Abstract
Although whole-genome sequencing has uncovered a large number of mutations that drive tumorigenesis, functional ratification for most mutations remains sparse. Here, we present an approach to test functional relevance of tumor mutations employing CRISPR/Cas9. Combining comprehensive sgRNA design and an efficient reporter assay to nominate efficient and selective sgRNAs, we establish a pipeline to dissect roles of cancer mutations with potential applicability to personalized medicine and future therapeutic use.
Genetic mutations are a hallmark of cancer development, and more than 140 cancer driver genes have been described to date (1,2). Identification of all mutations in an actual tumor of a patient by whole-genome sequencing is rapidly emerging as the method of choice for precision diagnostics (3). However, detailed knowledge of the functional roles and relevance of most mutations arising during tumorigenesis are still lacking.
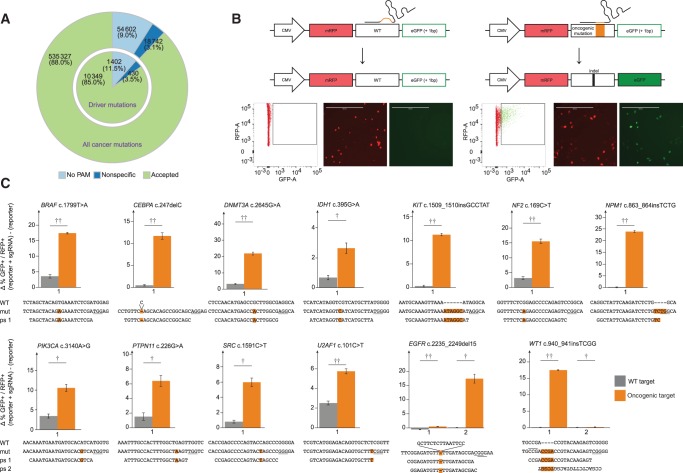
We set out to test whether the CRISPR/Cas9 system (4) can aid the functional investigation of mutations detected in cancer cells. To first investigate how many cancer mutations could theoretically be targeted by Streptococcus pyogenes (sp)Cas9, we performed a comprehensive bioinformatics analysis of published cancer mutations (2). From the reported 608 671 unique mutations, we were able to design 1 909 172 sgRNAs that cover 554069 mutations (91.0%) and 20 756 out of 20 948 mutated genes. We then performed an analysis to avoid off-target cleavage and discarded all sgRNAs having additional perfect matches to sequences in the reference genome, in addition to prioritizing sgRNAs with the highest divergence to homologous sequences elsewhere in the genome (5). Based on these criteria, we nominated 1 701 813 sgRNAs that could theoretically target 535 327 (88.0%) of known cancer mutations encompassing 10 349 (85.0%) of known cancer driver mutations (Figure 1A;Supplementary Table 1, available online).
Figure 1.
sgRNA design and evaluation of sgRNA efficacy and selectivity. A) Bioinformatics analysis and sgRNA design for cancer mutations. A pie chart for reported cancer mutations and for cancer driver mutations for Streptococcus pyogenes sgRNAs is shown. B) Overview of “traffic light” reporter assay. Important elements are indicated. Representative examples of fluorescence-activated cell sorting plots and microscopy images are shown (scale bars = 400 µm). C) Activity and selectivity of employed sgRNAs. The targeted mutations are indicated above each graph, with the wild-type, mutant, and protospacer sequences illustrated below each graph. Error bars represent SD from experiments performed in triplicates. Two-sided Student’s t test *P<.05; **P<.01; ***P<.001. mut = mutant; ps = protospacer; WT = wild-type.
We next established a “traffic-light” reporter system (6), where Cas9 cleavage activates GFP expression in transiently transfected mammalian cells in culture to rapidly evaluate efficacy and selectivity of designed sgRNAs (Figure 1B). Sequences bearing 13 different cancer mutations or the corresponding WT sequences were cloned into the reporter construct and subsequently cotransfected into HeLa cells with a Cas9 expression plasmid (7) that also expressed the cancer mutation–specific sgRNA (Figure 1, B and C). Efficient cleavage was observed for most constructs bearing cancer mutations, with 10 out of 13 sgRNAs also showing a higher than 4-fold target site selectivity over the wild-type (WT) sequence, with the remaining three still showing 2.7- to 3.8-fold selectivity of mutant over WT. In particular, insertion and deletion mutations reported in the genes KIT, NPM1, CEBPA, EGFR, and WT1 showed little to no appearance of green cells when combined with the WT reporters (Figure 1C), reflecting that the WT sequences were not cleaved efficiently. In contrast, 10% to 25% of GFP-positive cells were detected when the cancer mutation reporters were used in combination with matching sgRNAs. Hence, these sgRNAs created indels in the reporter plasmids that brought the GFP sequence into the correct reading frame, demonstrating their potency to cleave the cancer mutation sequence. Overall, we observed a descent correlation between the sgRNA prediction score and the actual activity in the traffic light reporter assay. However, we detected considerable differences in cleavage efficacy for some sgRNAs targeting the identical cancer mutation, despite the fact that their prediction scores (8,9) were similar (Supplementary Table 2, available online). For instance, sgRNA#1 with a score of 0.42 for the EGFR 2235_2249del15 mutation only produced 0.5% (+/-0.1%) of GFP-positive cells, whereas the related sgRNA#2 with a score of 0.33 that is only shifted by one base pair was highly efficient and resulted in more than 17.4% (+/-1.5%) of GFP-positive cells. Hence, the current prediction algorithms provide a guideline for the design of efficient sgRNAs, but experimental testing of the actual sequences seems recommendable (Supplementary Figure 1, available online). Interestingly, it was recently shown that nucleosome occupancy impedes Cas9 function (10), possibly explaining the discrepancy between score and activity for some sgRNAs. Remarkably, many point mutations, such as the DNMT3A c.2645G>A mutation, were efficiently cleaved by the cancer mutation sgRNA (21.9% [+/-0.8%] GFP-positive cells) without appreciably cleaving the WT sequence (3% [+/-0.2%] GFP-positive cells), demonstrating that the CRISPR/Cas9 system can be sensitive enough to distinguish single base pair alterations. Taken together, these results show that the CRISPR/Cas9 traffic-light reporter system is a valuable method to classify efficient and selective sgRNAs that can cleave cancer mutations.
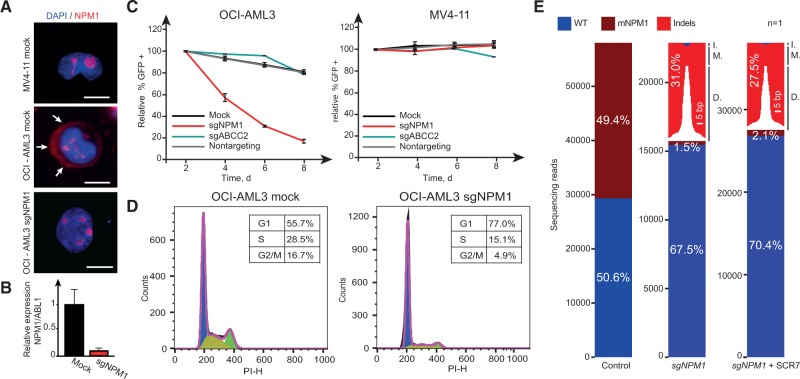
We next investigated the functional relevance of two common cancer mutations in tumor cells. The nucleophosmin gene (NPM1) is mutated in about 30% of patients suffering from acute myeloid leukemia (AML) (11). Mutant NPM1 is thought to play an important role in AML proliferation, indicating that a direct way to inactivate the mutation could affect malignant growth (12). We cloned the tested sgRNA sequence targeting mutant NPM1 (Figure 1C) into a lentiviral vector (13) expressing Cas9 in conjunction with EGFP and transduced NPM1 mutant OCI-AML3 cells and NPM1 WT MV4-11 cells with the virus. Efficient cleavage of mutant NPM1 in OCI-AML3 cells was evident in employing multiple assays (Figure 2). Strikingly, transduced OCI-AML3, but not the MV4-11 cells, were successively depleted over time (Figure 2C), signifying that the mutant NPM1 protein is required for efficient cell proliferation in OCI-AML3. Cell cycle analyses revealed that OCI-AML3 cells treated with the NPM1 sgRNA arrested in G1 without markedly altering the subG1 fraction (Figure 2D), suggesting that mutant NPM1 expression in these cells is required for cell cycle progression. To investigate the mutational spectrum at the site of cleavage, we performed deep sequencing of the NPM1 locus in control- and sgNPM1-treated cells. As expected, cells treated with a control sgRNA revealed a 50:50 ratio for the WT and mutant allele, reflecting the heterozygous nature of the NPM1 mutation. In contrast, cells treated with the sgRNA-targeting mutant NPM1 showed efficient cleavage and repair of the mutant allele. Remarkably, the WT:NPM1 ratio in this sample increased to around 70:30, indicating that a substantial fraction (34.2%) of the cells had repaired the mutation back to the WT sequence (Figure 2E), likely through homologous recombination utilizing the WT allele as a template. The fraction of indel mutations was further reduced (from 62.8% to 55.7%) when the cells were treated with the DNA ligase IV inhibitor SCR7 (14), indicating that enhanced HR-mediated repair can be achieved when the NHEJ pathway is inhibited. Hence, expression of a cancer-specific sgRNA can act in a gene drive fashion to push selection toward the WT sequence. Similar results were obtained targeting a second common cancer mutation (BRAF c.1799T>A) in the colon carcinoma cell line RKO (Supplementary Figure 2, available online), demonstrating that the approach can pinpoint cancer mutation dependencies in cell lines of different origins. Overall, we conclude that mutant NPM1 and mutant BRAF are required for OCI-AML3 and RKO proliferation, respectively, and that the CRISPR/Cas9 system is a powerful tool to dissect the relevance of cancer mutations in tumor cells.
Figure 2.
Effects of mutant NPM1 inactivation. A) Localization of NPM1 in MV4-11 and OCI-AML3 cells under indicated treatment conditions (scale bars = 10 µm). Arrows highlight the cytoplasmatic localization of mutant NPM1 in mock-treated OCI-AML3 cells. OCI-AML3 sgNPM1 images were taken 96 hours after sgRNA treatment. B) Relative mRNA expression level of mutated NPM1 after sgRNA treatment. mRNA was extracted 96 hours after sgRNA treatment. C) Relative abundance of cells treated with indicated sgRNAs in MV4-11 and OCI-AML3 over time. Error bars show SD from experiments performed in triplicates. D) Cell-cycle profile of OCI-AML3 cells after indicated treatments. Fluorescence-activated cell sorting analyses were performed eight days after sgRNA treatment. E) Graphical representation of NPM1 sequencing reads under indicated conditions (one biological replicate each). Size of deletions and insertions are indicated in white and black, respectively. sgNPM1–OCI-AML3 cells treated with sgRNA-targeting mutant NPM1 (mNPM1). sgNPM1 + SCR7–OCI-AML3 cells treated with sgRNA-targeting mutant NPM1 in the presence of the ligase IV inhibitor SCR7. Sequencing was performed on genomic DNA isolated eight days after sgRNA treatment. Sixty-four point six percent of the indels resulted in reading frame shifts. D. = deletions; I. = insertions; M. = mutations; WT = wild-type.
With the spCas9, we were able to design sgRNAs for 88% of reported cancer mutations. Orthogonal CRISPR/Cas9 systems (15) and/or the engineering of Cas9 proteins to recognize alternative PAMs (16) will increase the spectrum of cancer mutations that can be targeted (Supplementary Figure 3, available online). By generating cell line–specific sgRNA libraries, it might be possible to rapidly identify the most important driver mutations in this setting. Furthermore, we envision that this approach is transferable to primary patient samples (Supplementary Figure 4, available online), and, in the long run, CRISPR/Cas9 could potentially be considered a therapeutic approach to target patient-specific mutations in affected individuals. Delivery of Cas9 and mutation-specific sgRNAs into tumor cells by, eg, oncolytic viruses (17) could form a potent, individualized therapy that could complement current treatment strategies. In particular, combination therapy, where two or more cancer mutations are targeted at the same time, is relatively straightforward in this setting, when several specific sgRNAs can be provided simultaneously (Supplementary Figure 4, available online).
Limitations to our study include that even with additional orthogonal CRISPR/Cas systems it will be impossible to design unique sgRNAs for every cancer mutation. Furthermore, control over the repair mechanism after Cas9-mediated DNA cleavage is limited. It is therefore likely that sgRNA-resistant clones may emerge that maintain the oncogenic phenotype. In addition, off-target cleavage has to be considered a potential risk factor in a therapeutic setting.
Nevertheless, considering current cancer treatment regimes employing DNA-damaging drugs and/or radiation, the CRISPR/Cas9 system is conceivable to be less genotoxic and cause less undesired DNA lesions in cells. Given the prominent gene drive effect we observed to repair the cancer mutation back to the WT sequence, installing the CRISPR/Cas9 system as a “tumor protection system” is also worth contemplating (Supplementary Figure 5, available online). This way, the system would act as a “cancer mutation immune system,” eliminating or repairing malignant lesions when they occur and before cells become cancerous.
Funding
This work was supported by grants from the Mildred-Scheel Program of the German cancer aid, the Else Kröner-Promotionskolleg, the Excellence Initiative by the German Federal and State Governments (Institutional Strategy, measure “support the best ZUK 64”), and the German Cancer Consortium joint funding program “novel tools for dissection of oncogenic pathways.”
Notes
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication. The authors declare no competing financial interest. Sequencing data will be deposited into the National Center for Biotechnology Information Short Read Archive. The authors thank Juliane Reh and Marika Karger for technical assistance, Stanislava Popova for help with cell cycle analyses, and Martin Schneider for support with cloning and fluorescence-activated cell sorting analyses.
Supplementary Material
References
- 1. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dewey FE, Grove ME, Pan C, et al. Clinical interpretation and implications of whole-genome sequencing. JAMA. 2014;311(10):1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karimova M, Beschorner N, Dammermann W, et al. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci Rep. 2015;5:13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doench JG, Hartenian E, Graham DB, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32(12):1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doench JG, Fusi N, Sullender M, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horlbeck MA, Witkowsky LB, Guglielmi B, et al. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. [DOI] [PubMed] [Google Scholar]
- 12. Federici L, Falini B. Nucleophosmin mutations in acute myeloid leukemia: a tale of protein unfolding and mislocalization. Protein Sci. 2013;22(5):545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32(9):941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Srivastava M, Nambiar M, Sharma S, et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell. 2012;151(7):1474–1487. [DOI] [PubMed] [Google Scholar]
- 15. Esvelt KM, Mali P, Braff JL, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleinstiver BP, Prew MS, Tsai SQ, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14(9):642–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.