Abstract
Background
The aim of this study was to explore the influence of ubiquitin associated protein 2-like (UBAP2L) on the growth and metastasis of hepatocellular carcinoma (HCC) and its potential underlying mechanism.
Material/Methods
UBAP2L gene was knocked down in SMMC-7721 by RNA interference and cell function experiments were performed. A subcutaneous xenograft tumor model was constructed to examine the effect of UBAP2L silence on HCC growth. Finally, the whole genomic microarrays were used to screen the potential mechanism of UBAP2L in regulating the biological function of HCC.
Results
Compared with those in the control group, the cell proliferation and clone formation were significantly reduced, cell cycle was arrested in G2/M phase, the number of apoptotic cells was remarkably increased, and the abilities of vascular formation and cell migration and metastasis were dramatically weakened in the shUBAP2L group (all P<0.05). UBAP2L knockdown significantly suppressed the tumor growth of HCC in vivo. Moreover, a total of 320 genes changed significantly after UBAP2L knockdown, among which, 159 genes were upregulated and 161 genes were downregulated. Then, gene enrichment analysis revealed that PI3K/AKT and P53 signal pathway were the most significant in the top 10 enrichments. Finally, Western blot analysis verified that UBAP2L knockdown caused the increase of P21 and PTEN and decrease of CDK1, CCNB1, p-PI3K, and p-AKT.
Conclusions
UBAP2L plays an oncogenic role in HCC, and knockdown of its expression significantly inhibits HCC growth and metastasis, which may be related to the regulation of PI3K/AKT and P53 signaling pathways by UBAP2L.
MeSH Keywords: Carcinoma, Hepatocellular; Molecular Biology; Ubiquitins
Background
Primary liver cancer (PLC) is one of the most common malignant tumors of the digestive tract, characterized by high recurrence and metastasis. In China, the incidence and mortality rates of this cancer rank fourth and third, respectively [1,2]. Hepatectomy and liver transplantation are the first choice for radical treatment of liver cancer patients [3–6]. However, most patients are advanced at the time of diagnosis and have lost the chance to benefit from radical surgery. Even though some of these patients receive systemic therapy such as chemotherapy or molecular targeted therapy, the curative effect is still poor and the survival is very short [7,8]. Therefore, it is especially necessary and urgent to explore new biomarkers or therapeutic targets for the early diagnosis or effective treatment for liver cancer.
Ubiquitin-associated protein 2-like (UBAP2L) is a ubiquitin-binding molecule [9] that participates in a variety of cellular pathophysiological processes [10–13]. Recent studies have found that UBAP2L is highly expressed in various malignancies, such as prostate cancer, glioma, colorectal cancer, lung adenocarcinoma, and liver cancer, and is closely related to tumor proliferation, cell cycle, apoptosis, invasion, and metastasis [14–18]. In our previous study [19], we confirmed that UBAP2L was highly expressed in hepatocellular carcinoma (HCC), and UBAP2L overexpression could serve as an independent predictor of poor prognosis in HCC patients. However, the detailed molecular biological role and potential mechanism of UBAP2L in HCC remains unclear.
Therefore, in the present study, we mainly focused on exploring the biological role of UBAP2L in HCC and its potential underlying mechanism. First, UBAP2L-overexpressed hepatoma cell line SMMC-7721 was selected as the target cell line and then the UBAP2L gene expression was knocked down by RNA interference. Second, we assessed the effect of UBAP2L knockdown on hepatoma cell function (including cell proliferation, cell cycle, apoptosis, colony formation, wound healing, migration and invasion, and angiogenesis). Third, a subcutaneous xenograft tumor model was established in nude mice, and the effect of UBAP2L knockdown on HCC growth was observed. Finally, the whole genomic microarrays were used to screen and validate the potential mechanism of UBAP2L in regulating the biological function of HCC.
Material and Methods
Cell culture
HCC cell line SMMC-7721 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). Cell were cultured based on the manufacturer’s protocol and our previously reported method [20, 21]. The study was authorized by the Ethics Committee of our hospital.
Quantitative real-time PCR (qRT-PCR)
TRIzol® reagent (Invitrogen, Carlsbad, CA) was used to isolate the total RNA from SMMC-7721 cells based on the manufacturer’s protocol. Then, reverse transcription was performed using the PrimeScript™ RT reagent kit (Takara, Dalian, P.R. China). The mRNA levels were normalized against those of the GAPDH housekeeping gene. UBAP2L primer sequence was: forward primer: 5′-ATTCGCCTCACTCTCCACAC-3′, reverse primer: 5′-TACCACCACACAACA CAGCA-3′. GAPDH primer sequence was listed below: sense primer: 5′-TGACTT CAACAGCGACACCCA-3′, antisense primer: 5′-CACCCTGTTGCTGTAGCCAA A-3′. Finally, qRT-PCR was done by using SYBR Premix Ex Taq™ (Takara, Dalian, P.R. China) according to the experimental protocol.
Western blot
SMMC-7721 cells were used as the original sample to extract the protein. Then, Western blot analysis was performed as previously described [19–21]. Equivalent amounts of extracted proteins were used in Western blot analysis. Anti-UBAP2L (Abcam, UK) and anti-GAPDH antibodies (Abcam, UK) were used.
Construction and infection of lentivirus
To silence the expression of UBAP2L gene in SMMC-7721, a recombinant lentivirus expression vector containing green fluorescent protein (GFP) tag (pGSIL-sh UBAP2L) was constructed. In order to produce lentiviral particles, the recombinant expression plasmid was co-transfected into SMMC-7721 cells by packaging plasmid system (psPAX2 and pMD2G), and the virus particles were collected 48 h later. SMMC-7721 was infected with shUBAP2L lentiviral vector or empty vector (NC) for 96 h. The infection efficiency was initially evaluated under a fluorescent microscope in each experiment, and then GFP-positive cells were sorted and measured by flow cytometry (BD, USA). The stable infected cells were amplified and harvested for further experiments. The shUBAP2L target sequences (5′-3′) were as follows: Sense: CACCGCCAA TACTGATGATAACGAATTATCAGTATTGGCTGGC; Antisense: AAAAGCCAGC CAAT ACTGATGAT AATTCGTTATCATC AGTATTGGCTGGC.
Cell proliferation assay
SMMC-7721 cells infected with shUBAP2L or shCtrl were inoculated in 96-well plates at a density of 2000 cells per pore of 100 ml and placed in incubators at 37°C and 5% CO2. Cellomics Array Scan VTI (Thermo, Rockford, IL, MA, USA) was used to quantify the number of cells automatically once a day at 488 nm for 5 days. Then, the cell growth curve was generated for each condition.
Clonogenic formation assay
Cells infected with UBAP2L-shRNA lentiviruses or NC lentiviruses were inoculated into 6-well plates at a density of 300–500 cells/pores and further cultured in complete medium for 14 days. After removing the medium and rinsing it twice with PBS, the colony was immobilized with methanol for 30–60 min, stained with GIMSA solution for 10–20 min, and photographed with a digital camera (Leica, Germany). The experiment was repeated 3 times.
Cell cycle and apoptosis
The infected SMMC-7721 cells were harvested at 48 h. Following the manufacturer’s protocol, the Cell Quest software (BD Biosciences) was used to analyze the cells by flow cytometry (FACScan; BD Biosciences) after double staining with FITC-annexin V and propidium iodide (PI). The percentages of G0/G1, S, and G2/M cells were calculated and compared.
Transwell assay
Digestive and re-suspended cells were collected 48 h after plasmid was transfected and adjusted. Matrigel was coated on the polycarbonate microporous membrane of the Transwell system. The upper chamber of the Transwell system was loaded with 100 μl of the digested and re-suspended cells, and the lower chamber was loaded with 600 μl of the corresponding culture supernatant per well for incubation for 24 h. Testing was performed in triplicate in each group.
Wound-healing assay
Wound-healing migration was assayed on confluent cells and monitored using time-lapse microscopy. Data acquisitions were performed every 5 min on a mean of 24-h time course using multisite microscopy. Videos were analyzed using ImageJ (National Institutes of Health), and quantifications were performed with Photoshop (Adobe) software.
Tube formation assay
HUVECs (2×104/well) were cultured in a 96-well plate coated with 70 μL Matrigel (BD Biosciences). Tube formation was defined as a tube-like structure exhibiting a length 4 times its width. Tube morphology was quantified after 24 h in 5 random microscopic fields with a computer-assisted microscope.
Construction of a subcutaneous xenograft model of HCC in nude mice
A 6-week-old female BALB/c nude mice were acquired from the animal center of the Cancer Institute of the Chinese Academy of Medical Science. As described previously [20,21], a subcutaneous xenograft model of HCC in nude mice was constructed by injection of SMMC-7721 infected cells. We randomly divided the nude mice into 2 groups: the UBAP2L silence group (shUBAP2L) and the control group (NC), and each group had 6 nude mice. After 13 days, the mice were euthanized. The tumor volume was calculated as follows: volume=length×width2/2.
Gene expression profiling after UBAP2L knockdown
Whole-genome gene expression profiles were performed after UBAP2L knockdown in SMMC-7721 by using Affymetrix GeneChip Microarray. Differential expressed genes were acquired by comparison between the shUBAP2L and control group (we set the criterion as P<0.05 and fold change >1.5). The DAVID online tool (https://david.ncifcrf.gov/) was used to do the pathway enrichment analysis. The protein levels of some of the key differential expressed genes in classical pathways were validated by Western blot.
Statistical analysis of data
SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) was used to do statistical analysis. Quantitative experimental data are expressed as mean ± standard deviation (SD). The 2 independent samples t test was performed to compare the differences between 2 groups. GraphPad Prism software (Version 5.0) was used to draw statistical graphics. A P value of less than 0.05 was considered to be statistically different.
Results
Knockdown of UBAP2L in SMMC-7721 by lentiviral-mediated RNA interference and efficiency validation
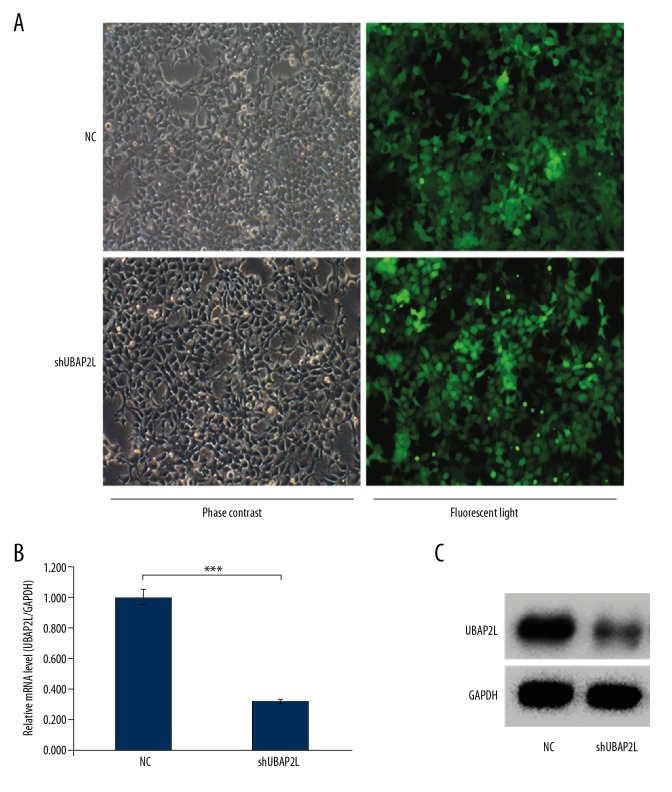
A lentivirus-mediated RNAi technique was used to knock down the UBAP2L gene expression in SMMC-7721 cells. As shown in Figure 1A, the infection efficiency of SMMC-7721 cells after lentivirus infection for 96 h was 94.5%. Then, the knockdown efficiency of UBAP2L was validated by qRT-PCR and Western blot, respectively. The results revealed that compared with the control group, mRNA and protein expression levels of UBAP2L were both significantly reduced after knockdown by RNA interference (Figure 1B, 1C).
Figure 1.
Efficiency of lentivirus infection and the protein and mRNA expression levels of UBAP2L in HCC cells. (A) In SMMC-7721 cells were selected by puromycin for 96 h after infection, and examined by fluorescence and light microscopy, more than 85% of the cells expressed GFP (magnified 100×). (B) qRT-PCR examination of UBAP2L mRNA expression. (C) Western blot examination of UBAP2L protein expression. NC – SMMC-7721 infected with control shRNA lentivirus; shUBAP2L – SMMC-7721 cells infected with UBAP2L shRNA lentivirus. Data are shown as means ±SD, *** P<0.001.
Knockdown of UBAP2L inhibited cell proliferation and colony formation and promoted cell cycle arrest and apoptosis of HCC
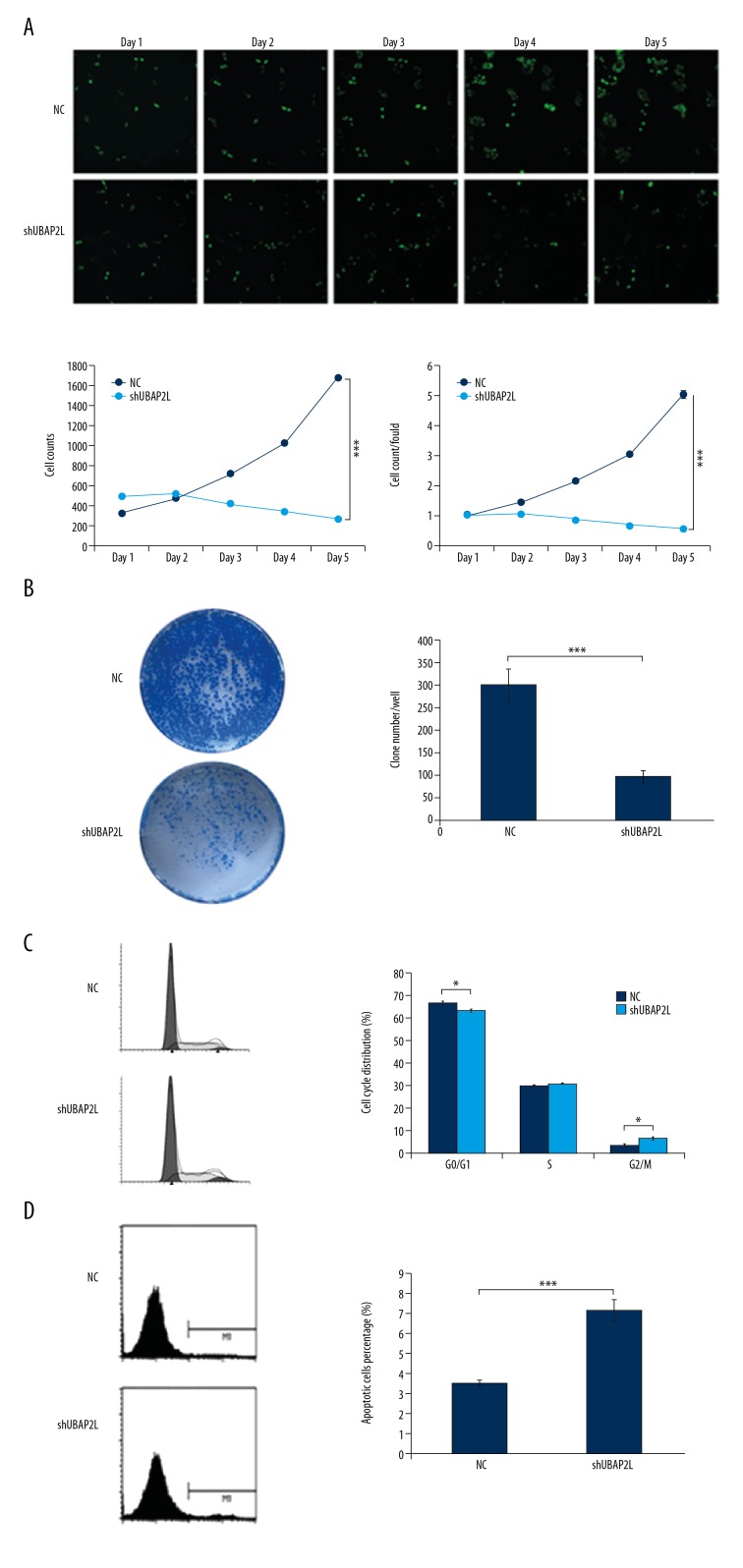
Subsequently, we performed various experiments to observe the influence of UBAP2L knockdown on the cell function of HCC. As shown in Figure 2A, compared with those in the control group, cell count and cell proliferation fold of SMMC-7721 in the shUBAP2L group tended to decrease and reached the maximum on the 5th day, with a statistically significant difference (both P values <0.001). The number of clones in the UBAP2L knockdown group was dramatically reduced compared to that in the control group (P<0.001, Figure 2B). The cell cycle assay showed that the number of SMMC-7721 cells decreased at G0/G1 phase and increased at G2/M phase after UBAP2L knockdown (P<0.05, Figure 2C). Additionally, the proportion of apoptosis in the shUBAP2L group was remarkably increased compared with that in the control group (7.2% vs. 3.5%, P<0.001, Figure 2D). These results suggest that UBAP2L can act as an oncogene and is involved in the occurrence and development of HCC.
Figure 2.
Knockdown of UBAP2L inhibited cell proliferation and colony formation, induced cell cycle arrest and apoptosis, and retarded cell migration and invasion the ability and angiogenic formation in HCC. (A) Cell growth was measured via multiparametric high-content screening (HCS) every day for 5 days after lentivirus infection. Then, SMMC-7721 cell numbers were quantified by Cellomics ArrayScan VTI every day and the cell proliferation rate was analyzed. (B) UBAP2L knockdown reduced colony formation in SMMC-7721 cells. (C) Flowcytometry results showed that the percentage of SMMC-7721 cells transfected with shUBAP2L in the G2/M phase was higher than that in the control group. (D) Flowcytometry showed that the proportion of apoptosis in the shUBAP2L group was remarkably increased compared with that in the control group. Data are shown as means ±SD, *P<0.05, *** P<0.001.
Knockdown of UBAP2L suppressed the ability of cell migration and invasion and angiogenic formation in HCC
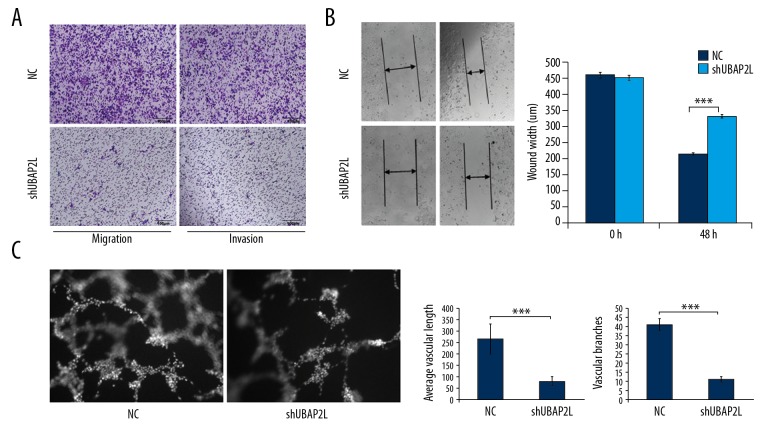
Transwell experiments showed that the cell migration and invasion abilities in the shUBAP2L group were dramatically lower than that of the control group (Figure 3A). As shown in Figure 3B, the shUBAP2L group had significantly wider wound width than that in the control group (P<0.001) at 48 h. Besides, the average vascular length and number of vascular branches were remarkably decreased compared to the control group (P<0.001, Figure 3C). These data suggest that UBAP2L knockdown retarded the ability of cell migration and invasion and angiogenic formation in HCC.
Figure 3.
Knockdown of UBAP2L retarded the ability of cell migration and invasion and angiogenic formation in HCC. (A) Transwell migration and invasion assays. Representative photographs (right) and quantification (left) are shown (Crystal violet stain, ×100). (B) Wound-healing assay. UBAP2L knockdown reduced the migration ability of SMMC-7721 cells. (C) Tube formation assay. UBAP2L knockdown decreased tube formation in SMMC-7721 cells; NC – cells infected with non-targeting shRNA lentivirus, shUBAP2L – cells infected with UBAP2L-targeting shRNA. Data are shown as means ±SD, *** P<0.001.
Knockdown of UBAP2L retarded tumor growth in the HCC nude model in vivo
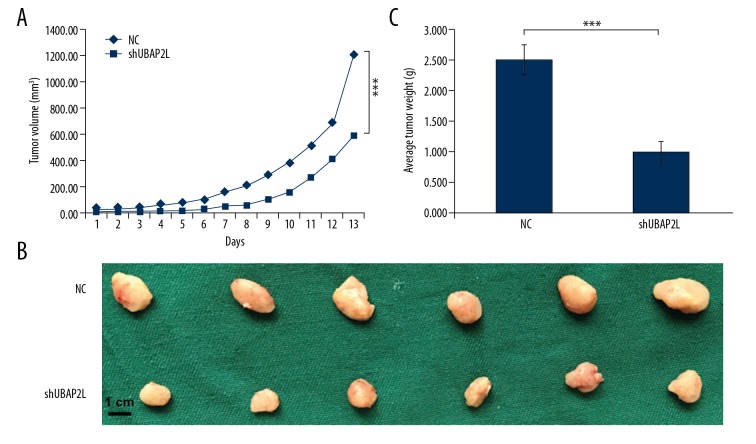
Lentivirus-infected SMMC-7721 cells silenced by UBAP2L were injected subcutaneously into nude mice to construct a subcutaneous HCC xenograft model. The tumor sizes in the 2 groups was recorded daily during the growth phase. All animals were sacrificed after 13 days of continuous observation, and the tumor weight was recorded. As shown in Figure 4, the tumor volume decreased continuously in the shUBAP2L group, and the tumor volume and the average tumor weight in the shUBAP2L group were remarkably lower than those in the control group at the 13th day (All P values <0.001).
Figure 4.
UBAP2L knockdown retarded tumor growth of HCC. (A) Tumor volumes were measured on the indicated days. Knockdown of UBAP2L decreased the volumes of xenografts significantly. (B) UBAP2L knockdown reduced the weight of xenografts. (C) UBAP2L knockdown significantly suppressed the formation and growth of xenografts. Data are shown as means ±SD, bar=1 cm, *** P<0.001.
Gene expression profiling after UBAP2L silence in SMMC-7721
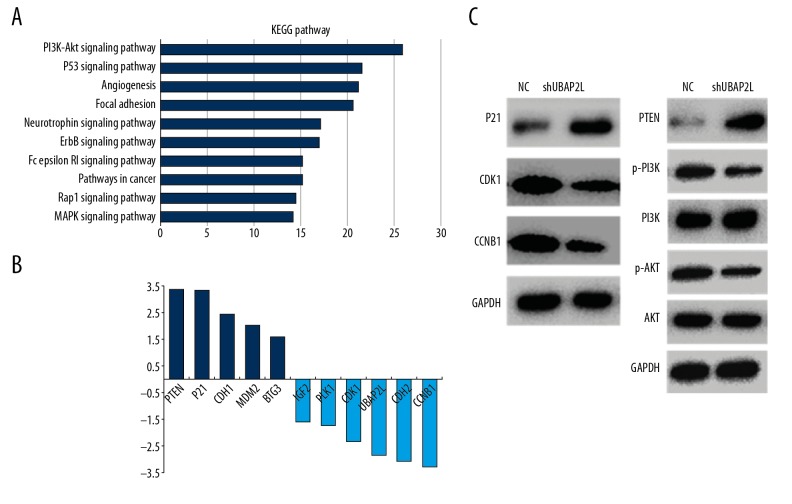
We used gene expression microarray to screen the differentially expressed genes between UBAP2L-silence SMMC-7721 cells and NC cells. Compared to control cells, a total of 320 genes were significantly changed after UBAP2L knockdown, of which 159 genes were upregulated and 161 genes were downregulated. Pathway enrichment analysis of the differential genes in the DAVID online website showed that among the top 10 enrichment pathways, PI3K/AKT and P53 signaling pathways were the most enriched (Figure 5A). Moreover, as shown in Figure 5B, the highest upregulated genes were PTEN, P21, CDH1, MDM2, and BTG3, and the highest downregulated genes were CCNB1 (namely Cyclin B1), CDH2, UBAP2L, CDK1, PLK1, and IGF2. Subsequent Western blot analysis was performed to verify that knockdown of UBAP2L caused the increase of P21 and PTEN, decrease of CDK1 and CCNB1, and phosphorylated PI3K and AKT, while the changes in PI3K and AKT were not obvious (Figure 5C).
Figure 5.
Differential genes and pathway enrichment after UBAP2L knockdown (A) Pathway enrichment analysis was performed using the DAVID online tool. (B) The most significantly differentially expressed genes. (C) The protein levels of part of the key differentially expressed genes in classical pathways were validated by Western blot.
Discussion
Related research on UBAP2L in tumors has gradually increased in recent years. All the existing reports suggest that UBAP2L was highly expressed in malignant tumors and played an oncogenic role in promoting cancer growth and metastasis. For instance, Li et al. [14] found that UBAP2L was overexpressed in PC-3 and DU145 prostate cancer cell lines. Knockdown of UBAP2L significantly inhibited the proliferation and colony formation of both cell lines and arrested the cell cycle in S phase and G2/M phase, respectively, and the migration ability was significantly weakened, which might be associated with the blockade of AMPKα, Bad, and PRAS40 signaling after UBAP2L depletion. Zhao et al. [15] found that UBAP2L was overexpressed in human glioma cells, and knockdown of UBAP2L significantly inhibited cell proliferation and clone formation ability and caused cell cycle arrest in S phase in A172 and G0/G1 phase in U251 and U373. Similar results were found in colorectal cancer cell lines HCT116, SW1116, and RKO [16]. Knockdown of UBAP2L significantly decreased the colony formation and cell proliferation, and promoted cell cycle arrest and apoptosis, by inhibition of P38 phosphorylation and activation of PRAS40, Bad, Bax, cleaved PARP, and Caspase 3. Recent studies have also revealed that UBAP2L can promote tumor metastasis by inducing EMT in lung adenocarcinoma [17] and HCC [18]. However, there are no studies reporting the effects of UBPA2L on other biological functions of HCC, such as cell proliferation, apoptosis, and angiogenesis.
Previously, we confirmed that UBAP2L was overexpressed in HCC, and found that high expression of UBAP2L could be used as independent factor for predicting the poor prognosis of HCC patients [19]. On this basis, in the present study we first used the lentivirus-mediated RNA interference technique to successfully knock down the expression of UBAP2L in the SMMC-7721 cell line, and cell function experiments were performed in vitro. The results showed that compared to those in the control group, cell proliferation and clone formation were significantly reduced, cell cycle was arrested in G2/M phase, the number of apoptotic cells was remarkably increased, and the abilities of vascular formation and cell migration and metastasis were dramatically weakened in the UBAP2L knockdown group. Knockdown of UBAP2L significantly suppressed the tumor growth of HCCs in the xenograft nude model in vivo. These findings are consistent with previous reports of UBAP2L in malignancies [15–18]. Therefore, the above data suggest that UBAP2L acts as an oncogene and plays a key role in HCC occurrence and development.
Subsequently, we made a preliminary exploration of the related molecular mechanisms. First of all, gene expression profiling showed that a total of 320 genes changed significantly after UBAP2L knockdown, among which, 159 genes were upregulated and 161 genes were downregulated. Then, gene enrichment analysis in the DAVID website was performed and revealed that PI3K/AKT and P53 signal pathway was most significant in the top 10 enrichments. Finally, verification by Western blot showed that UBAP2L knockdown caused the increase of P21 and PTEN, and decrease of CDK1, CCNB1, p-PI3K, and p-AKT. The PI3K/AKT signaling pathway and P53 signaling pathway are closely related to the occurrence and development of tumors [22–33], and are involved in apoptosis, cell cycle, angiogenesis, invasion, and metastasis. Thus, together with the KEGG search results and our own findings, we speculated that the potential mechanism was as follows: knockdown of UBAP2L can induce the expression of tumor suppressor gene PTEN, which can activate PI3K/AKT signaling pathway, and AKT can upregulate the expression of P21 and then induce the down-regulation of CDK1/Cyclin B1 (CCNB1) expression, leading to cell cycle arrest in G2/M phase. In fact, recent research has demonstrated that Rab27B can modulate PI3K/AKT/P21 signaling to promote the HCC proliferation [34], and inhibition of P21/CDK1/Cyclin B1 signaling by UBAP2L knockdown can induce cell cycle arrest in G2/M phase in breast cancer cells [35]. These findings are similar to ours and could provide some theoretical support for the possible mechanism detailed above.
There are some limitations in this study. First of all, only 1 strain of hepatoma cell line (SMMC-7721) was used, and the target of RNA interference of UBAP2L was a single sequence. It was difficult to circumvent the possible off-target effect and interfere with the experimental results. Second, the subcutaneously transplanted tumor model in mice was mainly used to observe the effect of UBAP2L on the growth of HCC, and its effect on metastasis could not be evaluated. Additionally, speculation about the downstream mechanism was only based on the preliminary experimental results and previous studies, and this speculated mechanism needs to be confirmed by experiments on the rescue or protein interaction. These deficiencies will be improved and validated in our future research.
Conclusions
Taken together, our preliminary findings show that UBAP2L plays an oncogenic role in HCC, and knockdown of its expression significantly inhibited HCC growth and metastasis, the potential mechanism of which might be related to the regulation of PI3K/AKT and P53 signaling pathways by UBAP2L. UBAP2L may become a potential new therapeutic target for HCC and this warrants further exploration.
Footnotes
Source of support: This research was partly supported by the Natural Science Foundation of Anhui Province (No. 1708085QH177)
Conflicts of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics. Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bureau of Medical Administration, National Health and Family Planning Commission of the PRC. Standardization of diagnosis and treatment for hepatocellular carcinoma (2017 edition) Chin J Dig Surg. 2017;16(7):635–47. [Google Scholar]
- 4.Erstad DJ, Tanabe KK. Hepatocellular carcinoma: Early-stage management challenges. J Hepatocell Carcinoma. 2017;4:81–92. doi: 10.2147/JHC.S107370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie DY, Ren ZG, Zhou J, et al. Critical appraisal of Chinese 2017 guideline on the management of hepatocellular carcinoma. Hepatobiliary Surg Nutr. 2017;6(6):387–96. doi: 10.21037/hbsn.2017.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudo M. Systemic therapy for hepatocellular carcinoma: 2017 update. Oncology. 2017;93(Suppl 1):135–46. doi: 10.1159/000481244. [DOI] [PubMed] [Google Scholar]
- 7.Le Grazie M, Biagini MR, Tarocchi M, et al. Chemotherapy for hepatocellular carcinoma: The present and the future. World J Hepatol. 2017;9(21):907–20. doi: 10.4254/wjh.v9.i21.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taketomi A. Clinical trials of antiangiogenic therapy for hepatocellular carcinoma. Int J Clin Oncol. 2016;21(2):213–18. doi: 10.1007/s10147-016-0966-0. [DOI] [PubMed] [Google Scholar]
- 9.Wilde IB, Brack M, Winget JM, et al. Proteomic characterization of aggregating proteins after the inhibition of the ubiquitin proteasome system. J Proteome Res. 2011;10(3):1062–72. doi: 10.1021/pr1008543. [DOI] [PubMed] [Google Scholar]
- 10.Naz RK, Dhandapani L. Identification of human sperm proteins that interact with human zona pellucida3 (ZP3) using yeast two-hybrid system. J Reprod Immunol. 2010;84(1):24–31. doi: 10.1016/j.jri.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel AK, Schilling M, Comte-Walters S, et al. Identification of O-linked N-acetylglucosamine (O-GlcNAc)-modified osteoblast proteins by electron transfer dissociation tandem mass spectrometry reveals proteins critical for bone formation. Mol Cell Proteomics. 2013;12(4):945–55. doi: 10.1074/mcp.M112.026633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bordeleau ME, Aucagne R, Chagraoui J, et al. UBAP2L is a novel BMI1-interacting protein essential for hematopoietic stem cell activity. Blood. 2014;124(15):2362–69. doi: 10.1182/blood-2014-01-548651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda M, Hasegawa H, Sugiyama M, et al. Arginine methylation of ubiquitin-associated protein 2-like is required for the accurate distribution of chromosomes. FASEB J. 2016;30(1):312–23. doi: 10.1096/fj.14-268987. [DOI] [PubMed] [Google Scholar]
- 14.Li D, Huang Y. Knockdown of ubiquitin associated protein 2-like inhibits the growth and migration of prostate cancer cells. Oncol Rep. 2014;32(4):1578–84. doi: 10.3892/or.2014.3360. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Zong G, Xie Y, et al. Downregulation of ubiquitin-associated protein 2-like with a short hairpin RNA inhibits human glioma cell growth in vitro. Int J Mol Med. 2015;36(4):1012–18. doi: 10.3892/ijmm.2015.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chai R, Yu X, Tu S, et al. Depletion of UBA protein 2-like protein inhibits growth and induces apoptosis of human colorectal carcinoma cells. Tumour Biol. 2016;37(10):13225–35. doi: 10.1007/s13277-016-5159-y. [DOI] [PubMed] [Google Scholar]
- 17.Aucagne R, Girard S, Mayotte N, et al. UBAP2L is amplified in a large subset of human lung adenocarcinoma and is critical for epithelial lung cell identity and tumor metastasis. FASEB J. 2017;31(11):5012–18. doi: 10.1096/fj.201601219RRR. [DOI] [PubMed] [Google Scholar]
- 18.Ye T, Xu J, Du L, et al. Downregulation of UBAP2L inhibits the epithelial-mesenchymal transition via SNAIL1 regulation in hepatocellular carcinoma cells. Cell Physiol Biochem. 2017;41(4):1584–95. doi: 10.1159/000470824. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, Zhang M, Peng Y, et al. Ubiquitin associated protein 2-like (UBAP2L) overexpression in patients with hepatocellular carcinoma and its clinical significance. Med Sci Monit. 2017;23:4779–88. doi: 10.12659/MSM.907071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Jia WD, Hu B, et al. RAB10 overexpression promotes tumor growth and indicates poor prognosis of hepatocellular carcinoma. Oncotarget. 2017;8(16):26434–47. doi: 10.18632/oncotarget.15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu WB, Jia WD, Ma JL, et al. Knockdown of GTPBP4 inhibits cell growth and survival in human hepatocellular carcinoma and its prognostic significance. Oncotarget. 2017;8(55):93984–97. doi: 10.18632/oncotarget.21500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8(35):59950–64. doi: 10.18632/oncotarget.19048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular carcinoma: Implications for genomic and immune therapies. Mol Cancer. 2017;16(1):149. doi: 10.1186/s12943-017-0712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pons-Tostivint E, Thibault B, Guillermet-Guibert J. Targeting PI3K signaling in combination cancer therapy. Trends Cancer. 2017;3(6):454–69. doi: 10.1016/j.trecan.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Spangle JM, Roberts TM, Zhao JJ. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim Biophys Acta. 2017;1868(1):123–31. doi: 10.1016/j.bbcan.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duran I, Lambea J, Maroto P, et al. Resistance to targeted therapies in renal cancer: The importance of changing the mechanism of action. Target Oncol. 2017;12(1):19–35. doi: 10.1007/s11523-016-0463-4. [DOI] [PubMed] [Google Scholar]
- 27.Tazawa H, Kagawa S, Fujiwara T. p53 replacement therapy for cancer. Recent Results Cancer Res. 2016;209:1–15. doi: 10.1007/978-3-319-42934-2_1. [DOI] [PubMed] [Google Scholar]
- 28.Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2017;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: What we know, what we need to know and the path forward. Nat Rev Cancer. 2017;17(12):725–37. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- 30.Ferraz da Costa DC, Fialho E, Silva JL. Cancer chemoprevention by resveratrol: The p53 tumor suppressor protein as a promising molecular target. Molecules. 2017;22(6) doi: 10.3390/molecules22061014. pii: E1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prokocimer M, Molchadsky A, Rotter V. Dysfunctional diversity of p53 proteins in adult acute myeloid leukemia: Projections on diagnostic workup and therapy. Blood. 2017;130(6):699–712. doi: 10.1182/blood-2017-02-763086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheok CF, Lane DP. Exploiting the p53 pathway for therapy. Cold Spring Harb Perspect Med. 2017;7(3) doi: 10.1101/cshperspect.a026310. pii: a026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirzayans R, Andrais B, Kumar P, et al. Significance of wild-type p53 signaling in suppressing apoptosis in response to chemical genotoxic agents: Impact on chemotherapy outcome. Int J Mol Sci. 2017;18(5) doi: 10.3390/ijms18050928. pii: E928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, Ye X, Sun L, et al. Downregulation of serum RAB27B confers improved prognosis and is associated with hepatocellular carcinoma progression through PI3K-AKT-P21 signaling. Oncotarget. 2017;8(37):61118–32. doi: 10.18632/oncotarget.18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He J, Chen Y, Cai L, et al. UBAP2L silencing inhibits cell proliferation and G2/M phase transition in breast cancer. Breast Cancer. 2018;25(2):224–32. doi: 10.1007/s12282-017-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]