Abstract
The information characterizing key events in an Adverse Outcome Pathway (AOP) can be generated from in silico, in chemico, in vitro and in vivo approaches. Integration of this information and interpretation for decision making are known as integrated approaches to testing and assessment (IATA). One such IATA was published by Jaworska et al (2013) which describes a Bayesian network model known as ITS-2. The current work evaluated the performance of ITS-2 using a stratified cross validation approach. We also characterized the impact of replacing the most significant component of the network, output from the expert system TIMES-SS with structural alert information from the OECD Toolbox and Toxtree. Lack of structural alerts or TIMES-SS predictions, yielded a sensitization potential prediction of 79%. If the TIMES-SS prediction was replaced by a structural alert indicator, the network predictivity increased up to 87%. The original network’s predictivity was 89%. The local applicability domain of the original ITS-2 network was also evaluated using reaction mechanistic domains to understand what types of chemicals ITS-2 was able to make the best predictions for. We found that the original network was successful at predicting which chemicals would be sensitizers, but not at predicting their potency.
Keywords: skin sensitization, adverse outcome pathway (AOP), IATA, ITS, QSAR, TIMES-SS, reaction domains, OECD QSAR Toolbox, Toxtree
Introduction
The last decade has seen a major surge in global chemical regulation. Notable has been the Registration Evaluation Authorization restriction of CHemicals (REACH) regulation within Europe (EC 2006), similar programmes within China and Korea [1–4], and Toxic Substances Control Act (TSCA) reform within the US [5]. These regulations require large numbers of chemicals to be assessed for their potential human health and environmental impacts. Some regulations, such as REACH [1–2], stipulate that animal testing should be used only as a last resort and call for greater use of non-animal approaches such as (Q)SARs or in vitro methods.
The development of the Adverse Outcome Pathway (AOP) framework, which provides information on the causal links between a molecular initiating event (MIE), key events (KEs) and an adverse outcome (AO) of regulatory concern, offers the biological context to generate and interpret mechanistically relevant information from in vitro and QSAR studies [6,7]. The first AOP developed and endorsed by OECD was that for skin sensitization [8]. It was chosen since skin sensitization is an important endpoint in chemical legislation, has been well studied over several decades and the mechanistic understanding could be readily structured in the AOP framework. The skin sensitization AOP also served as the pilot case study to inform the development and application of integrated approaches to testing and assessment (IATA), new and revised test methods and chemical categories [9].
In brief, for skin sensitization to be induced, a chemical needs to gain access to the viable epidermis, be electrophilic either directly or upon transformation in order to bind covalently with skin proteins, and the ensuing hapten complex formed needs to mature and migrate to the draining lymph nodes activating keratinocytes and dendritic cells in the process [8]. At the draining lymph node, the complex is presented to the naïve T cells to cause the immune response thus resulting in the proliferation of memory T cells. Test methods notably the Direct Reactivity Peptide Assay (DRPA) (OECD TG 442c) exist to measure the covalent binding (the MIE), the KeratinoSens™ (OECD TG 442d) to measure the keratinocyte activation (KE2) and the dendritic activation by the human cell line activation test (h-CLAT) (KE3) [10–12]. The current animal test method typically conducted, the local lymph node assay (LLNA) (OECD TG 429) measures the T-cell proliferation as a cumulative impact of the preceding events [13].
The availability of data from these non-animal test methods has prompted much study in exploring ways and means of efficiently integrating these different information sources together for regulatory decision making. To address this need, a number of IATAs have been developed and published in the literature for skin sensitization. Examples include the ‘2 out of 3’ prediction model by researchers at BASF (this relies on a majority vote based on the outcomes from the KeratinoSens™ (or LuSens), Myeloid U937 skin sensitization test (MUSST) or h-CLAT and DRPA) (Urbisch et al.), artificial neural network approaches by researchers in Japan [14–15], and a Bayesian prediction model by researchers at RIVM [16]. One of the IATA developed was a Bayesian network known as ITS-2 that was developed by researchers at P&G [17]. This ITS-2 network was also implemented into an open source tool [18] accessible on the NICEATM website [see http://ntp.niehs.nih.gov/pubhealth/evalatm/test-method-evaluations/immunotoxicity/nonanimal/index.html#NICEATM-Collaboration-With-P-G-to-Develop-an-Open-source-Integrated-Testing-Strategy].
In this current study, we evaluated the performance of ITS-2 globally (using cross validation) and locally (using reaction mechanistic domains). ITS-2 [17] was selected as a case study since the network is freely available in an open source format. An update to this network, ITS-3 has since been published by Jaworska et al. [19], but a non-proprietary version is not currently available.
The ITS-2 network predicts the sensitizing potency category as would be derived from the LLNA using a number of inputs which can be mapped to the corresponding AOP. The LLNA provides a quantitative measure of sensitizing potency, known as the EC3. This is the test concentration causing a threefold increase of lymph node cell proliferation compared to the vehicle control. Determinations of potency based on EC3 values have been shown to correlate closely with what is known of the relative ability of contact allergens to cause skin sensitization among humans. As such proposals have been made to categorize contact allergens according to their skin sensitizing potency [25,26]. Kimber et al., (2003) identified four sub-categories: ‘extreme’, ‘strong’, ‘moderate’ and ‘weak’ based on thresholds defined by specific, derived EC3 values [25]. In the ITS-2 network, strong and extreme potency categories were collapsed into one category. The four potency categories chosen in the ITS-2 network: non-sensitizing, weak, moderate and strong/extreme, are labelled as numbers from 1 to 4. Non-sensitizers with no EC3 value calculated are denoted category 1. Weak sensitizers (category 2) correspond to an EC3 value greater than 10%. Moderate sensitizers (category 3) have an EC3 value between 1 and 10% and strong/extreme sensitizers (category 4) correspond to an EC3 value less than 1%.
ITS-2 aims to predict the sensitization potency as measured in the LLNA [17]. The network inputs cover each of the events described in the AOP. Skin bioavailability is addressed by parameters used to model skin penetration although it should be noted that current evidence suggests that skin penetration is not a determining factor for inducing skin sensitization [27–29]. The DRPA is used to characterize the MIE, the covalent binding to skin proteins. The KeratinoSens™ and MUSST are used to characterise KEs 2 and 3, the activation of keratinocytes and dendritic cells respectively. A prediction from the commercial expert system TIMES-SS is also considered, to account for the inherent reactivity of a chemical in terms of whether it can react directly or requires activation either enzymatic or chemical in nature. TIMES-SS is a module within the TIssue MEtabolism Simulator platform which relies on structure-metabolism and structure-activity rules for the prediction of skin sensitization potency. The development and performance of the TIMES-SS expert system has been discussed in much more detail elsewhere [20,21].
Our study sought to evaluate the global performance of the ITS-2 model using cross validation, explore freely available alternatives to TIMES-SS and assess their impact on predictive performance. The way in which TIMES-SS works is as follows: a chemical is introduced into TIMES-SS and matched against a list of all its hierarchical transformations (these comprise both structure-activity and structure-metabolism relationships). For all the matches identified, the reactive species or metabolic species and their respective protein adducts are then generated. Some pathways are underpinned by 3D-QSAR models which assign potency. These same structure-activity relationships (or structural alerts) are also implemented in the OECD QSAR Toolbox (version 3.3), a freely available tool, as a protein binding for skin sensitization profiler. A second freely available tool, Toxtree (Ideaconsult Ltd) includes a module to assign reaction mechanistic domains as described by Roberts and Aptula [22] using SMiles ARbitrary Target Specification (SMARTS) derived by Enoch et al. [30]. It should be noted that the Toxtree module does not incorporate any autoxidation or metabolism simulators. These profilers were used as surrogates for TIMES-SS. Alerts identified by these profilers were categorized into one of the 5 reaction mechanistic domains described by Roberts and Aptula [22]. These organic chemistry principles have been used to evaluate many other compilations of skin sensitization data to rationalize their skin sensitizing behaviour (see references [23,24]). The five domains are Schiff base formers, Acylating agents, Michael acceptors, SN2 and SNAr. Chemicals which do not fit into these domains are either non-reactive or have special considerations such as acting via a free radical route or by a SN1 reaction scheme. The reaction domains, as identified using the OECD QSAR Toolbox, were then used to evaluate the practical utility of the ITS-2 model in terms of its local chemistry domain – i.e. whether specific reaction domains were better represented, rendering the model more predictive for one or other reaction domain. This evaluation was termed a ‘local validity analysis’.
Materials and Methods
Although our study intended to evaluate the predictive performance of ITS-2 as published, the open source version of this network as coded in the R programming language made available by Pirone et al. was used in practice [18]. This was a re-derivation of the same network, and used the same training and test sets as referenced in Jaworska et al. [17]. There were some minor differences in the initial settings that could not be recreated in the open source version of the network which are discussed in more detail in Pirone et al., [18] but the R version still yielded similar results to the original ITS-2 network on both the test and training sets.
The ITS-2 network predicts the probability of a LLNA potency category from conditional probability tables (CPTs) generated using discretized values of the input variables (Table 1). The conditional probability tables are based on the location of the nodes in a network. Table 2 provides an example of a conditional probability table for two of the input parameters in the network; bioavailability and LogKow. The conditional probability tables for the entire network calculated based on the entire data set are given in the supplemental information. The conditional probability tables were generated with the R package gRain, which uses a variant of the junction tree algorithm to estimate the parameters for the conditional probability tables based on the training data [31]. To determine the CPTs for the latent variables of cysteine and bioavailability, the R package poLCA was used [32].
Table 1.
Input variables used in the ITS-2 network.
| Input Nodes | Description |
|---|---|
| LogKow, Cfree, and AUC120 | LogKow is the log of the octanol-water partition coefficient, AUC120 the area under the flux curve at 120 hrs as a percentage of the applied dose and Cfree the free test chemical concentration in the mid epidermis multiplied by the thickness of viable epidermis expressed as a percentage of applied dose. |
| DPRACys, DPRALys | Percentage of peptide remaining after reaction in the DPRA cysteine and lysine assay respectively |
| KEC1.5, KEC3, IC50 | KEC1.5 and KEC3 are the amounts needed to see a 1.5 and 3 fold increase of luciferase activity in the KeratinoSens™ assay. IC50 is used to control for cell viability in the assay |
| CD86 | EC150 (uM) Amount needed for 150% cell surface activation in the U937 assay |
| TIMES-SS | Skin sensitization potency as predicted by the TIMES-SS module – non-sensitizer, weak and moderate/strong/extreme |
Table 2:
Example of a conditional probability table.
| Conditional Probability Table for LogKo/w and Bioavailability | |||
|---|---|---|---|
| Bioavailability | |||
| LogKow | Cluster 1 | Cluster 2 | Cluster 3 |
| [-Inf, 0.094] | 22.6 % | 16.0 % | 0.4 % |
| [0.094, 1.92] | 39.5 % | 31.9 % | 47.0 % |
| [1.92, 3.83] | 30.2 % | 31.9 % | 52.2 % |
| [3.83, Inf] | 7.7 % | 20.3 % | 0.4 % |
Global Performance Assessment of the ITS-2 network
The test set used for external validation purposes in Jaworska et al. comprised 21 chemicals [17]. This limited a robust assessment of true predictivity. Here, we sought to evaluate the performance of the ITS-2 using a stratified 10-fold cross-validation. Stratified cross-validation is a process where a single dataset is broken up into different groups of the same size, in this case 10. The stratification refers to the fact that the data is distributed so that each group has the same number of items with a given proportion of properties. The procedure allowed for a more robust evaluation of the ITS-2 network to be performed. The training and test sets reported in Jaworska et al. [17] were combined together to form a dataset of 145 chemicals. Four chemicals namely 5910–85-0, 3326–32-7, 2785–87-7, 26172–55-4 were removed from further consideration as they did not contain a complete dataset of information. Groups were stratified based on their LLNA classification, so that a typical group had 4 non-sensitizers, 3 weak sensitizers, 4 moderate sensitizers, and 3 strong/extreme sensitizers.
The cross validation was performed using modifications to the R scripts from Pirone, et al. [18] and the instructions for their use are provided in the supplementary information. Figure 1 outlines the procedure followed for performing the cross validation.
Figure 1.
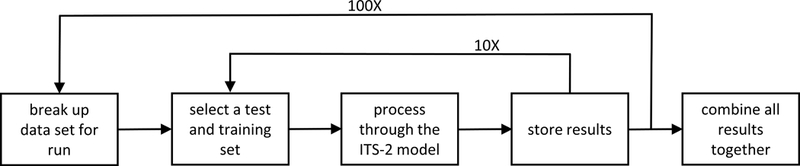
Diagram of the different steps in running the stratified cross-validation.
The first step of our code allows each individual subset of 10 runs to occur on multiple processors using the DoMC package for R [33]. The second step splits the dataset into the 10 separate folds for each run. The third step uses the original R code from Pirone, et al. [18]. All but one of the sets were combined into the training set. The remaining set was used as the test set looping through so that each of the 10 sets becomes the test set once, while the remainder comprise the training set. The results were stored for each run and the test completed 10 times before moving on to complete another set. Lastly, the results were combined using Python scripts and output into a csv file for subsequent evaluation. An example is presented in the supplemental information.
The convergence in distance weighted error was used to judge whether an optimal number of cross validation runs had been performed [34]. The distance weighted error was calculated using equation 1 [34].
| (1) |
Where pi is the probability that a given compound will be in a particular class and d(class,i) is the absolute distance the prediction is from the correct class when the classes are numbered sequentially 1 through 4. To determine the uncertainty of each run, an interval was computed using the quantiles function in R to simply select the 5% and 95% percentile cut-offs and subtract those from the mean to determine the uncertainty. For all values the upper and lower bounds are given as a super and subscript respectively example, .
Evaluation of global network performance using two freely available alternatives to TIMES-SS
The most significant input to the ITS-2 network was the information arising from TIMES-SS. Jaworska, et al. [17] reported a relative mutual information between the LLNA result and the TIMES-SS prediction of 36%, significantly higher than any other node. Given TIMES-SS is a commercial expert system, we sought to identify a surrogate for the sort of information TIMES-SS provided and evaluate the effect this had on the performance characteristics. Of the 145 chemicals used in the ITS-2 dataset, 95 were part of the training set underpinning TIMES-SS. This could mean that the ITS-2 model is over-fitted due to its high reliance on the TIMES-SS prediction. TIMES-SS identifies electrophilic features within chemical structures either directly or upon activation based on a set of structure-activity and structure-metabolism rules.
Alerts were identified using the protein binding profiler by OASIS version 1.3, which is freely available in the OECD QSAR Toolbox v3.3 and the SMARTS module within Toxtree v2.6.13. Alert predictions were then made using both tools on all of the autoxidation products and metabolites generated in the OECD QSAR Toolbox using the OASIS autoxidation simulator v3.3 and the OASIS skin metabolism simulator v3.2. The profiling outcomes were summarized and converted into a binary score, 1 to signify presence of an alert and 0 to signify absence of an alert(s). The node was renamed “reaction (rxn) alert” for all four sets of alert outcomes, i.e. OECD QSAR Toolbox alerts with and without metabolism and autoxidation as well as the Toxtree alerts with and without metabolism and autoxidation. A control network was also constructed to provide a measure of the baseline performance relative to the original ITS-2 with TIMES-SS and the modified network which included the profiling alert as its alternative. Both networks were tested using the same stratified cross-validation procedure and the distance weighted error values checked for convergence.
Local Validity Analysis using the OECD QSAR Toolbox protein binding alerts
An analysis of the local performance of the ITS-2 network on the basis of the reaction mechanistic domains as described by Roberts and Aptula [22] was also undertaken. The results for each chemical in the cross validation procedure had to be separated out on the basis of its presumed reaction mechanistic domain to enable the analysis. The OASIS v1.3 protein binding alerts, previously used to replace the TIMES-SS node (see above), were also used to determine one of the 5 main reaction mechanistic domains as previously discussed. A sdf (structure data file) file was generated by matching CAS numbers listed in the original dataset against the DSStox inventory [35]. The addition of the reaction domain enabled the mean and standard deviation of each result to be determined. All code/scripts are available in supplemental file 1.
Results
Evaluation of global network performance
The modified network where TIMES-SS is replaced with the reaction alert node is presented in figure 2. The performance of the network was assessed with each of the 4 different reaction alerts i.e. the OECD QSAR Toolbox and Toxtree with and without activation. Figure 3 shows the baseline performance network with the TIMES-SS node removed.
Figure 2.
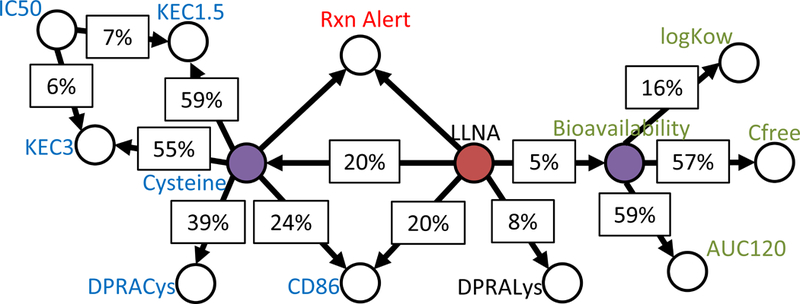
The original ITS-2 network modified to contain a reaction alert (Rxn Alert) node in place of the TIMES-SS node. The reaction alert node is based upon the outcome of the OASIS version 1.3 protein binding for skin sensitization tool in the OECD Toolbox.
Figure 3.
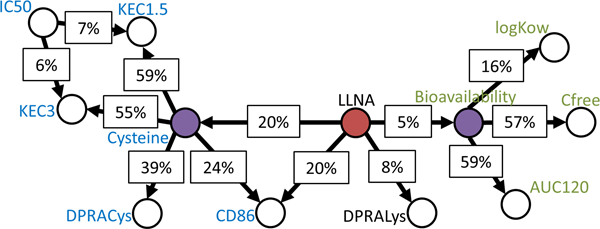
The original ITS-2 network with the TIMES-SS node removed, used to establish a baseline for prediction without input from TIMES-SS or the reaction alerts.
Plotting the distance weighted error over 100 runs showed that convergence had been reached - a negligible change in slope of the cumulative average distance weighted error was found when 100 runs of 10X stratified cross validation was selected for the original ITS-2 network and the 2 modified networks (see Figure 1 in the supplemental information).
Table 3 provides the overall average results of all three networks for both their ability to distinguish sensitizers from non-sensitizers and their prediction of the correct LLNA potency class from the 10-fold cross validation. This comparison revealed that the original ITS-2 network gave the most accurate results, distinguishing sensitizers from non-sensitizers of the time and LLNA potency class . If the OECD QSAR Toolbox predictions for reaction alerts with metabolism and autoxidation was used, the network’s ability to distinguish sensitizers from non-sensitizers was , almost the same as the original network with TIMES-SS itself. The prediction of the LLNA potency class was examined in more detail in table 4 to determine if an over or under prediction was more likely. This analysis revealed that the original ITS-2 network provides the best accuracy for both under and over prediction (table 4). The modified network never performed as well as the original network for the prediction of the LLNA potency, regardless of the approach used to derive the reaction alerts.
Table 3:
Overall averages of the results of 100 runs of stratified 10-fold cross validation on all six networks based on the 141 compound dataset.
| Stratified 10-fold Cross Validation | ||
|---|---|---|
| Test Set | Correct for Sensitization | Correct LLNA Potency Class |
| Original Network | ||
| No TIMES-SS | ||
| Toolbox Alerts | ||
| Toolbox Alerts with Metabolism and Autoxidation | ||
| Toxtree Alerts | ||
| Toxtree Alerts with Metabolism and Autoxidation | ||
Where Toolbox denotes the OECD QSAR Toolbox
Table 4:
How often a network predicted the LLNA potency class correctly, for 141 compounds as well as how often it over or under predicted the LLNA potency class.
| Over and Under Prediction of LLNA Potency class | |||
|---|---|---|---|
| Test Set | Correct LLNA Potency Class | Over predicted | Under predicted |
| Original Network | |||
| No TIMES-SS | |||
| Toolbox Alerts | |||
| Toolbox Alerts with Metabolism and Autoxidation | |||
| Toxtree Alerts | |||
| Toxtree Alerts with Metabolism and Autoxidation | |||
Where Toolbox denotes the OECD QSAR Toolbox
Twelve chemicals out of a total of 141 from the dataset were found to have their sensitization potential predicted correctly by all three networks over the 100 runs (regardless of the approach used to derive the reaction alerts). Two chemicals were predicted incorrectly on every run by the three networks. (Interestingly all of the chemicals predicted correctly and incorrectly every time were part of the TIMES training set.) The chemicals and their structures are shown in figure 4. (Note: Oxalic acid [144–62-7] is a known false positive in the LLNA and should be disregarded [36,37]). A list of all chemicals with their prediction percentages in the original network is provided in the supplementary information.
Figure 4.
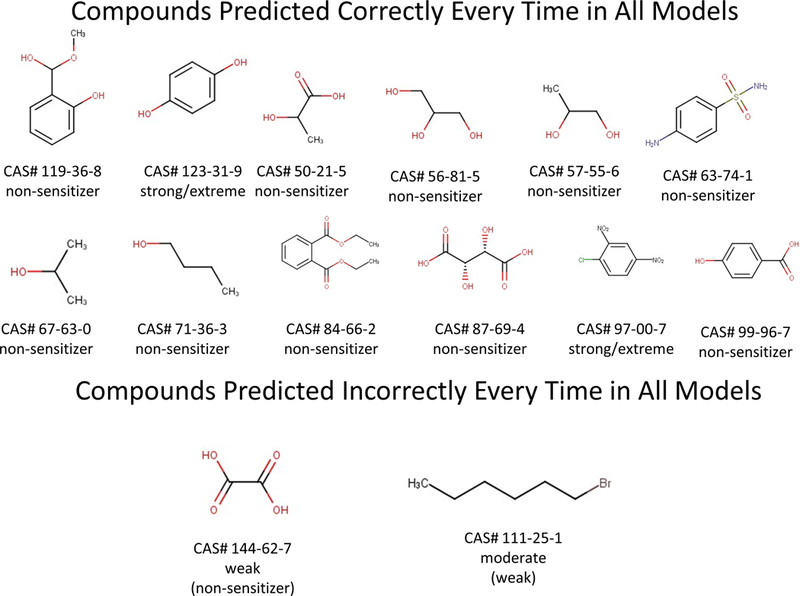
The 14 chemicals that were predicted correctly during all runs in all three networks as well as the five that were predicted incorrectly every time in all three models. The results of the actual LLNA tests are given below the chemical figure; those predicted incorrectly also include the most common prediction in parentheses.
All three networks were better at predicting non-sensitizers. These represent 10 of the 12 (83%) chemicals predicted correctly every time despite representing only 42 out of 141 (30%) all chemicals in the dataset. This could be because if a chemical does not show any positive results in any test, it can be readily rationalized to be a non-sensitizer, whereas a chemical with several positive results is likely to be a sensitizer even if its specific potency class may be more difficult to predict.
Local validity analysis
We compared the results of the original network to the reaction domain of each chemical provided by the OECD QSAR Toolbox. We chose to make our analysis without the use of metabolism and autoxidation given we found no significant difference in the global performance of the network. Chemicals with a Michael addition alert were the most likely to have their LLNA potency class predicted correctly, even without applying the Michael addition correction which the original authors suggested. The correction accounts for the fact that when predicted incorrectly, chemicals with a Michael addition alert tend to be over predicted. The results of all chemicals run in the original ITS-2 network grouped by reaction alert are given in table 5. A complete list of the reaction alerts reported for each chemical can be found in the supplemental information.
Table 5:
Results for sensitizing and non-sensitizing predictions for 141 chemicals as well as the LLNA potency class accuracy for all chemicals predicted in the original network, grouped by reaction domain. (Four chemicals are missing from this table, 2 had alerts for nucleophilic addition and the other 2 had alerts for SNAr.)
| Alert | Number of Compounds |
Sensitization Accuracy |
LLNA Potency Class Accuracy |
Over Prediction of Class |
Under Prediction of Class |
|---|---|---|---|---|---|
| Acylation | 15 | ||||
| Michael Acceptor | 21 | ||||
| No Alert Found | 64 | ||||
| Schiff Base Former | 23 | ||||
| SN2 | 14 | ||||
Discussion
Our work suggest that the ITS-2 network is well suited to the prediction of skin sensitization potential. However, it is unlikely to give correct results for LLNA potency with the exception of select local cases based on reaction domain analysis. Currently the largest problem with the network is overfitting, due to its heavy reliance on the results of predictions made using TIMES-SS. We have however shown that even when this node is removed the network can still predict sensitization potential correctly and that the TIMES-SS node may be replaced with predictions from the associated profiler in the OECD QSAR Toolbox.
Global Network Performance
The original test of the ITS-2 network by Jaworska et al. [17] reported a 95% correct sensitization potential and a 86% LLNA class prediction. Our cross validation analysis showed a sensitization potential and a LLNA potency class prediction. The original evaluation used a training set of 124 and a test set of only 21 chemicals. Using the technique of cross-validation, we were able to apply a much more rigorous evaluation of the network, using the same limited amount of data. The results showed that LLNA potency was much more difficult to predict, which we would expect given the wide variability within the LLNA [26].
The results of the original network may also be overstated due to the fact that the TIMES-SS training set contains 89 of the 124 chemicals in the data set. Eight of the 21 chemicals used in the evaluation of the network by Jaworska, et al. [17] also were part of the TIMES-SS training set. When the network was run without TIMES-SS, i.e. the baseline network, the performance for predicting sensitization potential was still reasonable at of the time. The performance of predicting LLNA potency class fared less favourably showing a drop in accuracy to only , thus while the ITS-2 network may be unable to predict the LLNA potency class without information from TIMES-SS, it is still performs well in predicting sensitization potential.
The performance of the network improved when the reaction alert node in lieu of TIMES-SS were included, for sensitization potential, when using the reaction alerts from the OECD QSAR Toolbox with metabolism and autoxidation. A for LLNA potential class, when the alerts from the OECD QSAR Toolbox without autoxidation were used. Using any other prediction of reaction alerts from the OECD QSAR Toolbox or Toxtree, with or without metabolism yielded better results than the network with no reaction node. This improvement is not surprising given that the reaction alerts and the simulators for autoxidation and metabolism contained within the OECD QSAR Toolbox arise from the TIMES-SS model itself. Given the marginal difference in performance across the different reaction alerts when metabolism/autoxidation was considered, reaction alerts alone as derived from the OECD QSAR Toolbox could be practically used in place of TIMES-SS, for predicting sensitization potential.
Local validity analysis
The local domain analysis identifies chemicals where the network would be most effective at predicting LLNA potency. Although the number of chemicals with a particular reaction domain is small and the confidence intervals for their predictions are high when compared to the overall performance of the network, the results are still clearly significant as can be seen in figure 5. Chemicals identified as Michael acceptors or SN2 electrophiles have an excellent probability of having their sensitization potential predicted correctly. This fits with what is currently known about the ability of the non-animal assays leading into the network mainly that the DPRACys and KeratinoSens™ assays tend to work best with soft electrophiles [38]. While LLNA class prediction was poor for those with an SN2 domain, it was excellent for Michael acceptors at , i.e. significantly above the overall class prediction of (This could be a reflection on the assays however, which may be good for assessing whether or not a chemical may be a sensitizer, but not its potency.) It also follows that hard electrophiles like acylating agents would have the most difficult time being predicted correctly. We see that this is true, with acylating agents not only having the worst results of all reaction types for predicting sensitization potential but also for LLNA potency class prediction.
Figure 5.
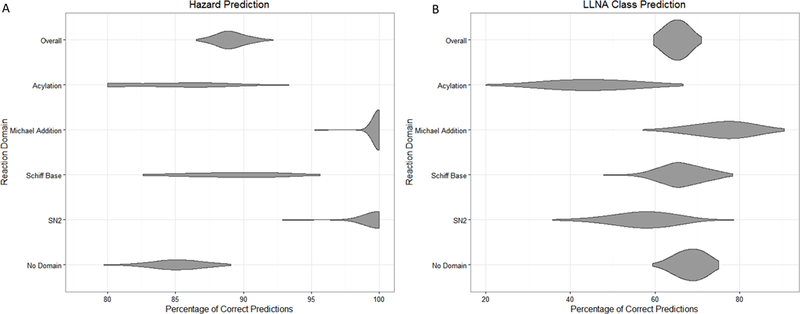
Violin plots showing the probability density for a given prediction. (a) Hazard confidence intervals based on reaction domains; (b) LLNA confidence intervals based on reaction domains.
Conclusions
The cross validation analysis performed herein confirms the previous finding [17] that the ITS-2 model predicts skin sensitization potential with reasonable accuracy. This validation also demonstrated that the ITS-2 network was generally unsuitable for correctly predicting LLNA potency. The strong performance found compared with the original network when using alerts identified by the OECD QSAR Toolbox demonstrated that these can be used in place of TIMES-SS to predict skin sensitization potential.
Supplementary Material
Footnotes
Disclaimer
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. The authors alone are responsible for the content and writing of this article.
References
- 1.The European Parliament and the Council of the European Union, Corrigendum to Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC, Official Journal of the European Union. L136 (2007), pp. 3–280. [Google Scholar]
- 2.The European Parliament and the Council of the European Union, Regulation (EC) No 1907/2006 of the European Parliament and of the council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC, Official Journal of the European Union. R1907 (2006), pp. 1–101. [Google Scholar]
- 3.National Assembly in Korea, Revised Korea REACH - The Act on the Registration and Evaluation of Chemicals, Chemical Inspection and Regulation Service; (2016). Available at http://www.cirs-reach.com/news-and-articles/revised-korea-reach---the-act-on-the-registration-and-evaluation-of-chemicals.html [Google Scholar]
- 4.Chinese Ministry of Environmental Protection (MEP), The Measures for Environmental Administration of New Chemical Substances (China MEP Order 7), 2010. Available at http://www.chemsafetypro.com/Topics/China/China_REACH_MEP_Order_7_New_Substance_Notification.html. [Google Scholar]
- 5.United States Government, Frank R Lautenberg Chemical Safety for the 21st Century Act, 114th Congress, Washington, District of Columbia, USA, 2016. [Google Scholar]
- 6.Ankley G, Bennett R, Erickson R, Hoff D, Hornung M, Johnson R, Mount D, Nichols J, Russom C, Schmieder P, Serrano J, Tietge J, and Villeneuve D, Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment, Environ. Toxicol. Chem. 29 (2010), pp. 730–41. [DOI] [PubMed] [Google Scholar]
- 7.Villeneuve D, In Response: The path forward for the adverse outcome pathway framework—A regulatory perspective, Environ. Toxicol. Chem. 34 (2015), pp. 1938–40. [DOI] [PubMed] [Google Scholar]
- 8.OECD, The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins, OECD Publishing; (2014). [Google Scholar]
- 9.Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts D, Mekenyan O, Towards AOP application – Implementation of an integrated approach to testing and assessment (IATA) into a pipeline tool for skin sensitization, Regul. Toxicol. Pharmacol. 69 (2014), pp. 529–545. [DOI] [PubMed] [Google Scholar]
- 10.OECD. OECD GUIDELINE FOR THE TESTING OF CHEMICALS In Chemico Skin Sensitisation: Direct Peptide Reactivity Assay (DPRA) TG 442C. 2015. [cited 2016 Jul 8]; Available from: http://ntp.niehs.nih.gov/iccvam/suppdocs/feddocs/oecd/oecd-tg442c-508.pdf [Google Scholar]
- 11.OECD. OECD GUIDELINE FOR THE TESTING OF CHEMICALS In Vitro Skin Sensitisation: ARE-Nrf2 Luciferase Test Method TG 442D. 2015 [Google Scholar]
- 12.OECD. OECD GUIDELINE FOR THE TESTING OF CHEMICALS 1 DRAFT PROPOSAL FOR A NEW TEST GUIDELINE 2 In Vitro Skin Sensitisation: human Cell Line Activation Test (h-CLAT). 2014 [Google Scholar]
- 13.Urbisch D, Mehling A, Guth K, Ramirez T, Honarvar N, Kolle S, Landsiedel R, Jaworska J, Kern P, Gerberick F, Natsch A, Emter R, Ashikaga T, Miyazawa M, Sakaguchi H, Assessing skin sensitization hazard in mice and men using non-animal test methods, Regul. Toxicol. Pharmacol. 71 (2015), pp. 337–351. [DOI] [PubMed] [Google Scholar]
- 14.Hirota M, Kouzuki H, Ashikaga T, Sono S, Tsujita K, Sasa H, Aiba S, Artificial neural network analysis of data from multiple in vitro assays for prediction of skin sensitization potency of chemicals, Toxicol In Vitro 27 (2013), pp. 1233–1246. [DOI] [PubMed] [Google Scholar]
- 15.Hirotaa M, Fukuib S, Okamotob K, Kurotanic S, Imaic N, Fujishirod M, Kyotanid D, Katoe Y, Kasaharaf T, Fujitaf M, Toyodag A, Sekiyah D, Watanabeh S, Setoi H, Takenouchij O, Ashikagaa T, and Miyazawaj M, Evaluation of combinations of in vitro sensitization test descriptors for the artificial neural network-based risk assessment model of skin sensitization: Evaluation of the descriptor for ANN risk assessment of skin sensitization, J. Appl. Toxicol. 35 (2015), pp. 1333–1347. [DOI] [PubMed] [Google Scholar]
- 16.van der Veen J, Rorije E, Emter R, Natsch A, van Loveren H, Ezendam J, Evaluating the performance of integrated approaches for hazard identification of skin sensitizing chemicals, Regul. Toxicol. Pharmacol 69 (2014), pp. 371–379. [DOI] [PubMed] [Google Scholar]
- 17.Jaworska J, Dancik Y, Kern P, Gerberick F, Natsch A, Bayesian integrated testing strategy to assess skin sensitization potency: from theory to practice, J. Appl. Toxicol. 33 (2013), pp. 1353–1364. [DOI] [PubMed] [Google Scholar]
- 18.Pirone J, Smith M, Kleinstreuer N, Burns T, Strickland J, Dancik Y, Morris R, Rinckel L, Casey W, and Jaworska J, Reproducing the ITS-2 model using R, (2014) Available from: http://ntp.niehs.nih.gov/iccvam/methods/immunotox/ITS-OS/ITS2_R_version-508.pdf [DOI] [PubMed]
- 19.Jaworska J, Natsch A, Ryan C, Strickland J, Ashikaga T, Miyazawa M, Bayesian integrated testing strategy (ITS) for skin sensitization potency assessment: a decision support system for quantitative weight of evidence and adaptive testing strategy, Arch. Toxicol. 89 (2015), pp. 2355–2383. [DOI] [PubMed] [Google Scholar]
- 20.Dimitrov S, Low L, Patlewicz G, Kern P, Dimitrova G, Comber M, Phillips R, Niemela J, Bailey P, Mekenyan O, Skin sensitization: Modelling based on skin metabolism simulation, Int. J. Toxicol. 24 (2005), pp. 189–204. [DOI] [PubMed] [Google Scholar]
- 21.Patlewicz G, Kuseva C, Mehmed A, Popova Y, Dimitrova G, Ellis G, Hunziker R, Kern P, Low L, Ringeissen S, Roberts D, Mekenyan O, TIMES-SS--recent refinements resulting from an industrial skin sensitisation consortium, SAR QSAR Environ Res. 25 (2014), pp. 367–391. [DOI] [PubMed] [Google Scholar]
- 22.Roberts D, Aptula A, Determinants of skin sensitisation potential, J. Appl. Toxicol. 28 (2008), pp. 377–387. [DOI] [PubMed] [Google Scholar]
- 23.Roberts D, Aptula A, Patlewicz G, Electrophilic Chemistry Related to Skin Sensitization. Reaction Mechanistic Applicability Domain Classification for a Published Data Set of 106 Chemicals Tested in the Mouse Local Lymph Node Assay, Chem Res Toxicol. 20 (2007), pp. 44–60. [DOI] [PubMed] [Google Scholar]
- 24.Roberts D, Patlewicz G, Kern P, Gerberick F, Kimber I, Dearman R, Ryan C, Basketter D, and Aptula A, Mechanistic Applicability Domain Classification of a Local Lymph Node Assay Dataset for Skin Sensitization, Chem. Res. Toxicol. 20 (2007), pp. 1019–1030. [DOI] [PubMed] [Google Scholar]
- 25.Kimber I, Basketter D, Butler M, Gamer A, Garrigue J, Gerberick G, Newsome C, Steiling W, Vohr H, Classification of contact allergens according to potency: proposals, Food Chem. Toxicol. 41 (2003), pp. 1799–1809. [DOI] [PubMed] [Google Scholar]
- 26.Basketter D, Clapp C, Jefferies D, Safford B, Ryan C, Gerberick F, Dearman R, Kimber I, Predictive identification of human skin sensitization thresholds, Contact Dermatitis. 53 (2005), pp. 260–267. [DOI] [PubMed] [Google Scholar]
- 27.Roberts D, Mekenyan O, Dimitrov S, Dimitrova G, What determines skin sensitization potency-myths, maybes and realities. Part 1. The 500 molecular weight cut-off, Contact Dermatitis. 68 (2012), pp. 32–41. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick J, Roberts D, Patlewicz G, What determines skin sensitization potency: Myths, maybes and realities. The 500 molecular weight cut-off: An updated analysis, J. Appl. Toxicol. 37 (2017), pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 29.Fitzpatrick J, Roberts D, Patlewicz G, Is skin penetration a determining factor in skin sensitization potential and potency? Refuting the notion of a LogKow threshold for skin sensitization, J. Appl. Toxicol. 37 (2017), pp. 117–127. [DOI] [PubMed] [Google Scholar]
- 30.Enoch S, Madden J, Cronin M, Identification of mechanisms of toxic action for skin sensitisation using a SMARTS pattern based approach, SAR QSAR Environ. Res. 19 (2008), pp. 555–578. [DOI] [PubMed] [Google Scholar]
- 31.Højsgaard S, Graphical independence networks with the gRain package for R. J Stat Softw 46 (2012), pp. 1–26.22837731 [Google Scholar]
- 32.Linzer D, Lewis J, poLCA: An R package for polytomous variable latent class analysis. J Stat Softw. 42 (2011) pp. 1–29. [Google Scholar]
- 33.Weston S, Getting Started with doMC and foreach, CRAN (2015) pp. 1–6 [Google Scholar]
- 34.Luechtefeld T, Maertens A, McKim J, Hartung T, Kleensang A, Sá-Rocha V, Probabilistic hazard assessment for skin sensitization potency by dose-response modeling using feature elimination instead of quantitative structure-activity relationships: Probabilistic hazard assessment for skin sensitization potency, J. Appl. Toxicol. 35 (2015), pp. 1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.personal communication with Grulke C, EPA-NCCT.
- 36.Roberts D, Schultz T, Api A, Chemical applicability domain of the Local Lymph Node Assay (LLNA) for skin sensitization potency. Part 3. Apparent discrepancies between LLNA and GPMT sensitization potential: False positives or differences in sensitivity?, Regul. Toxicol. Pharmacol. 80 (2016), pp. 260–267. [DOI] [PubMed] [Google Scholar]
- 37.Anderson S, Siegel P, Meade B, The LLNA: A Brief Review of Recent Advances and Limitations, J. Allergy. 2011 (2011), pp. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urbisch D, Becker M, Honarvar N, Kolle S, Mehling A, Teubner W, Wareing B, and Landsiedel R, Assessment of Pre- and Pro-haptens Using Nonanimal Test Methods for Skin Sensitization, Chem. Res. Toxicol. 29 (2016), pp. 901–913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


