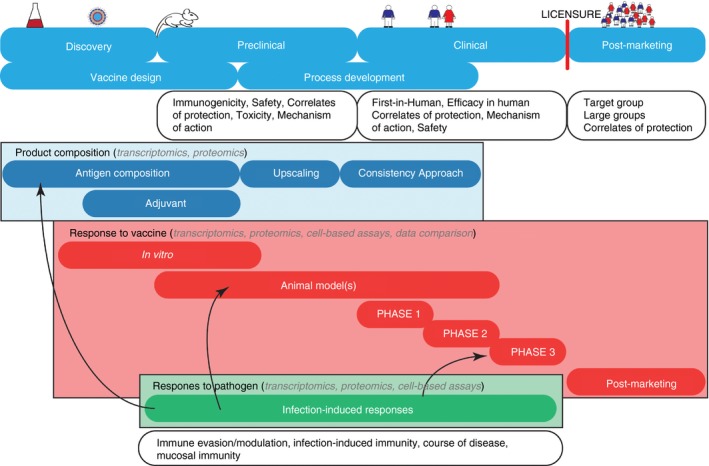
Vaccine development and application of systems vaccinology. Systems vaccinology can be applied to all phases implementing different techniques (e.g. transcriptomics, proteomics, cell‐based assays) to provide detailed insight in different research objectives, such as composition, immunogenicity and safety of the vaccine. The host–pathogen responses are not essential for vaccine development but since protection after infection is often superior in terms of efficacy – but not safety – this knowledge can be useful throughout the vaccine development chain, as illustrated by arrows.
Table 2.
Applications of systems vaccinology
| Aim, deliverables | Benefits | Examples |
|---|---|---|
| Predicting responses | Clinical development, post‐marketing surveillance | 2, 3, 14, 120, 121, 122, 123, 124, 125, 126, 129, 175 |
| Understand mode of action of vaccines | Risk mitigation: less late‐stage failure | 127, 129, 176 |
| Identify universal vaccine signatures | Improve vaccine development | 79, 118, 133 |
| Select new immune modulators and delivery systems |
Risk mitigation: less early‐stage failure Better vaccines |
13, 111, 113, 128 |
| Assess vaccine safety and adverse effects | Clinical development, post‐marketing surveillance | 117, 118, 119 |
| Understand mode of action of infection | Host–pathogen interaction | 100, 102, 104, 105 |
| Rational vaccine design |
Risk mitigation: less early‐stage failure Facilitates regulatory acceptance |
N.A. |
| Development of animal models |
Improved early development Risk mitigation: facilitates preclinical to clinical decision. |
118 |
Exploratory studies in vaccine development
During the discovery phase a systems approach can be implemented to investigate vaccine composition as is especially interesting for complex multi‐protein vaccines (Fig. 2). For example, transcriptomics and/or proteomics approaches can be applied during strain generation for an inactivated vaccine to provide detailed information on protein composition and, more specifically, the proportion of immunogenic proteins. These techniques can subsequently be used for process development to verify whether or not the product is similar when produced at a larger scale. This was, for instance, demonstrated for pertussis whole‐cell vaccines.97 Information on the relationship between cultivation conditions and the composition of the final vaccine, considerably enhances the process of optimizing vaccine efficacy. In addition, it reveals key process parameters that can subsequently be tightly monitored, making the process more consistent and thereby eventual vaccine release easier.
To study the vaccine responses in the discovery phase, systems vaccinology methods can be used to study the mechanism of action of vaccines or adjuvants, both in vitro as well as in experimental animals (Fig. 2). In vitro cell‐based assays allow the gain of information on the magnitude and direction of the innate response, but they lack the possibility to study coherent interactions between innate and adaptive immune responses. Especially in the transition from innate to adaptive responses, a lot of immunological information is still missing.
While the main focus in systems vaccinology is on vaccine‐induced responses, pathogen‐induced responses during infection are also of great interest, as these can enable the discovery of potential markers of, or mechanisms involved in, protective immunity, or unwanted immunological effects such as immune evasion (Fig. 2). Systems biology can assist to investigate host–pathogen interactions by looking at pathogen behavior and clearance after infection. A natural infection sometimes induces effective protection, e.g. mumps, that may serve as a benchmark for a vaccine, whereas other pathogens may display immune evasive or suppressive responses, which one would like to avoid in a vaccine‐induced response. A systems‐based approach was for instance used to investigate infection‐induced responses during sepsis.98 In terms of vaccine‐preventable diseases, investigators have looked at infection‐induced responses for pathogens such as RSV,99, 100, 101 M. tuberculosis,102 B. pertussis 12, 78, 103 and influenza.104, 105 The course of a B. pertussis infection was investigated in mice to unravel molecular and cellular signatures of the effective infection‐induced immunity.12, 106 Results indicated that a B. pertussis infection induced T helper type 17 (Th17) cells and mucosal IgA responses that were preceded by a prolonged acute‐phase response, broad pathogen recognition and early gene signatures of subsequent T‐cell recruitment in the lungs.12 In addition, the infection led to enhanced activation of pulmonary innate immune cells and markers indicating recruitment of CCR6+ B‐cells and Th17 cells.106 These promising markers of infection‐induced responses should ideally be confirmed in human B. pertussis challenge models.107 In a comparative network analysis of two data sets, the pulmonary molecular signatures that preceded the effector cells in B. pertussis infection‐induced‐immunity12 were compared with signatures induced upon pulmonary immunization with a novel Outer Membrane Vesicle Pertussis vaccine (omvPV).15 Network analysis demonstrated that similar genes and pathways were involved in both responses but that the intensity and kinetics between infection‐induced and vaccine‐induced responses were different. Both responses did, however, result in induction of mucosal immunity and better protection compared with subcutaneous vaccine administration.
Systems approaches have also been used to investigate innate responses, for example responses induced by adjuvants.108 In vitro studies were performed to investigate monocyte‐specific responses induced by alum salts using a proteomics approach109 and to study subset‐specific dendritic cell responses following lipopolysaccharide stimulation using a transcriptomics approach.110 These extensive in vitro studies provide insight into novel markers, pathways and cell subsets activated by adjuvants. Yet the complexity of cell‐to‐cell interaction during immune responses requires in vivo studies. This was also addressed by Mosca et al.,13 who conducted one of the first systems‐based studies in mice for better understanding of adjuvants, such as MF59, alum, CpG ODN and α‐GalCer. Adjuvant‐induced responses were profiled in muscle at the site of injection. This provided comparative information on plain adjuvant responses such as the fact that CpG elicited stronger systemic innate responses compared with MF59 and alum. Others have studied the responses of adjuvants in combination with antigen, that might also influence the immune response. Olafsdottir et al.111 determined molecular signatures induced in mice of four clinically tested vaccine adjuvants (CAF01, IC31 GLA‐SE and Alum) using a tuberculosis vaccine candidate (H56) as model antigen. With a transcriptomics analysis, signatures of T follicular helper (Tfh) and germinal center (GC) B‐cell responses were found to be enhanced in adjuvated vaccines compared with those of the H56 antigen alone. Using a similar combination of H56 adjuvanted with CAF01, Santoro et al.112 applied systems vaccinology to demonstrate that recall innate responses following boosting were modulated by the use of adjuvant in the primary immunization. Lindqvist et al.113 focused on specifying the molecular signatures in mice induced by mucosal adjuvants for vaccination against sexually transmitted infections.
Relevant animal models to study infection are not always available. In such cases, the application of systems approaches in vaccination studies can provide valuable information on the mechanism of action of candidate vaccines. Gómez et al. unraveled transcriptomic activation markers of human myeloid and plasmacytoid dendritic cells upon stimulation with a vaccine candidate against HIV/AIDS, based on an MVA‐based vector. Subsequently, HIV‐1‐specific CD4+ and CD8+ T‐cell memory responses were demonstrated in vivo.114 Strouts et al.115 were able to link early specific gene transcripts related to T‐cell activation and type I interferon response to the magnitude of neutralizing antibody titers after immunization with a live‐attenuated tetravalent dengue vaccine in non‐human primates. As part of the development of a novel Outer Membrane Vesicle Pertussis vaccine (omvPV), vaccine‐induced responses were investigated in mice. An extensive proteomics approach for antibody profiling was used to determine the distinct antibody specificity and subclass distribution induced by different pertussis vaccines.59 Transcriptome and cytokine profiling demonstrated reduced pro‐inflammatory responses, potentially leading to less adverse effects by the omvPV compared with a classic whole‐cell pertussis vaccine while maintaining the vaccine efficacy.116 To investigate the effect of the route of immunization on protective immunity, the responses induced by subcutaneous and pulmonary administration of omvPV in mice were compared.15 The study demonstrated improved mucosal immunity, and therefore protection, against B. pertussis after pulmonary immunization. Finally, in terms of vaccine safety, Mizukami et al.117 applied the systems‐approach in a rat model to predict safety and batch‐to‐batch consistency of influenza vaccines. Wang et al.118 investigated blood gene signatures in non‐human primates immunized with seven marketed or experimental vaccines to isolate markers of vaccine efficacy and safety. Whereas overlapping trends in markers of efficacy were found between some vaccines, no markers were found correlating to adverse effects.
Clinical studies
Predictive markers that have been found in the exploratory studies (described in the previous section) can be applied in subsequent clinical studies. Moreover, systems vaccinology analyses are also useful in a clinical study itself to obtain better insight into safety and efficacy (Fig. 2). However, to our knowledge in only a few clinical studies has systems vaccinology been performed to assess signatures that correlate to adverse effects, such as fever and seizures. In the public–private project BIOVACSAFE, systems approaches are applied to investigate markers correlating to vaccine safety in human and animal models potentially providing novel markers.119 Investigators have conducted systems‐based studies in humans investigating vaccine candidates against Ebola (rVSV‐ZEBOV),120 HIV‐1,121 malaria (RTS,S)122, 123 and M. tuberculosis (M72/AS01).124 For Ebola (rVSV‐ZEBOV), IP‐10 and CXCR6+ natural killer (NK) cells were independently correlating with antibody titers, suggesting that IP‐10 could be a potential target to influence vaccine‐induced responses.120 Anderson et al.121 investigated transcriptome profiles in the blood of healthy volunteers following immunization with a novel HIV‐1 vaccine adjuvanted with a Toll‐like receptor 4 agonist. High responders, based on serum antibodies, contained modules of genes expressed in NK cells, whereas modules of genes related to myeloid cells, monocytes and integrin cell surface interactions were detected in low responders. These signatures enable vaccine responses to be distinguished in an early phase and perhaps even allows steering of immune responses in low responders by adjustment of the vaccine. In a phase I randomized controlled trial using AS03‐adjuvanted and unadjuvanted inactivated split‐virus H5N1 influenza vaccines, the effect of adjuvant on vaccine‐induced responses, such as serum cytokines, antibody titers, and gene expression levels, was investigated. This led to novel insights into antigen processing, presentation markers in neutrophils and revealed the correlation of serum IP‐10 levels with proliferation of NK cells.125 Interestingly, for the prediction of protection induced by malaria (RTS,S) immunization, NK cells correlated negatively whereas positive correlations were detected for molecular signatures of B‐cells and plasma cells122 and the nuclear factor‐κB and interferon‐γ pathways.123 These findings indicate that different vaccine–adjuvant combinations may induce different immune (non)effective responses.
Post‐marketing studies
After vaccines have been licensed and introduced on the market, post‐marketing surveillance studies are performed to monitor vaccine safety and efficacy in larger populations (Fig. 2). At this point in the vaccine life cycle, correlates of protection, or surrogates thereof, are sometimes known, however the mechanism of action is often underexposed. Systems approaches in this phase are not routinely applied, as far as we know. The validation of the methods is complex and expensive, with the added value not yet proven convincingly. However, pioneering studies have demonstrated that vaccine efficacy after yellow fever and influenza vaccination could be predicted in an early stage after immunization by analyzing molecular signatures.14, 126, 127 For the yellow fever vaccine, the enhanced levels of specific gene expression profiles in blood obtained 1 to 7 days after vaccination were predictive for antigen‐specific CD8+ T‐cell levels and antibody titers usually obtained 60 days after immunization, thus inducing a significant time advantage. In a clinical phase, regulators might want to see positive classical end‐points but for post‐marketing purposes this approach of looking at early correlates may be valuable. Furthermore, these studies are of value in comparative studies of different vaccine formulations. Nakaya et al.128 compared two different vaccine formulations, trivalent inactivated influenza vaccine (TIV) and MF59‐adjuvanted TIV (ATIV), in children and predicted influenza antibody titers 1 month after vaccination with a seasonal influenza vaccine across five consecutive seasons by analyzing specific signatures of innate immunity and plasma blasts.129 In addition, a systems biology approach could also be applied to investigate the differences in vaccine responsiveness in different target groups. For example, researchers predicted age‐related vaccine hypo‐responses against a hepatitis B vaccine in the elderly with a gene marker130 and exposed transcriptomic signatures of vaccine‐induced immunity, both cellular and humoral immune responses, after seasonal influenza vaccination specifically in older adults.131 Obviously, these post‐marketing surveillance studies facilitate further clinical development of improved vaccines or provide insight into general mechanisms that will enhance future vaccine development, even if correlates of protection are unknown.132
Finally, a great benefit of systems vaccinology studies is the ability to combine data sets from vaccine responses against different pathogens in integrative network modeling to reveal detailed insight into universal signatures of vaccine responsiveness.78, 79, 118, 133 Li et al.79 compared molecular signatures, induced by five different human vaccines against specific bacterial or viral infections, which could predict antibody responses. This indicates that investigating responses against a single pathogen can lead to universal markers that serve as benchmark for vaccine development against new emerging diseases. This also supports the desire for data obtained in systems‐approaches to be publicly available (open access) for future large meta‐analyses.
Future outlook
Systems vaccinology has been implemented in the different phases of the vaccine development chain and has become a relevant approach in the field of vaccinology. The recent progression in systems vaccinology has led to successful initiatives such as the ADITEC project134 that provided a novel gene expression method to analyze biomarkers of tuberculosis pathogenesis135 as well as a study where safety and immunogenicity of a novel vaccine against tuberculosis (VPM1002) were evaluated.136 To achieve maximum vaccine efficacy and safety, future vaccine research will have to focus on target groups such as neonates137, 138 or even on the development of personalized vaccines.139 Systems vaccinology approaches can be further applied to investigate the inter‐group differences in vaccine responsiveness between healthy adults and specific target groups such as infants, and elderly or immune‐deficient individuals.140
Investigating vaccine‐induced responses with systems vaccinology has led to a trend of gathering massive amounts of data going as deep as investigating every molecule in single cells. Acquiring large data sets is far easier than extracting novel relevant information from those data. Furthermore, the amount of data is constantly increasing, especially when combining data sets of different origin (genes versus proteins versus cells). This requires equally advanced tools for (i) data visualization and (ii) translation of content to knowledge and understanding, forcing a dialogue between vaccine developers and bioinformatics experts. In the future, implementation of artificial intelligence could assist in data processing and for data visualization online publications may become interactive allowing readers to scroll through, for example, the immune responses induced by immunization over time.
In summary, systems vaccinology is becoming a valuable tool in each phase of the vaccine development chain. The link with predicting immune responses beforehand is, however, still difficult. Small but important steps are currently taken towards application of the insights created by systems vaccinology. Recent studies show that the application of systems vaccinology is especially interesting for investigating complex or dangerous pathogens such as Ebola virus, HIV, malaria‐causing Plasmodium spp. and M. tuberculosis where phase 3 studies are challenging or even impossible,120, 121, 122, 123, 124 or even for investigating the heterologous or non‐specific effects that vaccines induce against other diseases.141 These promising changes call for the inclusion of systems vaccinology as early as possible in the vaccine development chain to better understand why some vaccines work and others do not. This will enable efficiency of vaccine development proportionally in the design phase and will lead to improved vaccine evaluation in early phases, thereby reducing time and costs.
Author contribution
R.R., E.R., H.M., B.M. and G.K. wrote the review.
Disclosures
The authors declare no competing interest.
References
- 1. Wack A, Rappuoli R. Vaccinology at the beginning of the 21st century. Curr Opin Immunol 2005; 17:411–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oberg AL, Kennedy RB, Li P, Ovsyannikova IG, Poland GA. Systems biology approaches to new vaccine development. Curr Opin Immunol 2011; 23:436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity 2010; 33:516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aderem A. Systems biology: its practice and challenges. Cell 2005; 121:511–3. [DOI] [PubMed] [Google Scholar]
- 5. Rappuoli R. Reverse vaccinology. Curr Opin Microbiol 2000; 3:445–50. [DOI] [PubMed] [Google Scholar]
- 6. Flanagan KL, Noho‐Konteh F, Ghazal P, Dickinson P. Transcriptional profiling technology for studying vaccine responses: an untapped goldmine. Methods 2013; 60:269–74. [DOI] [PubMed] [Google Scholar]
- 7. Touzot M, Dahirel A, Cappuccio A, Segura E, Hupe P, Soumelis V. Using transcriptional signatures to assess immune cell function: from basic mechanisms to immune‐related disease. J Mol Biol 2015; 427:3356–67. [DOI] [PubMed] [Google Scholar]
- 8. Brummelman J, Raeven RHM, Helm K, Pennings JL, Metz B, van Eden W et al Transcriptome signature for dampened Th2 dominance in acellular pertussis vaccine‐induced CD4(+) T cell responses through TLR4 ligation. Sci Rep 2016; 6:25064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldstein LD, Chen YJ, Dunne J, Mir A, Hubschle H, Guillory J et al Massively parallel nanowell‐based single‐cell gene expression profiling. BMC Genom 2017; 18:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stubbington MJT, Rozenblatt‐Rosen O, Regev A, Teichmann SA. Single‐cell transcriptomics to explore the immune system in health and disease. Science 2017; 358:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, Fisher S et al Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods 2014; 11:190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raeven RHM, Brummelman J, Pennings JL, Nijst OE, Kuipers B, Blok LE et al Molecular signatures of the evolving immune response in mice following a bordetella pertussis infection. PLoS ONE 2014; 9:e104548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C et al Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A 2008; 105:10501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie‐Kunze S, Haining WN et al Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 2011; 12:786–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raeven RHM, Brummelman J, Pennings J, van der Maas L, Helm K, Tilstra W et al Molecular and cellular signatures underlying superior immunity against Bordetella pertussis upon pulmonary vaccination. Mucosal Immunol 2018; 11:979–93. [DOI] [PubMed] [Google Scholar]
- 16. Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN et al Comparative analysis of proteome and transcriptome variation in mouse. PLoS Genet 2011; 7:e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stoeckius M, Hafemeister C, Stephenson W, Houck‐Loomis B, Chattopadhyay PK, Swerdlow H et al Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017; 14:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post‐transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9:102–14. [DOI] [PubMed] [Google Scholar]
- 19. Purcell AW, Gorman JJ. Immunoproteomics: Mass spectrometry‐based methods to study the targets of the immune response. Mol Cell Proteomics 2004; 3:193–208. [DOI] [PubMed] [Google Scholar]
- 20. Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N et al Characterization of peptides bound to the class I MHC molecule HLA‐A2.1 by mass spectrometry. Science 1992; 255:1261–3. [DOI] [PubMed] [Google Scholar]
- 21. Schellens IM, Meiring HD, Hoof I, Spijkers SN, Poelen MC, van Gaans‐van den Brink JA et al Measles virus epitope presentation by HLA: novel insights into epitope selection, dominance, and microvariation. Front Immunol 2015; 6:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spencer CT, Bezbradica JS, Ramos MG, Arico CD, Conant SB, Gilchuk P et al Viral infection causes a shift in the self peptide repertoire presented by human MHC class I molecules. Proteomics Clin Appl 2015; 9:1035–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marcilla M, Alvarez I, Ramos‐Fernandez A, Lombardia M, Paradela A, Albar JP. Comparative analysis of the endogenous peptidomes displayed by HLA‐B*27 and Mamu‐B*08: two MHC class I alleles associated with elite control of HIV/SIV infection. J Proteome Res 2016; 15:1059–69. [DOI] [PubMed] [Google Scholar]
- 24. Draheim M, Wlodarczyk MF, Crozat K, Saliou JM, Alayi TD, Tomavo S et al Profiling MHC II immunopeptidome of blood‐stage malaria reveals that cDC1 control the functionality of parasite‐specific CD4 T cells. EMBO Mol Med 2017; 9:1605–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Creech AL, Ting YS, Goulding SP, Sauld JFK, Barthelme D, Rooney MS et al The role of mass spectrometry and proteogenomics in the advancement of HLA epitope prediction. Proteomics 2018; 18:e1700259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ebstein F, Textoris‐Taube K, Keller C, Golnik R, Vigneron N, Van den Eynde BJ et al Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non‐spliced epitopes. Sci Rep 2016; 6:24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lim WC, Khan AM. Mapping HLA‐A2, ‐A3 and ‐B7 supertype‐restricted T‐cell epitopes in the ebolavirus proteome. BMC Genom 2018; 19(Suppl 1):42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berkers CR, de Jong A, Schuurman KG, Linnemann C, Meiring HD, Janssen L et al Definition of proteasomal peptide splicing rules for high‐efficiency spliced peptide presentation by MHC class I molecules. J Immunol 2015; 195:4085–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liepe J, Marino F, Sidney J, Jeko A, Bunting DE, Sette A et al A large fraction of HLA class I ligands are proteasome‐generated spliced peptides. Science 2016; 354:354–8. [DOI] [PubMed] [Google Scholar]
- 30. Galassie AC, Link AJ. Proteomic contributions to our understanding of vaccine and immune responses. Proteomics Clin Appl 2015; 9:972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donnarumma D, Faleri A, Costantino P, Rappuoli R, Norais N. The role of structural proteomics in vaccine development: recent advances and future prospects. Expert Rev Proteomics 2016; 13:55–68. [DOI] [PubMed] [Google Scholar]
- 32. Guy AJ, Irani V, Beeson JG, Webb B, Sali A, Richards JS et al Proteome‐wide mapping of immune features onto Plasmodium protein three‐dimensional structures. Sci Rep 2018; 8:4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Opuni KFM, Al‐Majdoub M, Yefremova Y, El‐Kased RF, Koy C, Glocker MO. Mass spectrometric epitope mapping. Mass Spectrom Rev 2018; 37:229–41. [DOI] [PubMed] [Google Scholar]
- 34. Kao A, Chiu CL, Vellucci D, Yang Y, Patel VR, Guan S et al Development of a novel cross‐linking strategy for fast and accurate identification of cross‐linked peptides of protein complexes. Mol Cell Proteomics 2011; 10:M110 002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barysz HM, Malmstroem J. Development of large‐scale cross‐linking mass spectrometry. Mol Cell Proteomics 2017; 17:1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sinz A. Divide and conquer: cleavable cross‐linkers to study protein conformation and protein–protein interactions. Anal Bioanal Chem 2017; 409:33–44. [DOI] [PubMed] [Google Scholar]
- 37. Liu F, Rijkers DT, Post H, Heck AJ. Proteome‐wide profiling of protein assemblies by cross‐linking mass spectrometry. Nat Methods 2015; 12:1179–84. [DOI] [PubMed] [Google Scholar]
- 38. Nessen MA, Kramer G, Back J, Baskin JM, Smeenk LE, de Koning LJ et al Selective enrichment of azide‐containing peptides from complex mixtures. J Proteome Res 2009; 8:3702–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eichelbaum K, Winter M, Berriel Diaz M, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol 2012; 30:984–90. [DOI] [PubMed] [Google Scholar]
- 40. Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc Natl Acad Sci U S A 2006; 103:9482–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eichelbaum K, Krijgsveld J. Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol Cell Proteomics 2014; 13:792–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frost DC, Greer T, Li L. High‐resolution enabled 12‐plex DiLeu isobaric tags for quantitative proteomics. Anal Chem 2015; 87:1646–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Potts GK, Voigt EA, Bailey DJ, Rose CM, Westphall MS, Hebert AS et al Neucode labels for multiplexed, absolute protein quantification. Anal Chem 2016; 88:3295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nyman TA, Lorey MB, Cypryk W, Matikainen S. Mass spectrometry‐based proteomic exploration of the human immune system: focus on the inflammasome, global protein secretion, and T cells. Expert Rev Proteomics 2017; 14:395–407. [DOI] [PubMed] [Google Scholar]
- 45. Hagan T, Pulendran B. Will systems biology deliver its promise and contribute to the development of new or improved vaccines? From data to understanding through systems biology. Cold Spring Harb Perspect Biol 2018; 10:pii: a028894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS et al Metabolic phenotypes of response to vaccination in humans. Cell 2017; 169:862–77. e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dunn WB, Broadhurst DI, Atherton HJ, Goodacre R, Griffin JL. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 2011; 40:387–426. [DOI] [PubMed] [Google Scholar]
- 48. Bousso P, Moreau HD. Functional immunoimaging: the revolution continues. Nat Rev Immunol 2012; 12:858–64. [DOI] [PubMed] [Google Scholar]
- 49. Chattopadhyay PK, Gierahn TM, Roederer M, Love JC. Single‐cell technologies for monitoring immune systems. Nat Immunol 2014; 15:128–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Proserpio V, Lonnberg T. Single‐cell technologies are revolutionizing the approach to rare cells. Immunol Cell Biol 2016; 94:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J Jr. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B‐cell differentiation. Blood 2003; 102:592–600. [DOI] [PubMed] [Google Scholar]
- 52. Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O'Rourke P, de Silva AD et al Precursors of human CD4+ cytotoxic T lymphocytes identified by single‐cell transcriptome analysis. Sci Immunol 2018; 3:pii: eaan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brodie TM, Tosevski V. Broad immune monitoring and profiling of T cell subsets with mass cytometry. Methods Mol Biol 2018; 1745:67–82. [DOI] [PubMed] [Google Scholar]
- 54. van Unen V, Li N, Molendijk I, Temurhan M, Hollt T, van der Meulen‐de Jong AE et al Mass cytometry of the human mucosal immune system identifies tissue‐ and disease‐associated immune subsets. Immunity 2016; 44:1227–39. [DOI] [PubMed] [Google Scholar]
- 55. Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high‐throughput sequencing of the antibody repertoire. Nat Biotechnol 2014; 32:158–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Corti D, Sallusto F, Lanzavecchia A. High throughput cellular screens to interrogate the human T and B cell repertoires. Curr Opin Immunol 2011; 23:430–5. [DOI] [PubMed] [Google Scholar]
- 57. Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A et al T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 2015; 347:400–6. [DOI] [PubMed] [Google Scholar]
- 58. Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS. In silico‐accelerated identification of conserved and immunogenic variola/vaccinia T‐cell epitopes. Vaccine 2009; 27:6471–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Raeven RHM, van der Maas L, Tilstra W, Uittenbogaard JP, Bindels TH, Kuipers B et al Immunoproteomic profiling of Bordetella pertussis outer membrane vesicle vaccine reveals broad and balanced humoral immunogenicity. J Proteome Res 2015; 14:2929–42. [DOI] [PubMed] [Google Scholar]
- 60. Legutki JB, Johnston SA. Immunosignatures can predict vaccine efficacy. Proc Natl Acad Sci U S A 2013; 110:18614–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M et al Broadly cross‐reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 2011; 208:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY et al Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross‐reactive memory B cells. Proc Natl Acad Sci U S A 2012; 109:9047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D et al A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850–6. [DOI] [PubMed] [Google Scholar]
- 64. Pappas L, Foglierini M, Piccoli L, Kallewaard NL, Turrini F, Silacci C et al Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014; 516:418–22. [DOI] [PubMed] [Google Scholar]
- 65. Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 2013; 31:705–42. [DOI] [PubMed] [Google Scholar]
- 66. Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol 2010; 17:1055–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol 2018; 18:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW et al Antigen‐specific antibody glycosylation is regulated via vaccination. PLoS Pathog 2016; 12:e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Selman MH, de Jong SE, Soonawala D, Kroon FP, Adegnika AA, Deelder AM et al Changes in antigen‐specific IgG1 Fc N‐glycosylation upon influenza and tetanus vaccination. Mol Cell Proteomics 2012; 11:M111 014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang J, Balog CI, Stavenhagen K, Koeleman CA, Scherer HU, Selman MH et al Fc‐glycosylation of IgG1 is modulated by B‐cell stimuli. Mol Cell Proteomics 2011; 10:M110 004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ackerman ME, Barouch DH, Alter G. Systems serology for evaluation of HIV vaccine trials. Immunol Rev 2017; 275:262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Arnold KB, Chung AW. Prospects from systems serology research. Immunology 2018; 153:279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ et al Dissecting polyclonal vaccine‐induced humoral immunity against HIV using systems serology. Cell 2015; 163:988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Weiner J 3rd, Kaufmann SH, Maertzdorf J. High‐throughput data analysis and data integration for vaccine trials. Vaccine 2015; 33:5249–55. [DOI] [PubMed] [Google Scholar]
- 75. Nakaya HI, Li S, Pulendran B. Systems vaccinology: learning to compute the behavior of vaccine induced immunity. Wiley Interdiscip Rev Syst Biol Med 2012; 4:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ren S, Hinzman AA, Kang EL, Szczesniak RD, Lu LJ. Computational and statistical analysis of metabolomics data. Metabolomics 2015; 11:1492–513. [Google Scholar]
- 77. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Raeven RHM, Pennings JL, van Riet E, Kersten GF, Metz B. Meta‐Analysis of pulmonary transcriptomes from differently primed mice identifies molecular signatures to differentiate immune responses following bordetella pertussis challenge. J Immunol Res 2017; 2017:8512847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li S, Rouphael N, Duraisingham S, Romero‐Steiner S, Presnell S, Davis C et al Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 2014; 15:195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. van Unen V, Hollt T, Pezzotti N, Li N, Reinders MJT, Eisemann E et al Visual analysis of mass cytometry data by hierarchical stochastic neighbour embedding reveals rare cell types. Nat Commun 2017; 8:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kidd BA, Peters LA, Schadt EE, Dudley JT. Unifying immunology with informatics and multiscale biology. Nat Immunol 2014; 15:118–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 83. Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C et al A systems biology approach for pathway level analysis. Genome Res 2007; 17:1537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM et al Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 2000; 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 1999; 27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S et al BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009; 10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A et al InnateDB: facilitating systems‐level analyses of the mammalian innate immune response. Mol Syst Biol 2008; 4:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res 2009; 37(Database issue):D852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Russo PST, Ferreira GR, Cardozo LE, Burger MC, Arias‐Carrasco R, Maruyama SR et al CEMiTool: a Bioconductor package for performing comprehensive modular co‐expression analyses. BMC Bioinformatics 2018; 19:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bagaev DV, Zvyagin IV, Putintseva EV, Izraelson M, Britanova OV, Chudakov DM et al VDJviz: a versatile browser for immunogenomics data. BMC Genom 2016; 17:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G et al VDJdb: a curated database of T‐cell receptor sequences with known antigen specificity. Nucleic Acids Res 2018; 46(D1):D419–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wooden SL, Koff WC. The Human Vaccines Project: towards a comprehensive understanding of the human immune response to immunization. Hum Vaccin Immunother 2018; 14:2214–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kitano H. Computational systems biology. Nature 2002; 420:206–10. [DOI] [PubMed] [Google Scholar]
- 94. Wang HW, Pai TW. Machine learning‐based methods for prediction of linear B‐cell epitopes. Methods Mol Biol 2014; 1184:217–36. [DOI] [PubMed] [Google Scholar]
- 95. Desai DV, Kulkarni‐Kale U. T‐cell epitope prediction methods: an overview. Methods Mol Biol 2014; 1184:333–64. [DOI] [PubMed] [Google Scholar]
- 96. Gupta S, Mittal P, Madhu MK, Sharma VK. IL17eScan: a Tool for the Identification of Peptides Inducing IL‐17 Response. Front Immunol 2017; 8:1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Metz B, Hoonakker M, Uittenbogaard JP, Weyts M, Mommen GP, Meiring HD et al Proteome analysis is a valuable tool to monitor antigen expression during upstream processing of whole‐cell pertussis vaccines. J Proteome Res 2017; 16:528–37. [DOI] [PubMed] [Google Scholar]
- 98. Smith CL, Dickinson P, Forster T, Craigon M, Ross A, Khondoker MR et al Identification of a human neonatal immune‐metabolic network associated with bacterial infection. Nat Commun 2014; 5:4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pennings JL, Schuurhof A, Hodemaekers HM, Buisman A, de Rond LC, Widjojoatmodjo MN et al Systemic signature of the lung response to respiratory syncytial virus infection. PLoS ONE 2011; 6:e21461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mejias A, Dimo B, Suarez NM, Garcia C, Suarez‐Arrabal MC, Jartti T et al Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med 2013; 10:e1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhao Y, Jamaluddin M, Zhang Y, Sun H, Ivanciuc T, Garofalo RP et al Systematic analysis of cell‐type differences in the epithelial secretome reveals insights into the pathogenesis of respiratory syncytial virus‐induced lower respiratory tract infections. J Immunol 2017; 198:3345–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T et al An interferon‐inducible neutrophil‐driven blood transcriptional signature in human tuberculosis. Nature 2010; 466:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Banus S, Pennings J, Vandebriel R, Wester P, Breit T, Mooi F et al Lung response to Bordetella pertussis infection in mice identified by gene‐expression profiling. Immunogenetics 2007; 59:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 2013; 154:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tam VC, Quehenberger O, Oshansky CM, Suen R, Armando AM, Treuting PM et al Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 2013; 154:213–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Raeven RHM, Brummelman J, van der Maas L, Tilstra W, Pennings JL, Han WG et al Immunological signatures after Bordetella pertussis infection demonstrate importance of pulmonary innate immune cells. PLoS ONE 2016; 11:e0164027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Merkel TJ, Halperin SA. Nonhuman primate and human challenge models of pertussis. J Infect Dis 2014; 209(Suppl 1):S20–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Harandi AM. Systems analysis of human vaccine adjuvants. Semin Immunol 2018; pii: S1044‐5323(18)30069‐1. 10.1016/j.smim.2018.08.001. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 109. Kooijman S, Brummelman J, van Els C, Marino F, Heck AJR, Mommen GPM et al Novel identified aluminum hydroxide‐induced pathways prove monocyte activation and pro‐inflammatory preparedness. J Proteomics 2018; 175:144–55. [DOI] [PubMed] [Google Scholar]
- 110. Hancock DG, Shklovskaya E, Guy TV, Falsafi R, Fjell CD, Ritchie W et al A systems biology approach to the analysis of subset‐specific responses to lipopolysaccharide in dendritic cells. PLoS ONE 2014; 9:e100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Olafsdottir TA, Lindqvist M, Nookaew I, Andersen P, Maertzdorf J, Persson J et al Comparative systems analyses reveal molecular signatures of clinically tested vaccine adjuvants. Sci Rep 2016; 6:39097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Santoro F, Pettini E, Kazmin D, Ciabattini A, Fiorino F, Gilfillan GD et al Transcriptomics of the vaccine immune response: priming with adjuvant modulates recall innate responses after boosting. Front Immunol 2018; 9:1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lindqvist M, Nookaew I, Brinkenberg I, Samuelson E, Thorn K, Nielsen J et al Unraveling molecular signatures of immunostimulatory adjuvants in the female genital tract through systems biology. PLoS ONE 2011; 6:e20448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gomez CE, Perdiguero B, Jimenez V, Filali‐Mouhim A, Ghneim K, Haddad EK et al Systems analysis of MVA‐C induced immune response reveals its significance as a vaccine candidate against HIV/AIDS of clade C. PLoS ONE 2012; 7:e35485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Strouts FR, Popper SJ, Partidos CD, Stinchcomb DT, Osorio JE, Relman DA. Early transcriptional signatures of the immune response to a live attenuated tetravalent dengue vaccine candidate in non‐human primates. PLoS Negl Trop Dis 2016; 10:e0004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Raeven RHM, Brummelman J, Pennings JL, van der Maas L, Tilstra W, Helm K et al Bordetella pertussis outer membrane vesicle vaccine confers equal efficacy in mice with milder inflammatory responses compared to a whole‐cell vaccine. Sci Rep 2016; 6:38240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mizukami T, Momose H, Kuramitsu M, Takizawa K, Araki K, Furuhata K et al System vaccinology for the evaluation of influenza vaccine safety by multiplex gene detection of novel biomarkers in a preclinical study and batch release test. PLoS ONE 2014; 9:e101835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wang IM, Bett AJ, Cristescu R, Loboda A, ter Meulen J. Transcriptional profiling of vaccine‐induced immune responses in humans and non‐human primates. Microb Biotechnol 2012; 5:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lewis DJ, Lythgoe MP. Application of “systems vaccinology” to evaluate inflammation and reactogenicity of adjuvanted preventative vaccines. J Immunol Res 2015; 2015:909406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Rechtien A, Richert L, Lorenzo H, Martrus G, Hejblum B, Dahlke C et al Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the Ebola vaccine rVSV‐ZEBOV. Cell Rep 2017; 20:2251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Anderson J, Olafsdottir TA, Kratochvil S, McKay PF, Ostensson M, Persson J et al Molecular signatures of a TLR4 agonist‐adjuvanted HIV‐1 vaccine candidate in humans. Front Immunol 2018; 9:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kazmin D, Nakaya HI, Lee EK, Johnson MJ, van der Most R, van den Berg RA et al Systems analysis of protective immune responses to RTS, S malaria vaccination in humans. Proc Natl Acad Sci U S A 2017; 114:2425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. van den Berg RA, Coccia M, Ballou WR, Kester KE, Ockenhouse CF, Vekemans J et al Predicting RTS, S vaccine‐mediated protection from transcriptomes in a malaria‐challenge clinical trial. Front Immunol 2017; 8:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. van den Berg RA, De Mot L, Leroux‐Roels G, Bechtold V, Clement F, Coccia M et al Adjuvant‐associated peripheral blood mRNA profiles and kinetics induced by the adjuvanted recombinant protein candidate tuberculosis vaccine M72/AS01 in bacillus Calmette–Guérin‐vaccinated adults. Front Immunol 2018; 9:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Howard LM, Hoek KL, Goll JB, Samir P, Galassie A, Allos TM et al Cell‐based systems biology analysis of human AS03‐adjuvanted H5N1 avian influenza vaccine responses: a phase I randomized controlled trial. PLoS ONE 2017; 12:e0167488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D et al Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 2009; 10:116–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali‐Mouhim A et al Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 2008; 205:3119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE et al Systems biology of immunity to MF59‐adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc Natl Acad Sci U S A 2016; 113:1853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N et al Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity 2015; 43:1186–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Fourati S, Cristescu R, Loboda A, Talla A, Filali A, Railkar R et al Pre‐vaccination inflammation and B‐cell signalling predict age‐related hyporesponse to hepatitis B vaccination. Nat Commun 2016; 7:10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Voigt EA, Grill DE, Zimmermann MT, Simon WL, Ovsyannikova IG, Kennedy RB et al Transcriptomic signatures of cellular and humoral immune responses in older adults after seasonal influenza vaccination identified by data‐driven clustering. Sci Rep 2018; 8:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Plotkin SA. Complex correlates of protection after vaccination. Clin Infect Dis 2013; 56:1458–65. [DOI] [PubMed] [Google Scholar]
- 133. Obermoser G, Presnell S, Domico K, Xu H, Wang Y, Anguiano E et al Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity 2013; 38:831–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Haks MC, Bottazzi B, Cecchinato V, De Gregorio C, Del Giudice G, Kaufmann SHE et al Molecular signatures of immunity and immunogenicity in infection and vaccination. Front Immunol 2017; 8:1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, Jacobsen M et al Identification of biomarkers for tuberculosis disease using a novel dual‐color RT‐MLPA assay. Genes Immun 2012; 13:71–82. [DOI] [PubMed] [Google Scholar]
- 136. Loxton AG, Knaul JK, Grode L, Gutschmidt A, Meller C, Eisele B et al Safety and immunogenicity of the recombinant Mycobacterium bovis BCG vaccine VPM1002 in HIV‐unexposed newborn infants in South Africa. Clin Vaccine Immunol 2017; 24:pii: e00439‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Olin A, Henckel E, Chen Y, Lakshmikanth T, Pou C, Mikes J et al Stereotypic immune system development in newborn children. Cell 2018; 174:1277–92. e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Jennewein MF, Butler AL, Alter G. Neonate‐omics: charting the unknown immune response in early life. Cell 2018; 174:1051–3. [DOI] [PubMed] [Google Scholar]
- 139. Delhalle S, Bode SFN, Balling R, Ollert M, He FQ. A roadmap towards personalized immunology. NPJ Syst Biol Appl 2018; 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Pulendran B. Systems vaccinology: probing humanity's diverse immune systems with vaccines. Proc Natl Acad Sci U S A 2014; 111:12300–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. de Bree LCJ, Koeken V, Joosten LAB, Aaby P, Benn CS, van Crevel R et al Non‐specific effects of vaccines: current evidence and potential implications. Semin Immunol 2018; pii: S1044‐5323(18)30011‐3. 10.1016/j.smim.2018.06.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 142. Seib KL, Dougan G, Rappuoli R. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet 2009; 5:e1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Grover A, Sharma PC. Development and use of molecular markers: past and present. Crit Rev Biotechnol 2016; 36:290–302. [DOI] [PubMed] [Google Scholar]
- 144. Ovsyannikova IG, Salk HM, Larrabee BR, Pankratz VS, Poland GA. Single‐nucleotide polymorphism associations in common with immune responses to measles and rubella vaccines. Immunogenetics 2014; 66:663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Linnik JE, Egli A. Impact of host genetic polymorphisms on vaccine induced antibody response. Hum Vaccin Immunother 2016; 12:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Tserel L, Kolde R, Limbach M, Tretyakov K, Kasela S, Kisand K et al Age‐related profiling of DNA methylation in CD8+ T cells reveals changes in immune response and transcriptional regulator genes. Sci Rep 2015; 5:13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zimmermann MT, Oberg AL, Grill DE, Ovsyannikova IG, Haralambieva IH, Kennedy RB et al System‐wide associations between DNA‐methylation, gene expression, and humoral immune response to influenza vaccination. PLoS ONE 2016; 11:e0152034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Youngblood B, Hale JS, Akondy R. Using epigenetics to define vaccine‐induced memory T cells. Curr Opin Virol 2013; 3:371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Poland GA, Ovsyannikova IG, Kennedy RB. Personalized vaccinology: a review. Vaccine 2018; 36:5350–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Poland GA, Kennedy RB, McKinney BA, Ovsyannikova IG, Lambert ND, Jacobson RM et al Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century. Semin Immunol 2013; 25:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Castiblanco J, Anaya JM. Genetics and vaccines in the era of personalized medicine. Curr Genomics 2015; 16:47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol 2017; 13:e1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Barton AJ, Hill J, Pollard AJ, Blohmke CJ. Transcriptomics in human challenge models. Front Immunol 2017; 8:1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bakken T, Cowell L, Aevermann BD, Novotny M, Hodge R, Miller JA et al Cell type discovery and representation in the era of high‐content single cell phenotyping. BMC Bioinformatics 2017; 18(Suppl 17):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Westermann AJ, Vogel J. Host‐pathogen transcriptomics by dual RNA‐Seq. Methods Mol Biol 2018; 1737:59–75. [DOI] [PubMed] [Google Scholar]
- 156. Westermann AJ, Barquist L, Vogel J. Resolving host–pathogen interactions by dual RNA‐seq. PLoS Pathog 2017; 13:e1006033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Ontoria E, Hernandez‐Santana YE, Gonzalez‐Garcia AC, Lopez MC, Valladares B, Carmelo E. Transcriptional profiling of immune‐related genes in Leishmania infantum‐infected mice: identification of potential biomarkers of infection and progression of disease. Front Cell Infect Microbiol 2018; 8:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Sharon J, Rynkiewicz MJ, Lu Z, Yang CY. Discovery of protective B‐cell epitopes for development of antimicrobial vaccines and antibody therapeutics. Immunology 2014; 142:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Behrendt I, Pradzinska M, Spodzieja M, Kolodziejczyk AS, Rodziewicz‐Motowidlo S, Szymanska A et al Epitope location for two monoclonal antibodies against human cystatin C, representing opposite aggregation inhibitory properties. Amino Acids 2016; 48:1717–29. [DOI] [PubMed] [Google Scholar]
- 160. Hager‐Braun C, Tomer KB. Determination of protein‐derived epitopes by mass spectrometry. Expert Rev Proteomics 2005; 2:745–56. [DOI] [PubMed] [Google Scholar]
- 161. Vaughan K, Xu X, Caron E, Peters B, Sette A. Deciphering the MHC‐associated peptidome: a review of naturally processed ligand data. Expert Rev Proteomics 2017; 14:729–36. [DOI] [PubMed] [Google Scholar]
- 162. Weiner J 3rd, Mohney RP, Kaufmann SHE. The potential of metabolic profiling for vaccine development. Semin Immunol 2018; pii: S1044‐5323(18)30027‐7. 10.1016/j.smim.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 163. Gardinassi LG, Arevalo‐Herrera M, Herrera S, Cordy RJ, Tran V, Smith MR et al Integrative metabolomics and transcriptomics signatures of clinical tolerance to Plasmodium vivax reveal activation of innate cell immunity and T cell signaling. Redox Biol 2018; 17:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wood PL, Steinman M, Erol E, Carter C, Christmann U, Verma A. Lipidomic analysis of immune activation in equine leptospirosis and Leptospira‐vaccinated horses. PLoS ONE 2018; 13:e0193424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Tisoncik‐Go J, Gasper DJ, Kyle JE, Eisfeld AJ, Selinger C, Hatta M et al Integrated omics analysis of pathogenic host responses during pandemic H1N1 influenza virus infection: the crucial role of lipid metabolism. Cell Host Microbe 2016; 19:254–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Mahan AE, Tedesco J, Dionne K, Baruah K, Cheng HD, De Jager PL et al A method for high‐throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. J Immunol Methods 2015; 417:34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL Jr et al Effect of antibiotic‐mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized‐control proof‐of‐concept trial. Cell Host Microbe 2018; 24:197–207. e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Barrila J, Crabbe A, Yang J, Franco K, Nydam SD, Forsyth RJ et al Modeling host–pathogen interactions in the context of the microenvironment: 3‐D cell culture comes of age. Infect Immun 2018; 86 (11) e00282–18; 10.1128/IAI.00282-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Harris VC. The significance of the intestinal microbiome for vaccinology: from correlations to therapeutic applications. Drugs 2018; 78:1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J et al Best practices for analysing microbiomes. Nat Rev Microbiol 2018; 16:410–22. [DOI] [PubMed] [Google Scholar]
- 171. Marsland BJ, Gollwitzer ES. Host–microorganism interactions in lung diseases. Nat Rev Immunol 2014; 14:827–35. [DOI] [PubMed] [Google Scholar]
- 172. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Schnupf P, Gaboriau‐Routhiau V, Cerf‐Bensussan N. Modulation of the gut microbiota to improve innate resistance. Curr Opin Immunol 2018; 54:137–44. [DOI] [PubMed] [Google Scholar]
- 174. Zimmermann P, Curtis N. The influence of the intestinal microbiome on vaccine responses. Vaccine 2018; 36:4433–9. [DOI] [PubMed] [Google Scholar]
- 175. Furman D, Jojic V, Kidd B, Shen‐Orr S, Price J, Jarrell J et al Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol 2013; 9:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Nino D et al Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis 2011; 203:921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]


