Abstract
Lysosomes degrade cellular proteins and organelles and regulate cell signaling by providing a surface for the formation of critical protein complexes, notably molecular target of rapamycin (mTOR) complex 1 (mTORC1). Striking differences in the lysosomes of cancer versus normal cells suggest that they could be targets for drug development. Although the lysomotropic drugs chloroquine (CQ) and hydroxychloroquine (HCQ) have been widely investigated, studies have focused on their ability to inhibit autophagy. We synthesized a novel compound, called EAD1, which is structurally related to CQ but is a 14-fold more potent inhibitor of cell proliferation. Here we find that EAD1 causes rapid relocation, membrane permeabilization (LMP), and deacidification of lysosomes, and it induces apoptosis and irreversibly blocks proliferation of human lung cancer H460, H520, H1299, HCC827, and H1703 cells. EAD1 causes dissociation of mTOR from lysosomes and increases mTOR’s perinuclear versus cytoplasmic localization, changes previously shown to inactivate mTORC1. The effect on mTOR was not seen with HCQ, even at >10-fold greater concentrations. Phosphorylation of a downstream target of mTORC1, ribosomal protein S6, was inhibited by EAD1. Although EAD1 also inhibited autophagy, it retained full antiproliferative activity in autophagy-deficient H1650 lung cancer cells, which have a biallelic deletion of Atg7, and in H460 Atg7-knockout cells. As Atg7 is critical for the canonical autophagy pathway, it is likely that inhibition of autophagy is not how EAD1 inhibits cell proliferation. Further studies are needed to determine the relationship of LMP to mTORC1 disruption and their relative contributions to drug-induced cell death. These studies support the lysosome as an underexplored target for new drug development.
Introduction
The treatment of non–small cell lung cancer (NSCLC) has rapidly advanced in recent years, with the incorporation of genomic sequencing of tumors and the availability of molecularly targeted drugs. Although genome-driven therapy has demonstrated significant benefits, most NSCLC patients either do not have a mutation for which a matched drug is available, will not have a meaningful response to the drug, or will develop resistance to an initially effective agent (Jordan et al., 2017). Consequently, there is a need for additional approaches to therapy, including those that are based on cancer cell biology, rather than on a specific genetic alteration. These could be used alone or to complement the actions of molecularly matched drugs.
The regulation of programmed cell death and related pathways is often altered in cancer cells and has been an attractive target for drug development, with drugs targeting apoptosis the most advanced and those affecting autophagy only more recently being evaluated clinically. The development of autophagy-modulating approaches has been hampered by several factors: uncertainty as to which tumor types and/or genetic alterations are most likely to be responsive; the low potency and lack of specificity of the only two Food and Drug Administration–approved drugs, chloroquine (CQ) and hydroxychloroquine (HCQ), known to inhibit autophagy; some uncertainty as to whether autophagy inhibition versus autophagy stimulation would be beneficial for a particular tumor; and reports that the antitumor actions of CQ and HCQ may not be dependent on their effects on autophagy (Maycotte et al., 2012; Gewirtz, 2014; Amaravadi et al., 2016; Eng et al., 2016; Rebecca and Amaravadi, 2016; Chen et al., 2017; Levy et al., 2017; Rebecca et al., 2017). Although they were initially recognized as lysomotropic and to have the ability to disrupt lysosome function, studies over the years of CQ and HCQ have focused predominantly on their ability to inhibit autophagy (Weissmann, 1964; Boya and Kroemer, 2008). Despite extensive investigation, the precise mechanism(s) of CQ action remains a long-standing question.
Lysosomal function is closely intertwined with vesicular trafficking, including macroautophagy, microautophagy, and chaperone-mediated autophagy (Kroemer and Jäättelä, 2005). In addition to their central role in the degradation of cellular macromolecules, proteins, and organelles, lysosomes also have more complex biologic functions. They play a critical role in the integration of death signals in programmed cell death and regulate other cell signaling pathways by providing a surface for the formation of protein complexes, notably for the molecular target of rapamycin (mTOR)-containing mTORC1 (Carroll and Dunlop, 2017), which is a central regulator of multiple signaling pathways, coordinating aspects of nutrient sensing, cell metabolism, and cell proliferation, among other biologic functions (Saxton and Sabatini, 2017). Interestingly, chaperone-mediated autophagy is regulated by a mTORC2-containing lysosomal-associated complex (Arias et al., 2015).
Lysosomes and lysosome-associated proteins are being increasingly seen as useful targets for drug development (Kallunki et al., 2013; Piao and Amaravadi, 2016; Rebecca et al., 2017). There are striking changes in lysosomal volume, composition, cellular distribution, and enzyme activity seen during cancer progression and metastasis (Fehrenbacher et al., 2004, 2008; Boya and Kroemer, 2008; Kallunki et al., 2013; Piao and Amaravadi, 2016). Increased expression and activity of lysosomal cysteine cathepsins correlate with the metastatic capacity and aggressiveness of tumors. Immortalization and oncogene-driven transformation lead to increased sensitivity to the lysosomal cell death pathways, and this was due to changes in lysosomes themselves rather than to changes in signaling pathways that lead to lysosomal permeabilization (Fehrenbacher et al., 2008). Lysosomal membranes are less stable in cancer cells compared with normal cells, a difference that could be exploited for the development of agents whose primary site of action would be the lysosome (Piao and Amaravadi, 2016; Rebecca et al., 2017; Towers and Thorburn, 2017). Several classes of drugs that cause lysosomal membrane permeabilization (LMP) have been identified, and LMP inducers may preferentially target lysosomes of cancer cells compared with normal cells (Fehrenbacher et al., 2004). Some of the known lysosome-targeting agents can trigger cell death, even in apoptosis-resistant and drug-resistant cancer cells (Wiedmer et al., 2017). Numerous other anticancer drugs, including several targeted multityrosine kinase inhibitors that are not thought to act through lysosomes, are weak bases, are lysomotropic, and can cause LMP (Piao and Amaravadi, 2016; Wiedmer et al., 2017).
Whereas CQ and HCQ are considered relatively safe drugs, their low potency necessitates the use of high doses (up to 1200 mg daily) that are at the upper range of tolerability. Even these doses produce blood drug concentrations that are below those generally required for their antiproliferative effects in vitro (Goldberg et al., 2012; Rangwala et al., 2014). To address these limitations and as part of our medicinal chemistry program to identify new aminochloroquinoline inhibitors, we synthesized a compound, EAD1 (Fig. 1), that is structurally related to CQ but up to 14-fold more potent than CQ and HCQ as an inhibitor of proliferation of NSCLC cells (Nordstrøm et al., 2015). We report here that EAD1 disrupts multiple lysosomal functions, including mTORC1 localization, and although it also potently blocks autophagy, the data presented suggest that inhibition of autophagy is not the mechanism by which EAD1 inhibits cell proliferation. Given the interest in the use of lysosomal-modulating drugs as anticancer agents, a better understanding of their mechanism(s) of action would help to optimize their use and suggest avenues for new drug development.
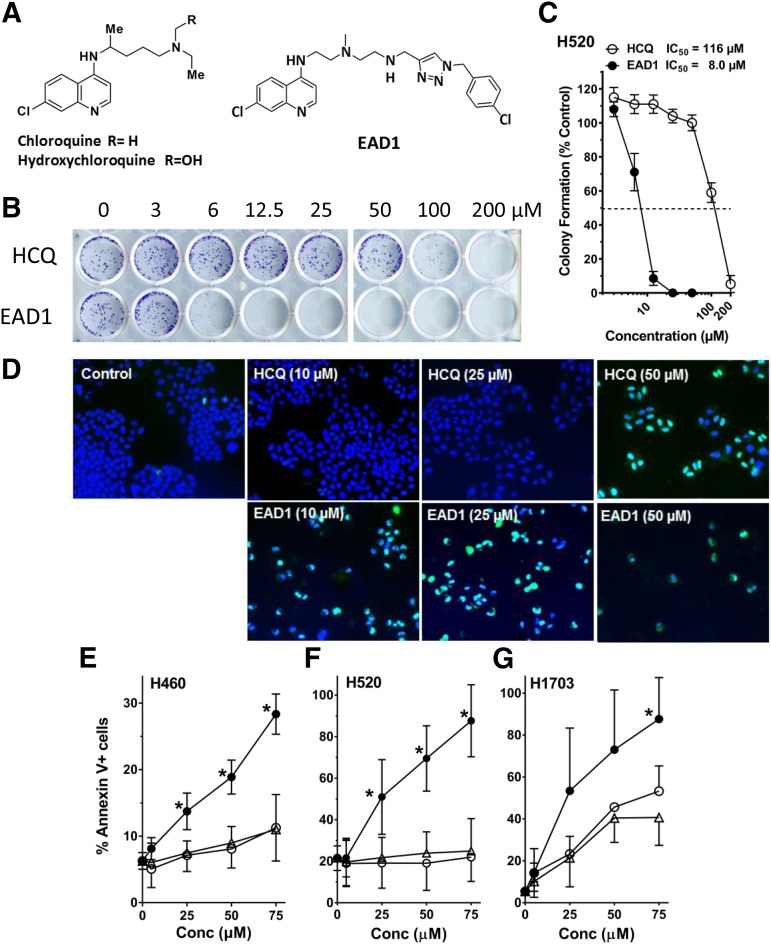
Fig. 1.
Effect of EAD1 on cell proliferation and apoptosis. (A) Structure of EAD1, HCQ, and CQ. (B) H520 NSCLC cells were treated with HCQ or EAD1 for 24 hours. Cells were washed, and drug-free medium was added for an additional 10 days. Colonies were stained with crystal violet. (C) Quantification of colonies shows EAD1 was 14-fold more potent in killing cells than was HCQ. (D–G) EAD1 induces apoptosis in cells treated with the indicated concentrations of HCQ and EAD1 for 24 hours. (D) H460 cells were fixed and stained using a TUNEL assay; apoptotic cells stain aqua/green. (E–G) H460, H520, and H1703 cells were treated with the indicated concentrations of CQ (∆), HCQ (○), and EAD1 (●) for 24 hours and analyzed by flow cytometry for annexin-V staining (means ± S.D. for three experiments). *Significantly different from CQ and HCQ, P < 0.05.
Materials and Methods
Cell Lines and Materials
Human NSCLC cell lines H460, H1299, H520, HCC827, H1650, and H1703 cell lines were from American Type Culture Collection (Manassas, VA), which confirmed their identity by short tandem repeat profiling, and were maintained in RPMI-1640 medium with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere with 5% CO2. Chloroquine and hydroxychloroquine sulfate were from Spectrum Chemicals (New Brunswick, NJ) and Sigma-Aldrich (St. Louis, MO), respectively. Chloroquine analogs were synthesized as previously described, dissolved in H2O, and stored at −20°C (Nordstrøm et al., 2015).
Cell Proliferation
Cells were seeded in 96-well plates in triplicate (2–4 × 103/well) and allowed to attach overnight before different concentrations of the drugs were added for an additional 72 hours. Cell proliferation was quantified by a sulforhodamine B (SRB) assay. Attached cells were fixed with 10% trichloroacetic acid and incubated for 1 hour at 5°C. The cells were stained with SRB (0.4% in 1% acetic acid) by incubating at room temperature for 30 minutes. The plate was rinsed 4× with 1% acetic acid and dried. The SRB was dissolved by adding 10 mM Tris-base. Absorbances were read at 510 nm on plate reader. Cell numbers were expressed relative to untreated controls, and IC50 levels were calculated from concentration-response graphs.
Colony-Formation Assay
H460 cells were plated at 250 cells/well in 24-well plates. After allowing for cell attachment overnight, HCQ and EAD1 were added at the indicated concentrations. After 24 hours, drug-containing medium was removed, and drug-free medium was added. Cell colonies were stained with crystal violet and counted after an additional 10-day growth in the absence of drug.
Apoptosis
Cells were cultured in medium containing HCQ, CQ, or EAD1 for 24 hours, and apoptosis was assessed by using APC-conjugated annexin V (eBioscience, San Diego, CA) to determine phosphatidylserine exposure. For quantitative determination, cells were trypsinized, stained with annexin V for 15 minutes at room temperature, and analyzed by flow cytometry. Each experiment was performed in triplicate.
Neutral Red Uptake
Cells were plated in 96-well plates and incubated with HCQ or EAD1. After 1 hour, neutral red was added to a final concentration of 40 µg/ml (Repetto et al., 2008). After an additional 2 hours, cells were washed briefly in phosphate-buffered saline, lysed, and the neutral red dye extracted with 0.1 ml of acidified ethanol (50% ethanol, 1% glacial acetic acid in water). Absorbance at 540 nm was determined in a plate reader.
Electron Microscopy
Monolayer cells were fixed with 2.5% glutaraldehyde, in 0.1 M sodium cacodylate buffer, postfixed with 1% osmium tetroxide, followed by 2% uranyl acetate, and dehydrated through a graded series of ethanol. Cells were lifted from the monolayer with propylene oxide and embedded as a loose pellet in LX112 resin (LADD Research Industries, Burlington, VT). Ultrathin sections were cut on a Leica Ultracut UC7, stained with uranyl acetate, followed by lead citrate, and viewed on a JEOL 1200EX transmission electron microscope at 80 kV.
Immunoblot Analysis
Cells were scraped from culture dishes, cell lysates were prepared, and immunoblot analysis was performed. Cell extracts were prepared in cold lysis buffer [50 mM Tris pH 7.5, 100 mM NaCl, 50 mM NaF, 5 mM EDTA, 1% Triton X-100, 200 µM Na orthovanadate, and protease inhibitor cocktail (HALT; Thermo Scientific)] Protein concentrations were determined using the Lowry Reagent B (Bio-Rad, Hercules, CA), and normalized cell lysates were mixed with sample buffer (Bio-Rad) containing 2-mercaptoethanol and boiled for 5 minutes. The samples were run on SDS-PAGE gels and transferred to nitrocellulose or polyvinylidene fluoride membranes. The membranes were incubated overnight with primary antibodies in TBS-T buffer containing 5% nonfat milk. Antibodies used were LC3 A/B (cs12741), mTOR and phospho-ser2448-mTOR (cs2983 and cs5536), ULK1 (cs80541), phospho-Ser555-ULK1 (cs5869), phospho-Ser757-ULK1 (cs6888), p70S6K1 (cs2708), phospho-Thr389-p70S6K (cs9234), phospho-Thr421/Ser424-p70S6K1 (cs9204), p90 RSK (cs9355), phospho-Thr359-p90RSK (cs8753), phospho-ThrSer380-p90RSK (cs11989), ribosomal protein S6 (rpS6) (cs2217), phospho-Ser240/244 rpS6 (cs5364), phospho-Ser235 rpS6 (cs4858) from Cell Signaling (Danvers, MA); Atg7 (ab6251) from Abcam (p62/SQSTM1; Cambridge, UK) (P0067) from Sigma; and Atg5 (cs2630). After washing, the membranes were incubated with horseradish peroxidase HRP-conjugated secondary antibody for 1 hour. The bands were detected with enhanced chemiluminescence reagent (Pierce Biotechnology, Waltham, MA).
Immunofluorescence
Immunofluorescence was done on cells treated with EAD1 or HCQ for 24 hours. Cells were washed with phosphate-buffered saline-Tween 20 and fixed for 15 minutes in 4% paraformaldehyde at room temperature. Antigen retrieval was done in 100 mM Tris-HCl, 5% urea (pH 9.5) for 10 minutes at 95°, and the cells were permeabilized and blocked with 0.3% Triton X-100, 5% goat serum, glycine, and 1% BSA for 1 hour. Incubation with primary antibodies was done overnight at 5°C as follows: lysosome-associated membrane protein (LAMP)-2 (sc-18822, 1:100), galectin-3 (BD556904, 1:100), and mTOR (cs2983, 1:400). Appropriate secondary antibodies were used at 1:500, and cells were counterstained with Hoechst 33342.
CRISPR-Based Atg7 Knockout Cells
H460 cells were sequentially transduced with a two-vector lentiviral system: a doxycycline-inducible Cas9 lentivirus with puromycin resistance and a human Atg7 sgRNA-PAM-TGG vector with blasticidin resistance. The sgRNA, targeting exon 3 of Atg7, was GCTGCCAGCTCGCTTAACAT. Clones grown from single cells were isolated and screened by Western blot for Atg7 protein expression.
Cell-Starvation Assay
H460 cells (wild-type or Atg7-minus clones) were plated in 12-well plates and allowed to attach overnight. Medium was removed, and the cells were washed three times and resuspended with serum-free Hanks’ balanced salt solution (HBSS). After 12 hours, the HBSS was removed and replaced with complete media (RPMI 1640 + 10% FBS). Cells were maintained in complete medium for 10 days and colonies were then stained with crystal violet.
Cell-Size Determination
Cells were treated with HCQ or EAD1, stained with propidium iodide, and analyzed by flow cytometry. The size distribution of live G1-phase cells was determined, and the median cell size, determined by FSC-A, was calculated for treated cells relative to untreated controls.
Statistical Analysis
Two-way ANOVA, followed by Tukey’s multiple comparisons test, was performed using GraphPad Prism version 7.01 for Windows (GraphPad Software, La Jolla, CA).
Results
A Novel Chloroquine Analog Inhibits Cell Growth and Disrupts Lysosomes.
We previously synthesized a series of 4-aminoquinoline analogs that inhibited lung cancer cell proliferation with an increased potency compared with chloroquine (Nordstrøm et al., 2015). The lead compound, EAD1, retains the 4-aminoquinoline subunit of CQ and HCQ and is coupled to a 4-chlorophenyl triazole unit via a polyamine linker (Fig. 1A). EAD1 inhibited proliferation in a colony formation assay in which H520 cells were exposed to the compound for 24 hours, after which the cells were washed, and drug-free medium added for an additional 14 days (Fig. 1B). The IC50 for EAD1 in this assay was 8 µM, 14-fold lower than that of HCQ (Fig. 1C). This is comparable to the IC50 we previously reported for H460 cells in the same assay (Nordstrøm et al., 2015). To focus on early drug-induced changes, in this report, all cellular and molecular assays used cells treated with drugs for 24 hours or less. Using both a TUNEL assay and flow cytometry for annexin-V, 24-hour treatment with EAD1 induced a concentration-dependent increase in apoptosis in multiple NSCLC cell lines (Fig. 1, D–G). In all cases, EAD1 had the same increased potency, compared with HCQ and CQ, seen in the cell proliferation assay.
The inhibition of autophagy by CQ and HCQ is thought to be due to their accumulation in lysosomes, causing lysosomal deacidification, which prevents them from fusing with autophagosomes to form autophagolysosomes. We sought to determine whether EAD1 was also affecting lysosome function; to do this, we used several assays of lysosomal integrity and structure. Lysosomes comprise more than 25 membrane proteins, with lysosomal-associated proteins 1 and 2 (LAMP-1, LAMP-2) by far the most abundant (together, >50% of total) (Piao and Amaravadi, 2016). The LAMPs are glycoproteins located on the lysosome membrane, where they are involved in the fusion of lysosomes and autophagosomes (LAMP-2) and are receptors for chaperone-mediated autophagy (LAMP-2A); they are well characterized markers for lysosomes.
LAMP-2 was diffusely expressed throughout the cytoplasm in control H460 cells with lightly staining puncta present (Fig. 2A). EAD1 treatment caused a dramatic increase in the intensity of the staining of the puncta, with a corresponding decrease in the more diffuse cytoplasmic staining. This change was observed in some cells at the lowest EAD1 concentration tested (5 µM) and increased to a near uniform pattern in a concentration-dependent manner. Interestingly, puncta were observed throughout the cytoplasm at 5 µM EAD1, but they were increasingly localized to the perinuclear region at higher concentrations. These changes were not seen in the cells treated with HCQ (Fig. 2A). Although there was a modest increase in puncta intensity with 25 and 50 µM HCQ, these remained distributed throughout the cytoplasm, with no evidence of a shift to perinuclear localization.
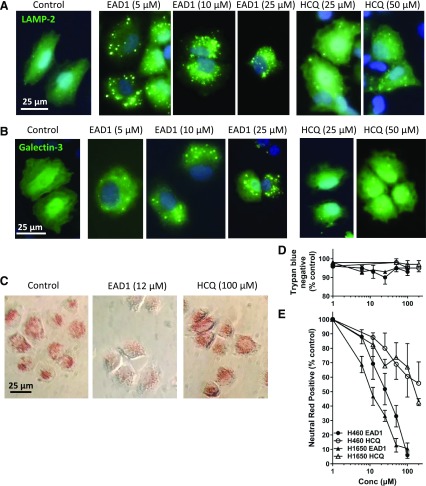
Fig. 2.
Effect of EAD1 on lysosomes. Immunofluorescence microscopy of H460 cells treated for 24 hours with EAD1 or HCQ at the indicated concentrations and stained for LAMP-2 (green) and Hoechst 33342 (blue) (A) or galectin-3 (green) and Hoechst 33342 (blue) (B). (C) H460 and H1650 cells were plated in 96-well plates and incubated with HCQ or EAD1. After 1 hour, neutral red was added to a final concentration of 40 µg/ml. After an additional 2 hours, cells were washed briefly in phosphate-buffered saline and photographed. (D) HCQ and EAD1 did not disrupt the membrane integrity of the cells as determined by the lack of uptake of trypan blue. (E) Quantification of neutral red levels in the cells after the treatment described in (C) and extraction of the dye with 0.1 ml of acidified ethanol (50% ethanol, 1% glacial acetic acid in water). Absorbance at 540 nm was determined in a plate reader. Results are means of three experiments each done in triplicate.
To characterize more fully the EAD1-induced changes in the lysosomes, we used a highly sensitive measure of lysosomal membrane permeabilization. The galectins are carbohydrate-binding lectins that bind β–galactoside sugars with specific carbohydrate-recognition domains (Aits et al., 2015). They are normally found in the cytoplasm, in the nucleus, and on the cell surface, but they localize to the lysosomes during LMP as a consequence of their gaining access to the inside of the lysosomal membrane. Galectin staining specifically marks individual leaky lysosomes and, as such, is a highly sensitive probe for early LMP, detecting LMP before and in the absence of cell death (Aits et al., 2015). Staining with galectins has been superior to other methods to detect LMP. Treatment with EAD1 caused a change in cell staining for galectin-3 from a diffuse pattern throughout the cell to one with prominent, well defined puncta (Fig. 2B). As for LAMP-2, puncta were observed at the lowest concentration of EAD1 tested (5 µM) and progressively increased in abundance and in a perinuclear localization with increasing concentrations. In contrast, with HCQ treatment, there were fewer and less-intensely stained puncta, and these were observed only at the highest concentration (50 µM) of HCQ.
EAD1-treated cells undergoing LMP would not be anticipated to be able to maintain the pH gradient that keeps lysosomes at the acidic environment for optimal lysosomal enzyme function. To assess lysosomal pH, we adapted a previously reported assay to develop a rapid and readily quantifiable measure of the uptake and retention of the dye neutral red (Repetto et al., 2008). Neutral red is weakly cationic and readily crosses lipid membranes in a nonionized form at neutral pH. Inside the low pH environment of the lysosome, it becomes charged and accumulates. We treated cells for 1 hour with increasing concentrations of EAD1 or HCQ, added neutral red for 2 hours, briefly washed the cells to remove nonionized dye, and then lysed the cells and measured the levels of neutral red (Fig. 2C). As shown in Fig. 2E, both compounds caused a concentration-dependent reduction of neutral red retention in two NSCLC cell lines, with EAD1 having much greater potency than HCQ. Strikingly, the curves and the IC50 levels for inhibition of neutral red retention by EAD1 closely overlapped those for its cytotoxic actions. The lack of retention of neutral red was not due to a loss of plasma membrane integrity of the cells, as EAD1 had no effect on the trypan-blue staining of the cells at this time point (Fig. 2D).
Transmission electron microscopy was used to further understand the cellular changes. Cells treated with EAD1 showed a pronounced formation and accumulation of large clumps of 0.1–1 µm diameter “empty” vesicles (Fig. 3A). These electron-lucent vesicles were mostly single membraned and resembled dilated lysosomes, as previously reported with CQ treatment, albeit at much higher (120 µM) CQ concentrations (Yoon et al., 2010). Some of the vesicles appeared to be double-membraned and resembled the autophagic vacuoles with clear content, previously reported to result from cargo loading failure and inefficient autophagy (Chen et al., 2014b). Other more electron-dense structures (0.5 µm diameter) contained osmiophilic inclusions and resembled classic lysosomes.
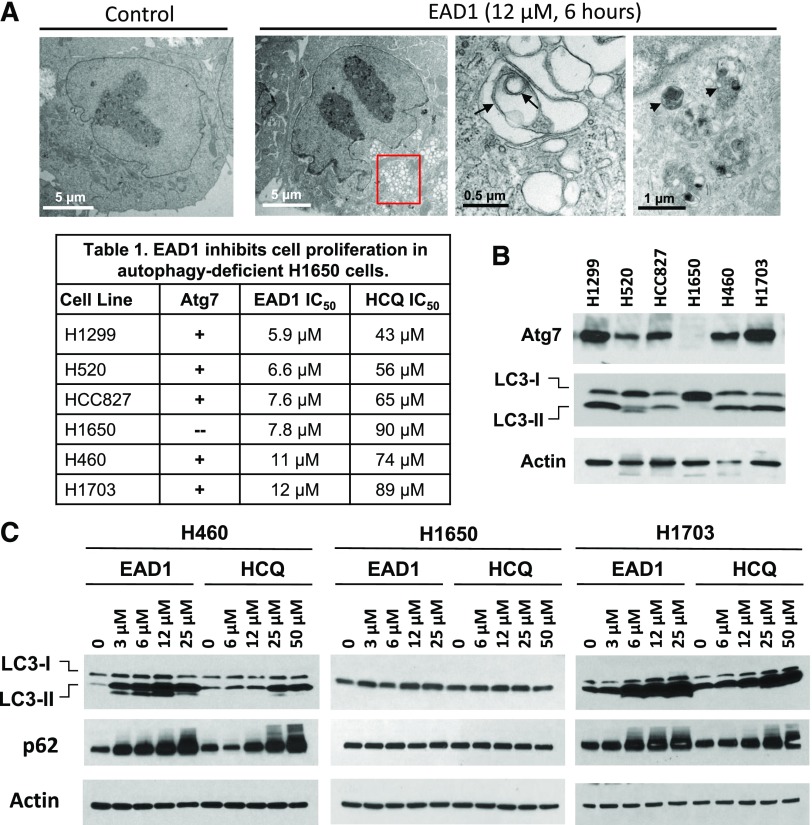
Fig. 3.
Effect of EAD1 on autophagy and cell proliferation in cells lacking the autophagic pathway. (A) Electron micrographs showing large accumulation of autophagosomes (red box) in H460 cells treated with EAD1 (12 µM for 6 hours). Double-membrane vesicles (black arrows) and lysosomes (black arrowheads) are indicated. (Table 1) Cells were treated with EAD1 or HCQ for 72 hours, and antiproliferative activity was measured using SRB assays. Results are means of at least three experiments, each done in triplicate. (B) Total lysates of untreated NSCLC cells analyzed by Western blot and probed for Atg7, LC3-I, and LC3-II. Atg7 and LC3-II were completely absent from the H1650 cells but were present in the other NSCLC cell lines. (C) EAD1 and HCQ caused large, concentration-dependent accumulations of LC3-II and p62 in H460 and H1703 cells but not in H1650 cells.
Inhibition of Cell Proliferation by EAD1 is Autophagy-Independent.
We next extended the growth inhibitory experiments with EAD1 to additional NSCLC cell lines and found inhibition occurred in a small range of IC50s (5.9–12 µM), which in all cases were lower than for HCQ (Table 1). No apparent differences were found between cell lines that were derived from adenocarcinomas (H1299, HCC827, H1650, H460) compared with those derived from squamous tumors (H520, H1703), nor did the presence of a mutant Ras (H460, H1299) versus wild-type Ras (all other cells) appear to affect sensitivity in this small panel. What was striking was the observation that both EAD1 and HCQ effectively inhibited proliferation of the H1650 cells, a human NSCLC cell line that has a biallelic deletion within the Atg7 locus (Mandelbaum et al., 2015). Atg7 is an E1 ligase that has several functions, including promoting Atg12-Atg5 conjugation during elongation of the autophagosome isolation membrane; it is required for the functioning of the canonical autophagy pathway (Mizushima et al., 2011). We confirmed that the H1650 cells lacked Atg7 protein and lacked the autophagosomal form of LC3, LC3-II (Fig. 3B). LC3 (also known as Atg8) is a ubiquitin-like protein that exists in a cytoplasmic form (LC3-I) and a PE-conjugated form (LC3-II) that is localized in autophagosomes. LC3 is involved in the biogenesis of, and cargo recruitment into, autophagosomes; thus, the absence of LC3-II indicates that the H1650 cells are autophagy-deficient.
TABLE 1.
EAD1 inhibits cell proliferation in autophagy-deficient H1650 cells
| Cell Line | Atg7 | EAD1 IC50 | HCQ IC50 (μM) |
|---|---|---|---|
| μM | μM | ||
| H1299 | + | 5.9 | 43 |
| H520 | + | 6.6 | 56 |
| HCC827 | + | 7.6 | 65 |
| H1650 | − | 7.8 | 90 |
| H460 | + | 11 | 74 |
| H1703 | + | 12 | 89 |
HCQ, hydroxychloroquine.
HCQ prevents the fusion of autophagosomes with lysosomes, with the subsequent accumulation of large number of autophagosomes, and we have previously found that EAD1 has a similar effect (Nordstrøm et al., 2015). We confirmed that there was no accumulation of LC3-II in the H1650 cells, in contrast to the large increases seen in H460 and H1703 cells in the presence of the drugs (Fig. 3C). Similarly, there was a drug-mediated accumulation of SQSTM1/p62, a cargo adaptor protein that is incorporated into autophagosomes, binds to LC3, and is itself selectively degraded by the autophagic pathway (Fig. 3C). To confirm the lack of a role of autophagy as a determinant of sensitivity to EAD1 and HCQ, we used CRISPR to disrupt exon 3 in the Atg7 gene in H460 cells. Two clones (2B4 and 3F9) were selected and are shown in Fig. 4A, documenting the lack of Atg7 and LC3-II in these cells. To demonstrate more clearly the absence of the autophagy pathway in these cells, we examined the Atg7-mediated conjugation of Atg12 to Atg5, which subsequently catalyzes LC3-I to LC3-II (Fig. 4B). Both the H1650 cells and the H460/Atg7-minus clones lacked the Atg12+Atg5 complex, with the latter expressing unconjugated Atg5 (Fig. 4A).
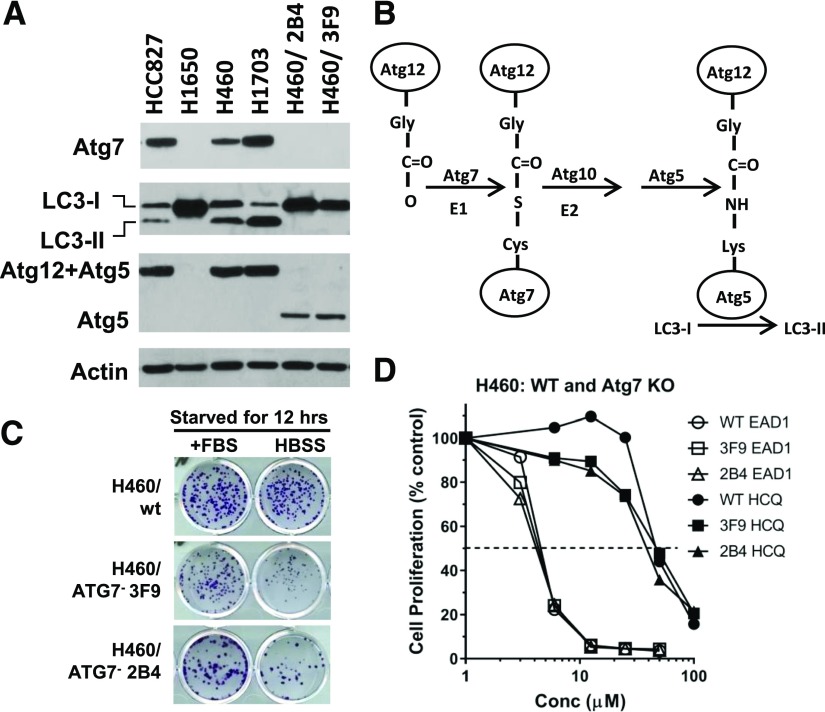
Fig. 4.
EAD1-induced cell death in cells lacking Atg7 expression and autophagy. (A) Lysates from NSCLC cells lines HCC827, H460, H1703 (with wild-type Atg7), H1650 (deleted Atg7), and two clones of H460 cells with Atg7-KO (2B4 and 3F9) were analyzed by Western blot. The H1650 cells and the H460-KO cells lack Atg7 protein expression and cannot form LC3-II. These cells also were unable to conjugate Atg12 to Atg5 (B), a critical step in the canonical autophagy pathway. (C) Whereas the H460-Atg7-KO cells showed no change in proliferation rate in serum-containing medium, they were more susceptible to cell death in serum-free HBSS medium. H460 Atg7-wt and ATG7-KO cells (3F9 and 2B4) were incubated with serum-containing medium (+FBS) or serum-free HBSS for 12 hours. Serum-containing medium was then restored, and colony formation was assessed after an additional 10–14 days. (D) Despite lacking the canonical autophagy pathway, the H460-Atg7-KO cells retained full sensitivity to the cytotoxic actions of EAD1 in a 72-hour SRB proliferation assay.
Although these data confirm the lack of the canonical autophagy in the Atg7-deficient cells, there have been reports of alternative autophagy pathways that are independent of Atg7 (Nishida et al., 2009; Honda et al., 2014). To test for this possibility, we used a functional assay, the ability of the cells to survive brief starvation conditions, to demonstrate the lack of autophagy in the H460/Atg7-minus cell. The standard growth medium was replaced with serum-free HBSS for 12 hours, after which the standard medium was returned. H460 wild-type cells can survive in serum-free HBSS for 12 hours, as illustrated by colony formation after 14 days (Fig. 4C). In contrast, both H460-Atg7-minus clones (3F9 and 2B4) had substantially fewer colonies, indicating that cells were less able to survive the starvation stress. Despite the absence of the autophagy pathway, the sensitivity of these cells to EAD1 and HCQ was identical to that of the wild- type cells with functioning autophagy (Fig. 4D). These data suggest that whereas both EAD1 and HCQ can inhibit autophagy, this inhibition is not the mechanism of their growth inhibitory actions.
EAD1 Disrupts the Cellular Distribution of mTOR and Its Association with Lysosomes.
In addition to their role in autophagy and macromolecule degradation and recycling, lysosomes are involved in the integration of intracellular signaling cascades that are mediated by mTOR, whereby the positioning of the mTOR complex 1 (mTORC1) on lysosomes can lead to its activation (Korolchuk et al., 2011). The recruitment of mTORC1 to the lysosomal membrane brings it into close proximity to its master regulator, Rheb, which is found on the lysosome, a process that is also dependent on a group of small GTPases called Rag GTPases and a complex called Ragulator (Sancak et al., 2008, 2010; Carroll and Dunlop, 2017). The direct interaction between Rheb and mTOR can activate the latter’s kinase activity and cause the phosphorylation of its downstream targets (Carroll and Dunlop, 2017). It is noteworthy that mTOR activity can also be solely regulated by changes in the intraluminal pH of the lysosome (Carroll and Dunlop, 2017). Thus, the lysosome creates a signaling hub that tightly controls mTORC1 activity.
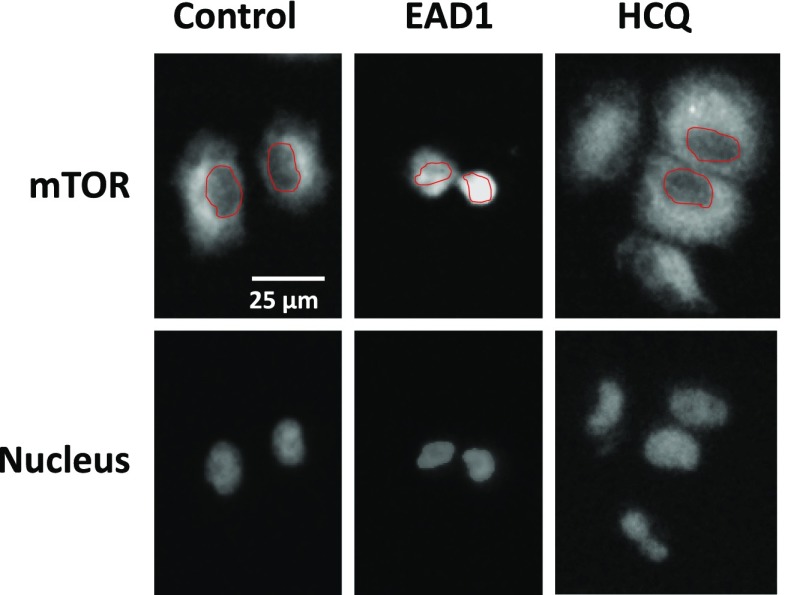
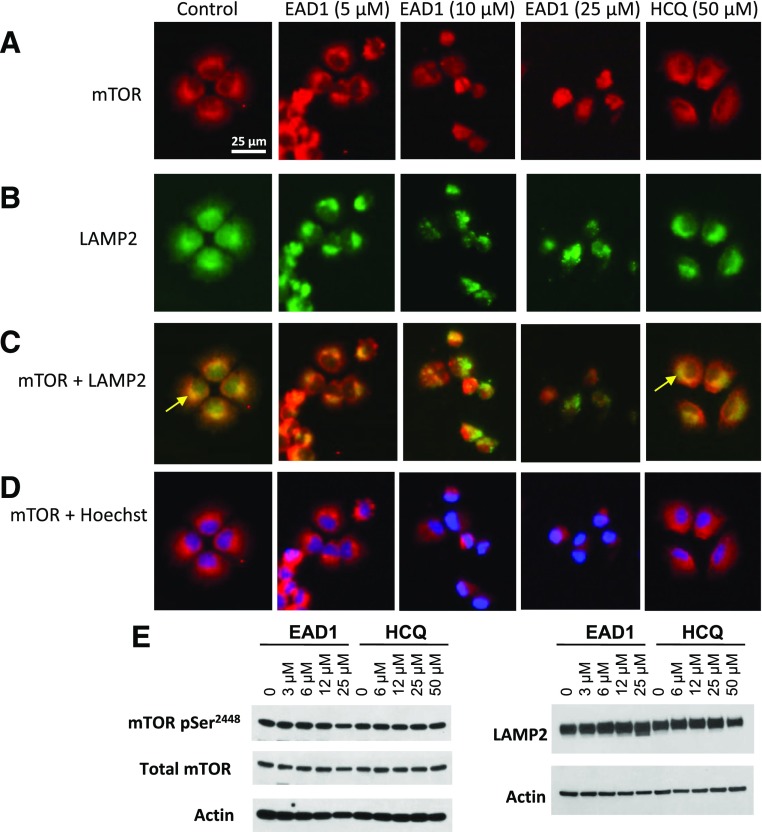
We first examined the intracellular distribution of mTOR by immunofluorescence. In control cells, nearly all the mTOR is localized in the cytoplasm, with little staining in the nucleus (as defined with Hoechst 33342) (Fig. 5). In contrast, treatment with 10 µM EAD1 caused a relocation of mTOR to the nuclear and perinuclear regions, with a nearly complete loss of cytoplasmic mTOR. Comparison of the effect of EAD1 with that of HCQ is striking, as there was no change in mTOR intracellular localization seen with the latter agent. We then costained cells with antibodies to mTOR and LAMP-2 and observed a substantial degree of colocalization (yellow arrows in Fig. 6C). EAD1 caused a progressive loss of yellow fluorescence with increasing concentrations, indicative of reduced colocalization, with a corresponding increase in red mTOR fluorescence. In contrast, cells treated with 50 µM HCQ appeared virtually identical to the control untreated cells (Fig. 6C). Interestingly, although mTOR did not colocalize with lysosomes in cells treated with EAD1 at the highest concentration (10 and 25 µM), it did appear to be present in only the perinuclear region (Fig. 6D). Again, there was no change in the cellular localization of mTOR in the HCQ-treated cells. It was difficult to determine whether EAD1 was affecting the cellular levels of mTOR or LAMP-2 when assessed by immunofluorescence (Fig. 6A), but immunoblots of drug-treated cells clearly showed no difference in total mTOR or in the levels of serine 2448-phosphorylated mTOR and a modest increase in the cellular levels of LAMP2; the latter was not seen with HCQ (Fig. 6E).
Fig. 5.
Intracellular localization of mTOR after EAD1 treatment. H460 cells attached overnight to multichamber slides and then treated with EAD1 (10 µM) or HCQ (50 µM) for 24 hours. Cells were fixed, permeabilized, and labeled with a rabbit monoclonal to mTOR (upper images) or Hoechst 33342 (lower images). Red shapes delineate the nucleus, as defined in the images after Hoechst staining.
Fig. 6.
EAD1 affects lysosome-associated mTOR. (A–D) H460 cells attached overnight to multichamber slides and then treated with the indicated concentrations of EAD1 or HCQ for 24 hours. Cells were fixed, permeabilized, and colabeled with a rabbit monoclonal to mTOR (A), a mouse monoclonal antibody to LAMP-2 (B), and Hoechst 33342. (C) Dual staining of cells for mTOR and LAMP2 or mTOR and Hoechst (D). Arrows in (C) indicate colocalization (yellow) of mTOR with lysosomes. (E) Western blot of H460 cells treated for 24 hours with the indicated concentrations of EAD1 and HCQ and probed for pSer2448-mTOR, total mTOR, and LAMP-2.
EAD1 Inhibits Downstream mTORC1 Signaling.
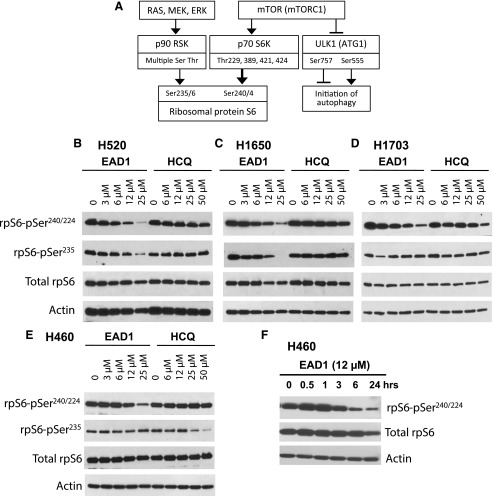
The phosphorylation of ribosomal protein S6 (rpS6) is often measured as a surrogate marker for mTOR activity, as it is downstream of the mTORC1 signaling pathway (Fig. 7A). In contrast to its minimal effect of mTOR phosphorylation, EAD1 produced a large and consistent reduction in rpS6 phosphorylation in a concentration (Fig. 7, B–E) and time-dependent (Fig. 7F) manner. Reduced rpS6 phosphorylation occurred in all four NSCLC cell lines examined and was seen at a low concentration of 3 µM EAD1 in some of these cell lines; it also caused a >90% reduction at the highest concentration tested. Phosphorylation of serines 240 and 244, which are downstream of mTORC1, occurred in all four cells lines, whereas decreases in serine 235, downstream of the RAS/MEK/ERK pathway were observed in two of the cell lines. Decreased phospho-rpS6 could be seen at 3 hours, with maximal effect by 6–24 hours of EAD1 treatment (Fig. 7F). Note that a strong reduction of phospho-rpS6 by EAD1 was also observed in the ATG7-minus and autophagy-deficient H1650 cells, which shows that this reduction was independent of any effect EAD1 has on autophagy. HCQ had a more modest effect on rpS6 phosphorylation, and only at the highest concentration tested (50 µM).
Fig. 7.
Effect of EAD1 on rpS6 phosphorylation at concentrations that inhibit cell proliferation and in H1650 cells that are autophagy-deficient. (A) Schematic diagram illustrating selected downstream signaling from mTOR. (B–E) Four NSCLC cell lines were treated with the indicated concentrations of EAD1 or HCQ for 24 hours, and the levels of phospho-rpS6 and total rpS6 were determined. H1650 are Atg7-and lack the canonical autophagy pathway. (F) Time course for the loss of pSer-rpS6 in H460 cells treated with 12 µM EAD1.
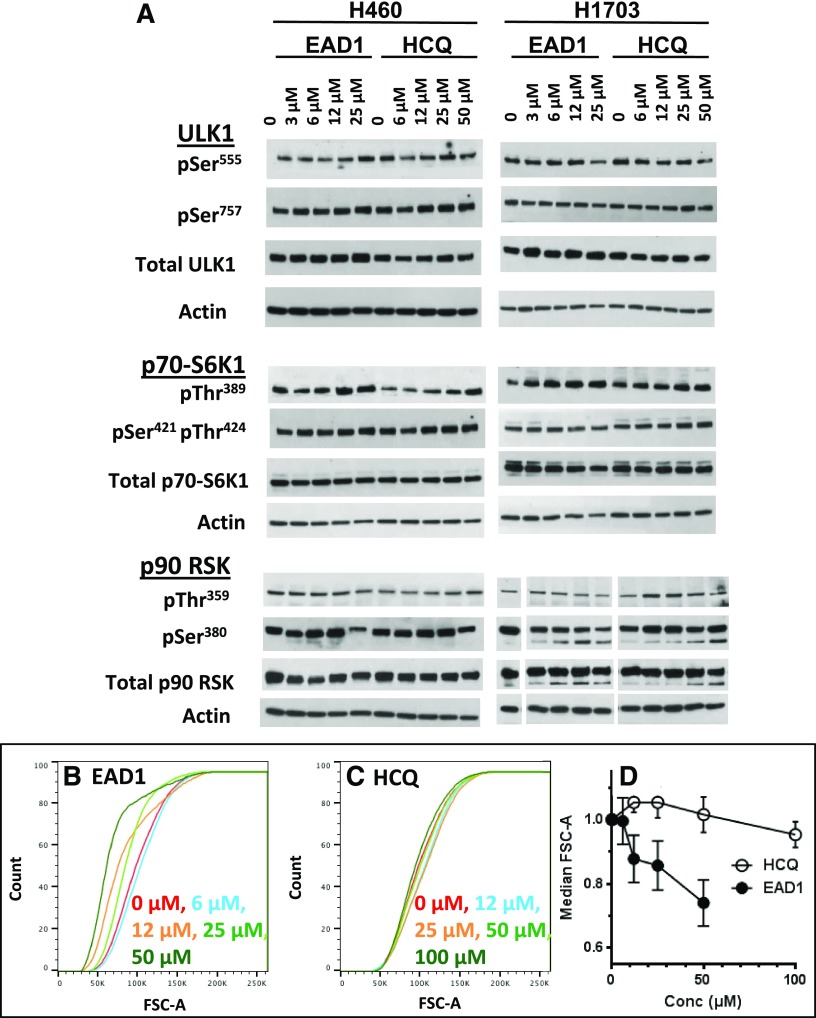
rpS6 phosphorylation can be modulated by a variety of physiologic and pathologic stimuli, and as rpS6 is downstream of two central cellular signaling pathways, multiple potential upstream sites could be responsible for the effects seen with EAD1. rpS6 is phosphorylated predominantly by two known family of kinases: p70 S6 kinases (S6Ks), which are downstream effectors of the PI3K/AKT/mTOR pathway and are the major kinases responsible for rpS6 phosphorylation, and the p90 S6Ks, which are downstream of the RAS/RAF/ERK pathway and play a lesser role in rpS6 phosphorylation (Fig. 7A). Both the p70 and p90 S6 kinases can be activated by phosphorylation, and we looked for changes in their phosphorylation as a possible explanation for the decrease in rpS6 phosphorylation. Rather than a decrease in activation, however, EAD1 caused a modest increase in p70-S6K1 phospho-Thr389 (H1703 and H460 cells) and p70-S6K1 phospho-Ser421/Thr389 (H1703 cells) (Fig. 8A). A modest decrease in the levels of phospho-p90 RSK were seen, although only at pSer380 and not at pThr359.
Fig. 8.
Effect of EAD1 on cell size and signaling components upstream of rpS6. (A) H460 and H1703 cells were treated with the indicated concentrations of EAD1 or HCQ for 24 hours. Cell lysates were analyzed by immunoblots for phospho- and total-ULK1 and the S6 kinases p70-S6K1 and p90 RSK. (B–D) H460 cells were treated for 24 hours with the indicated concentrations of EAD1 (B) or HCQ (C), stained with propidium iodide, and analyzed by flow cytometry. The size distribution of live G1-phase cells was determined (B and C). (D) Median cell size, as determined by FSC-A, for treated cells relative to control. Significant differences were found between HCQ and EAD1 at all concentrations (mean ± S.D., n = 3 experiments).
We also examined the effect of EAD1 on the phosphorylation of ULK1, as it also is downstream of mTORC1 in a pathway parallel to that of rpS6. Furthermore, phosphorylation of ULK1 on serine residues can lead to the initiation or inhibition of the canonical autophagy (Ser555 and Ser757, respectively), and it has also been implicated in noncanonical autophagy pathways (Nishida et al., 2009; Honda et al., 2014); however, neither EAD1 nor HCQ had a consistent effect on ULK1 phosphorylation in the NSCLC cell lines examined (Fig. 8A). Taken together, these data show that although EAD1 has a dramatic effect on mTOR localization and rpS6 phosphorylation, there were no obvious changes in intermediate steps in the pathway.
The schematic in Fig. 7A greatly oversimplifies the signaling inputs, outputs, and feedback regulation of the pathways in which mTOR participates. Rather than attempt to evaluate all possible changes, we decided to focus on the one change that was observed, rpS6 phosphorylation, and to ask whether there was a biologic consequence of this reduction. The downstream molecular and cellular effects of rpS6 phosphorylation remain somewhat obscure, despite extensive investigations (Meyuhas, 2015). One phenotypic change that is consistently observed, however, is cell-size regulation; thus, a wide variety of cell types derived from rpS6P−/− mice are significantly smaller than their wild-type counterparts (Meyuhas, 2015). We used flow cytometry to measure cell size in H460 cells treated for 24 hours with EAD1 (Fig. 8B) or HCQ (Fig. 8C) and found that the cumulative size distribution of viable G1-phase cells was significantly reduced in a concentration-dependent manner by EAD1, but not by HCQ (Fig. 8D). The decrease in cell size was consistent with the concentrations of EAD1 that reduced rpS6 phosphorylation and inhibited cell proliferation.
Discussion
Autophagy is a catabolic recycling process that can serve as an intracellular self-defense mechanism, allowing tumor cells to overcome stress and survive during oncogenesis and after treatment with chemotherapy (Mizushima et al., 2011). The inhibition of autophagy by CQ and HCQ has been shown to sensitize cancer cells to a range of cytotoxic and targeted drugs in preclinical models and is the basis for several ongoing clinical trials (Rebecca and Amaravadi, 2016). CQ and HCQ are weak bases that diffuse into lysosomes and neutralize their acidic pH, thereby preventing the fusion of the lysosome to autophagosomes and blocking the autophagic process. Although CQ and HCQ are known to have other effects on cells that could contribute to their anticancer actions, the predominant focus in recent years has been on their ability to block autophagy (Amaravadi et al., 2016). This focus has been called into question, however, by recent studies showing that the inhibitory effects of CQ on autophagy can be dissociated from its inhibitory effect on cell proliferation and its ability to overcome drug resistance (Maycotte et al., 2012; Eng et al., 2016; Chen et al., 2017). In these studies, leukemia and breast, colon, and pancreatic cancer cells retained full sensitivity to CQ after the complete abrogation of the canonical macroautophagy pathway by deletion of the autophagy regulatory proteins Atg7, Atg12, or beclin1. Furthermore, the ability of CQ to sensitize the cells to several anticancer drugs—including cisplatin, erlotinib, and sunitinib—occurred independent of the suppressive effects of CQ on autophagy. At the same time, Atg7 deficiency alone failed to sensitize cells to chemotherapy (Eng et al., 2016). These studies, however, did not identify any alternative, autophagy-independent mechanisms for CQ’s actions.
The importance of lysosomes in the control of cell death has prompted the search for agents that target this organelle (Kroemer and Jäättelä, 2005; Piao and Amaravadi, 2016). In a direct mechanism, leakage of cathepsins from lysosomes into the cytoplasm initiates both caspase-dependent and -independent apoptosis and necroptosis. Numerous stimuli that initiate this process have been identified, many of which are relevant to cancer cells (Kroemer and Jäättelä, 2005). Acting in part by effects on BID, BAX, Atgs, and other proteins, lysosomes also have indirect effects on programmed cell-death pathways, including apoptosis and all forms of autophagy. Besides the chloroquine derivatives, agents that have been investigated as targeting lysosomes include inhibitors of the lysosome-associated proteins vacuolar H+-ATPase, acid sphingomyelinase, cathepsins, and Hsp70 (Piao and Amaravadi, 2016). A recent medicinal chemistry approach identified novel dimeric quinacrines that can be targeted to lysosomes, can induce LMP, disrupt mTOR at the lysosomal membrane, inhibit autophagy, and have antitumor activity (Rebecca et al., 2017). Photoaffinity analysis in this report identified the lysosomal protein palmitoyl-protein thioesterase (PPT1) as a potential molecular target of the quinacrine agents.
Our data demonstrate multiple effects of EAD1 on lysosomes, suggesting that the organelle could be its site of action for killing lung cancer cells. EAD1 might mediate cell death via numerous mechanisms that are lysosome-dependent but autophagy-independent. For example, when CQ and HCQ become protonated and trapped in the lysosome, they can cause lysosomal swelling, cathepsin release, and autophagy-independent lysosomal cell death (LCD) (Boya et al., 2003; King et al., 2016). LCD is mediated by the cleavage of the BH3-only protein BID and subsequent caspase-3 activation and has been shown to be the mechanism for the synergistic cell death seen when CQ is combined with some anticancer drugs (King et al., 2016). Whereas LCD is usually initiated by LMP, less extensive permeabilization can also occur and does not necessarily lead to cell death. Thus, the degree of LMP can lead to different outcomes, suggesting that lysosomes and LMP have other cellular functions (Kroemer and Jäättelä, 2005; Maejima et al., 2013; Hämälistö and Jäättelä, 2016; Piao and Amaravadi, 2016). The role of the lysosome in the regulation of mTORC1 activity is particularly intriguing, as mTORC1 plays a central role in tightly controlling and coordinating cell growth and proliferation with nutritional status of the cell, receiving upstream signals from growth factors, intracellular energy levels, and amino acid availability (Sancak et al., 2008, 2010).
mTORC1 activity is regulated at the lysosome when Rag GTPases bring mTORC1 close to Rheb, allowing mTORC1 activation and subsequent phosphorylation of its downstream substrates (Sancak et al., 2008, 2010). Nutrient deprivation releases mTORC1 from lysosomes, thereby reducing its activity, whereas amino-acid replenishment restores lysosomal localization of mTORC1 and its activity. In addition to affecting the association of mTORC1 with lysosomes, another level of regulation is a function of the intracellular localization of mTORC1-lysosome complex, whereby it is active when located on lysosomes at the cell periphery but inactive when located on perinuclear lysosomes (Korolchuk et al., 2011). It is also possible that lysosomes located close to the cell periphery would promote activation of mTORC1 via signaling cascades originating at the plasma membrane, an action relevant to cancer cells with altered receptor tyrosine kinases (Carroll and Dunlop, 2017). We found that EAD1 potently decreases both the association of mTOR with lysosomes and its localization within the cells, both of which would be anticipated to inhibit or otherwise modulate its activity. It was noteworthy that, in contrast to the comparative effects of EAD1 and HCQ on autophagy inhibition, where the difference was mainly on the relative potency of the two, HCQ did not cause an apparent disruption of mTOR at the concentrations tested, suggesting that some of EAD1’s actions may be qualitatively different from that of HCQ.
Our experiments show that EAD1 decreases the phosphorylation of rpS6, an indispensable component of the 40S ribosome subunit and only one of two ribosomal proteins known to be phosphorylated in a regulated manner (Meyuhas, 2015). Changes in its phosphorylation occur in response to a variety of stimuli, including the presence or lack of growth factors, alterations in levels of amino acids, and changes in energy balance and oxygen supply (Meyuhas, 2015). These physiologic and pharmacologic stimuli affect rpS6 phosphorylation on five evolutionarily conserved serines in the C terminus. Loss of rpS6 phosphorylation does not affect the rate of total protein synthesis, but rpS6 phosphorylation does regulate the translation of selected mRNAs. It also regulates cell size, cell survival, and cell migration, including in lung cancer cells (Ruvinsky et al., 2005; Chen et al., 2015; Meyuhas, 2015). Phosphorylation may regulate other, yet-to-be identified functions of rpS6; however, as numerous proteins can bind specifically to unphosphorylated rpS6, but not to phosphorylated rpS6, suggesting that rpS6 can have biologic activity even in the unphosphorylated state (Wittenberg et al., 2016). Overexpression of rpS6 in low-expressing bronchial epithelial cells increases their rate of proliferation, whereas knockdown of rpS6 in high-expressing NSCLC cell lines inhibits their proliferation (Ruvinsky et al., 2005; Chen et al., 2014a). An interesting report found that a viral kinase identified in the Kaposi sarcoma-associated herpesvirus mimicked the S6 kinase’s activity and enhanced cell proliferation in infected cells, suggesting that in some instances, S6K can have oncogene-like activity (Bhatt et al., 2016). Reports studying rpS6 in human cancer found increased rpS6 phosphorylation levels in human sarcomas, leukemias, and lung and esophageal cancers (Iwenofu et al., 2008; Perl et al., 2012; Kim et al., 2013; Sun et al., 2014). A study of human NSCLC tumors found a significant correlation between high phospho-rpS6 and poor survival in patients with early stage lung cancer, supporting the hypothesis that phospho-rpS6 plays a significant role in cancer (Chen et al., 2015). Based on these and other studies, the S6Ks have been recognized as promising therapeutic targets and have led to the search for small molecule inhibitors (Byun et al., 2015; Jain et al., 2015).
EAD1 caused a modest and unexpected increase in phosphorylation of p70-S6K1. One explanation is that this increase could be a feedback response to the decrease in phospho-rpS6, analogous to that reported by Pearce et al. (2010) whereby a concentration of a specific p70-S6K1 inhibitor that completely suppressed phosphorylation of rpS6 also induced the rapid phosphorylation of S6K1. These authors suggest that there is a feedback loop by which S6K1 might regulate its own phosphorylation, although they did not identify the components of this loop. Alternatively, EAD1 may be somehow uncoupling the mTOR-S6K1-rpS6 signaling pathway.
In conclusion, a new chloroquinoline derivative, EAD1, inhibits the proliferation of lung cancer cells. Experiments to identify possible sites of action of EAD1 showed disruption of multiple properties of lysosomes, which correlated with its inhibition of cell proliferation, which were not seen with HCQ treatment and were independent of the compound’s effect on autophagy. Although the site of action of EAD1 is not yet precisely defined, the data are consistent with a direct effect on lysosomal function.
Acknowledgments
We thank Xheni Nishku and Frank Macaluso for assistance with the electron microscopy.
Abbreviations
- CQ
chloroquine
- FBS
fetal bovine serum
- HBSS
Hanks’ balanced salt solution
- HCQ
hydroxychloroquine
- LAMP
lysosome-associated membrane protein
- LCD
lysosomal mediated cell death
- LMP
lysosomal membrane permeabilization
- mTOR
molecular target of rapamycin
- mTORC1
mTOR complex 1
- NSCLC
non–small cell lung cancer
- rpS6
ribosomal protein S6
- S6K
S6 kinase
- SRB
sulforhodamine B
Authorship Contributions
Participated in research design: Sironi, Aranda, Schwartz.
Conducted experiments: Sironi, Aranda, Nordstrøm, Schwartz.
Performed data analysis: Schwartz.
Wrote or contributed to the writing of the manuscript: Schwartz.
Footnotes
This work was supported by National Institutes of Health, National Cancer Institute [Grants P30 CA013330], and the National Center for Advancing Translational Sciences [Grants UL1 TR001073, TL1 TR001072, and KL2 TR001071].
References
- Aits S, Kricker J, Liu B, Ellegaard AM, Hämälistö S, Tvingsholm S, Corcelle-Termeau E, Høgh S, Farkas T, Holm Jonassen A, et al. (2015) Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11:1408–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R, Kimmelman AC, White E. (2016) Recent insights into the function of autophagy in cancer. Genes Dev 30:1913–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. (2015) Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol Cell 59:270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AP, Wong JP, Weinberg MS, Host KM, Giffin LC, Buijnink J, van Dijk E, Izumiya Y, Kung HJ, Temple BR, et al. (2016) A viral kinase mimics S6 kinase to enhance cell proliferation. Proc Natl Acad Sci USA 113:7876–7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T, Perfettini JL, Kroemer G. (2003) Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 22:3927–3936. [DOI] [PubMed] [Google Scholar]
- Boya P, Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27:6434–6451. [DOI] [PubMed] [Google Scholar]
- Byun S, Lim S, Mun JY, Kim KH, Ramadhar TR, Farrand L, Shin SH, Thimmegowda NR, Lee HJ, Frank DA, et al. (2015) Identification of a dual inhibitor of Janus kinase 2 (JAK2) and p70 ribosomal S6 kinase1 (S6K1) pathways. J Biol Chem 290:23553–23562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Dunlop EA. (2017) The lysosome: a crucial hub for AMPK and mTORC1 signalling. Biochem J 474:1453–1466. [DOI] [PubMed] [Google Scholar]
- Chen B, Tan Z, Gao J, Wu W, Liu L, Jin W, Cao Y, Zhao S, Zhang W, Qiu Z, et al. (2015) Hyperphosphorylation of ribosomal protein S6 predicts unfavorable clinical survival in non-small cell lung cancer. J Exp Clin Cancer Res 34:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zhang W, Gao J, Chen H, Jiang L, Liu D, Cao Y, Zhao S, Qiu Z, Zeng J, et al. (2014a) Downregulation of ribosomal protein S6 inhibits the growth of non-small cell lung cancer by inducing cell cycle arrest, rather than apoptosis. Cancer Lett 354:378–389. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou L, Zhang Y, Leng Y, Pei XY, Lin H, Jones R, Orlowski RZ, Dai Y, Grant S. (2014b) Targeting SQSTM1/p62 induces cargo loading failure and converts autophagy to apoptosis via NBK/Bik. Mol Cell Biol 34:3435–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Clark J, Wunderlich M, Fan C, Davis A, Chen S, Guan JL, Mulloy JC, Kumar A, Zheng Y. (2017) Autophagy is dispensable for Kmt2a/Mll-Mllt3/Af9 AML maintenance and anti-leukemic effect of chloroquine. Autophagy 13:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CH, Wang Z, Tkach D, Toral-Barza L, Ugwonali S, Liu S, Fitzgerald SL, George E, Frias E, Cochran N, et al. (2016) Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci USA 113:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrenbacher N, Bastholm L, Kirkegaard-Sørensen T, Rafn B, Bøttzauw T, Nielsen C, Weber E, Shirasawa S, Kallunki T, Jäättelä M. (2008) Sensitization to the lysosomal cell death pathway by oncogene-induced down-regulation of lysosome-associated membrane proteins 1 and 2. Cancer Res 68:6623–6633. [DOI] [PubMed] [Google Scholar]
- Fehrenbacher N, Gyrd-Hansen M, Poulsen B, Felbor U, Kallunki T, Boes M, Weber E, Leist M, Jäättelä M. (2004) Sensitization to the lysosomal cell death pathway upon immortalization and transformation. Cancer Res 64:5301–5310. [DOI] [PubMed] [Google Scholar]
- Gewirtz DA. (2014) The four faces of autophagy: implications for cancer therapy. Cancer Res 74:647–651. [DOI] [PubMed] [Google Scholar]
- Goldberg SB, Supko JG, Neal JW, Muzikansky A, Digumarthy S, Fidias P, Temel JS, Heist RS, Shaw AT, McCarthy PO, et al. (2012) A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J Thorac Oncol 7:1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämälistö S, Jäättelä M. (2016) Lysosomes in cancer-living on the edge (of the cell). Curr Opin Cell Biol 39:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Arakawa S, Nishida Y, Yamaguchi H, Ishii E, Shimizu S. (2014) Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun 5:4004. [DOI] [PubMed] [Google Scholar]
- Iwenofu OH, Lackman RD, Staddon AP, Goodwin DG, Haupt HM, Brooks JS. (2008) Phospho-S6 ribosomal protein: a potential new predictive sarcoma marker for targeted mTOR therapy. Mod Pathol 21:231–237. [DOI] [PubMed] [Google Scholar]
- Jain R, Mathur M, Lan J, Costales A, Atallah G, Ramurthy S, Subramanian S, Setti L, Feucht P, Warne B, et al. (2015) Discovery of potent and selective RSK inhibitors as biological probes. J Med Chem 58:6766–6783. [DOI] [PubMed] [Google Scholar]
- Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, Chang MT, Ni A, Kundra R, Jonsson P, et al. (2017) Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov 7:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallunki T, Olsen OD, Jäättelä M. (2013) Cancer-associated lysosomal changes: friends or foes? Oncogene 32:1995–2004. [DOI] [PubMed] [Google Scholar]
- Kim SH, Jang YH, Chau GC, Pyo S, Um SH. (2013) Prognostic significance and function of phosphorylated ribosomal protein S6 in esophageal squamous cell carcinoma. Mod Pathol 26:327–335. [DOI] [PubMed] [Google Scholar]
- King MA, Ganley IG, Flemington V. (2016) Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene 35:4518–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13:453–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Jäättelä M. (2005) Lysosomes and autophagy in cell death control. Nat Rev Cancer 5:886–897. [DOI] [PubMed] [Google Scholar]
- Levy JMM, Towers CG, Thorburn A. (2017) Targeting autophagy in cancer. Nat Rev Cancer 17:528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. (2013) Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32:2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbaum J, Rollins N, Shah P, Bowman D, Lee JY, Tayber O, Bernard H, LeRoy P, Li P, Koenig, et al. (2015) Identification of a lung cancer cell line deficient in atg7-dependent autophagy. Autophagy DOI: 10.1080/15548627.2015.1056966 [published ahead of print]. [DOI] [PubMed]
- Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. (2012) Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8:200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyuhas O. (2015) Ribosomal protein S6 phosphorylation: four decades of research. Int Rev Cell Mol Biol 320:41–73. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27:107–132. [DOI] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. [DOI] [PubMed] [Google Scholar]
- Nordstrøm LU, Sironi J, Aranda E, Maisonet J, Perez-Soler R, Wu P, Schwartz EL. (2015) Discovery of autophagy inhibitors with antiproliferative activity in lung and pancreatic cancer cells. ACS Med Chem Lett 6:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce LR, Alton GR, Richter DT, Kath JC, Lingardo L, Chapman J, Hwang C, Alessi DR. (2010) Characterization of PF-4708671, a novel and highly specific inhibitor of p70 ribosomal S6 kinase (S6K1). Biochem J 431:245–255. [DOI] [PubMed] [Google Scholar]
- Perl AE, Kasner MT, Shank D, Luger SM, Carroll M. (2012) Single-cell pharmacodynamic monitoring of S6 ribosomal protein phosphorylation in AML blasts during a clinical trial combining the mTOR inhibitor sirolimus and intensive chemotherapy. Clin Cancer Res 18:1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao S, Amaravadi RK. (2016) Targeting the lysosome in cancer. Ann N Y Acad Sci 1371:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, et al. (2014) Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 10:1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecca VW, Amaravadi RK. (2016) Emerging strategies to effectively target autophagy in cancer. Oncogene 35:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecca VW, Nicastri MC, McLaughlin N, Fennelly C, McAfee Q, Ronghe A, Nofal M, Lim CY, Witze E, Chude CI, et al. (2017) A unified approach to targeting the lysosome’s degradative and growth signaling roles. Cancer Discov 7:1266–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto G, del Peso A, Zurita JL. (2008) Neutral red uptake assay for the estimation of cell viability/cytotoxicity. Nat Protoc 3:1125–1131. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. (2005) Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19:2199–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM. (2017) mTOR signaling in growth, metabolism, and disease [published correction appears in Cell (2017) 169:361–371]. Cell 168:960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CK, Zhang F, Xiang T, Chen Q, Pandita TK, Huang Y, Hu MC, Yang Q. (2014) Phosphorylation of ribosomal protein S6 confers PARP inhibitor resistance in BRCA1-deficient cancers. Oncotarget 5:3375–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers CG, Thorburn A. (2017) Targeting the lysosome for cancer therapy. Cancer Discov 7:1218–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann G. (1964) Labilization and stabilization of lysosomes. Fed Proc 23:1038–1044. [PubMed] [Google Scholar]
- Wiedmer T, Blank A, Pantasis S, Normand L, Bill R, Krebs P, Tschan MP, Marinoni I, Perren A. (2017) Autophagy inhibition improves sunitinib efficacy in pancreatic neuroendocrine tumors via a lysosome-dependent mechanism. Mol Cancer Ther 16:2502–2515. [DOI] [PubMed] [Google Scholar]
- Wittenberg AD, Azar S, Klochendler A, Stolovich-Rain M, Avraham S, Birnbaum L, Binder Gallimidi A, Katz M, Dor Y, Meyuhas O. (2016) Phosphorylated ribosomal protein S6 is required for Akt-driven hyperplasia and malignant transformation, but not for hypertrophy, aneuploidy and hyperfunction of pancreatic β-cells. PLoS One 11:e0149995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YH, Cho KS, Hwang JJ, Lee SJ, Choi JA, Koh JY. (2010) Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci 51:6030–6037. [DOI] [PubMed] [Google Scholar]