Abstract
Background
Effective targeting therapies for common chronic liver disease nonalcoholic steatohepatitis (NASH) are in urgent need. MicroRNA-targeted therapeutics would be potentially an effective treatment strategy of hepatic diseases. Here we investigated the functional role of miR-221/222 and the therapeutic effects of antimiRs-221/222 in NASH mouse models.
Methods
We generated the miR-221/222flox/flox mice on a C57BL/6 J background and the hepatic miR-221/222 knockout (miR-221/222-LKO) mice. The mice were challenged with the methionine and choline deficient diet (MCDD) or chronic carbon tetrachloride (CCl4) treatment to generate experimental steatohepatitis models. Adenovirus-mediated re-expression of miR-221/222 was performed on the MCDD-fed miR-221/222-LKO mice. The MCDD and control diet-fed mice were treated with locked nucleic acid (LNA)-based antimiRs of miR-221/222 to evaluate the therapeutic effects. Histological analysis, RNA-seq, quantitative PCR and Western blot of liver tissues were carried out to study the hepatic lipid accumulation, inflammation and collagen deposition in mouse models.
Findings
Hepatic deletion of miR-221/222 resulted in significant reduction of liver fibrosis, lipid deposition and inflammatory infiltration in the MCDD-fed and CCl4-treated mouse models. The hepatic steatosis and fibrosis were dramatically aggravated by miR-221/222 re-expression in MCDD-fed miR-221/222-LKO mice. AntimiRs of miR-221/222 could effectively reduce the MCDD-mediated hepatic steatosis and fibrosis. Systematically mechanistic study revealed that hepatic miR-221/222 controlled the expression of target gene Timp3 and promoted the progression of NASH.
Interpretation
Our findings demonstrate that miR-221/222 are crucial for the regulation of lipid metabolism, inflammation and fibrosis in the liver. LNA-antimiRs targeted miR-221/222 could reduce steatohepatitis with prominent antifibrotic effect in NASH mice.
Fund
This work is supported by the Natural Science Foundation of China (81530020, 81390352 to Dr. Ning and 81522032 to Dr. Cao and 81670793 to Dr. Jiang); National Key Research and Development Program (No. 2016YFC0905001 and 2017YFC0909703 to Dr. Cao); the Shanghai Rising-Star Program (15QA1402900 to Dr. Cao); Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant (20171905 to Dr. Jiang).
Keywords: miR-221/222, NASH, Liver fibrosis, miRNA-targeted therapeutics
Research in context.
Evidence before this study
The upregulated expression of highly conserved cluster miR-221 and 222 has been observed in HCC, liver of NASH patients and biliary atresia induced liver fibrosis in mice. In vivo delivery of anti-miR-221 oligonucleotides could reduce tumor size in HCC mice. The mechanistic role of hepatic miR-221/222 and the importance of miR-221/222 antimiRs in NASH treatment are still lack of investigation.
Added value of this study
Our findings demonstrate that miR-221/222 are crucial for the regulation of lipid metabolism, inflammation and fibrosis in the liver. LNA-antimiRs targeted miR-221/222 could reduce steatohepatitis with prominent antifibrotic effect in NASH mice.
Implications of all the available evidence
Our findings provided evidences that miR-221/222 are an efficient and potent target for hepatic steatosis, inflammation and liver fibrosis in NASH mouse models. Our data and previous reports revealed upregulated expression of hepatic miR-221/222 in NASH mouse models and patients. Our work indicated that antimiR-221/222 could be an effective approach for NASH treatment in vivo. The combined therapies contain antimiR-221/222 and other small molecules or monoclonal antibodies which targeting multiple pathways in NASH might be of great interest.
Alt-text: Unlabelled Box
1. Introduction
Nonalcoholic steatohepatitis (NASH), a severe form of nonalcoholic fatty liver disease (NAFLD), is a worldwide common form of progressive chronic liver disease result in liver cirrhosis, hepatic decompensation and failure or hepatocellular carcinoma (HCC). The prevalence of NAFLD and NASH has dramatically increased with the rising incidence of obesity, type 2 diabetes mellitus and metabolic syndrome in recent decades. The clinical characteristics of NASH include hepatic steatosis, lobular inflammation, hepatocyte ballooning and progressive pericellular fibrosis [1]. Liver fibrosis is the determinant stage of NASH, which predicts high-risk outcome, such as liver transplantation or liver-related mortality [2,3]. Given the limitations and difficulties of the individual therapeutic lifestyle change interventions, safe and effective pharmacological therapeutics are in urgent need for NASH treatment. However, the approved drug for the treatment of NASH is still absent currently.
Therapeutic intervention of NASH should integrate antifibrotics and reduction of fibrosis triggers, including hepatic inflammation, cell stress and insulin resistance. Insulin sensitizers such as Thiazolidinediones (TZDs) for diabetes treatment have proven evidence of benefit for off-label therapeutic use in NASH, with the limitations of adverse effects and safety issues [4,5]. Recent clinical trials of target therapies for NAFLD or NASH have shed light on the encouraging progress of NASH treatment. The effective and competitive drugs include proliferator-activator receptor agonists (elafibranor), farnesoid X receptor agonist (obeticholic acid), antifibrotic monoclonal antibody (simtuzumab), bile acid and farnesoid X receptor agonist (obeticholic acid) and ASK1 inhibitor (selonsertib) [[5], [6], [7], [8]]. Considering the pathophysiological complexity of NASH, continuous discovery of crucial potential therapeutic targets and antifibrotic agents is essential for ideal combination therapy and personalized therapy for the majority of NASH patients in the future.
MiRNA-based treatments for many diseases are under rapid clinical development with the advantages of multiple targets and feasible chemical modification over small molecule drugs. AntimiRs which directly and specifically inhibit miRNA function in vivo could be most efficiently taken up by the liver. Liver diseases are the most promising models for the development of practical miRNA-based gene therapies [[9], [10], [11]]. One of the most important cases is LNA-modified antimiR targeting the liver-specific miR-122 (miravirsen) is effective in prevention of HCV replication [[12], [13], [14]]. MiR-103/107 are demonstrated to modulate insulin sensitivity in animal models, therefore antimiRs targeting miR-103/107 (RG-125) are developed as an effective insulin sensitizer for the treatment of NASH patients with type 2 diabetes [11,15]. In addition, antimiRs of miR-21 reduced hepatic inflammation and liver steatosis through PPARα [16].
The upregulated expression of highly conserved cluster miR-221 and 222 has been reported in HCC, liver of NASH patients and biliary atresia induced liver fibrosis in mice [[17], [18], [19], [20], [21]]. miR-221/222 were shown to be the most upregulated miRNAs in HCC samples. miR-221 could stimulate the growth and tumorigenesis of murine hepatic progenitor cells in vivo [17]. In vivo delivery of anti-miR-221 oligonucleotides leads to a significant reduction of tumor size in HCC mice [18]. The mechanistic role of hepatic miR-221/222 and the importance of miR-221/222 antimiRs in NASH treatment are still lack of investigation. In this study, we generated the hepatocyte-specific miR-221/222 knockout mice and performed targeted inhibition of miR-221/222 by locked nucleic acid (LNA)-antimiR in vivo. Our results demonstrated that the oncogenic and profibrotic miR-221/222 were effective therapeutic targets for NASH.
2. Methods
2.1. Animals
All of the animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. To generate the miR-221/222 targeting vector, a mouse genomic DNA corresponding to miR-221 and miR-222 was cloned. A cassette expressing the neomycin resistant gene (Neo) and thymidine kinase (TK) flanked by two loxP sites was placed 500 bp downstream of miR-221 and another loxP site was inserted 3000 bp upstream of miR-222. The resulting 20.7-kb transgene was electroporated into ES cells and targeted ES clones were identified by Southern blotting. The targeted ES clones were microinjected into the pronuclei of fertilized oocytes from C57BL/6J mice to get chimeric mice. The chimeric mice were crossed with Flp-expressing transgenic mice to obtain the floxed allele with deletion of the NeoTK cassette. Genotypes were confirmed by PCR using the following primers: 5′-AGGTGGACAGTTTGATGAGTT-3′ and 5′-TTGAAGTAGCAAGCATAGGG-3′. MiR-221/222flox/flox mice in which miR-221 and 222 were flanked by loxP sites were bred with mice expressing Cre recombinase driven by the albumin promoter (Alb-Cre). Heterozygous (miR-221/222flox/+-Cre) mice were crossed to generate hepatocyte-specific miR-221/222 knockout mice (miR-221/222flox/flox-Cre, miR-221/222-LKO) and their age-matched littermate controls (miR-221/222flox/flox). The C57BL/6J mice were purchased from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences (SLAC, CAS). Alb-Cre mice were purchased from Jackson Laboratory. All of the mice were housed in pathogen-free facilities with a 12 h light/dark cycle and had free access to water and food. Mice were subjected to two different fibrosis models. The miR-221/222-LKO and control mice were fed a methionine or choline deficient diet (MCDD) or control diet (Research Diets) for 6 weeks to generate the dietary model of NASH. In the second model, the miR-221/222-LKO and control mice were injected intraperitoneally (i.p.) with carbon tetrachloride (CCl4) or vehicle for 6 weeks (0.5 ml/kg, twice weekly). Mice were sacrificed 3 days after the last injection. All mice used for the experiments were males.
2.2. Adenovirus infection of miR-221/222 and adeno-associated virus infection of Timp3 shRNA
The adenovirus expressing miR-221 and miR-222 was constructed and packaged using Ad5CMV K-NpA vector. Ad-GFP was used as a control. Mice were injected with adenoviruses through the tail vein at 1 × 1011 plaque-forming units in 0.2 ml PBS. Injection of Ad-miR-221/222/GFP did not affect food consumption compared to that of untreated animals. Mice were sacrificed at the indicated days after the adenovirus injection. The adeno-associated virus (AAV) expressing Timp3 shRNA was constructed and packaged using AAV8 vector. Mice were injected with AAV through the tail vein at 1 × 1012 plaque-forming units in 0.2 ml PBS. Mice were sacrificed 4 weeks after the AAV injection and MCDD feeding.
2.3. LNA-antimiRs injection in vivo
Custom LNA-modified oligonucleotides were purchased from Exiqon (Vedbaek, Denmark). AntimiRs LNA-i-miR-221 (sequence: CAGCAGACAATGTAGC) and LNA-i-miR-222 (sequence: AGTAGCCAGATGTAGC) are 16-mer DNA/LNA oligonucleotides. A mismatch 15-mer oligonucleotide LNA-i-miR-NC (sequence: ACGTCTATACGCCCA) is served as negative control. All these oligonucleotides have a phosphorothioate-modified backbone and are purified by HPLC followed by Na+ salt exchange and lyophilization. The control mice were randomized into four groups and treated i.p. with a dose of 25 mg/kg LNA-i-miR-Ctrl, LNA-i-miR-221, LNA-i-miR-222 or 12.5 mg/kg LNA-i-miR-221 and 12.5 mg/kg LNA-i-miR-222 (LNA-i-miR-221 + 222) on days 1, 4, 8, 15, and 22. The mice were fed with MCDD on the day of first dose and sacrificed for serum and tissue collection after 1 week from the last dose.
2.4. Cell culture and transfection
Mouse HCC hepa1–6 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum at 37 °C in 5% CO2 infusion and humidified air. Hepa1–6 cells were transfected with LNA-antimiRs or control at the concentration of 50 nM and 100 nM by Lipofectamine 2000 (Invitrogen).
2.5. Hepatic histological analysis
The liver tissues were fixed in 4% paraformaldehyde, dehydrated and paraffin embedded. The liver sections were stained with hematoxylin-eosin, Masson's trichrome solution or Sirius Red (saturated picric acid containing 0.1% (wt/vol) Direct Red 80). The Sirius red–positive area or masson-positive area was quantitated by digital image analysis. The results were presented as percentage area positively stained for Sirius Red or Masson. For the detection of neutral lipids accumulation, liver cryosections were stained using Oil Red O (Sigma) according to standard procedures. The images were acquired using an Olympus microscopy system.
2.6. Biochemical analysis
Serum and liver total cholesterol (TC) and triglycerides (TG) were measured using commercially available kits (BioVision) following the manufacturer's instructions. Serum alanine aminotransferase (ALT) was measured commercially available kits (BioVision) following the manufacturer's instructions. The glucose measurements were taken by tail-blood analysis using One-Touch Ultra glucometers (LifeScan).
2.7. Hydroxyproline measurement
The colorimetric measurement of the collagen-specific amino acid hydroxyproline was performed using Hydroxyproline Assay Kit (Sigma) following the manufacturer's instructions. Hydroxyproline content was expressed as nanogram (ng) of hydroxyproline per milligram (mg) liver.
2.8. Ultrastructural analysis
For transmission electron microscopic (TEM) analysis, fresh liver samples (1 mm3 volume) were fixed in 2.5% glutaraldehyde/formaldehyde. Subsequently, the specimens were postfixed in 2% osmium tetroxide (OsO4) for 1 h. Following fixation, the tissues were dehydrated through a graded series of ethanols and propylene oxide, embedded in Epon 812, and sectioned on a Reichert ultramicrotome (Reichert Ultracut S) to obtain semithin sections. The sections were stained with methylene blue in sodium borate, contrasted with uranyl acetate and lead citrate, and photographed using an Opton EM 900 transmission electron microscope (Zeiss, Oberkochen, Germany).
2.9. Quantitative RT–PCR
The total RNA, containing the miRNA, was extracted using a miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. For the miRNA reverse transcription, 1 μg of the total RNA was reverse transcribed using a reverse transcription kit (Promega) following the manufacturer's instruction with the following primers: miR-221: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAAACCC-3′; miR-222: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCCAGT-3′. Quantitative PCR (qPCR) of miR-221/222 was performed using a QuantStudio Dx Real-Time instrument (Thermo Fisher) with a SYBR Premix Ex Taq Kit (Takara) using the following primers: miR-221: F: GGGAAGCTACATTGTCTGC R: CAGTGCGTGTCGTGGAGT; miR-222: F: GGGGAGCTACATCTGGCT R: TGCGTGTCGTGGAGTC and normalized to U6 (small nuclear RNA) as the endogenous control. The U6 primers were: F: GCTTCGGCAGCACATATACTAAAAT R: CGCTTCACGAATTTGCGTGTCAT. For the RNA reverse transcription, 1 μg of the total RNA was converted into cDNA using a reverse transcription kit (Promega) following the manufacturer's instruction. qPCR was performed using a QuantStudio Dx Real-Time instrument (Thermo Fisher) with a SYBR Premix Ex Taq Kit (Takara). The gene expression levels were normalized to the housekeeping gene Gapdh. For the primers used, please refer to Supplemental Experimental Procedures.
2.10. Western blot
Protein preparation and Western blots were performed as described previously [22]. The following primary antibodies were used: anti-F4/80 (1:1000; Abcam), anti-α-SMA (1:1000; Sigma), anti-TIMP3 (1:1000; Cell Signaling Technology), anti-α-tubulin (1:1000; Cell Signaling Technology), anti-PGC-1β (1:1000; abcam), anti-G6PC (1:1000; Invitrogen).
2.11. Luciferase assays
The pRL-NULL and pGL3-control vectors were obtained from Professor Mofang Liu (Shanghai Institutes for Biological Sciences). The hepa1-6 cells were seeded at 5 × 105 cells per well in 24-well plates and transfected with 10 ng pRL-NULL-3′-UTR (untranslated region) plasmids with 100 ng of pGL3-control vector. The cells were harvested at 24 h after transfection, and luciferase activities were measured. The values were normalized using the dual-luciferase reporter assay system according to the manufacturer's protocol (Promega). To generate the wild-type and mutant Timp3 luciferase reporter constructs, the 3′-UTRs were cloned and inserted into pRL-NULL vectors with the appropriate restriction enzymes.
2.12. RNA sequencing
Library construction and sequencing were performed on the BGISEQ-500 sequencer by Beijing Genomic Institution (BGI Genomics, BGI-Shenzhen, China). Briefly, the core steps in preparing RNA for sequencing are: (1) enrich poly(A) + RNAs by oligo (dT) selection; (2) fragment the target RNA and reverse transcription to double-stranded DNA; (3) end repair the dscDNA and generate bubble adapter ligation; (4) denature the PCR product and cyclize the single strand DNA for sequencing. Sequencing reads were mapped to reference genome and transcripts available at the UCSC hg19 (http://genome.ucsc.edu/) using Bowtie2 and HISAT, respectively [23,24]. Gene expression was determined using RSEM [25]. The R/Bioconductor package “DEGSeq” and Wilcoxon were used to identify differentially expressed genes.
2.13. Statistical analysis
Significant differences were analyzed using two-tail unpaired Student's t-test or one-way analysis of variance (ANOVA) for multiple group comparison. The error bars in the graphs represent standard deviations (SD). Differences were considered significant if P < .05.
3. Results
3.1. Hepatocyte specific miR-221/222 knockout mice
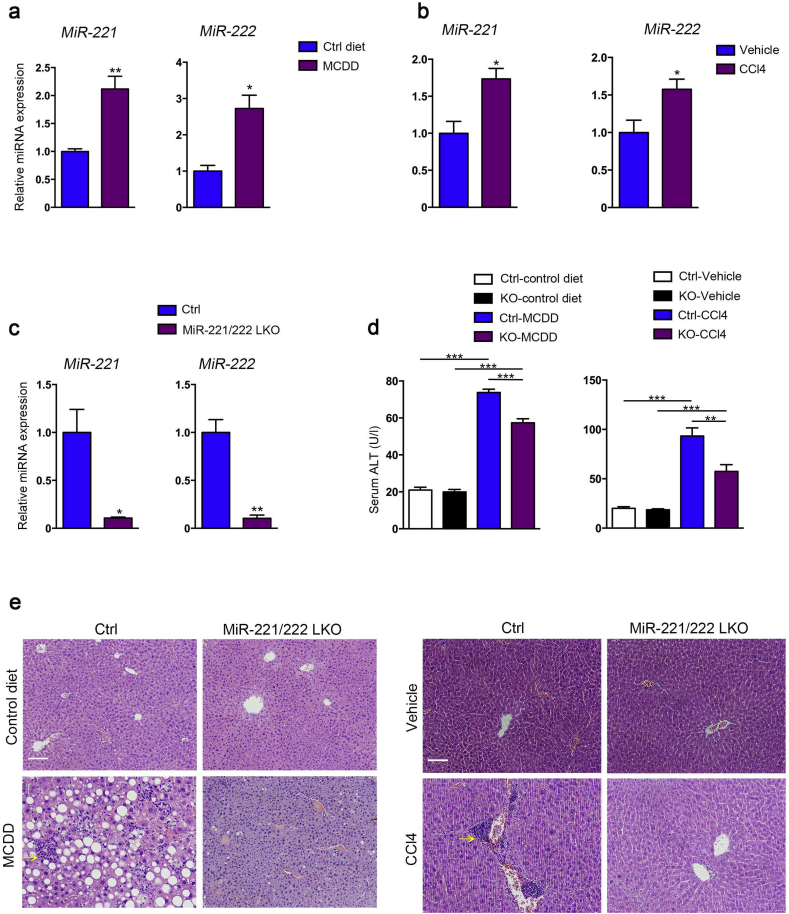
The correlation of miR-221/222 expression with development of NASH was investigated in two experimental models of liver fibrosis: MCDD and chronic CCl4 treatment in C57/BL6 mice. Our data showed the expression of liver miR-221/222 was significantly increased in both fibrosis models compared to control mice (Fig. 1a, b). These results and the consistent previous reports [20,21] indicated the upregulation of miR-221/222 was an important profibrotic factor response to liver inflammation and injury in both human and mice.
Fig. 1.
MiR-221/222-LKO mice under the challenge of MCDD or CCl4. (a) The expression of miR-221 and 222 in the livers of control mice treated with MCDD or control diet for 6 weeks (n = 4–6). (b) The expression of miR-221 and 222 in the livers of control mice treated with CCl4 or vehicle for 6 weeks (n = 3–7). (c) The expression of miR-221 and 222 in the livers of 8-week-old miR-221/222-LKO and control mice (n = 4 for each group). The data represent the mean ± SD, Student's t-test. (d) Serum ALT levels of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks (n = 3–9), and CCl4 or vehicle for 6 weeks (n = 3–5). The data represent the mean ± SD, one-way ANOVA. (e) H&E staining of the liver sections from miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks, and CCl4 or vehicle for 6 weeks. Diffuse inflammatory foci containing mononucleated cells (arrowheads) apparent in the livers of control mice under MCDD or CCl4 treatment. *P < .05, **P < .01, ***P < .001. Scale bars, 100 μm.
To elucidate the precise functions of miR-221/222 in NASH, we generated hepatocyte specific miR-221/222 knockout mice (miR-221/222-LKO) harboring floxed miR-221/222 alleles (miR-221/222flox/flox mice) and Alb-Cre (Fig. S1a, b). The expression levels of miR-221/222 in liver were significantly abolished in miR-221/222-LKO mice relative to control mice (Fig. 1c). Meanwhile the miR-221/222 expression in other tissues were indistinguishable between miR-221/222-LKO and control mice (Fig. S2). The 2 months old miR-221/222-LKO mice exhibited a normal metabolic phenotype under normal diet, including body and liver weight, fasting blood glucose, serum and hepatic triglyceride (TG) and total cholesterol (TC) (Fig. S3).
3.2. Hepatic miR-221/222 deletion reduced hepatic steatosis and fibrosis
To evaluate the effects of miR-221/222 expression in hepatic steatosis and fibrosis, we challenged the miR-221/222-LKO and control mice with MCDD or chronic CCl4 treatment. Our results showed hepatocyte miR-221/222 knockout significantly reduced the serum level of ALT in MCDD-fed and CCl4-treated miR-221/222-LKO mice compared to the control mice (Fig. 1d). Hematoxylin and eosin (H&E) staining showed the MCDD-fed control mice developed severe hepatic steatosis and hepatocyte ballooning, while the MCDD-fed miR-221/222-LKO mice showed slight steatosis (Fig. 1e). Moreover, the CCl4-induced liver injury was markedly alleviated in miR-221/222-LKO mice (Fig. 1e). Altogether, hepatocyte specific miR-221/222 knockout could reduce hepatosteatosis in the NASH mouse models.
3.3. MiR-221/222 ablation ameliorated hepatic lipid deposition and inflammatory infiltration
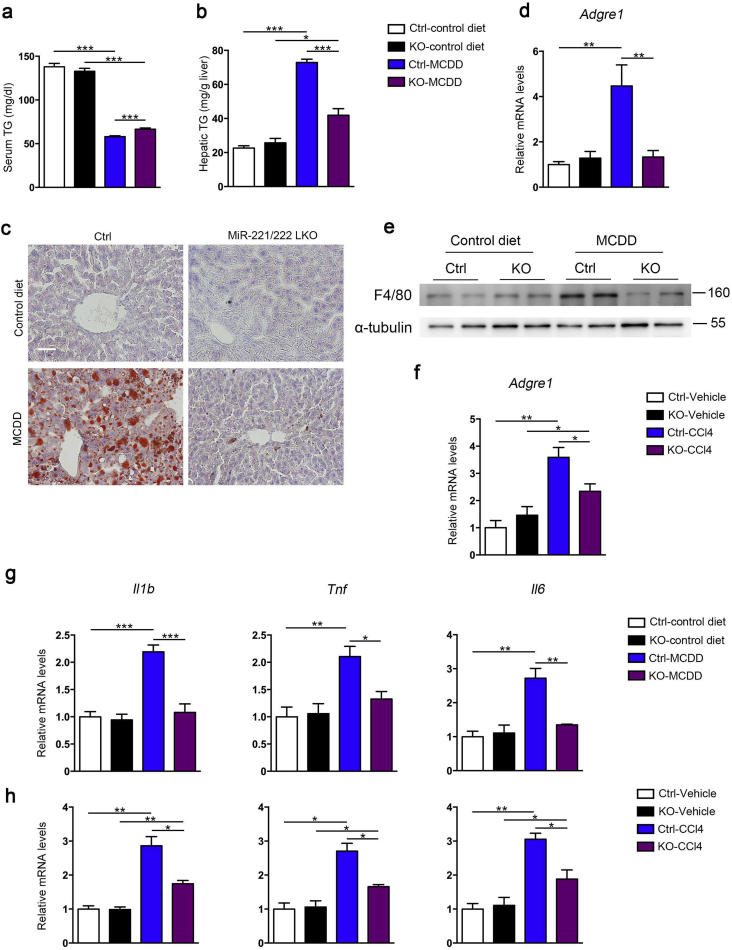
To determine the effects of miR-221/222 on liver lipid metabolism, we analyzed the lipid circulation and deposition in MCDD-fed mice. The MCDD challenged miR-221/222-LKO mice had significantly higher serum TG and similar serum TC levels compared to control mice (Fig. 2a and S4a). Meanwhile, MCDD challenging significantly increased hepatic lipid accumulation in mice. The miR-221/222-LKO mice exhibited much lower hepatic TG and similar hepatic TC compared to control mice (Fig. 2b and S4b). Moreover, Oil Red O staining analysis of liver sections showed that miR-221/222-LKO mice had markedly reduced hepatic fat accumulation compared to control mice under MCDD challenging (Fig. 2c). TEM analysis of liver sections revealed that MCDD-fed miR-221/222-LKO mice only had slight lipid droplets deposition compared to the severe circumstances in control mice (Fig. S4c). Taken together, these data support that miR-221/222 ablation suppressed MCDD-induced hepatic fat deposition in mice.
Fig. 2.
Hepatic deletion of miR-221/222 reduces hepatic inflammation and lipid deposition in NASH mice. (a and b) Triglyceride (TG) levels in the serum (a) and liver (b) of control and miR-221/222-LKO mice treated with MCDD or control diet for 6 weeks (n = 3–5). (c) Oil Red O staining of liver sections from miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks. Scale bars, 100 μm. (d) The expression of Argre1 in the livers of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks (n = 4 for each group). (e) Western blot analyses of F4/80 in the livers of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks (n = 2 for each group). The data shown represent three independent experiments. (f) The expression of Argre1 in the livers of miR-221/222-LKO and control mice treated with CCl4 or vehicle for 6 weeks (n = 3–5). (g and h) The expression of Il1b, Tnf and Il6 in the livers of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks (n = 3 for each group) (g), and (h) CCl4 or vehicle for 6 weeks (n = 3 for each group). The data represent the mean ± SD, *P < .05, **P < .01, ***P < .001, one-way ANOVA.
Furthermore, knockout of miR-221/222 significantly reduced the hepatic inflammatory infiltration in keeping with the reduction of lipid deposition. The mRNA and protein levels of macrophage marker F4/80 were significantly lower in MCDD-fed miR-221/222-LKO mice compared to control mice (Fig. 2d, e and S4d). The similar effects were also observed in the CCl4-treated mouse model (Fig. 2f). In addition, the production of key pro-inflammatory chemokines and cytokines, including tumor necrosis factor-α (TNF-α), interleukin (IL)-1β and IL-6, were significantly decreased by hepatic miR-221/222 ablation in the MCDD-fed and CCl4-treated mouse models (Fig. 2g, h). NASH was characterized by diffuse inflammatory foci containing mononucleated cells in the mice under the challenge of MCDD or CCl4, whereas the hepatic inflammation was much improved in miR-221/222-LKO mice (Fig. 1e). In summary, miR-221/222 deficiency could efficiently antagonize the hepatic inflammatory.
3.4. MiR-221/222 ablation induced antifibrotics in NASH models
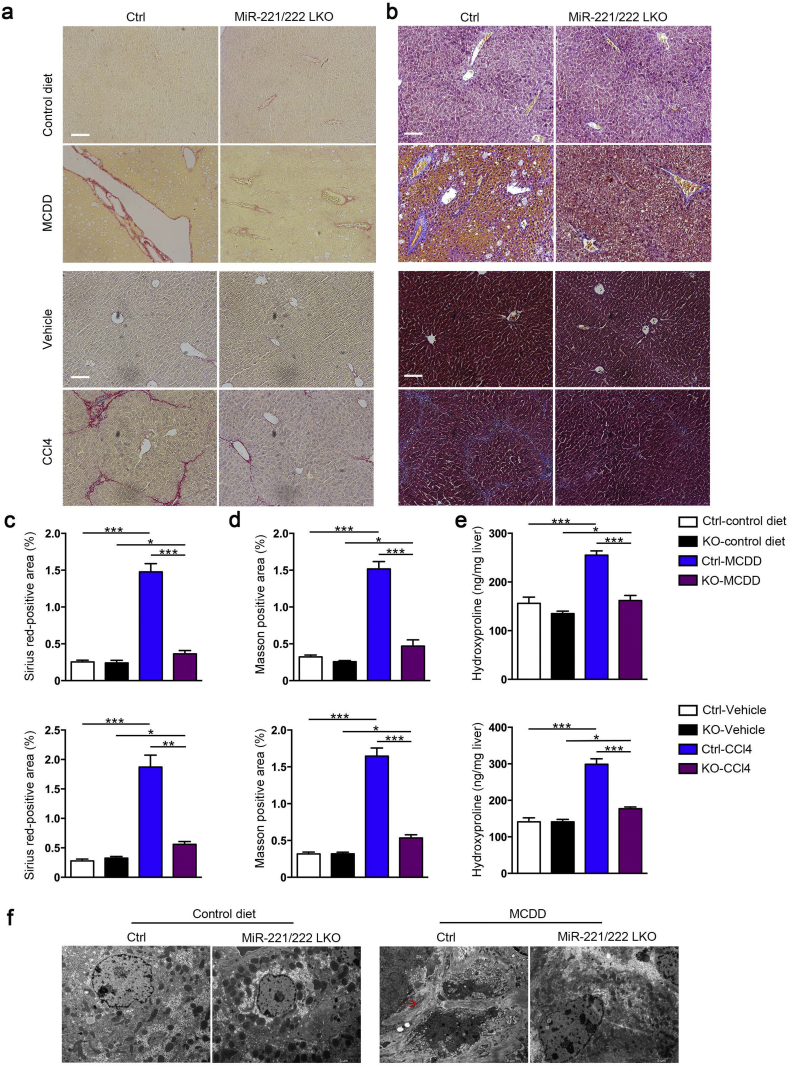
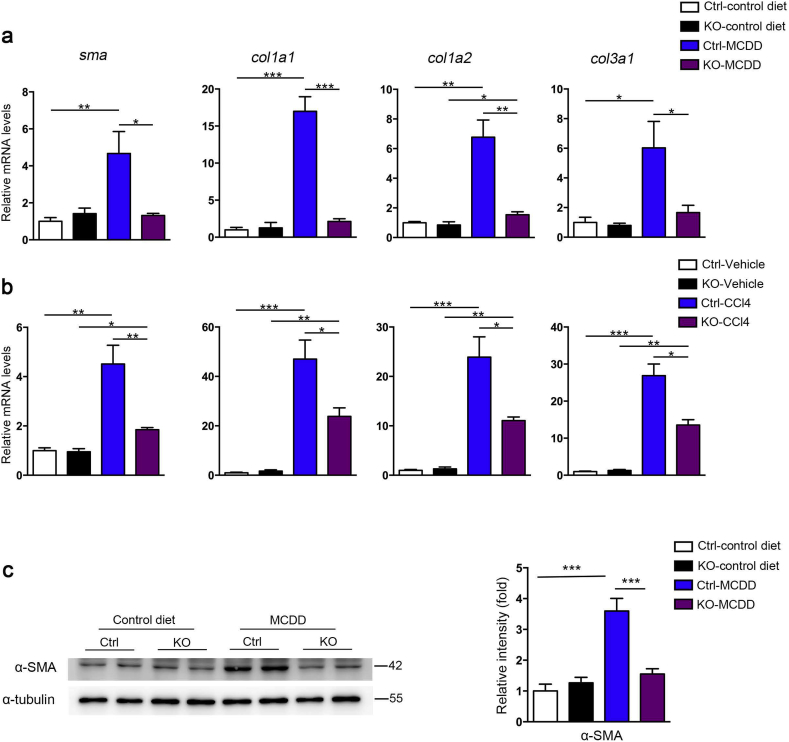
The antifibrotic effect of hepatic miR-221/222 ablation is one of the fundamental evaluation indicators in our investigation. Notably, the hepatic collagen deposition could be much ameliorated in miR-221/222-LKO mice response to MCDD and CCl4 challenge. The morphometric assessments of hepatic fibrosis by Sirius Red and Masson's trichrome staining showed reduced positive area in miR-221/222-LKO mice (Fig. 3a–d). The hepatic content of collagen-specific amino acid hydroxyproline was significantly reduced by miR-221/222 knockout under the treatment of MCDD or CCl4 (Fig. 3e). TEM analysis of liver sections showed that MCDD-fed miR-221/222-LKO mice had improved collagen deposition and fibrogenesis (Fig. 3f), accompanied with significantly decreased a-SMA, Col1a1, Col1a2 and Col3a1 expression levels and similar expression of Col5a1 and Col5a2 (Fig. 4a, b and S5). Correspondingly, the protein level of hepatic a-SMA in KO mice was lower than control mice (Fig. 4c). Taken together, these results indicated that liver fibrosis could be controlled through miR-221/222 in mouse models.
Fig. 3.
Hepatic deletion of miR-221/222 reduces liver fibrosis in NASH mice. (a and b) Representative Sirius red staining (a) and Masson staining (b) of the liver sections from miR-221/222-LKO and control mice treated with MCDD or control diet (upper part) and CCl4 or vehicle (bottom part). Scale bars, 100 μm. (c and d) Analysis of the Sirius red-positive area (c) and Masson-positive area (d) of the liver sections from miR-221/222-LKO and control mice treated with MCDD or control diet (upper part) and CCl4 or vehicle (bottom part) (n = 4–5). (e) Hepatic hydroxyproline concentration of miR-221/222-LKO and control mice treated with MCDD or control diet (upper part) and CCl4 or vehicle (bottom part) (n = 3–5). (f) TEM micrographs (arrowheads, collagen) of liver samples from miR-221/222-LKO and control mice treated with MCDD or control diet. Scale bars, 5 μm. Data represent mean ± SD. *P < .05, **P < .01, ***P < .001, one-way ANOVA.
Fig. 4.
Hepatic deletion of miR-221/222 reduces the expression of procollagen genes in NASH mice. (a and b) The expression of sma, Col1a1, Col1a2 and Col3a1 in the livers of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks (a) (n = 3–9), and (b) CCl4 or vehicle diet for 6 weeks (n = 3–5). (c) Western blot analyses of α-SMA in the livers of miR-221/222-LKO and control mice treated with MCDD or control diet for 6 weeks. The relative intensity of the bands was analyzed for three independent experiments. Data represent mean ± SD. *P < .05, **P < .01, ***P < .001, one-way ANOVA.
3.5. Adenovirus-mediated re-expression of miR-221/222 aggravated NASH in mice
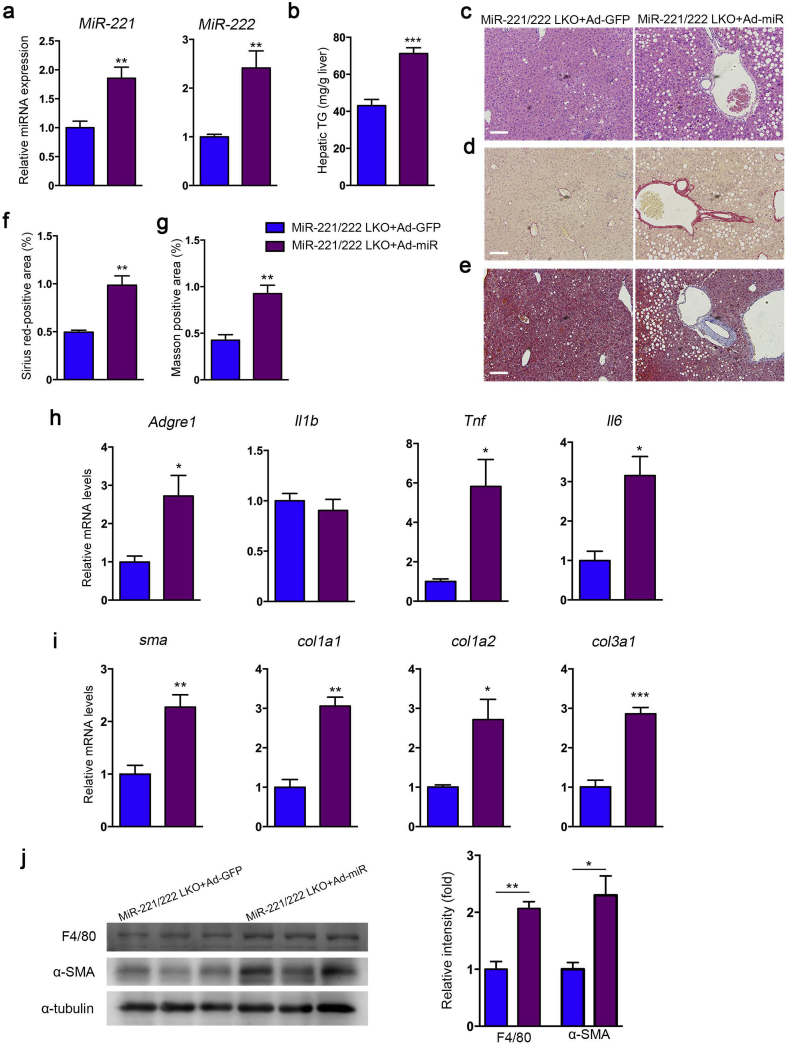
In order to determine the driver effects of miR-221/222 in the development of NASH, we overexpressed miR-221/222 in the liver of MCDD-fed miR-221/222-LKO mice by adenovirus (Ad-miR-221/222) injection (Fig. 5a). Re-expression of miR-221/222 significantly elevated the hepatic TG levels and promoted hepatic steatosis and hepatocyte ballooning in MCDD-fed mice (Fig. 5b, c). The sirius Red and Masson's trichrome staining indicated miR-221/222 overexpression resulted in aggravating liver fibrosis progress in mice (Fig. 5d-g). Moreover, the expression of miR-221/222 facilitated the upregulation of inflammation and fibrosis related genes, including Adgre1, TNF-α, IL-1β, IL-6 and a-SMA, Col1a1, Col1a2, Col3a1, while the levels of Col5a1 and Col5a2 were indistinguishable (Fig. 5h, i and S6). Correspondingly, the protein levels of hepatic F4/80 and a-SMA were also increased by miR-221/222 re-expression (Fig. 5j). Altogether, our data revealed that miR-221/222 overexpression deteriorate liver fibrosis.
Fig. 5.
Re-expression of hepatic miR-221/222 deteriorates liver steatosis and fibrosis in miR-221/222-LKO mice. (a) The expression of miR-221 and miR-222 in the livers of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 4 for each group). (b) The hepatic TG of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 4 for each group). (c) H&E staining of the liver sections from miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP. (d and e) Representative (d) Sirius red staining and (e) Masson staining of the liver sections from miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP. Scale bars, 100 μm. (f and g) Analysis of the (f) Sirius red-positive area and (g) Masson-positive area of the liver sections of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 4–5). (h and i) The expression of (h) Argre1, Il1b, Tnf, Il6 and (i) sma, Col1a1, Col1a2, Col3a1 in the livers of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 4 for each group). (j) Western blot analyses of F4/80 and α-SMA in the livers of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 3 for each group). The relative intensity of the bands was analyzed. Data represent mean ± SD. *P < .05, **P < .01, ***P < .001, Student's t-test.
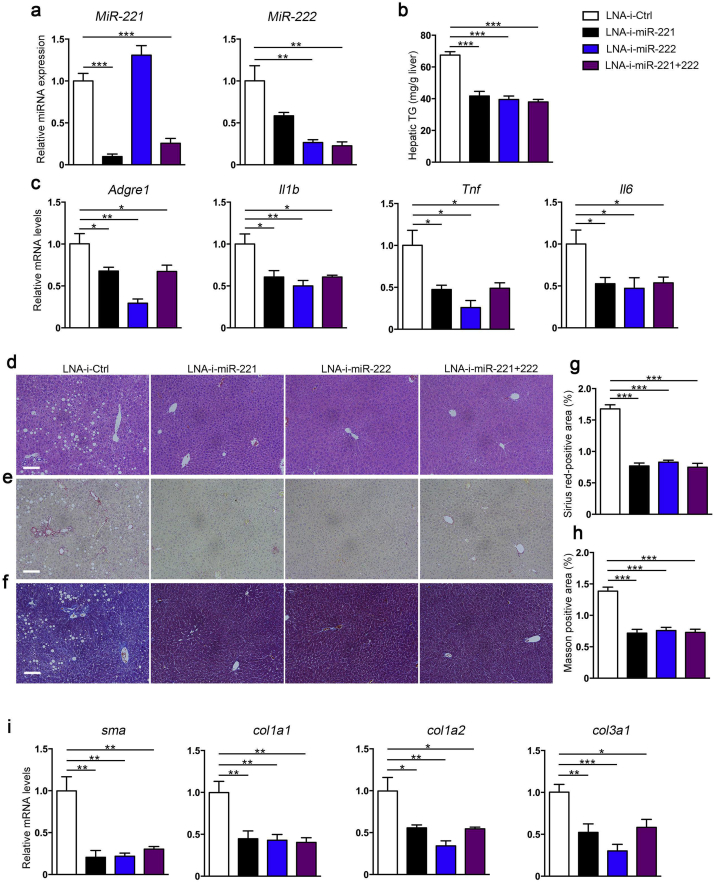
3.6. LNA-antimiR-221/222 treatment effectively reduced hepatic steatosis and fibrosis in vivo
Based on the findings from miR-221/222-LKO mice, we performed in vivo miR-221/222 inhibition by LNA-antimiRs in MCDD challenged mice to explore the therapeutic potential on NASH. The inhibition efficiency and concentration of LNA-antimiR-221/222 were pre-assessed in vitro on mouse HCC hepa1-6 cells. The expression of miR-221 and 222 was effectively inhibited by LNA-i-miR-221 and LNA-i-miR-222 at the concentration of 50 nM and 100 nM, respectively. The 100 nM concentration with solid effect was used for the following experiments (Fig. S7a). The mRNA and protein levels of p27, which was well-established target gene of miR-221/222, could be decreased by LNA-antimiR-221/222 at 100 nM in vitro (Fig. S7b, c). The effects of LNA-antimiR-221/222 in MCDD-fed mice were further evaluated. LNA-i-miR-221 and LNA-i-miR-222 efficiently and equivalently inhibited the expression of hepatic miR-221 and 222 (Fig. 6a). LNA-antimiR-221/222 significantly decreased hepatic TG and suppressed the expression of inflammation related genes (Adgre1, TNF-α, IL-1β and IL-6) in MCDD-fed mice compared to controls (Fig. 6b, c). In addition, LNA-antimiR-221/222 improved the hepatic steatosis and hepatocyte ballooning in NASH mice as depicted (Fig. 6d). Hepatic collagen deposition in MCDD challenged mice was significantly reduced by LNA-antimiR-221/222 treatment, as proved by sirius Red and Masson's trichrome staining analysis (Fig. 6e–h). The expression of collagen related genes, a-SMA, Col1a1, Col1a2 and Col3a1, were markedly suppressed by antimiRs correspondingly while Col5a1 and Col5a2 were unchanged (Fig. 6i and S8). Taken together, our results demonstrate that LNA-antimiR-221/222 treatment could effectively and simultaneously ameliorate hepatic steatosis, inflammation and fibrosis in vivo in NASH mice. MiR-221/222 inhibitors could be candidate in miRNA-based gene therapies for the intervention of NASH.
Fig. 6.
The effects of LNA-antimiR-221/222 in the MCDD-fed mice. (a) The expression of miR-221 and miR-222 in the livers from control mice treated with LNA-antimiRs (LNA-i-miR-221, LNA-i-miR-222, LNA-i-miR-221 + 222) or control (LNA-i-Ctrl) (n = 5–6). The mice were injected with the LNA-antimiRs on days 1, 4, 8,15 and 22 at the dose of 25 mg/kg. All the mice were fed the MCDD from day 1 to 29. (b) The hepatic TG of control mice treated with LNA-antimiRs or control (n = 6 for each group). (c) The expression of Argre1, Il1b, Tnf and Il6 in the livers of control mice treated with LNA-antimiRs or control (n = 5–6). (d) H&E staining of the liver sections from control mice treated with LNA-antimiRs or control. (e and f) Representative (e) Sirius red staining and (f) Masson staining of the liver sections from control mice treated with LNA-antimiRs or control. Scale bars, 100 μm. (g and h) Analysis of the (g) Sirius red-positive area and (h) Masson-positive area of the liver sections (n = 6 for each group). (i) The expression of sma, Col1a1, Col1a2 and Col3a1 in the livers of control mice treated with LNA-antimiRs or control (n = 5–6). Data represent mean ± SD. *P < .05, **P < .01, ***P < .001, one-way ANOVA.
3.7. Hepatic miR-221/222 targeted Timp3 in NASH mice
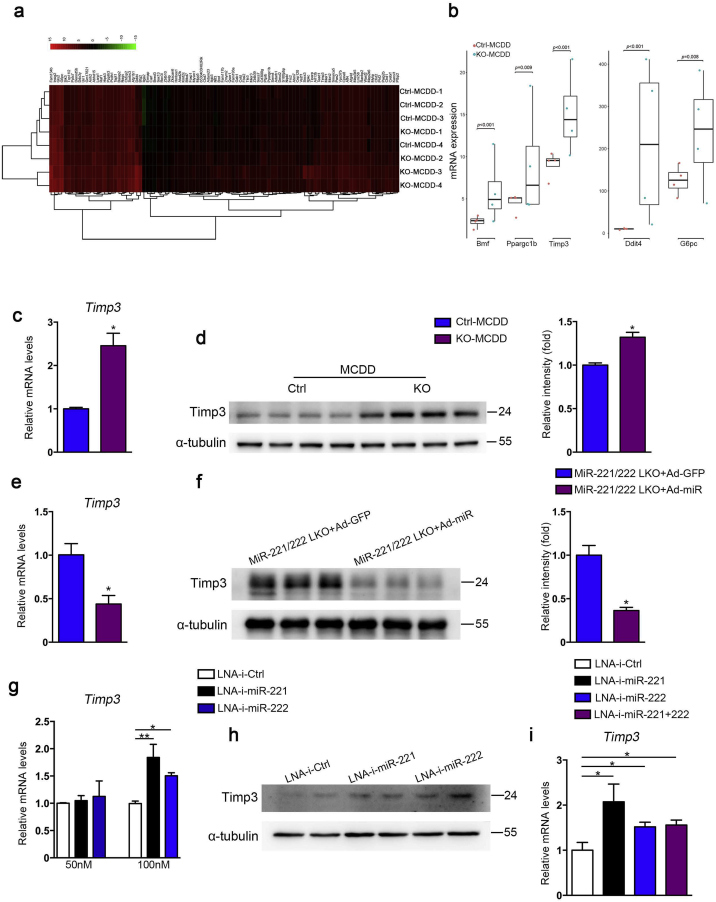
To investigate the mechanic functions of miR-221/222 in NASH, we performed target gene prediction of miR-221/222 by TargetScan (www.targetscan.org) and systematic gene expression analysis by RNA-seq of liver in the MCDD-fed miR-221/222-LKO mice. We identified a series of predicted target genes of miR-221/222 were up-regulated in knockout mice. These genes include confirmed target genes Ddit4, Bmf and Timp3 (Fig. 7a), and metabolic genes PGC-1b (Ppargc1b) and glucose-6-phosphatase-α (G6pc) (Fig. 7b, S9). Importantly, Timp3 (tissue inhibitor of metalloproteinase 3) was top of the most significant upregulated candidate genes (P = .0018; fold change = 1.97).
Fig. 7.
Hepatic miR-221/222 directly targeted Timp3 in NASH mice. (a) RNA-seq analysis and target gene prediction in the liver of MCDD-fed miR-221/222-LKO mice. Significantly upregulated (P < .05 and fold change >1.5) and predicted target genes of miR-221/222 are listed. (b) The expression of Bmf, Ppargc1b, Timp3, Ddit4 and G6pc in the liver of MCDD-fed miR-221/222-LKO mice by RNA-seq analysis. (c and d) The mRNA and protein levels of Timp3 in the liver of miR-221/222-LKO and control mice treated with MCDD for 6 weeks (n = 3–4). The relative intensity of the bands was analyzed. The protein levels of Timp3 were normalized to that of α-tubulin. (e and f) The mRNA (e) and protein levels (f) of Timp3 in the livers of miR-221/222-LKO mice injected with Ad-miR-221/222 or Ad-GFP (n = 3–4). The relative intensity of the bands was analyzed for Western blots. The data represent the mean ± SD, Student's t-test. (g) The expression of Timp3 in the hepa1–6 cells transfected with LNA-antimiRs or control at the concentration of 50 nM and 100 nM (n = 3 for each group). (h) Western blot analyses of Timp3 in the hepa1–6 cells transfected with LNA-antimiRs or control at the concentration of 100 nM. The data shown represent three independent experiments (n = 3 for each group). (i) The expression of Timp3 in the livers of control mice treated with LNA-antimiRs or control (n = 5–6). The data represent the mean ± SD, *P < .05, **P < .01, one-way ANOVA.
Timp3, the main natural regulator of TACE (TNF-α-converting enzyme) activity [26], is a crucial regulator in inflammation, fibrosis and progression of NAFLD and HCC [[27], [28], [29]]. Mice lacking Timp3 result in liver inflammation caused by increased TNF-α activity and defective response to liver injury [27]. Hepatocyte-specific overexpression of Timp3 could prevents NAFLD and tumorigenesis through the regulation of ADAM17 activity [28]. Macrophage-specific overexpression of Timp3 could protects from insulin resistance, NASH and metabolic inflammation in mice [29]. Therefore, Timp3 could be a crucial target gene for the miR-221/222 inhibition-mediated effects in NASH.
Sequence analysis showed the seed sequence of miR-221/222 and the 3′ untranslated region (UTR) of Timp3 had 2 perfect matches, suggesting that Timp3 could be direct target of miR-221/222 in liver (Fig. S10a). We generated reporter constructs with wild type or mutated Timp3 3′UTR cloned downstream of the luciferase gene. Overexpression of miR-221/222 significantly inhibited the 3′UTR luciferase activity of wild type Timp3 constructs in hepa1-6 cells, whereas the mutated 3′UTR at one binding site maintained partial inhibition and mutation at 2 binding sites lost inhibition (Fig. S10b). The mRNA and protein levels of Timp3 were significantly increased in the liver of MCDD-fed miR-221/222-LKO mice (Fig. 7c, d). Consistently, the mRNA and protein levels of Timp3 were decreased by adenovirus-mediated re-expression of miR-221/222 in the liver of knockout mice (Fig. 7e, f). Finally, LNA-antimiR-221/222 significantly increased the mRNA and protein levels of timp3 at the concentration of 100 nM in vitro and in vivo (Fig. 7g–i). In summary, our findings and previous studies indicate that Timp3 is the crucial target gene of miR-221/222 in the pathological processes of liver disease and act as an important regulator in NASH.
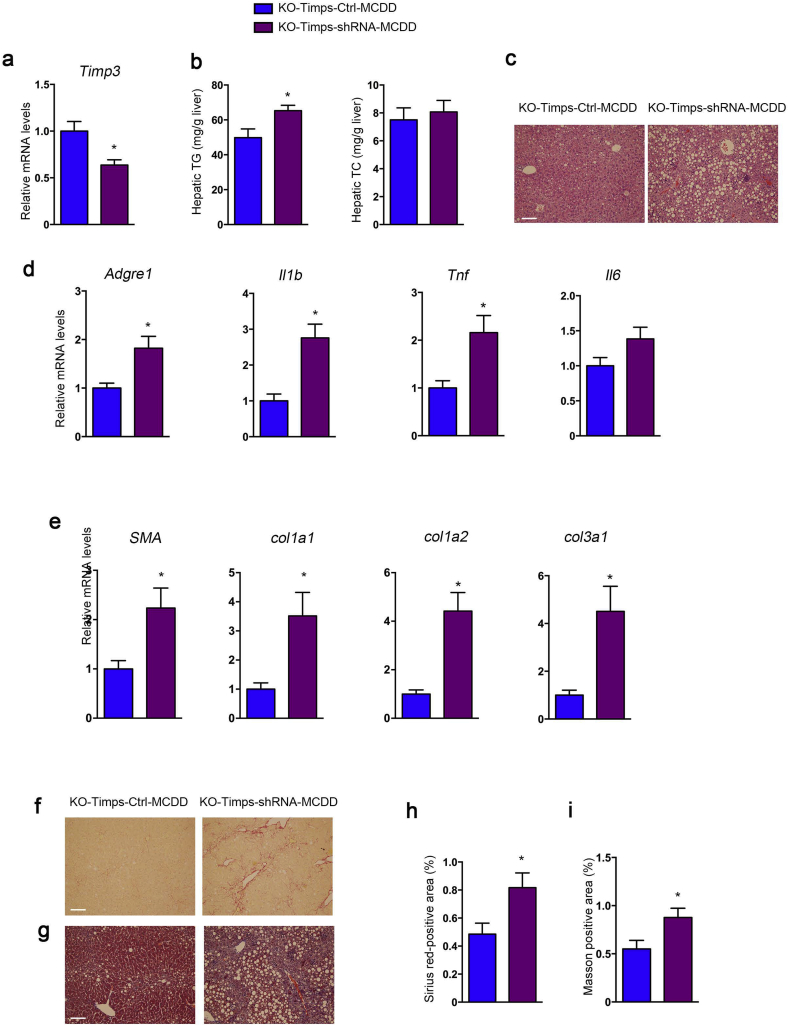
3.8. Inhibition of Timp3 aggravated NASH in miR-221/222-LKO mice
In order to investigate whether Timp3 mediates the effect of miR-221/222 in hepatic steatosis and fibrosis, Timp3 was knocked-down in the liver of MCDD-fed miR-221/222-LKO mice by adeno-associated virus (AAV8-Timp3 shRNA) injection (Fig. 8a). Inhibition of Timp3 significantly elevated the hepatic TG levels and promoted hepatic steatosis and hepatocyte ballooning in MCDD-fed miR-221/222-LKO mice (Fig. 8b, c). The hepatic TC levels were indistinguishable between the two groups. Moreover, the decreased expression of Timp3 facilitated the upregulation of inflammation and fibrosis related genes, including Adgre1, TNF-α, IL-1β, IL-6 and a-SMA, Col1a1, Col1a2, Col3a1, while the levels of Col5a1 and Col5a2 were unchanged (Fig. 8d, e and S10c). Correspondingly, the sirius Red and Masson's trichrome staining indicated Timp3 inhibition resulted in aggravating liver fibrosis progress in MCDD-fed miR-221/222-LKO mice (Fig. 8f–i). Altogether, our data revealed that Timp3 inhibition deteriorate liver fibrosis in MCDD-fed miR-221/222-LKO mice.
Fig. 8.
Inhibition of hepatic Timp3 deteriorates liver steatosis and fibrosis in miR-221/222-LKO mice. (a) The expression of Timp3 in the livers of MCDD-fed miR-221/222-LKO mice injected with adeno-associated virus (AAV8) AAV-Timp3-shRNA or AAV-Ctrl (n = 4 for each group). (b) The hepatic TG and TC of MCDD-fed miR-221/222-LKO mice injected with AAV-Timp3-shRNA or AAV-Ctrl (n = 4 for each group). (c) H&E staining of the liver sections from MCDD-fed miR-221/222-LKO mice injected with AAV-Timp3-shRNA or AAV-Ctrl. (d and e) The expression of (d) Argre1, Il1b, Tnf, Il6 and (e) sma, Col1a1, Col1a2, Col3a1 in the livers of MCDD-fed miR-221/222-LKO mice injected with AAV-Timp3-shRNA or AAV-Ctrl (n = 4 for each group). (f and g) Representative (f) Sirius red staining and (g) Masson staining of the liver sections from MCDD-fed miR-221/222-LKO mice injected with AAV-Timp3-shRNA or AAV-Ctrl. Scale bars, 100 μm. (h and i) Analysis of the (h) Sirius red-positive area and (i) Masson-positive area of the liver sections of MCDD-fed miR-221/222-LKO mice injected with AAV-Timp3-shRNA or AAV-Ctrl (n = 4). Data represent mean ± SD. *P < .05, Student's t-test.
4. Discussion
In this study, our findings provided evidences that miR-221/222 are an efficient and potent targets for hepatic steatosis, inflammation and liver fibrosis in NASH mouse models. Our data and previous reports revealed conservative upregulation of hepatic miR-221/222 in NASH mouse models and patients [20,21]. Overexpression of miR-221/222 promoted the progression of hepatic steatosis, fibrosis and inflammatory infiltration. Meanwhile, the well-known oncogenic functions of miR-221/222 cluster have been extensively investigated in HCC [[17], [18], [19]] and many other cancers, including glioblastomas, breast cancer, prostate cancer, breast cancer, leukemia, colorectal cancer and multiple myeloma [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39]]. The upregulation of miR-221/222 expression might be induced by growth factors or NF-kB activation during the pathogenesis of liver diseases [20,32]. Interestingly, pilot studies for therapeutic applications and pharmacokinetics and pharmacodynamics characteristics of miR-221/222 inhibitors in cancers have shed light on the promising future of antimiR-221/222 in drug therapies [18,39,40]. Importantly, miR-221/222 inhibitors significantly shrink tumors and prolong survival of HCC mouse models [18]. Our work and previous studies indicated that antimiR-221/222 might be effective for NAFLD, NASH and HCC treatment in vivo at the same time, which is an ideal solution for the complicated situation in different stages of chronic liver diseases. The combined therapies contain antimiR-221/222 and other small molecules or monoclonal antibodies which targeting multiple pathways in NASH might be of great interest in the future.
Take in advantage of the nucleotide modification technologies, targeting mir-221/222 may provide innovative practical therapeutics for NASH patients. LNA-based oligonucleotides have been demonstrated high affinity and specificity for the specific complementary strand, such as mature miRNA. Therefore, we used LNA-modified antimiRs for intervention experiments. Intravenous administration of LNA-miR-221 inhibitor in mice and monkeys showed long-lasting detectable LNA-i-miR-221 in tissues, plasma and urine. The safety profile evaluation demonstrated no acute toxicity nor immune stimulation was induced by LNA-i-miR-221 treatment at 10-folds increase of therapeutic dose in monkeys [40]. Here, our data showed LNA-antimiR-221/222 and LNA-i-miR-222 treatment was effective in alleviating NASH characterized by reduced collagen deposition, lipid deposition and inflammatory infiltration in vivo. Taken together, these data support the considerable suitability of LNA-antimiR-221/222 for preclinical and clinical trial use of liver diseases.
Majority of the functional studies of miR-221/222 identified many tumor suppressors as miR-221/222 targets in cancers, such as p27, p57 and PUMA [[30], [31], [32], [33], [34], [35], [36]]. As one of the most increased miRNAs in HCC, hepatic miR-221/222 targets includes DDIT4, p27, p57, PTEN and TIMP3 [17,19,32]. In human and mouse NASH liver, upregulation of miR-221/222 was found to associate with liver fibrosis, stellate cell activation and increase of Col1A1 and α-SMA [20]. These reports indicate the conserved miR-221/222 cluster could regulate hepatic pathological processes through multiple factors and signaling pathways. In our study, systematic target gene screening by RNA-seq and bioinformatics analysis revealed the consistent key genes Ddit4 and Timp3. DDIT4 (encode DNA damage response 1, REDD1) is a negative regulator of mTOR through activation of the tuberous sclerosis tumor suppressor TSC1/2 complex, which is associated with cancer, insulin resistance [17,[41], [42], [43]]. Timp3 is a crucial regulator of inflammation and fibrosis in multiple organ systems. Timp3 knockout promotes the development of fibrosis in kidney and heart [44,45]. Timp3 deficiency increased TACE activity and promotes hepatic steatosis and inflammation in high-fat-diet-fed insulin resistance mouse model [46]. In addition, the expression of Timp3 is down-regulated in livers of NASH patients. Decreasing of Timp3 mediate the increased fibrogenic activity and up-regulation of fibrosis related genes a-SMA, Col1a1 and TGF-β in NASH [47]. These reports indicated that the maintain of Timp3 level is benefit and protective for NASH. However, the mechanisms of Timp3 regulation and intervention are still lack of investigation. In this work, we showed that miR-221/222 directly targeted and inhibited Timp3 during the progression of NASH. MiR-221/222 knockout promoted Timp3 expression whereas miR-221/222 overexpression reduced Timp3 expression. LNA-antimiR-221/222 efficiently rescued the hepatic Timp3 levels and reduced a-SMA and procollagen levels in mouse model. Based on these results, targeting miR-221/222 may ameliorate liver inflammation and fibrosis partially via restoration of Timp3.
In summary, our data show that hepatic miR-221/222 deficiency improved the hepatic steatosis, inflammation and fibrosis in NASH mouse models. Targeting miR-221/222 by LNA-based antimiRs significantly inhibited the progression of steatohepatitis. Liver fibrosis could be markedly ameliorated by the inhibition of miR-221/222 in MCDD-fed mice. Our findings might provide a potent therapeutic approach for treatment of NASH and enhanced the importance of miRNA-based gene therapy in future treatment strategies for liver diseases.
Author contributions
X.J., L.J., A.S. and Y.Ca. contributed to research data, data analysis and wrote the manuscript. Y.S., Y. Ch., D.S., and H.J. contributed to the mouse model experiments. W. W. contributed to discussion and revision of the manuscript; Y.Ca. and G.N. conceived and designed the study, reviewed and edited the manuscript.
Conflict of interests
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.09.051.
Contributor Information
Guang Ning, Email: guangning@medmail.com.cn.
Weiqing Wang, Email: wqingw61@163.com.
Yanan Cao, Email: caoyanan@vip.sina.com.
Appendix A. Supplementary data
Supplementary material
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Angulo P., Kleiner D.E., Dam-Larsen S. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlton M.R., Burns J.M., Pedersen R.A., Watt K.D., Heimbach J.K., Dierkhising R.A. Frequency and outcomes of liver transplantation for nonalcoholic steatohepatitis in the United States. Gastroenterology. 2011;141:1249–1253. doi: 10.1053/j.gastro.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 4.Ratziu V., Goodman Z., Sanyal A. Current efforts and trends in the treatment of NASH. J Hepatol. 2015;62:S65–S75. doi: 10.1016/j.jhep.2015.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Rotman Y., Sanyal A.J. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66:180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 6.Ratziu V., Harrison S.A., Francque S. Elafibranor, an agonist of the peroxisome proliferator-activated receptor-α and -δ, induces resolution of nonalcoholic steatohepatitis without fibrosis worsening. Gastroenterology. 2016;150:1147–1159. doi: 10.1053/j.gastro.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 7.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loomba R., Lawitz E., Mantry P.S., Jayakumar S., Caldwell S.H., Arnold H. The ASK1 inhibitor selonsertib in patients with nonalcoholic steatohepatitis: a randomized, phase 2 trial. Hepatology. 2017 doi: 10.1002/hep.29514. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X.W., Heegaard N.H., Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431–1443. doi: 10.1053/j.gastro.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabo G., Sarnow P., Bala S. MicroRNA silencing and the development of novel therapies for liver disease. J Hepatol. 2012;57:462–466. doi: 10.1016/j.jhep.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 12.Esau C., Davis S., Murray S.F. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 14.Janssen H.L.A., Reesink H.W., Lawitz E.J. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 15.Trajkovski M., Hausser J., Soutschek J. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 16.Loyer X., Paradis V., Hénique C. Liver microRNA-21 is overexpressed in non-alcoholic steatohepatitis and contributes to the disease in experimental models by inhibiting PPARα expression. Gut. 2016;65:1882–1894. doi: 10.1136/gutjnl-2014-308883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pineau P., Volinia S., McJunkin K. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2011;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park J.K., Kogure T., Nuovo G.J. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011;71:7608–7616. doi: 10.1158/0008-5472.CAN-11-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callegari E., Elamin B.K., Giannone F. Liver tumorigenicity promoted by microRNA-221 in a mouse transgenic model. Hepatology. 2012;56:1025–1033. doi: 10.1002/hep.25747. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T., Enomoto M., Fujii H. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600–1609. doi: 10.1136/gutjnl-2011-300717. [DOI] [PubMed] [Google Scholar]
- 21.Shen W.J., Dong R., Chen G., Zheng S. MicroRNA-222 modulates liver fibrosis in a murine model of biliary atresia. Biochem Biophys Res Commun. 2014;446:155–159. doi: 10.1016/j.bbrc.2014.02.065. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y., Liu R., Jiang X. Nuclear-cytoplasmic shuttling of menin regulates nuclear translocation of beta-catenin. Mol Cell Biol. 2009;29:5477–5487. doi: 10.1128/MCB.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Federici M., Hribal M.L., Menghini R. Timp3 deficiency in insulin receptor-haploinsufficient mice promotes diabetes and vascular inflammation via increased TNF-alpha. J Clin Invest. 2005;115:3494–3505. doi: 10.1172/JCI26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohammed F.F., Smookler D.S., Taylor S.E. Abnormal TNF activity in Timp3−/− mice leads to chronic hepatic inflammation and failure of liver regeneration. Nat Genet. 2004;36(2004):969–977. doi: 10.1038/ng1413. [DOI] [PubMed] [Google Scholar]
- 28.Casagrande V., Mauriello A., Bischetti S., Mavilio M., Federici M., Menghini R. Hepatocyte specific TIMP3 expression prevents diet dependent fatty liver disease and hepatocellular carcinoma. Sci Rep. 2017;7:6747. doi: 10.1038/s41598-017-06439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menghini R., Casagrande V., Menini S. TIMP3 overexpression in macrophages protects from insulin resistance, adipose inflammation, and nonalcoholic fatty liver disease in mice. Diabetes. 2012;61:454–462. doi: 10.2337/db11-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fornari F., Gramantieri L., Ferracin M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 31.le Sage C., Nagel R., Egan D.A. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garofalo M., Di Leva G., Romano G. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Sun T., Wang Q., Balk S., Brown M., Lee G.S., Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Leva G., Gasparini P., Piovan C. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frenquelli M., Muzio M., Scielzo C. MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood. 2010;115 doi: 10.1182/blood-2009-11-254656. 3949–3959. [DOI] [PubMed] [Google Scholar]
- 36.Zhang C.Z., Zhang J.X., Zhang A.L. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintavalle C., Garofalo M., Zanca C. miR-221/222 overexpession in human glioblastoma increases invasiveness by targeting the protein phosphate PTPμ. Oncogene. 2012;31:858–868. doi: 10.1038/onc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., Sun X., Wang M. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gullà A., Di Martino M.T., Cantafio Gallo. A 13 mer LNA-i-miR-221 inhibitor restores drug sensitivity in melphalan-refractory multiple myeloma cells. Clin Cancer Res. 2016;22:1222–1233. doi: 10.1158/1078-0432.CCR-15-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallo Cantafio M.E., Nielsen B.S., Mignogna C. Pharmacokinetics and pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in mice and non-human primates. Mol Ther Nucleic Acids. 2016;5:e326. doi: 10.1038/mtna.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ben Sahra I., Regazzetti C., Robert G. Metformin, independent of AMPK, induces mTOR inhibition and cell-cycle arrest through REDD1. Cancer Res. 2011;71:4366–4372. doi: 10.1158/0008-5472.CAN-10-1769. [DOI] [PubMed] [Google Scholar]
- 42.Williamson D.L., Dungan C.M., Mahmoud A.M., Mey J.T., Blackburn B.K., Haus J.M. Aberrant REDD1-mTORC1 responses to insulin in skeletal muscle from Type 2 diabetics. Am J Physiol Regul Integr Comp Physiol. 2015;309:R855–R863. doi: 10.1152/ajpregu.00285.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pastor F., Dumas K., Barthélémy M.A. Implication of REDD1 in the activation of inflammatory pathways. Sci Rep. 2017;7:7023. doi: 10.1038/s41598-017-07182-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kassiri Z., Oudit G.Y., Kandalam V. Loss of TIMP3 enhances interstitial nephritis and fibrosis. J Am Soc Nephrol. 2009;20:1223–1235. doi: 10.1681/ASN.2008050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan D., Takawale A., Basu R. Differential role of TIMP2 and TIMP3 in cardiac hypertrophy, fibrosis, and diastolic dysfunction. Cardiovasc Res. 2014;103:268–280. doi: 10.1093/cvr/cvu072. [DOI] [PubMed] [Google Scholar]
- 46.Menghini R., Menini S., Amoruso R. Tissue inhibitor of metalloproteinase 3 deficiency causes hepatic steatosis and adipose tissue inflammation in mice. Gastroenterology. 2009;136:663–672. doi: 10.1053/j.gastro.2008.10.079. [DOI] [PubMed] [Google Scholar]
- 47.Jiang J.X., Chen X., Fukada H., Serizawa N., Devaraj S., Török N.J. Advanced glycation endproducts induce fibrogenic activity in nonalcoholic steatohepatitis by modulating TNF-α-converting enzyme activity in mice. Hepatology. 2013;58:1339–1348. doi: 10.1002/hep.26491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material