Abstract
Local inflammation in obese adipose tissue has been shown to contribute to insulin resistance; however, the role of macrophage infiltration within skeletal muscle is still debatable. This study aimed to evaluate the association of skeletal muscle macrophage gene expression with adiposity levels and insulin sensitivity in obese patients. Twenty-two nondiabetic obese patients and 23 healthy lean controls were included. Obese patients underwent a 3-month weight loss intervention. Macrophage gene expression in skeletal muscle (quantitative real-time polymerase chain reaction), body composition (dual-energy X-ray absorptiometry), and insulin sensitivity (homeostatic model assessment (HOMA) and oral glucose tolerance test) were compared between groups and their associations were analyzed. To validate skeletal muscle findings, we repeated the analyses with macrophage gene expression in adipose tissue. Expression levels of macrophage genes (CD68, CD11b, CD206, CD16, CD40, and CD163) were lower in skeletal muscle tissue of obese versus lean participants. Macrophage gene expression was also found to be inversely associated with adiposity, fasting insulin, and HOMA (r = −0.4 ~ −0.6, p < 0.05), as well as positively associated with insulin sensitivity (r = 0.4 ~ 0.8, p < 0.05). On the other hand, adipose tissue macrophage gene expression showed higher levels in obese versus lean participants, presenting a positive association with adiposity levels. Macrophage gene expression, in both skeletal and adipose tissue samples, was only minimally affected by the weight loss intervention. In contrast with the established positive relationship between adiposity and macrophage gene expression, an unexpected inverse correlation between these 2 variables was observed in skeletal muscle tissue. Additionally, muscle macrophage gene expression was inversely correlated with insulin resistance.
Keywords: M1/M2 macrophage, weight loss, inflammation, insulin sensitivity, skeletal muscle, adipose tissue
Résumé :
L’inflammation locale dans le tissu adipeux des personnes obèses contribue, étudesà l’appui,à l’insulinorésistance; toutefois, le rôle de l’infiltration des macrophages dans le muscle squelettique suscite encore des débats. Cette étude a pour objectif d’évaluer l’association entre l’expression génique des macrophages du muscle squelettique et le degré d’adiposité ainsi que la sensibilitéà l’insuline chez des patients obèses. L’étude comprend 22 patients obèses sans diabète et 23 témoins minces en bonne santé. Les patients obèses participentà un programme de perte de poids d’une durée de 3 mois. On compare les variables suivantes entre les groupes et on évalue leur degré d’association : expression génique des macrophages du tissu musculosquelettique (RT-PCR quantitative), composition corporelle (absorptiométrieà rayons X en double énergie) et sensibilitéà l’insuline (modèle HOMA et épreuve d’hyperglycémie provoquée par voie orale). Pour valider les observations dans le muscle squelettique, on reprend les analyses de l’expression génique des macrophages dans le tissu adipeux. Le taux d’expression génique des macrophages (CD68, CD11b, CD206, CD16, CD40 et CD163) est plus faible dans le tissu musculosquelettique des personnes obèses comparativement aux participants minces. L’expression génique des macrophages est inversement associéeà l’adiposité, au taux d’insulineà jeun età HOMA (r = −0,4 ~ −0,6, p < 0,05) et positivement associéeà la sensibilitéà l’insuline (r = 0,4 ~ 0,8, p < 0,05). D’autre part, l’expression génique des macrophages du tissu adipeux est plus élevée chez les personnes obèses comparativement aux sujets minces et présente une association positive avec le degré d’adiposité. L’expression génique des macrophages dans les échantillons de tissus adipeux et squelettique est minimalement influencée par le programme de perte de poids. En contraste avec l’association positive établie entre l’adiposité et l’expression génique des macrophages, on observe une corrélation inverse inattendue entre ces deux variables dans le tissu musculosquelettique. De plus, l’expression génique des macrophages dans le muscle est inversement corréléeà l’insulinorésistance. [Traduit par la Rédaction]
Keywords: macrophage M1/M2, perte de poids, inflammation, sensibilité à l’insuline, muscle squelettique, tissu adipeux
Introduction
Skeletal muscle contains a population of resident immune cells, mainly macrophages and dendritic cells, which play a fundamental role in muscle regeneration and repair (Summan et al. 2006; Chazaud 2016). During pathophysiological conditions such as obesity and diabetes, additional immune cells, such as monocytes, may infiltrate muscle and the macrophage activation state might shift (Pillon et al. 2013). Macrophages exhibit a wide range of activation states. Along the continuum in macrophage activation states, “classically activated” (designated as M1) and “alternatively activated” (designated as M2) macrophage represent 2 extremes of the spectrum. Clusters of differentiation (CD) antigens are available to classify macrophages according to their function. In previous studies, macrophages were largely determined using a single marker (i.e., CD68), and thus the macrophage cell type heterogeneity was not considered. To address this limitation, in the present study, we used multiple marker genes specific for each subtype as well as CD68, to evaluate the associations between adiposity, adipose, and skeletal muscle tissue macrophage content and insulin sensitivity in obese individuals. Markers for M1 macrophage included CD11b, CD16, and CD40, and M2 macrophages markers included CD163 and CD206 (or MRC1) (Aron-Wisnewsky et al. 2009; Przybyla et al. 2006). We also measured the gene expression of several inflammatory (tumor necrosis factor (TNF), MCP1, and interleukin (IL)-6) and anti-inflammatory (IL-10) cytokines to further characterize the inflammatory status of adipose tissue and skeletal muscle as a consequence of obesity and weight loss. We hypothesized that as with adipose tissue macrophages, the abundance and properties of skeletal muscle macrophages are influenced by obesity, and they are associated with insulin sensitivity.
It has been proposed that macrophage-muscle fiber cross-talk in obesity and diabetes plays a significant role in inducing local inflammation and muscle insulin resistance (Odegaard and Chawla 2008; Varma et al. 2009; Pillon et al. 2012; Khan et al. 2015), which precedes the development of systemic insulin resistance and the occurrence of overt diabetes. Studies using a mouse model of diet-induced obesity and insulin resistance demonstrated increased infiltration of macrophages, especially M1 macrophages, in skeletal muscle. However, studies on humans are limited and reports are inconsistent. While some authors report an association between obesity and increased infiltration of skeletal muscle macrophages (Torres et al. 2004; Varma et al. 2009), others report little evidence of macrophage accumulation in skeletal muscle in obesity with or without type 2 diabetes (Xu et al. 2003; Tam et al. 2012; Amouzou et al. 2016). Consequently, we conducted the present study in obese and lean humans with the goal of further clarifying if obese skeletal muscle becomes infiltrated with macrophages as it happens with obese adipose tissue. A secondary purpose of the study was to assess whether skeletal muscle macrophages are associated with insulin sensitivity.
Materials and methods
Participants
Participants were recruited from the University of Michigan Nutrition Obesity Research Consortium’s Investigational Weight Management Clinic. The University of Michigan Institutional Review Board approved the study. Twenty-two obese (body mass index (BMI) > 30 kg/m2) nondiabetic patients (13 men and 9 women, aged 49 ± 7 years) and 23 lean healthy controls (4 men and 19 women, aged 34 ± 12 years) were included in the study. Subjects’ characteristics are presented in Table 1. Written informed consent was obtained from each participant.
Table 1.
Body composition and insulin sensitivity in lean and obese subjects (pre- and post-weight loss).
| Obese subjects (n = 22) | |||||
|---|---|---|---|---|---|
| Variable | Lean control (n = 23) | Pre-weight loss | Post-weight loss | Difference | p |
| Weight (kg) | 62.0±3.5 | 120.7±2.9* | 102.8±2.6* | 21.0±4.8 | 0.0003 |
| BMI (kg/m2) | 20.6±0.9 | 40.7±0.8* | 34.8±0.8* | 6.9±1.1 | <0.0001 |
| Fat mass (kg) | 8.6±3.5 | 55.4±2.3* | 39.8±2.2* | 14.7±2.8 | <0.0001 |
| Fat% | 18.2±2.1 | 47.4±1.4* | 40.8±1.5* | 7.1±2.2 | 0.005 |
| Glucose (mg/dL) | 95.2±4.1 | 98.3±3.3 | 87.2±3.0 | 9.7±3.8 | 0.02 |
| Insulin (μU/mL) | 7.9±2.1 | 22.1±1.7* | 14.3±1.3* | 7.8±2.3 | 0.003 |
| HOMA | 1.9±0.6 | 5.6±0.5* | 3.1±0.3* | 2.5±0.6 | 0.0004 |
| ISI (n = 17) | 5.8±0.7 | 1.9±0.4* | 3.7±0.4 | −2.0±0.6 | 0.003 |
Note: Values are age- and sex- adjusted means ± SE. Difference was calculated as pre–post-weight-loss intervention in obese subjects. p values were derived from paired t test for the pre- and post-weight-loss differences. Seventeen patients undertook OGTT and had ISI available. BMI, body mass index; ISI, Insulin Sensitivity Index; OGTT, oral glucose tolerance test.
Significantly different from lean control (p < 0.001).
Obese patients participated in a weight-loss program, which was constituted by a low-calorie (~800–1200 kcal/day) high-protein liquid meal replacement (HMR, Boston, Mass., USA) along with healthy lifestyle education. The meal plan was designed for each individual patient with a goal of achieving 15% weight loss within 3 months. Individuals met with the dietitian in a weekly manner during the first month, followed by a monthly visit thereafter. The post-intervention assessments were conducted within 2 weeks after completion. Three days before the assessment, subjects were required to refrain from exercise and were prescribed a eucaloric diet consisting of 50%–60% carbohydrates, ~20% protein, and ~30% fat.
Body composition and insulin sensitivity
Body composition was determined using whole-body dual X-ray absorptiometry (GE Lunar Prodigy Advance Plus, GE Medical Systems, Madison, Wis., USA) and analyzed with Encore 2002 software (GE Medical Systems).
Following an overnight (12 h) fast, subjects completed a 75-g oral glucose tolerance test (OGTT). Blood samples were collected at 0, 30, 60, 90, and 120 min for the measurement of glucose and insulin. Insulin sensitivity was assessed using the Insulin Sensitivity Index (ISI) (Matsuda and DeFronzo 1999) and homeostasis model assessment (HOMA) methods when OGTT data were not available.
Tissue biopsy and quantitative real-time PCR (qRT-PCR)
At baseline (all subjects) and following weight loss (obese patients only), subcutaneous adipose tissue (abdominal) and skeletal muscle (vastus lateralis) biopsies were obtained. Total RNA was extracted from frozen tissue samples using Trizol reagent (Invitrogen, Rockville, Md., USA) and purified with RNAse kit (Qiagen, Santa Clara, Calif., USA). Total RNA concentrations and A260/A280 and A260/A230 ratios were determined using a NanoDrop ND1000 spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, De., USA) to ensure the integrity and purity of the messenger RNA (mRNA). Complementary DNA was synthesized using MMLV Reverse Transcriptase and cleaned using a QIAquick PCR purification kit (Qiagen, Carlsbad, Calif., USA). PCR was performed using an Applied Biosystems Taqman Gene Expression Assay on the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Life Technologies Corp., Carlsbad, Calif., USA). Taqman assays (Applied Biosystems) were used to measure the gene expression of CD68, encoding CD68 molecule (Hs00154355_m1); proinflammatory markers: CD16, encoding Fc fragment of IgG, low affinity IIIb, receptor (CD163 molecule) (Hs00275547_m1); CD40, encoding CD40 molecule (Hs01002913_g1); CD11b, encoding integrin, alpha M (ITGAM) (Hs00355885_m1); MCP1, encoding chemokine (C-C motif) ligand 2 (Hs00234140_m1); TNF, encoding tumor necrosis factor (Hs00174128_m1); IL-6, encoding interleukin 6 (Hs00985639_m1); and anti-inflammatory markers: CD163, encoding CD163 molecule (Hs00174705_m1); CD206, encoding mannose receptor (Hs00267207_m1); IL-10, encoding interleukin 10 (Hs00961622_m1). Eukaryotic 18S (Hs03928985_g1) and LRP10 (Hs01047362_m1) were used as the endogenous reference for gene expression in skeletal muscle and adipose tissue, respectively. The reaction was prepared according to standard Taqman gene expression assay protocol in a total volume of 20 μL (Applied Biosystems). The threshold cycle (Ct) values of reference genes were determined for each sample and used to normalize the mRNA expression of all target genes (ΔCt = Cttarget gene – Ctreference gene).
Statistical analysis
Body composition, insulin sensitivity, and gene expression were compared between lean and obese subjects via ANOVA, controlling for age and sex. The effect of weight loss following the intervention was tested using a paired t test. Multiple regression analysis was performed to test the associations between macrophage gene expression, body composition, and insulin sensitivity after controlling for age and sex. Results were considered significant at the p < 0.05 level.
Results
Macrophage gene expression in skeletal muscle was lower in obese patients versus lean controls
To fully characterize the phenotype of macrophages in skeletal muscle, we selected a panel of gene markers including CD68 and those representing M1 and M2 markers. Our data indicated that compared with lean controls, obese patients had lower mRNA levels of macrophage genes in skeletal muscle, including CD68, CD11b, CD16, CD40, CD163, and CD206, as well as TNF and MCP1. IL-6 and IL-10 were not detectable in most subjects (Fig. 1a). These results remained unchanged even after controlling for age and sex.
Fig. 1.
Macrophage-related gene expression in skeletal muscle and adipose tissue in obese patients at baseline and post-weight loss.
(a) Macrophage-related gene expression in skeletal muscle.
(b) Macrophage-related gene expression in adipose tissue. mRNA levels, expressed in arbitrary units (AUs), are relative to the mean levels of the lean controls after normalization with the reference gene. Dashed line represents the control level of the mRNA levels based on lean controls. *, Significantly different from the control level; †, significantly different from pre-weight loss. All genes in adipose tissue had mRNA levels below 5 AU except CD16 (25.00 ±7.50 pre-weight loss and 22.75 ± 4.25 post-weight loss). To better visualize the entire panel of genes, an up-limit of 10 was set for the y axis in panel b, and the mRNA levels for CD 16 were labeled. IL, interleukin; mRNA, messenger RNA; TNF, tumor necrosis factor.
Macrophage gene expression in skeletal muscle was negatively associated with fat mass and positively associated with insulin sensitivity
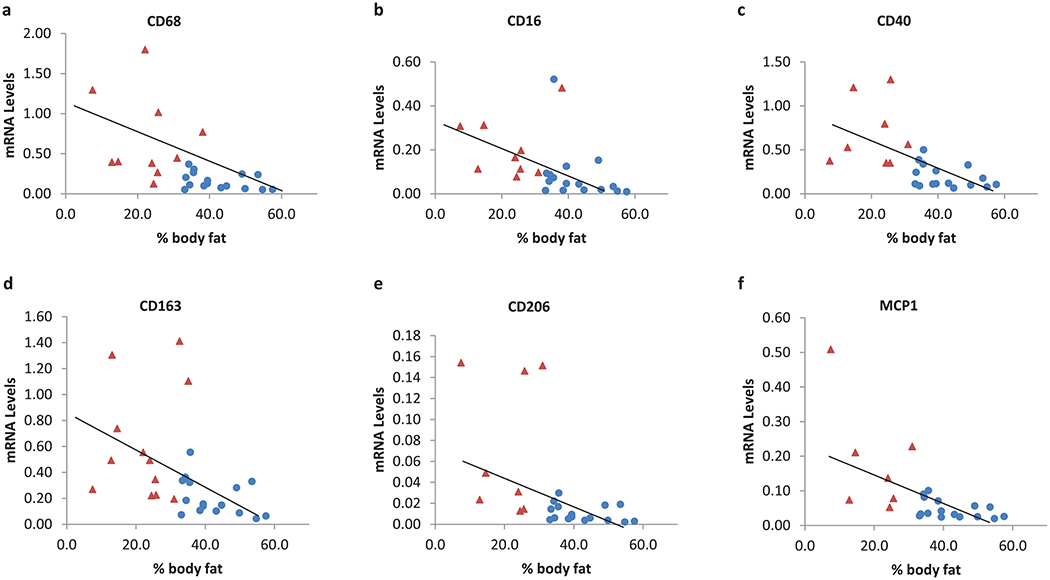
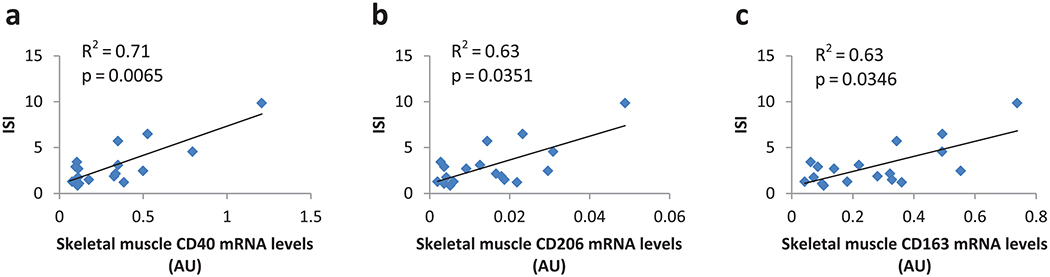
Combining obese patients and lean controls, we found that skeletal muscle mRNA levels of CD68, CD11b, CD16, CD40, CD163, CD206, and MCP1 were negatively correlated with body fat percentage in all subjects even after controlling for age and sex (Fig. 2). Similar results were obtained when excluding lean controls. The expression levels of these genes were negatively correlated with fasting insulin, glucose levels and HOMA, and positively correlated with ISI (Table 2). After controlling for BMI or fat mass, only the correlation between ISI and gene expression of CD40 (p = 0.007), CD206 (p = 0.04), and CD163 (p = 0.03) remained significant (Fig. 3).
Fig. 2.
Skeletal muscle macrophage gene expression was associated with body-fat percentage. Skeletal muscle macrophage gene expression was significantly lower in obese patients than in lean controls; to better visualize the relationship between adiposity and gene expression at low mRNA levels, a certain number of lean controls were not shown in the figure, as specified where appropriate. Red triangles represent lean control and blue circles represent obese patients. Sixteen obese patients were analyzed for each gene. Twelve lean controls were analyzed for CD68, CD16, and CD163, and 11 lean controls for CD40, CD206, and MCP1. (a) CD68, r = −0.30, p = 0.003; 2 lean controls with mRNA levels above2 were not shown in the figure. (b) CD16, r = −0.50, p = 0.0007; 3 lean controls with mRNA levels above 1 were now shown in the figure. (c) CD40, r = −0.50, p < 0.0001; 3 lean controls with mRNA levels above 2 were not shown. (d) CD163, r = −0.40, p = 0.0086. (e) CD206, r = −0.46, p = 0.013;3 lean controls with mRNA levels above 0.3 were not shown in the figure. (f) MCP1, r = −0.35, p = 0.014; 4 lean controls with mRNA levels above 1 were not shown. x axis represents percentage of body fat. y axis represents mRNA levels of each gene normalized to 18 s, calculated as 2−ΔCt ×1000. mRNA, messenger RNA. [Color online.]
Table 2.
Correlations between skeletal muscle macrophage gene expression, body composition and insulin sensitivity in obese patients at baseline (n = 16).
| Gene | Weight | BMI | Fat mass | FFM | FAT% | Glucose | Insulin | HOMA | ISI (n = 13) |
|---|---|---|---|---|---|---|---|---|---|
| CD68 | −0.39* | −0.43* | −0.46* | −0.19 | −0.46* | −0.29 | −0.43* | −0.41* | 0.58* |
| CD16 | −0.47* | −0.48* | −0.47* | −0.34 | −0.35 | −0.42* | −0.51* | −0.51* | 0.41 |
| CD40 | −0.60* | −0.60* | −0.60* | −0.42* | −0.50* | −0.51* | −0.60* | −0.59* | 0.81* |
| CD163 | −0.49* | −0.51* | −0.52* | −0.29* | −0.44* | −0.38* | −0.52* | −0.51* | 0.70* |
| CD206 | −0.40* | −0.39* | −0.41* | −0.27 | −0.34 | −0.33 | −0.42* | −0.41* | 0.69* |
| CD11b | −0.47* | −0.48* | −0.48* | −0.32 | −0.40* | −0.42* | −0.50* | −0.49* | 0.40* |
| MCP1 | −0.43* | −0.43* | −0.38* | −0.35 | −0.28 | −0.36 | −0.40* | −0.39* | 0.69* |
| TNF (n = 8) | −0.38 | −0.40 | −0.41 | −0.22 | −0.36 | −0.34 | −0.42 | −0.42 | 0.55 |
Note: Data are Pearson’s correlation coefficient;
, p < 0.05. Gene expression data were available in 16 obese patients, except for TNF, which was available in 8 patients. ISI was available for 13 patients. BMI, body mass index; FFM, fat free mass; HOMA, homeostasis model assessment; ISI, Insulin Sensitivity Index; TNF, tumor necrosis factor.
Fig. 3.
Macrophage gene expression in skeletal muscle positively related to insulin sensitivity independent of adiposity, age, and sex. Thirteen obese patients and 5 lean controls with both ISI and gene expression data available were included in these figures. (a) CD40 in skeletal muscle, model: ISI = 4.9595 + 4.6379 × CD40_M + 1.1418 × SEX −0.0426 × AGE −0.0591 × BMI, p = 0.0065, adjusted R2 = 0.71. (b) CD206in skeletal muscle, model: ISI = 6.6671 + 80.751 × CD206_M + 1.2613 × SEX −0.0473 × AGE −0.0968 × BMI, p = 0.0351, adjusted R2 = 0.63. (c) CD163 in skeletal muscle, model: ISI = 6.0885 + 5.3107 × CD163_M + 1.3702 × SEX −0.046 × AGE −0.0899 × BMI, p = 0.0346, adjusted R2 = 0.63. x axis represents gene expression levels in arbitrary units (normalized to 18 s, calculated as 2−ΔCt × 1000 in skeletal muscle, or normalized to LRP10, calculated as 2−ΔCt in adipose tissue); y axis represents Matsuda’s ISI. AU, arbitrary unit; ISI, Insulin Sensitivity Index; mRNA, messenger RNA. [Color online.]
Skeletal macrophage markers correlated with each other
Skeletal muscle macrophage markers measured in our study, CD68, CD11b, CD16, CD40, CD163, and CD206, were highly correlated with each other (r ≥ 0.8, p < 0.0001), suggesting that these macrophages comprise mainly 1 subtype that can express an array of markers (data not shown).
Macrophage gene expression in adipose tissue was higher in obese patients versus lean controls
We also analyzed adipose tissue macrophage gene expression from collected adipose tissue of obese and lean participants. Higher mRNA levels of CD68, CD206, CD11b, and TNF concentration was observed in the adipose tissue of obese patients versus lean controls. IL-6 expression levels tended to be higher in obese patients but not significantly (Fig. 1b).
Macrophage gene expression in adipose tissue was positively associated with fat mass and insulin resistance
The expression levels of macrophage genes (CD68, CD11b, MCP1, TNF, CD163, and CD206) in adipose tissue were positively correlated with adiposity levels in all participants irrespective of age and sex (Table 3). Similar results were obtained when excluding lean controls. The mRNA levels of CD68, CD206, CD11b, and TNF were positively correlated with fasting insulin and HOMA. However, this effect appeared to be mediated by adiposity as this relationship lost significance after adjusting for either BMI or fat mass. Intriguingly, the mRNA levels of CD40 were negatively associated with fasting glucose levels and positively with ISI, even after controlling for age, sex, and body fat (Table 3). A lack of correlation between CD40 and other macrophage markers was also observed.
Table 3.
Correlations between adipose tissue macrophage gene expression, body composition and insulin sensitivity in obese patients at baseline (n = 16).
| Gene | Weight | BMI | Fat mass | FFM | FAT% | Glucose | Insulin | HOMA | ISI (n = 13) |
|---|---|---|---|---|---|---|---|---|---|
| CD68 | 0.70* | 0.63* | 0.48* | 0.26 | 0.38 | 0.06 | 0.52* | 0.50* | −0.32 |
| CD40 | −0.27 | −0.27 | −0.10 | −0.15 | −0.13 | −0.44* | −0.24 | −0.28 | 0.58* |
| CD163 | 0.38* | 0.41* | 0.64* | 0.28 | 0.50* | 0.13 | 0.27 | 0.28 | −0.37 |
| CD206 | 0.55* | 0.52* | 0.63* | 0.14 | 0.54* | 0.09 | 0.53* | 0.51* | −0.25 |
| CD11b | 0.54* | 0.58* | 0.51* | −0.13 | 0.58* | 0.17 | 0.42* | 0.43* | −0.35 |
| MCP1 | 0.07 | 0.10 | 0.51* | 0.20 | 0.35 | 0.16 | −0.15 | −0.08 | −0.14 |
| TNF | 0.34 | 0.44* | 0.32 | −0.28 | 0.50* | 0.04 | 0.45* | 0.43* | −0.37 |
| IL-6 (n = 13) | 0.14 | 0.20 | 0.42 | 0.24 | 0.24 | 0.49* | 0.22 | 0.31 | −0.19 |
Note: Data are Pearson’s correlation coefficient;
, p < 0.05. Gene expression data were available in 16 obese patients, except for IL-6, which was available in 13 patients. ISI was available for 13 patients. BMI, body mass index; FFM, fat free mass; HOMA, homeostasis model assessment; IL, interleukin; ISI, Insulin Sensitivity Index; TNF, tumor necrosis factor.
Low-calorie diet minimally modified macrophage gene expression in both skeletal muscle and adipose tissue
To explore the causal relationship between adiposity and macrophage gene expression in skeletal muscle and adipose tissue, we obtained muscle and adipose tissue biopsies from obese patients before and after a 3-month weight-loss program and analyzed the association between changes in macrophage gene expression and metabolic health status. The supervised weight-loss program induced decreases in body weight (17.4% ± 4.0%) and total body fat(7.1% ± 2.2%), as well as improvements in glucose, insulin, HOMA, and ISI (Table 1). With exemption of CD206 in adipose tissue, which showed an increase after the weight-loss intervention (fold change, 1.85 ± 0.39, p < 0.05), there were minimal changes in the expression of CD11b, CD40, CD16, CD163, CD206, IL10, IL6, MCP1, and TNF in response to the weight-loss intervention in both adi-pose tissue and skeletal muscle (Fig. 1). Accordingly, no significant correlation was identified between weight-loss–induced improvement in insulin and changes in macrophage gene expression.
Discussion
Research evaluating the presence or absence of macrophage accumulation in the skeletal muscle of obese individuals is controversial (Xu et al. 2003; Torres et al. 2004; Varma et al. 2009). Our findings demonstrated that macrophage markers’ expression in human skeletal muscle were inversely associated with adiposity levels and positively associated with insulin sensitivity. Our results are consistent with a recent report in which Fink et al. (2013) found a positive association between gene expression levels of M2 markers in skeletal muscle (CD163 and CD206) and insulin sensitivity in overweight/obese individuals. However, Fink et al did not observe this relationship with M1 markers.
In agreement with our results, a recent study demonstrated an absence of skeletal muscle inflammation in both obese insulin-sensitive and insulin-resistant participants, thus suggesting an alternative pathway through which obesity alters insulin resistance. A muscle defect in the activation of the insulin meta bolic pathway (Akt phosphorylation) was identified as an early mechanism of insulin resistance development in these populations (Amouzou et al. 2016). Along with the study by Amouzou et al., recent animal data has also suggested the existence of alternative mechanisms, independent of previously proposed skeletal muscle inflammation, to generate insulin resistance (Evers-van Gogh et al. 2016; Rivas et al. 2016). If we consider that skeletal muscle macrophages have a critical role in promoting muscle recovery and regeneration (Chazaud 2016), we might speculate that there could be an impaired ability to mediate muscle regeneration among obese individuals. A delayed and weakened wound-healing response has already been observed in both obese and diabetic populations. Process that seems to take place because of an impaired inflammatory response within these populations (Pence and Woods 2014; Brown et al. 2015; Sinha et al. 2017). However, further research in the area is needed to verify this hypothesis.
However, not all studies are in agreement with our results. A recent study observed a significantly higher concentration of macrophage and T cell markers in human skeletal muscle of participants with obesity compared with a control group. This study also showed a significant increment on macrophages and T cells in skeletal muscle (which were localized in the extramyocelular adipose tissue) after mice were fed a high-fat diet (Khan et al. 2015). Similar results were observed by Varma et al. 2009, in which higher macrophage infiltration in skeletal muscle was observed in the group with obesity (Varma et al. 2009).
The high correlation between the macrophage markers measured in our study (CD68, CD11b, CD16, CD40, CD163, and CD206) suggests that these macrophages may comprise mainly 1 subtype that can express an array of markers. The independent positive association between macrophage gene expression and insulin sensitivity suggests that skeletal muscle macrophages may be functionally similar to M2, as it has been recognized that M2 macrophages have a beneficial effect on maintaining and enhancing insulin action (Hevener et al. 2007; Odegaard and Chawla 2011). Although, it is important to highlight that we observed this positive correlation between insulin sensitivity and both M1 and M2 markers.
Our adipose tissue macrophage gene expression findings are consistent with previous studies and indirectly confirm the validity of our methodology. Data on skeletal muscle macrophage phenotype in obesity is scarce, especially in humans. Differences in patients’ health status and methodology used in assessing macrophage content make it difficult to compare findings from different studies. In contrast, numerous studies have been conducted on human adipose tissue macrophages and they consistently report, as do we, a positive association between macrophage content and adiposity levels as well as with insulin resistance (Ortega Martinez de Victoria et al. 2009; Klimcakova et al. 2011). Nevertheless, the observed differential response between skeletal and adi-pose tissue macrophage infiltration is in agreement with previous animal research showing an increased inflammatory response in liver and adipose tissue with gradual suppression in skeletal muscle in response to weight gain (Kleemann et al. 2010).
The negative association between CD40 and fasting glucose levels as well as the positive association between CD40 and ISI suggests that CD40 might play a beneficial role in regulating insulin activity. This speculation is consistent with an earlier investigation revealing that the genetic deletion of CD40 aggravates adipose tissue inflammation and diet-induced insulin resistance in obese mice (Wolf et al. 2011). These results, along with our findings, suggest that the metabolic effect of CD40 warrants further investigation.
The nonsignificant change of macrophage gene expression in both skeletal and adipose tissues in response to weight loss might be due to increased lipolysis induced by the low-calorie diet, as it has been suggested that augmentation lipolysis can drive macrophage infiltration (Kosteli et al. 2010). It is possible that weight stabilization after weight-loss is required to observe a significant change in tissue macrophage gene expression levels, as has been previously found (Kovacikova et al. 2011).
A few limitations of the study need to be considered. First, owing to limited amounts of tissue samples, it was not possible to directly quantify macrophage counts using histological methods. Second, owing to difficulties involved in recruiting human subjects for tissue biopsies, we could not match lean subjects to obese patients by age and sex. However, in our analysis we did not find that age or sex significantly influenced the relationship between macrophage gene expression and metabolic variables. Finally, the study only analyzed subcutaneous adipose tissue, as this depot is more feasible to be measured in a lab setting. Potential differences in visceral adipose tissue versus subcutaneous adipose tissue are likely to occur as it has been previously demonstrated that visceral adipose tissue has higher pro-inflammatory characteristics than subcutaneous adipose tissue (Kralova Lesna et al. 2016). Nonetheless, we were still able to detect a significant correlation between macrophage gene expression in subcutaneous adipose tissue and total body fat mass, as well as insulin resistance. Despite these limitations, the novel findings from the present study provide preliminary evidence that macrophages may play different roles in regulating metabolism of adipose tissue and skeletal muscle.
In conclusion, expression of macrophage genes in skeletal muscle is associated with obesity and insulin sensitivity but in an opposite direction to the one observed in adipose tissue. Furthermore, these opposing relationships observed in adipose and skeletal muscle suggest a tissue-specific role for immune cells in regulating metabolism of parenchymal cells.
Further research is needed to clarify the discrepancies among studies in relation to the potential involvement of macrophages/inflammation in the skeletal muscle on the development of insulin resistance observed in obesity.
Acknowledgements
The authors would like to thank Tim Muth for his technical laboratory assistance. This project was funded in part from National Institutes of Health (NIH P30 DK089503).
Footnotes
Conflict of interest statement
The authors declare that they have no competing interests.
References
- Amouzou C, Breuker C, Fabre O, Bourret A, Lambert K, Birot O, et al. 2016. Skeletal muscle insulin resistance and absence of inflammation characterize insulin-resistant grade I obese women. PLoS ONE, 11: e0154119. doi:10.1371/journal.pone.0154119. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-Wisnewsky J, Tordjman J, Poitou C, Darakhshan F, Hugol D, Basdevant A, et al. 2009. Human adipose tissue macrophages: m1 and m2 cell surface markers in subcutaneous and omental depots and after weight loss. J. Clin. Endocrinol. Metab 94: 4619–4623. doi:10.1210/jc.2009-0925. PMID:. [DOI] [PubMed] [Google Scholar]
- Brown LA, Lee DE, Patton JF, Perry RA Jr., Brown JL, Baum JI, et al. 2015. Diet-induced obesity alters anabolic signalling in mice at the onset of skeletal muscle regeneration. Acta Physiol. (Oxf.), 215: 46–57. doi:10.1111/apha.12537. PMID:. [DOI] [PubMed] [Google Scholar]
- Chazaud B 2016. Inflammation during skeletal muscle regeneration and tissue remodeling: application to exercise-induced muscle damage management. Immunol. Cell Biol. 94: 140–145. doi:10.1038/icb.2015.97. PMID:. [DOI] [PubMed] [Google Scholar]
- Evers-van Gogh IJ, Oteng AB, Alex S, Hamers N, Catoire M, Stienstra R, et al. 2016. Muscle-specific inflammation induced by MCP-1 overexpression does not affect whole-body insulin sensitivity in mice. Diabetologia, 59: 624–633. doi:10.1007/s00125-015-3822-2. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink LN, Oberbach A, Costford SR, Chan KL, Sams A, Bluher M, and Klip A 2013. Expression of anti-inflammatory macrophage genes within skeletal muscle correlates with insulin sensitivity in human obesity and type 2 diabetes. Diabetologia, 56: 1623–1628. doi:10.1007/s00125-013-2897-x. PMID: . [DOI] [PubMed] [Google Scholar]
- Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, et al. 2007. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Invest 117: 1658–1669. doi:10.1172/JCI31561. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IM, Perrard XY, Brunner G, Lui H, Sparks LM, Smith SR, et al. 2015. Intermuscular and perimuscular fat expansion in obesity correlates with skeletal muscle T cell and macrophage infiltration and insulin resistance. Int. J. Obes. (Lond.), 39: 1607–1618. doi:10.1038/ijo.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann R, van Erk M, Verschuren L, van den Hoek AM, Koek M, Wielinga PY, et al. 2010. Time-resolved and tissue-specific systems analysis of the pathogenesis of insulin resistance. PLoS ONE, 5: e8817. doi:10.1371/journal.pone.0008817. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimcakova E, Roussel B, Kovacova Z, Kovacikova M, Siklova-Vitkova M, Combes M, et al. 2011. Macrophage gene expression is related to obesity and the metabolic syndrome in human subcutaneous fat as well as in visceral fat. Diabetologia, 54: 876–887. doi:10.1007/s00125-010-2014-3. PMID:. [DOI] [PubMed] [Google Scholar]
- Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, and Ferrante AW Jr. 2010. Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. J. Clin. Invest 120: 3466–3479. doi:10.1172/JCI42845. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova M, Sengenes C, Kovacova Z, Siklova-Vitkova M, Klimcakova E, Polak J, et al. 2011. Dietary intervention-induced weight loss decreases macrophage content in adipose tissue of obese women. Int. J. Obes. (Lond.), 35: 91–98. doi:10.1038/ijo.2010.112. [DOI] [PubMed] [Google Scholar]
- Kralova Lesna I, Kralova A, Cejkova S, Fronek J, Petras M, Sekerkova A, et al. 2016. Characterisation and comparison of adipose tissue macrophages from human subcutaneous, visceral and perivascular adipose tissue. J. Transl. Med 14: 208. doi:10.1186/s12967-016-0962-1. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, and DeFronzo RA 1999. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care, 22: 1462–1470. doi:10.2337/diacare.22.9.1462. PMID: . [DOI] [PubMed] [Google Scholar]
- Odegaard JI, and Chawla A 2008. Mechanisms of macrophage activation in obesity-induced insulin resistance. Nat. Clin. Pract. Endocrinol. Metab 4: 619–626. doi:10.1038/ncpendmet0976. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, and Chawla A 2011. Alternative macrophage activation and metabolism. Annu. Rev. Pathol 6: 275–297. doi:10.1146/annurev-pathol-011110-130138. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega Martinez, de Victoria E, Xu X, Koska J, Francisco AM, Scalise M, Ferrante AW Jr., and Krakoff J 2009. Macrophage content in subcutaneous adipose tissue: associations with adiposity, age, inflammatory markers, and whole-body insulin action in healthy Pima Indians. Diabetes, 58: 385–393. doi:10.2337/db08-0536. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence BD, and Woods JA 2014. Exercise, obesity, and cutaneous wound healing: evidence from rodent and human studies. Adv. Wound Care, (New Rochelle), 3: 71–79. doi:10.1089/wound.2012.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon NJ, Arane K, Bilan PJ, Chiu TT, and Klip A 2012. Muscle cells challenged with saturated fatty acids mount an autonomous inflammatory response that activates macrophages. Cell. Commun. Signal, 10: 30. doi:10.1186/1478-811X-10-30. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon NJ, Bilan PJ, Fink LN, and Klip A 2013. Cross-talk between skeletal muscle and immune cells: muscle-derived mediators and metabolic implications. Am. J. Physiol. Endocrinol. Metab 304: E453–E465. doi:10.1152/ajpendo. 00553.2012. PMID:. [DOI] [PubMed] [Google Scholar]
- Przybyla B, Gurley C, Harvey JF, Bearden E, Kortebein P, Evans WJ, et al. 2006. Aging alters macrophage properties in human skeletal muscle both at rest and in response to acute resistance exercise. Exp. Gerontol 41: 320–327. doi:10.1016/j.exger.2005.12.007. PMID:. [DOI] [PubMed] [Google Scholar]
- Rivas DA, McDonald DJ, Rice NP, Haran PH, Dolnikowski GG, and Fielding RA 2016. Diminished anabolic signaling response to insulin induced by intramuscular lipid accumulation is associated with inflammation in aging but not obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol 310: R561–R569. doi:10.1152/ajpregu.00198.2015. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I, Sakthivel D, and Varon DE 2017. Systemic regulators of skeletal muscle regeneration in obesity. Front. Endocrinol. (Lausanne), 8: 29. doi:10. 3389/fendo.2017.00029. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summan M, Warren GL, Mercer RR, Chapman R, Hulderman T, Van Rooijen N, and Simeonova PP 2006. Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. Am. J. Physiol. Regul. Integr. Comp. Physiol 290: R1488–R1495. doi:10.1152/ajpregu. 00465.2005. PMID:. [DOI] [PubMed] [Google Scholar]
- Tam CS, Sparks LM, Johannsen DL, Covington JD, Church TS, and Ravussin E 2012. Low macrophage accumulation in skeletal muscle of obese type 2 diabetics and elderly subjects. 20: 1530–1533. doi:10.1038/oby.2012.24. PMID: [Obesity, Silver Spring.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres SH, De Sanctis JB, de Briceno LM, Hernandez N, and Finol HJ 2004. Inflammation and nitric oxide production in skeletal muscle of type 2 diabetic patients. J. Endocrinol 181: 419–427. doi:10.1677/joe.0.1810419. PMID: . [DOI] [PubMed] [Google Scholar]
- Varma V, Yao-Borengasser A, Rasouli N, Nolen GT, Phanavanh B, Starks T, et al. 2009. Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am. J. Physiol. Endocrinol. Metab 296: E1300–E1310. doi:10.1152/ajpendo.90885.2008. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Jehle F, Lozhkin A, Hilgendorf I, Bode C, Peter K, and Zirlik A 2011. Abstract 15348: CD40 deficiency aggravates diet-induced obesity, adi-pose tissue inflammation, and insulin resistance in mice. Circulation, 124: A15348. [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. 2003. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest 112: 1821–1830. doi:10.1172/JCI200319451. PMID:. [DOI] [PMC free article] [PubMed] [Google Scholar]