SUMMARY
Distinct neuronal types connect in complex ways to generate functional neural circuits. The molecular diversity required to specify this connectivity could be supplied by multigene families of synaptic recognition molecules, but most studies to date have assessed just one or a few members at a time. Here, we analyze roles of cadherins (Cdhs) in formation of retinal circuits comprising eight neuronal types that inform the brain about motion in four directions. We show that at least 15 classical Cdhs are expressed by neurons in these circuits and at least 6 (Cdh 6-10 and 18) act individually or in combinations to promote specific connectivity among the cells. They act in part by directing the processes of output neurons and excitatory interneurons to a cellular scaffold formed by inhibitory interneurons. Because cadherins are expressed combinatorially by many central neurons, similar interactions could be involved in patterning circuits throughout the brain.
eTOC
Duan et al. show that 15 members of the classical cadherin family are expressed in retinal circuits that compute direction of motion. They reveal that 6 cadherins act combinatorially to regulate dendritic lamination and connectivity of circuit elements.
INTRODUCTION
As the central nervous system develops, neurons of many types match up to form complex circuits. A long-standing view is that selective expression of cell surface recognition molecules biases synapse formation in favor of appropriate partners; activity-dependent processes then fine-tune the initial choices (Sanes and Yamagata, 2009; Yogev and Shen, 2014). Some of the required molecular diversity is supplied by members of multigene families such as the cadherin (Cdh) and immunoglobulin superfamilies, semaphorins, and leucine rich repeat proteins(de Wit and Ghosh, 2016; Hirano and Takeichi, 2012; Kolodkin and Tessier-Lavigne, 2011; Koropouli and Kolodkin, 2014; Sanes and Yamagata, 2009; Yogev and Shen, 2014). With few exceptions, however, analyses of these families have assessed just one or a few members at a time.
Here, we analyze roles of “classical” cadherins (Cdhs), a family of ~20 related recognition molecules (Hirano and Takeichi, 2012; Hulpiau, 2016), in assembly of neural circuits. We use mouse retina as a model because, although its circuitry is arguably as complex as that of other brain regions, its genetic accessibility and the extensive knowledge about its structure and function enable detailed analysis (Hoon et al., 2014; Sanes and Masland, 2015; Sanes and Zipursky, 2010). In particular, we focus on circuits capable of reporting the direction in which objects are moving (Fig. 1A). The output neurons are called ON-OFF direction-selective RGCs (ooDSGCs), because they respond selectively to objects that are either brighter (ON stimuli) or darker than the background (OFF stimuli) if they are moving in a particular direction. There are four ooDSGC types, each tuned to motion along one of the cardinal axes of the retina (ventral, dorsal, nasal and temporal; V, D, N, and T) (Vaney et al., 2012). Photoreceptors synapse on two sets of bipolar cells (BCs) that in turn form excitatory synapses on ooDSGCs. Type 2 and Type 5BCs (BC2 and BC5) provide much of the OFF and ON input to the outer and inner strata of the ooDSGC arbor, respectively (Duan et al., 2014; Greene et al., 2016). The BCs also form synapses on ON and OFF starburst amacrine cells (SACs), which in turn form inhibitory synapses on all four types of ooDSGCs. SACs inhibit ooDSGCs most strongly when stimuli move from proximal to distal along their dendrites; the preferred direction of each ooDSGC type is therefore opposite to that of the SAC dendrites that innervate it (Briggman et al., 2011; Vaney et al., 2012).
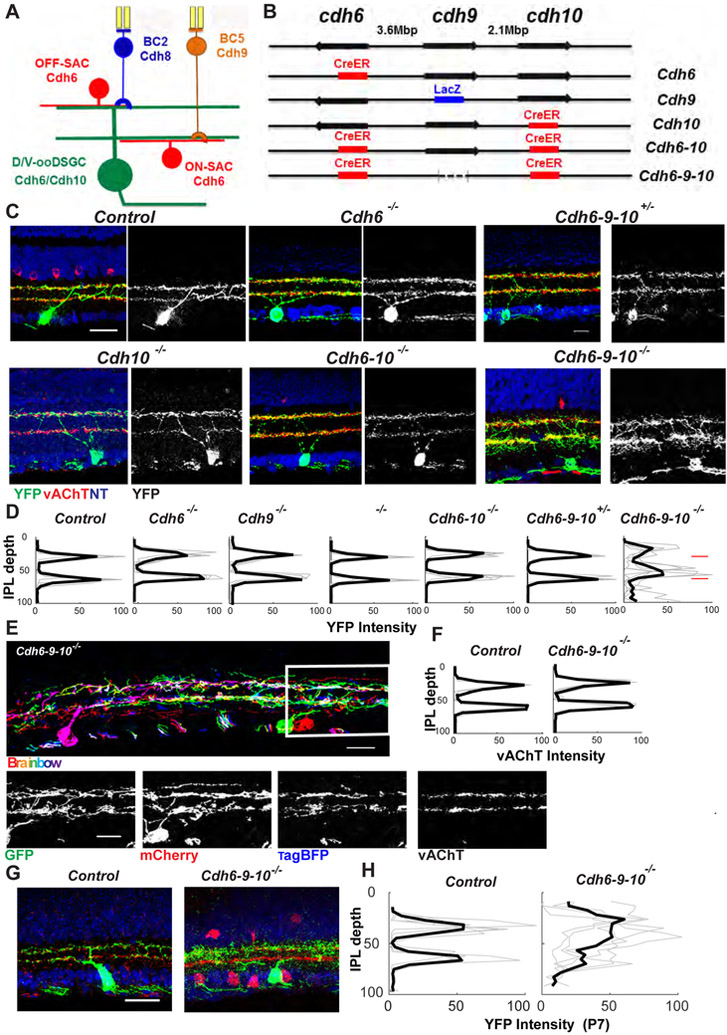
Figure 1. Cdh6, Cdh9 and Cdh10 pattern D/V-ooDSGC dendrites.
(A) Retinal ON-OFF direction-selective circuit, showing expression of Cdh6, Cdh8, Cdh9 and Cdh10 in bipolar cells (BCs), starburst amacrine cells (SACs) and dorsally and ventrally preferring ON-OFF direction selective retinal ganglion cells (D/V-ooDSGCs).
(B) The cdh6-cdh9-cdh10 locus on mouse Chromosome 15 and mutant alleles used in C-H. CreER, tamoxifen-inducible cre-recombinase; LacZ, beta-galactosidase; dotted line, indel deletion.
(C) ooDSGCs in control, Cdh6, Cdh10, Cdh6-10, Cdh6-9-10 mutants and Cdh6-9-10 heterozygotes at postnatal day (P) 21. ooDSGCs were labeled using a Cre-dependent reporter (YFP, green); sections were co-stained for vesicular acetylcholine transporter (vAChT, red) to label SAC dendrites and neurotrace (NT, blue) to visualize somata. Scale bar, 20 μm.
(D) Mean YFP intensity (± SEM) of ooDSGC dendrites across the inner plexiform layer (IPL) from indicated genotypes, derived from images such as those in C (n≥10 cells from each of ≥5 mice of each genotype; light lines show data from individual mice and heavy lines show means). A similarity index (see Methods), was used to tested differences in lamination pattern across genotypes. Cdh6-9-10 mutants differed from the other 5 genotypes (p<0.01), which did not differ significantly from each other.
(E) ooDSGCs in Cdh6-9-10 mutant retinas labeled using a Brainbow virus that marks individual cells in distinct colors. Separate channels are shown for the boxed region. Sections were also co-stained with anti-vAChT to label SAC dendrites (right panel).
(F) Mean vAChT level (± SEM) of SAC dendrites across the IPL in control and Cdh6-9-10 mutant retinas, measured as in “D” from images such as those in C (n as in Fig. 1D) for SACs only. SACs in control and Cdh6-9-10 mutants do not differ significantly in lamination, assessed as in D.
(G) D/V-ooDSGCs in control and Cdh6-9-10 mutant retinas at P7 labeled as in C Scale bar, 20 μm.
(H) Mean YFP intensity (± SEM) of P7 D/V-ooDSGC dendrites across the inner plexiform layer (IPL) from control and Cdh6-9-10 mutant retinas, measured from images such as those in E (n as in Fig. 1D). Similarity score indicates that lamination in mutants differs significantly from controls (p<0.05).
We show here that at least 15 Cdhs are expressed by cells of the direction-selective circuit and that at least 6 (Cdh6, 7, 8, 9, 10 and 18) function individually and in combinations to generate appropriate connectivity in these circuits. They act in part by directing ooDSGCs dendrites and BC axons to a cellular scaffold formed by dendrites of ON and OFF SACs. Because Cdhs are expressed combinatorially in central neurons (Hirano and Takeichi, 2012), and several have been implicated in hippocampal and cerebellar development (Basu et al., 2017; Hirano and Takeichi, 2012; Kuwako et al., 2014), we suggest that similar interactions could be involved in patterning circuits throughout the brain.
RESULTS
Cdh6, Cdh9 and Cdh10 pattern Dorsal/Ventral ooDSGC dendrites.
We showed previously that three Cdhs are selectively expressed by cells of the direction-selective circuit (Cdh6 by D-ooDSGCs, V-ooDSGCs and SACs, Cdh8 by BC2 and Cdh9 by BC5; Fig. 1A), and that Cdh8 and Cdh9 instruct the delivery of OFF and ON bipolar input, respectively, to ooDSGCs (Duan et al., 2014; Kay et al., 2011). To begin this study, we asked whether Cdh6 also plays a role in the DS circuit. We used a Cdh6 null allele in which a tamoxifen-dependent Cre recombinase (CreER) replaced the first coding exon (Fig. 1B, S1A). Administration of tamoxifen to Cdh6CreER mice that had been mated to a Cre-dependent reporter marked V- and D-ooDSGCs and SACs in heterozygotes (Cdh6CreER/+) and Cdh6 mutants (Cdh6CreER/CreER). We detected neither structural (Fig. 1C, D) nor physiological defects (see below) in mutant ooDSGCs or SACs.
Although cadherins are homophilic adhesion molecules, Cdh6 also binds heterophilically to its two closest relatives Cdh9 and Cdh10 (Shimoyama et al., 2000), and Cdh10 is expressed by V-ooDSGCs (Fig. S1E-H) albeit at lower levels than Cdh6 (see below). We therefore generated and analyzed Cdh10 mutants and Cdh6-10 double mutants but detected no defects in either mutant (Fig. 1C, D and S1B-D). Further analysis revealed, however, that Cdh9, which is not normally expressed by ooDSGCs or SACs (Duan et al., 2014), was upregulated in Cdh6-10 mutants (Fig. S1I), suggesting the existence of compensatory mechanisms. We therefore generated Cdh6-9-10 triple mutants using CRISPR-based genome editing; this was infeasible by mating single mutants as the three genes are closely linked (Fig. S1K).
Dendritic arbors of V- and D-ooDSGCs were strikingly abnormal in Cdh6-9-10 mutants. Whereas dendrites of control ooDSGCs co-stratify with SAC dendrites, those of Cdh6-9-10 mutant ooDSGCs were diffusely and variably distributed (Fig. 1C, D). Their variable arborization patterns were revealed clearly with a multi-color Brainbow strategy that marked ooDSGCs in different colors (Cai et al., 2013) (Fig. 1E). Most ooDSGCs (86%; 36 cells from 5 retinas) lost co-fasciculation with SACs. In that Cdh6 labels V-ooDSGCs and D-ooDSGC equally, we conclude that both V-ooDSGCs and D-ooDSGCs responded to Cdh6-9-10 mutants. Thus, Cdh6 and Cdh10 pattern ooDSGC arbors, but that the defects are revealed only when Cdh9 is also deleted. We speculate that Cdh6 may play the predominant role, with Cdh 10 acting in a redundant or compensatory fashion.
Defects were specific to ooDSGCs in that lamina-restricted arbors of other cell types, including SACs, were unaffected in Cdh6-9-10 mutants (Fig. 1E, F and S2A-C). Moreover, loss of Cdh6, 9 and 10 did not affect expression of the cell-type specific marker, Cart (Fig. S2A) (Kay et al., 2011), and we detected no significant change in the overall size or shape of ooDSGCs dendritic arbors or in the size of their somata (average soma size 26.2±4.2 μm2 in controls and 25.2±3.8 μm2 in Cdh6-9-10 mutants; average dendritic diameter 173±34 μm in controls and 162±23 μm in Cdh6-9-10 mutants; x±SEM from 4 animals, 7-20 cells per animal). Thus, Cdhs appear to act selectively on the laminar restriction of ooDSGC dendritic arbors.
Dendrites of ooDSGCs become tightly fasciculated with SACs at the end of the first postnatal (P) week (Peng et al., 2017). Cdhs could promote initial interactions between ooDSGC and SAC dendrites or maintain ooDSGC arbors following their patterning. Defects in ooDSGC arbors were apparent in Cdh6-9-10 mutants by P7, suggesting that Cdhs are required for initial patterning of ooDSGC (Fig. 1G, H).
Cadherins mediate interactions of V-ooDSGC dendrites with an interneuronal scaffold.
Based on the defects in Cdh6-9-10 mutants, we hypothesized that SAC dendrites, which stratify during the first few postnatal days (Ray et al., 2018), act as a scaffold to guide ooDSGC dendrites via cadherin-mediated interactions. This model predicts that eliminating SACs or deleting Cdh6, 9 and 10 in either SACs or ooDSGCs alone should phenocopy defects in global Cdh6-9-10 mutants. We tested these predictions.
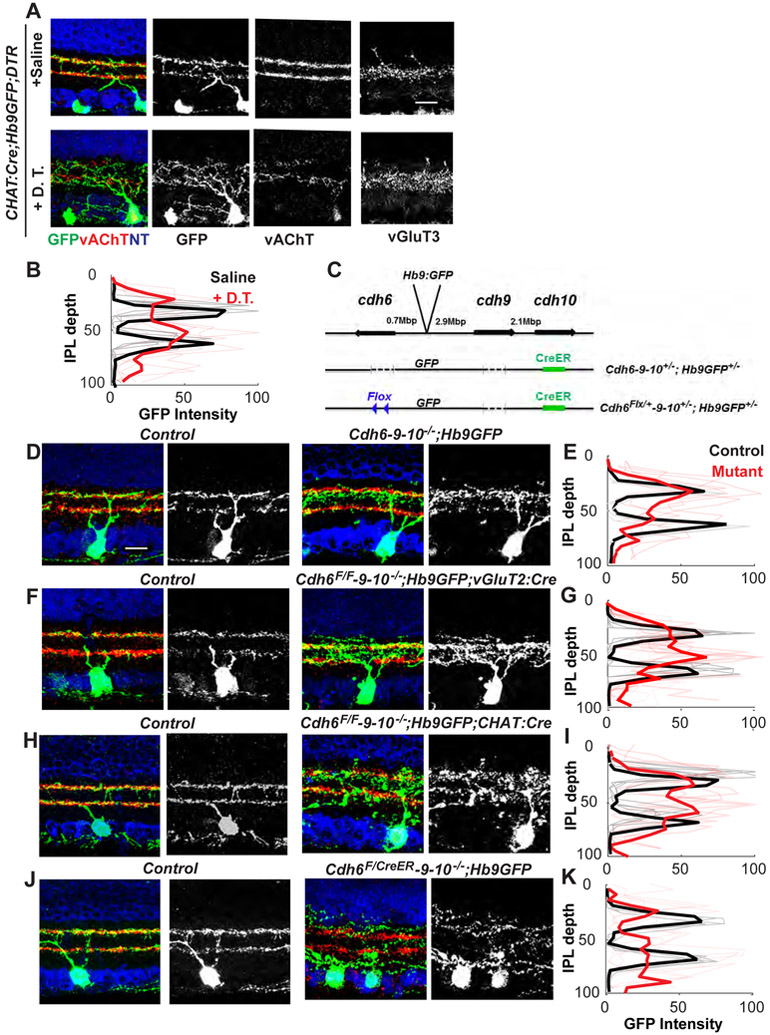
We eliminated SACs by expressing diphtheria toxin receptor in SACs (see Methods) and injecting diphtheria toxin at P0 to ablate SACs before ooDSGC dendrites arborize. We visualized V-ooDSGCs at P21 with the Hb9-GFP transgene, which selectively labels V-ooDSGCs (Trenholm et al., 2011). In regions with severe SAC depletion, V-ooDSGC dendrites arborized diffusely (Fig. 2A, B). The defect was specific to ooDSGCs, as cells that do not fasciculate with SACs, such as vGlut3 ACs, were unaffected, (Fig. 2A). Thus, SACs are required for arborization of V-ooDSGC dendrites.
Figure 2. Cadherins mediate interactions of V-ooDSGCs dendrites with an interneuronal scaffold.
(A) V-ooDSGCs in control retinas, and retinas from which SACs had been killed by diphtheria toxin (ChATcre;CAGS-stop-DTR;Hb9GFP). Sections were co-stained for vesicular acetylcholine transporter (vAChT, red) to label SAC dendrites and neurotrace (NT, blue) to visualize somata. Stratification of VG3 amacrine cells, marked with anti-VGlut3 in separate sections, is unaffected. Scale bar, 20 μm.
(B) Mean GFP intensity (± SEM) of V-ooDSGC dendrites across the inner plexiform layer (IPL), derived from images such as those in A (n as in Fig. 1D). Lamination pattern of Cdh6-9-10 mutants is significantly different from that of controls (p<0.05; see Fig. 1 legend).
(C) Cdh6-9-10;Hb9-GFP alleles generated using CRISPR/Cas9-based genome engineering (see also Fig. S1K, L).
(D,F,H,J) V-ooDSGCs in control and mutant retinas at P21. Staining as in A.
(E,G,I,K) Mean GFP intensity (± SEM) of ooDSGC dendrites across the IPL, derived from images such as those in D,F,H,J, respectively. Lamination pattern of mutants differ significantly from those of controls (p<0.05 for E,G,I and p<0.01 for K; see Fig. 1 legend). n as in Fig. 1D. Bar in D is 20μm.
We next asked whether Cdh6, 9 and 10 are required in ooDSGCs, SACs or both. We planned to use the Hb9-GFP transgenic line to mark V-ooDSGCs in combination with a conditional Cdh6 allele. Surprisingly, the chromosomal integration site of this transgene turned out to be 0.7MB from the Cdh6 locus (Fig. 2C), which may partially account for its expression pattern (ooDSGCs do not express Hb9 endogenously). We therefore used CRISPR/Cas9 to introduce constitutive and conditional Cdh6 alleles on an Hb9-GFP; Cdh9−; Cdh10− background (Fig. 2C and S1L, M). We then selectively deleted Cdh6 from SACs or RGCs with appropriate Cre drivers, and assessed dendritic arborization of Hb9-GFP-marked V-ooDSGCs. Defects in constitutive, SAC-specific and RGC-specific deletions were similar to each other and to in the constitutive Cdh6-9-10 allele (Fig. 2D-I). Thus Cdh6 plays a predominant role in both presynaptic SACs and postsynaptic ooDSGCs.
We also asked whether defects were cell-autonomous at the level of individual ooDSGCs by deleting Cdh6 from a sparse subset of D- and V-ooDSGCs using low doses of tamoxifen in Cdh6CreER/flox;Cdh9−/−;Cdh10−/− mice. Few SACs were mutated in this regimen. Dendritic defects were if anything more severe in isolated Cdh6-9-10-deficient ooDSGCs than when all ooDSGCs were Cdh6-9-10-deficient (Fig. 2J,K), raising the possibility that ooDSGCs dendrites compete for space on the SAC scaffold, with Cdh6-deficient arbors faring poorly.
Different cadherin combinations mediate V-ooDSGC and N-ooDSGC interactions with SACs.
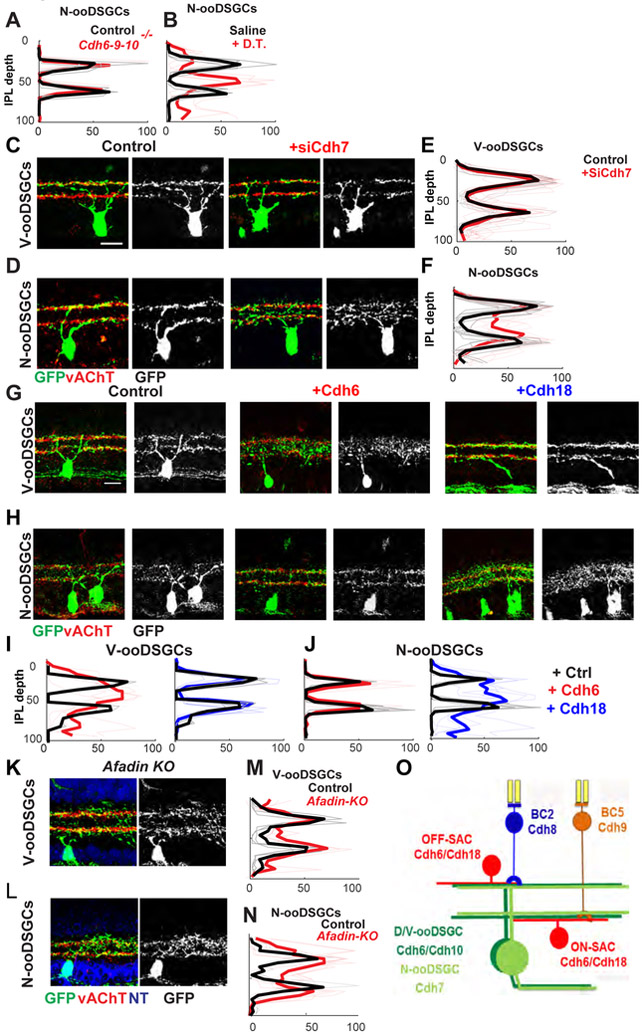
Since dendrites of all four ooDSGCs types fasciculate with SACs, we asked if ooDSGCs selective for other directions are patterned in the same way as D/V-ooDSGCs. Using the Drd4-GFP transgenic line to selectively mark N-ooDSGCs (Huberman et al., 2009), we found that their dendritic arbors were unperturbed in Cdh6-9-10 mutants (Fig. 3A, S3A),. On the other hand, dendrites of N-ooDSGCs, like those of V-ooDSGCs, were dispersed when SACs were ablated with diphtheria toxin (Fig. 3B, S3B). Thus, D-, V- and N-ooDSGCs all fasciculate on a SAC scaffold, but their interactions with the SAC scaffold are mediated by different molecules.
Figure 3. Different cadherins mediate connectivity of V-ooDSGCs and N-ooDSGCs.
(A) Mean Drd4-GFP (N-ooDSGC) intensity (± SEM) across the IPL in control (black) and Cdh6-9-10 mutants (red), calculated from micrographs such as those in Fig. S3A and plotted as in Fig. 1D (n as in Fig. 1D). Similarity score indicates that lamination in Cdh6-9-10 mutants do not differ significantly from those in controls (N.S.).
(B) Mean Drd4-GFP intensity (± SEM) across the IPL in control saline-injected (black) and diphtheria-toxin injected animals (red) calculated from micrographs such as those in Fig. S3B (n as in Fig. 1D). Difference in lamination between groups is significant (p<0.005).
(C, D) V-ooDSGC (Hb9-GFP; C), N-ooDSGC (Drd4-GFP; D) and SAC dendrites (vAChT, red) in control and Cdh7 knockdown retinas at P14.
(E, F) Hb9-GFP (E) or Dr4-GFP (F) intensity (± SEM) across the IPL in control (black) and Cdh7 knockdown (red) retinas (n≥10-12 cells from ≥3-4 mice of each group). Lamination in Cdh7 knockdown differs significantly from control for Drd4-GFP (p<0.005) but not Hb9-GFP.
(G, H) V-ooDSGC (Hb9-GFP; F) or N-ooDSGC (Dr4-GFP; G) and SAC dendrites (vAChT, red) in retinas electroporated with control (RFP), Cdh6, or Cdh18 vectors.
(I, J) Mean Hb9-GFP (H) and Dr4-GFP (I) intensity (± SEM) across the IPL in control (black), Cdh6 overexpression (red), and Cdh18 overexpression (blue) retinas (n as in Fig. 1D). Lamination of V-ooDGSC following overexpression of Cdh6 (p<0.05), and of N-ooDGSC following overexpression of Cdh18 differ from controls (p<0.05). Cdh6, Drd4-GFP and Cdh18, Hb9-GFP do not differ significantly from controls.
(K, L) V-ooDSGC (Hb9-GFP, green, K) or N-ooDSGC (Drd4-GFP, green, L) and SAC dendrites (vAChT, red) in conditional Afadin mutant retinas (Cdh6CreER/+;Thy1-Stop-YFP; AfadinFlx/Flx) at P21.
(M, N) Mean Hb9-GFP (M) and Drd4-GF) (N) intensity (± SEM) across the IPL in control (black) and Afadin mutants (red), calculated from micrographs such as those in K, L; n as in Fig. 1D). Lamination in mutant retinas differs significantly different from controls (p<0.05 for M, p<0.01 for N).
(O) Summary of the expression pattern of Type II Cdhs that wire up parallel direction-selective circuits. Light-green for N-ooDSGCs (Drd4-GFP), dark-green for V-ooDSGCs (Hb9-GFP).
See also Figure S3.
To identify potential mediators of ooDSGC-SAC interactions, we performed RNAseq on FACS-isolated V-ooDSGCs, N-ooDSGCs and SACs (Fig. S3D). Cdh7 was expressed at high levels in N-ooDSGCs but not V-ooDSGCs, and SACs were rich in its preferred heterophilic binding partner, Cdh18 [orthologous to CDH14 in humans; (Shimoyama et al., 2000)] (Fig. S3C), raising the possibility that roles of Cdh7 and Cdh18 in N-ooDSGCs are similar to those of Cdh6 and 10 in D/V-ooDSGCs.
To test this possibility, we first attenuated Cdh7 expression by RNA interference, using sequences previously shown to be effective in vivo (Kuwako et al., 2014). Cdh7 knockdown decreased the alignment of N-ooDSGC with SACs. (Fig. 3D, F), a phenotype similar to that observed in V-ooDSGCs following Cdh6-9-10 deletion. In contrast, Cdh7 knockdown had no effect on V-ooDSGC dendrites (Fig. 3C, E), just as Cdh6-9-10 deletion had no effect on N-ooDSGC dendrites.
We then used a gain-of function strategy to assess the differential sensitivity of N-ooDSGCs and V-ooDSGCs to Cdh6 and Cdh18 by expressing them ectopically in neonatal retina. Vectors encoding Cdh6 or Cdh18 plus a fluorescent protein were introduced by subretinal electroporation, which transduces bipolar, amacrine and Muller glia cells, which have processes in the IPL, but not RGCs. Dendrites of V-ooDSGCs (Hb9-GFP) but not N-ooDSGCs (Drd4-GFP) were disrupted by, and often grew along, processes of cells that ectopically expressed Cdh6. Conversely, Cdh18 disrupted arbors of N-ooDSGCs but not V-ooDSGCs (Fig. 3G-J). Arbors of SACs, were not detectably affected by either cadherin. Thus, Cdh7 and 18 play roles in wiring N-ooDSGC onto a SAC scaffold, similar to those that Cdh6, 9 and 10 play in wiring D/V-ooDSGC onto the same SAC scaffold.
We also deleted afadin, an intracellular signaling molecule that is required for localization and activation of multiple cadherins in several models, although its effects are not limited to cadherins (Beaudoin et al., 2012; Fujiwara et al., 2016). Conditional deletion of afadin from RGCs led to similar dendritic defects in both V-ooDSGCs and N-ooDSGCs (Fig. 3K-N), supporting the idea that different cadherins play similar roles in different populations of ooDSGCs.
Cdh6-9-10 regulate direction-selectivity of D/V-ooDSGCs.
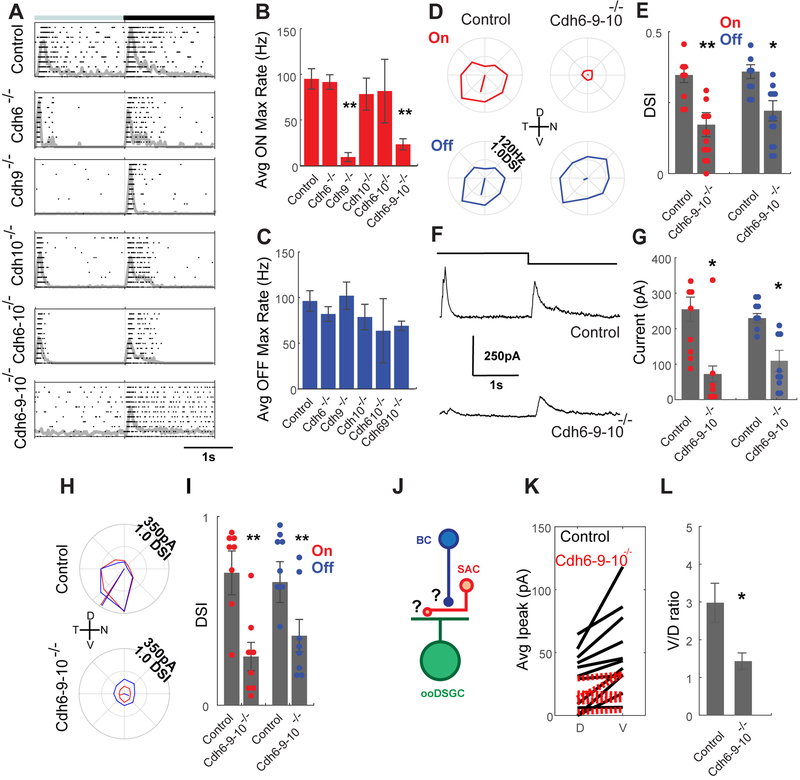
Finally, we investigated the consequences of cadherin deletion on ooDSGC function. Because germ-line reagents are unavailable for Cdhs7 and 18, we confined this analysis to roles of Cdh6, 9 and 10 in D/V-ooDSGCs. We marked D/V-ooDSGCs using fluorescent reporters as above, and targeted them for recording with patch electrodes (Krishnaswamy et al., 2015). Average peak responses of ooDSGCs in Cdh6, Cdh10, and Cdh6-10 mutant explants to spots of light were indistinguishable from those in controls, with robust ON and OFF responses at the beginning and end of the flash, respectively (Fig. 4A-C, S4A-D). OFF responses were also normal in Cdh9 and Cdh6-9-10 mutant ooDSGCs, although ON responses were greatly diminished in these genotypes, consistent with the previously reported loss of input from ON BCs in the absence of Cdh9 (Duan et al., 2014). Likewise, direction-selectivity, assessed by differential responses to bars moving in 8 directions, was normal in Cdh6, Cdh9, Cdh10, and Cdh6-10 mutants. In contrast, direction-selectivity was greatly reduced in Cdh6-9-10 mutant ooDSGCs (Fig. 4D-E, S4E-K). Thus, D/V-ooDSGCs respond robustly to light in the absence of Cdh6, Cdh9 and Cdh10, but their direction-selectivity was severely compromised.
Figure 4. Cdh6-9-10 selectively regulate D/V-ooDSGC direction-selectivity.
(A) Spike raster plots from D/V-ooDSGCs in control, single, double and triple mutants retinas in response to a ~200μm flashing spot centered on the receptive field from 10 trials. ON responses are strongly reduced in the absence of Cdh9.
(B, C) Average ON (B) and OFF (C) firing rates recorded from control (11 cells from 5 mice) and mutant (Cdh6 mutant, n=6; Cdh9 mutant, n=7; Cdh10 mutant, n=6; Cdh6-10 mutant, n=6; Cdh6-9-10 mutant, n=16 cells from 7 mice) D/V-ooDSGCs in response to stimulation as in (A). Cdh9 data are replotted from Duan et al. (2014).
(D) Polar plots of spike responses from D/V-ooDSGCs in control and Cdh6-9-10 mutant retinae in response to a bright bar moving in 8 different directions. Leading edge (ON, red) and trailing edge (OFF, blue) responses are shown separately. Leading edge (ON) responses are strongly reduced and trailing edge (OFF) responses lose direction selectivity in Cdh6-9-10 mutant retinae.
(E) Direction selective index (DSI) for experiments like those in (D) for controls (n=8 from 5 mice) and Cdh6-9-10 mutants (14 cells from 7 mice).
(F) Sample outward currents recorded from D/V-ooDSGCs in controls (top) and Cdh6-9-10 mutants (bottom) retinae to a ~200μm flashing spot.
(G) Average peak outward current for experiments like those in (F) for control (n=8 cells from 5 mice) and Cdh6-9-10 mutant (8 cells from 7 mice) in response to the onset (red) and offset (blue) of a flashing spot. Both ON and OFF inhibition are strongly reduced in Cdh6-9-10 mutant retinae.
(H) Polar plot of inhibitory currents on an V-ooDSGC evoked by a bar moving in 8 directions in control (top) and Cdh6-9-10 mutant (bottom) retinae. Leading (ON, red) and trailing (OFF, blue) edge responses are shown separately.
(I) Average DSI computed from experiments in (H) for control (n = 8 cells from 5 mice) and in Cdh6-9-10 mutant (8 cells from 7 mice) V-ooDSGCs. Outward currents are reduced in Cdh6-9-10 mutant retina and do not display directional tuning.
(J) Direction-selective outward currents might be reduced because of a loss of BC input to SACs or might be reduced because of a loss of SAC-ooDSGC synapses.
(K) Average SAC-evoked currents from stimulation of ChR2-positive SACs located dorsal (D) or ventral (V) of V-ooDSGCs in control (black, 11 cells from 5 mice) or Cdh6-9-10 mutant retinas (red, 5 cells from 5 mutants).
(L) Ventral/Dorsal ratio for data shown in in (K).
Bars in B,C,E,G,I and L show mean ± SEM. ** indicates p<0.01 and * indicated p<0.05 in B, E, G, I, L.
See also Figure S4.
The direction-selectivity of ooDSGCs is generated by inputs from SACs (Vaney et al., 2012; Wei and Feller, 2011). Defects in ooDSGC-SAC fasciculation documented above suggested that SAC-ooDSGC transmission might be compromised in Cdh6-9-10 mutants, which would explain the loss of direction-selectivity. We tested this possibility by recording inhibitory currents of ooDSGCs, which arise predominantly from SACs (Vaney et al., 2012; Wei and Feller, 2011). Inhibitory currents were drastically reduced in Cdh6-9-10 mutants, and residual inhibitory responses were not appreciably direction-selective, suggesting they arose from other sources (Fig. 4F-I). These deficits were specific to inhibitory ooDSGC inputs; excitatory OFF BC-ooDSGC responses in Cdh6-9-10 mutants were comparable to controls (Fig. S4L-O).
Loss of input from SACs, in turn, could result either from failure of BCs to excite SACs or from failure of SACs to form functional synapses on ooDSGCs (Fig. 4J). To distinguish these possibilities, we expressed channelrhodopsin-2 (ChR2) in SACs and used two-photon excitation to stimulate them directly (Krishnaswamy et al., 2015). Monosynaptic connections from SACs to V-ooDSGCs were greatly attenuated in Cdh6-9-10 mutants and asymmetric inhibition was markedly reduced (Fig. 4K, L, S4P-S), accounting for the loss of direction-selectivity. Together, these results demonstrate that the Cdh6-9-10 combination is required for the formation or function of the SAC-ooDSGC synapses that underlie direction selectivity.
DISCUSSION
We exploited advantageous features of the retina and prior knowledge of the direction-selective circuit (Vaney et al., 2012; Wei and Feller, 2011) to test the idea that multiple members of a gene family, in this case the classical cadherins, act in combination to promote the selective connectivity required for circuit function. Results reported here and previously (Duan et al., 2014) show that at least 6 cadherins (Cdh6, 7, 8, 9, 10, and 18) cooperate to pattern this circuit (Fig. 30).
Perhaps the most striking aspect of Cdh involvement is that different members of this multi-gene family restrict the arbors of distinct neuronal types to sublaminae within the IPL. Cdh8 is required to target OFF BC2 axons, Cdh9 to target ON BC5 axons, Cdh6, 9 and 10 to target D/V-ooDSGCs dendrites, and Cdh7 and 18 to target N-ooDSGCs dendrites. This division of labor is reflected in the physiological phenotypes of cadherin mutants. Thus, deleting Cdh8 dramatically decreases excitatory OFF responses in ooDSGCs, which are derived from OFF BCs, but leaves ON responses quantitatively intact and normally directional-selective. Likewise, deletion of Cdh9 decreases ON responses, delivered by ON BCs, with minimal effect on OFF responses. Deletion of Cdh6, 9 and 10 renders D/V-ooDSGCs largely direction non-selective with minimal effect on their bipolar-mediated responses to flashes. Germ-line mutants will be needed to assess functional roles of Cdh7 and 18, but morphological phenotypes suggest that these cadherins are required for direction-selectivity but not overall responsiveness of N-ooDSGCs. In short, there is a satisfying correspondence between the synapses specified by each cadherin or set of cadherins, and the functional consequences of manipulating cadherin expression.
Taken together, these results lead to two major conclusions. First, each Cdh or set of Cdhs specifies a unique synaptic type that subserves a unique function within a complex circuit. Second, circuit elements function with remarkable autonomy: loss of OFF inputs leaves ON inputs intact (and vice versa) and loss of direction-selectivity leaves light-sensitivity intact.
At a cellular level, the main structural consequence of cadherin mutation was to disrupt the the close association of BC axons and ooDGSC dendrites with SAC dendrites. Importantly, cadherin manipulation affected the laminar restriction of ooDSGCs dendrites with minimal perturbation of SAC dendrites, supporting a model in which SAC dendrites are patterned by cadherin-independent mechanisms and form a scaffold for cadherin-dependent patterning of ooDSGCs dendrites. The observations that SAC deletion phenocopies Cdh deletion (Cdh6, 9, 10 in D/V-ooDSGCs) or down-regulation (Cdh7 in N-ooDSGCs) supports this model, and the observation that Cdh6,9 and 10 are required in both SACs and D/V-ooDSGCs supports that idea that the interaction is based on homophilic interactions (or interactions among closely-related cadherins). As early-born retinal neurons, SACs are well placed to form a scaffold that patterns arbors of other neurons as they form. Recent observations from Kay and colleagues provide evidence that SACs also act as targets for the axonal arbors of BCs (Ray et al., 2018).
A major outstanding question is why D/V-ooDSGCs and N-ooDSGCs use different members of gene family to mediate the apparently similar intercellular interaction of associating their dendrites with those of SACs. One possibility is suggested by the way in which SACs synapse on ooDSGCs. SAC dendrites are themselves direction-selective, and ooDSGCs acquire direction selectivity because the “eastward-pointing” dendrites of many SACs connect selectively with “westward-preferring” ooDSGCs and so on (Briggman et al., 2011; Vaney et al., 2012; Wei and Feller, 2011). Cadherins could mediate this selective connectivity by interacting with ligands asymmetrically distributed across the SAC arbor. Available reagents do not permit a critical test of this idea, but do suggest strategies for seeking the hypothetical SAC ligands.
Finally, it is important to note that Type II cadherins do not act alone to pattern the direction-selective circuit; other recognition molecules including semaphorins, plexins, immunoglobulin superfamily molecules, Megf10/11, and protocadherins are also involved (Kay et al., 2012; Kostadinov and Sanes, 2015; Lefebvre et al., 2012; Peng et al., 2017; Sun et al., 2013). It is likely that similar combinatorial interactions underlie synaptic specificity throughout the brain, but at present, the complex perturbations of specific neuron needed to unravel the molecular logic of neural circuit assembly are particularly feasible in the retina.
STAR Methods
Contact Information for Reagent and Resource Sharing
Requests for reagents and further inquiries may be directed to the Lead Contact and corresponding author Joshua R. Sanes (sanesj@mcb.harvard.edu). Animal strain request will be fulfilled by Xin Duan (xin.duan@ucsf.edu)
Experimental Model and Subject Details
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) at Harvard and UCSF. Mice were maintained under regular housing conditions with standard access to food and drink in a pathogen-free facility. Immunohistochemistry experiments were carried out using P7-28 mice unless indicated otherwise. Retinal physiological recording was carried out on young adults (2-3 months). The RNA-Seq experiments were performed at postnatal age (P) 7. Male and female mice were used in roughly equal numbers; no sexual dimorphisms were observed. Animals with noticeable health problems or abnormalities were not used. Genotypes were determined by PCR of tail biopsy. The following mouse lines were used:
Cdh6CreER and Cdh10CreER mouse lines were established by targeted insertion of a frt-neo-frt cassette, a 6xmyc-tagged CreER-T2, and poly-adenylation signal at the translational start site of the cdh6 and cdh10 coding sequence. This removed their predicted signal sequences by deleting the rest of the exons encoding the N-terminal 76 amino acids of Cdh6 (MRTYRYFLLLFWVGQPYPTFSNPLSKRTSGFPAKRKALELSANSRNELSRSKRSWMWNQFFLLEEYTGSDYQYVGK) and the N-terminal 77 amino acids of Cdh10 (MTIYQFLRLFVLWACLPHFCCPELTFRRTPGIQQMTAESRAPRSDGKILHRQKRGWMWNQFFLLEEYTGSDYQYVGK). We generated targeting vectors by lambda phage-mediated recombineering . Mouse embryonic stem cells (V6.5) were electroporated and clones were screened for homologous recombination. Mouse chimeras were produced by the Harvard University Genome Modification Facility (GMF). High percentage chimeras transmitting the knock-in alleles were bred to animals expressing FLP recombinase to remove the Neo cassette. Indistinguishable expression patterns were confirmed for 2 independent founders for each knock-in allele, and we established a line from one of the two. Initial analysis of Cdh6CreER was reported (Kay et al., 2011). Both Cdh6CreER and Cdh10CreER are null alleles, but show no outward abnormality, and are viable and fertile. For sparse labeling of ooDSGCs, we used a low concentration of tamoxifen (50μg/kg, subcutaneously in the Cdh6CreER line), which we showed previously leads to preferential labeling of ooDSGCs with few SACs labeled (De la Huerta et al., 2012; Kay et al., 2011).
Cdh6-10 double mutant:
The strategy for generating Cdh6-10 double mutants took account of their close linkage (Figure. 1B). We first crossed Cdh6CreER/ CreER mice (Cdh6 mutants) to Cdh10CreER/ CreER mice (Cdh10 mutants) to obtain trans-heterozygotes. Transheterozygotes males were then mated to wild-type females, and progeny were screened to detect offspring carrying both cdh6 and cdh10 mutant alleles. We obtained one such cis-heterozygote from 320 offspring. This mouse was bred to establish the Cdh6-10 mutant line.
Cdh6-9-10 triple mutant:
Generating triple mutants by mating was infeasible, so we used CRISPR/Cas9 based genome engineering (Cong et al., 2013). Cas9 RNA and sgRNA against cdh9 were injected into fertilized zygotes from Cdh6-10 mutants, which were then implanted in pseudo-pregnant females. Pups carrying large indels in the first coding exon of cdh9 were identified by PCR. Of 19 such founders, we selected 8, which were bred to wildtype animals to determine whether the cdh9 indel and the Cdh6-10 mutant were in cis. Three lines were established: Line #7 (215bp indel), Line#34 (98bp indel) and Line #35 (38bp indel). All led to generation of short, truncated proteins with incomplete signal sequences
The sequence of the sgRNA was: GACUUACAGUUGUCUUCAACUGG
Indels detected in three Cdh6-9-10 alleles are as follows:
1) MRTYSCLQQHITRKG-QSLPEKDSESEKG-G-NAPSCQAWLDVESVLPLRRVYRYRHSVCRK Line #35 (38bp)
2) MRTYSCLQLVIWTCSIVPSVAGCGISSSS-KSIQVQTLSM- Line #34 (98bp)
3) MRTYSCLQLVIWnn Line #7 (215b)
Cdh6-9-10 mutant containing the Hb9-GFP transgene:
Attempts to generate Cdh6-9-10 mutant carrying the Hb9-GFP transgene failed. The failure suggested that the site of transgene insertion was in close proximity to the Cdh6-9-10 locus, a result that we confirmed by targeted locus amplification (X.D., M.A.L. and J.R.S. manuscript in preparation). While attempting to generate Cdh6-9-10; Hb9-GFP mice, we obtained a recombinant allele bearing a cdh9-10 mutant and the Hb9-GFP transgene. We then used CRISPR/Cas9 based genome engineering as described above to introduce a cdh6 mutation into this line. Out of 25 pups containing large indels in the first coding exon of cdh6, we used two to establish lines: Line #21 (40bp indel) and Line #7 (208bp indel). Both lines carry short, truncated proteins with incomplete signal sequences.
The sequence of the sgRNA was : GUUCGAAAAGGAGUUGGAUGUGG
Indels detected in two Cdh6-9-10;Hb9-GFP alleles are as follows:
1) MRTYRYFLLLFWVGQPYPTFSNPLSKRTSGFPAKRKALELSANSRNELSRWNTRDPIIST WA Line #21 (40bp deletion)
2) The entire exon2 was deleted - Line#7:215bp deletion.
Cdh6 conditional mutant containing Cdh9 and Cdh10 null alleles and Hb9-GFP transgene:
To generate a Cdh6 conditional mutant, we modified the targeting vector that had been used to generate the Cdh6CreER allele (Figure S1A). This vector contained 2.4kb upstream and 1.8kb downstream of the first coding exon of cdh6. LoxP sites flanking the first coding exon were synthesized and inserted between the two arms, and target sequence for the sgRNA was mutated to avoid cutting by Cas9. We used this vector to re-engineer the Cdh9-10;Hb9-GFP mutant. The sgRNA used for the Cdh6-9-10;Hb9-GFP allele was used for this purpose. The targeting vector, including a synthesized Floxed cdh6 Exon 2 (sgRNA-resistant) was as follows:
(Left- Arm,2.4kbp) GCATACAACGCCCACAGGGATCG.....TTCAAGTTTCGTAGCG(LoxP-Left) ataacttcgtataatgtatgctatacgaagttatAAGCATCTCTAAAAGTGCTTGATATGTTATTATTCTTTCC AGGTACCCTCTGAAAGCCAAGCAAAGAACATTAAGGAAGGAAGGAGGAATGAGCCTGGATTTGG TGCAGTGAAAAGAGGCGTATTAAGAAAAGGGGAGCTCACACCCAGACTCGACTGCCTGCCTTGCC AGCATCATGAGAACTTACCGGTACTTCTTGCTGCTCTTTTGGGTCGGCCAGCCCTACCCAACTTT CTCAAACCCATTATCTAAAAGGACTAGTGGCTTCCCAGCAAAGAGGAAAGCCCTGGAGCTCTCTG CAAACAGCAGGAATGAGCTGAGTCGTTCGAAAAGGAGTTGGATGTacAATCAGTTCTTCCTGTTG GAGGAATACACGGGATCCGATTATCAGTACGTGGGCAAGGTAGGCCTCCTTTGGGTGTTTCGACA GTCTAGGCTTataacttcgtataatgtatgctatacgaagttat (LoxP-right)
GAGAGAGAATGCTCTGGTGG.....CCGACAGTGAGAACTGGCGT (Right-Arm,1.8kbp)
Zygote injection was as described above. From 10 pups carrying the Cdh6Flox insert, we established one line carrying the targeted conditional Cdh6 allele.
Six3-Cre mice express Cre recombinase in all of the retina except its far periphery (Lefebvre et al., 2012).
Hb9-GFP transgenic mice express eGFP in V-ooDSGCs (Trenholm et al., 2011). Hb9 is not expressed endogenously in these cells.
Drd4-GFP transgenic mice express eGFP in N-ooDSGCs (Huberman et al., 2009). Drd4 is not expressed endogenously in these cells (Kay et al., 2011).
Thyl-stop-YFP Line #15 transgenic mice express eYFP driven by Cre-recombinase in many neuronal population (Buffelli et al., 2003), including the majority of retinal ganglion cells.
Cdh9lacZ “knock-in” mice express LacZ from the endogenous Cdh9 locus generating a null allele (Duan et al., 2014).
ChATCre mice express Cre recombinase from the endogenous choline acetyltransferase locus (Rossi et al., 2011). In retina, all and only SACs express ChAT.
vGlut2Cremice express Cre from the endogenous vGlut2 locus (Vong et al., 2011). In retina, all retinal ganglion cells express vGlut2.
To delete SACs, we generated triple-transgenic mice, combining a SAC-specific Cre-recombinase, choline acetyltransferase-cre (Rossi et al., 2011; SACs are the sole cholinergic cells in retina), a Cre-dependent diphtheria toxin receptor transgene (Buch et al., 2005), and the Hb9-GFP transgene, which selectively labels V-ooDSGCs (Trenholm et al., 2011). We injected diphtheria toxin intravitreally at P0; systemic injection was infeasible because motoneurons, which are cholinergic, were also receptor-positive
Method Details
Histology
Mice were euthanized by intraperitoneal injection of euthasol and enucleated. Eye cups were removed and fixed in 4% PFA in PBS on ice for 60 minutes, followed by retina dissection, post-fixation for 30 min, and rinsing with PBS. Retinas were analyzed as cryosections and/or wholemounts as previously described (Kim et al., 2010). Wholemount retina samples were incubated with blocking buffer (5% normal donkey serum, 0.5% Triton-X-100 in PBS for 1-2 hours), then incubated for 7 days at 4C with primary antibodies. For sectioning, fixed retinas were incubated with 30% sucrose in PBS for 2 hours, then quickly frozen and sectioned at 20μm in a cryostat. Sections were incubated with 0.3% Triton X-100, 3% donkey serum in PBS for 60mins, and then with primary antibodies overnight at 4C, and with secondary antibodies for 2 hours at room temperature. Retinas or sections were mounted onto glass slides using Vectashield (Vector Lab) or Prolong Gold Antifade Medium (Life Technology).
Antibodies used were as follows: rabbit and chicken anti-GFP (1:1000, Millipore; 1:500, Abcam); rabbit anti-DsRed (1:1000, Clontech); goat anti-choline acetyltransferase (ChAT) (1:500, Millipore); goat anti-VAChT (1:500, Santa Cruz Biotechnology); guinea pig anti-vGlut3 (1:2500, Millipore); sheep anti-tyrosine hydroxylase (TH) (1:2000, Millipore); mouse anti-PKCa (1:200, Abcam); rabbit anti-HCN4 (1:1000, Alomone); mouse anti-Syt2 (1:500, DSHB); rabbit anti-melanopsin (1:5000, Thermo Scientific); mouse anti-Kv4.2 (1:250, Rockland); goat anti-Sorcs3 (1:1,000, R&D Systems); guinea-pig anti-RBPMS (1:1000, PhosphoSolutions), guinea-pig anti-mKate2 (1:500, (Cai etal., 2013), rabbit anti-Cdh6 (1:1000, gift of G. Dressier, U. Michigan) (Cho et al., 1998), rabbit anti-Cdh10 (1:500, gift of M. Williams, U. Utah) (Basu et al., 2017). Nuclei were labeled with NeuroTrace Nissl 435/455 (1:500, Invitrogen). Secondary antibodies were conjugated to Alexa Fluor 488, Alexa Fluor 568 (Invitrogen), or Alexa Fluor 647 (Jackson ImmunoResearch) and used at 1:500.
In Situ Hybridization
In situ hybridization was performed as described (Duan et al., 2014; Kay et al., 2011). Mice were euthanized and the retina were fixed in 4% PFA/ PBS at 4C for 1 hour then incubated overnight in 30% sucrose/ PBS for cryopreservation, followed by quick-freezing. Retina sections (20μm) were mounted on Superfrost-Plus slides (VWR). Section hybridization was carried out at 65 C. Probes were detected using anti-digoxigenin (DIG) antibodies conjugated to horseradish peroxidase (HRP), followed by amplification with Cy3-tyramide (TSA-Plus System; Perkin-Elmer Life Sciences, MA) for 2hrs.
In vivo electroporation
A Cdh6 cDNA was reported from previous study (Yamagata et al., 2018) and transferred to an expression vector bearing the hEF1a promoter and an in-frame C-terminal mCherry-Tag (hEF1a-Cdh6-mCherry-WPRE). A mouse Cdh18 cDNA was synthesized (Genewiz Inc) based on the sequence from NCBI (NM_001081299.1). This cDNA was transferred to an expression vector bearing the hEF1a promoter and an in-frame C-terminal mKate2-Tag (hEF1a-Cdh18-mKate2-WPRE). In vivo electroporation was carried out as previously described (Matsuda and Cepko, 2004). Briefly, expression plasmids (~3mg/mL) were injected into the sub-retinal space of neonatal mice (P0/1), and current pulses (80 Volts) were applied across the head, using paddle electrodes (Harvard Apparatus, Size 7).
Adeno-Associated Virus
For Brainbow labeling (Cai et al., 2013), we used a mixture of rAAV9-hEF1a-lox-TagBFP-loxeYFPloxWPRE.hGH and rAAV9-hEF1a-lox-mCherry-lox-mTFP1-lox-WPRE-hGH. AAV was purchased from Penn Vector Core (1×10E13 titer, an equal titer mixture of the two AAVs, AV-9-PV2453 and AV-9-PV2454). In both cases, μl AAV was injected subretinally into Cre or CreER driver lines using a Hamilton syringe and 33G blunt-ended needle (Duan et al., 2015). Animals were euthanized and retinas were dissected 2+4 weeks following injection. AAV2-CAG-Cre (Park et al., 2008) was produced by the Childrens Hospital Boston AAV core.
siRNA
Customized siRNAs were synthesized by Dharmacon Inc based on previously validated sequences against mouse Cdh7 (Kuwako et al., 2014). Sequences were:
#1: 5’-GCCAUUACUAUACUGGAUAUU-3’
#2: 5’-GCCUCAAUACUCACGAGAAUU-3’
siRNAs were dissolved in RNase-free H2O to ~10μg/ul. 1μ1 of siRNA mixture containing both siRNAs and siGLO RISC-free control siRNA (Dharmacon) was injected intravitreally into P3~4 retinas using RNase-free glass pipettes. Control animals were either injected with siGLO RISC-free control siRNA only or uninjected. Eyes were collected at P9~10 for analysis.
Image Acquisition
Immunostained images were acquired from an Olympus-FV1000 Confocal Microscope, using 440, 488, 568, and 647 lasers with a step size of 0.5μm. We used ImageJ (NIH) software to analyze confocal stacks and generate maximum intensity projections.
RNAseq and Gene Expression Analysis
For RNAseq, V-ooDSGCs (Hb9-GFP), N-ooDSGCs (Drd4-GFP) and SACs (ChAT-cre; Thy1-stop-YFP) were FACS sorted at P6. Libraries were generated and sequenced as described in (Peng et al., 2017). RNAseq data were analyzed using Tuxedo tools (Trapnell et al., 2012).The gene expression level was calculated as transcripts per kilobase million (TPM). All data are shown as Mean ± SEM from at least three independent experimental replicates. Two-tailed Student’s t tests were used for two group comparisons, and one-way ANOVA followed by Bonferroni’s post-tests were used for multiple comparisons.
Electrophysiology
Mice were dark adapted for at least 2hrs prior to euthanasia. The retina was rapidly dissected under infrared illumination in oxygenated (95% O2; 5% CO2) Ames solution (Sigma). The ventral side of the retina was noted and three relaxing cuts were made and the retina was then placed in a recording chamber ganglion cells facing up on the stage of a custom built two-photon microscope and perfused with oxygenated Ames heated to 32-34 °C. Fluorescent ganglion cells were imaged using two-photon microscopy and targeted for recording. For loose cell-attached recordings, the patch electrodes (4-7MOhms) were filled with Ames Solution. For whole-cell recordings, patch electrodes of the same resistance were filled with a Celsium-based internal solution containing (in mM), 120 Cs-Methanesulfonate, 10 Na-Acetate, 0.2 CaCl2, 1 MgCl2, 10 EGTA, 5 CsCl, 2 Mg-GTP, and 0.5 Na2-GTP (pH 7.3). Intracellular recording solutions were supplemented with 5mM QX314-Br for V-ooDSGC voltage clamp recordings. This composition allowed for good separation of excitatory (Eglu ~ −10mV) and inhibitory (Ecl ~ −70mV) currents. Only cells with a Vm more negative than −50mV were used in this study. Signals from loose-patch and whole-cell recordings were acquired with a MultiClamp 700B amplifier (Molecular Devices) using custom software written in LabView (National Instruments). For spikes, the amplifier was put into I = 0 mode and signals were high pass filtered at 1Hz. For currents, signals were filtered at 3kHz and digitized at 20kHz.
Visual stimuli.
Light stimuli were delivered from a projector modified to project monochrome images centered on 410nm (frame rate 60Hz, magnification ~4μm/pixel; gray intensity = 1.5×104 Rstar/sec/rod). Visual stimuli were presented at 100:1 positive contrast and patterns generated using Psychophysics Toolbox in MATLAB. Before testing visual responses, the receptive field center was identified using a grid of flashing spots, and all subsequent stimuli were centered on this spot; Hb9-RGCs typically had receptive field centers that were ventrally offset from their soma position as previously described (Trenholm et al., 2011). Moving bars were presented as a bright long bar moving along its long axis that passed through the receptive field center; the bar was 300μm wide, 1000-1500μm, and moved with a velocity of 1000μm/sec to give good separation between the leading and trailing edges of the bar. Spikes and currents were analyzed as previously described (Kostadinov and Sanes, 2015; Krishnaswamy et al., 2015). Briefly, after chopping traces according to stimulus epochs, spikes were detected using the peak finder function in Matlab and spike counts used to calculate firing rate with 25ms bins; currents measurements were performed on an average of 6 stimulus presentations. Since leading edge response are severely attenuated in the absence of Cdh9, we used the midpoint of a moving bar epoch (bar movement in one direction) to standardize the measurement of leading and trailing edge current and spikes responses across genotypes (responses that preceded or followed this midpoint by ~500ms were considered leading and trailing respectively). A direction selectivity index (DSI) was calculated for spikes as previously described (Kim et al., 2010; Kostadinov and Sanes, 2015); for currents, we measured the peak current amplitude evoked by the leading and trailing edge of the moving bar and calculated the vector sum of these responses to measure DSI.
Optogenetic stimuli.
Methods for two-photon optogenetic stimulation have been described (Krishnaswamy et al., 2015). Briefly, Channelrhodopsin-tdTomato (ChR2) expressing starburst amacrine cells (SACs) in a ~300×300μm field centered on a voltage clamped Hb9-GFP RGC were imaged at low power (2-4mW at 920nm) and a stack of their cell body positions (for both INL and GCL SACs) were acquired. ChR2-positive SAC soma were highlighted with regions of interest until all available SACs were marked. Custom software written in LabView (National Instruments) used these ROIs to steer the two-photon laser to soma locations and activate ChR2 with either raster or spiral scan trajectories (~25-30mW at 920nm) that scanned through the soma in 1-2ms. Each soma was stimulated 6 times, responses were averaged across these repetitions, and stimulus-locked currents identified. For amplitude measurements, the average maximal response in a 40ms window following the stimulus was used; for latencies, stimulus locked currents had to be defined: as having a peak amplitude that was at least 1 standard deviation above the pre-stimulus average baseline and a variance of <15% to confirm that stimulus-locked currents were present on each trial. All analysis was performed in Matlab. Visualizations of connectivity as shown in Figure S4 were computed by computing a contour plot of current amplitude evoked by a field of SACs using the contour function in Matlab.
Quantification and Statistical Analysis
Data acquisition for images.
All images were acquired and processed as described in the method session above. In order to process samples in a systematic and random manner, a set of >15 retinal ganglion cells were sampled from consecutive sections of each retina. In practice, every eighth section was systematically sampled during cryostat preparation, thus ensuring coverage of the entire visual field. For the first cohort of Cdh6CreER, Cdh10CreEr, Cdh6-10 double mutants and Cdh6-9-10 triple mutants were analyzed in parallel, using the same imaging setting and analysis procedures. Different regions of the retinal (central and peripheral, dorsal and ventral regions) were randomized and analyzed. The second cohort of genetic experiments, alleles containing Hb9-GFP were analyzed in parallel in the same manner. Imaging experiments were not done in a blinded manner. Notably, the lamination assay was very robust and apparent to multiple co-authors. Statistical analysis was performed in MATLAB using the Anova1 function for ANOVA and multcomp function for pairwise testing. P values were reported individually throughout Figures 1-3, where the P values reflected the post-hoc pairwise testing results. All statistical tests, sample sizes (cell numbers and animal numbers) for each experiment were listed in the figure legends, accompanying Figures 1-3. No methods were used in the current data in order to determine whether the set of data met the assumptions of the statistical approach.
Dendritic lamination quantifications:
ooDSGC dendrites were quantified as previously described (Duan et al., 2014). Briefly images of ooDSGCs were acquired with a 40X Oil-Lens at a resolution of 1024×1024 pixels. Neurotrace counter-staining of nuclei was used to define the borders of the IPL. Intensity measurements were made in regions extending through the IPL; regions were ~40μm wide and chosen to avoid primary dendrites. The INL-IPL and IPL-GCL borders were assigned values of 0% and 100% respectively. Relative positions of YFP and vAChT signal within each image/cell were measured using the “Analyze/Plot Profile” function in ImageJ. “Plot Values” (X, depth within IPL; Y, cumulative signal intensity at any given X value) were obtained digitally from each image. The X value from the measurement was first normalized to the “total IPL depth” for each data point as “IPL depth %,” ranging between 0 and 100%. The Y value was normalized to the highest intensity pixel of each image as “YFP/VAChT Arbitrary Unit”. Normalized y values were then binned every five percent of IPL depth %, and averaged into one Y value. Thus each axonal arbor was transformed into a plot with 20 values (From 5%, 10%, to 95%, 100%) along the “IPL depth” Axis. Arbitrary Units were then calculated for each animal, and these were averaged by genotype or manipulation with “N” being the number of animals per genotype (>=5 animals with >=10 cells per animal). The IPL depth scores for the OFF and ON SAC dendrites were 30-35% and 60-65% respectively, as determined by vAChT staining.
Similarity index for control linescans was 0.85 ± 0.04 indicating that linescans from ooDSGC dendrites strongly resemble that for SACs. Similarity indices were pooled by genotyped and subjected to a one-way ANOVA to determine whether groups were significantly different. If differences were detected, posthoc pairwise tests were performed to determine the significance level reported in the figure legends.
Electrophysiology:
All statistics of currents were calculated in MATLAB. Pairwise comparisons were made using two-tailed t-test, and multiple samples were compared using one-way analysis of variance. Statistical tests and sample sizes (cell numbers and animal numbers) for the electrophysiology experiments are listed in relevant figure legends. Experimenter was blinded to genotype for electrophysiological experiments.
Data and Software Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE90673.
Supplementary Material
Highlights.
Cells of retinal direction-selective (DS) circuits express 15 cadherins (Cdhs)
Cdh 6, 9 and 10 regulate lamination of ventral motion-preferring DS cell dendrites
Cdh 7 and 18 regulate lamination of nasal motion-preferring DS cell dendrites
These Cdhs promote interactions of DS cell dendrites with an interneuronal scaffold
ACKNOWLEDGEMENTS
This work was supported by NIH (R37NS029169 and R01EY022073) to J.R.S; a Banting Fellowship to A.K; and Research to Prevent Blindness-CDA, That Man May See, Imaging-Morphology Core (P30EY002162) to X.D. Mutant mice were generated at the Harvard GMF. AAV was generated by Boston Childrens Hospital Viral-Core (P30EY012196.) We are grateful to M. Williams for Cdh10 antibody, G. Dressler for Cdh6 antibody, K. Kuwako for Cdh7 cDNA, L Reichardt and T. Jessell for afadin mutant mice.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Basu R, Duan X, Taylor MR, Martin EA, Muralidhar S, Wang Y, Gangi-Wellman L, Das SC, Yamagata M, West PJ, et al. (2017). Heterophilic Type II Cadherins Are Required for High-Magnitude Synaptic Potentiation in the Hippocampus. Neuron 96, 160–176 e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin GM 3rd, Schofield CM, Nuwal T, Zang K, Ullian EM, Huang B, and Reichardt LF (2012). Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci 32, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, and Denk W (2011). Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188. [DOI] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, and Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2, 419–426. [DOI] [PubMed] [Google Scholar]
- Buffelli M, Burgess RW, Feng G, Lobe CG, Lichtman JW, and Sanes JR (2003). Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature 424, 430–434. [DOI] [PubMed] [Google Scholar]
- Cai D, Cohen KB, Luo T, Lichtman JW, and Sanes JR (2013). Improved tools for the Brainbow toolbox. Nat Methods 10, 540–547. [PubMed] [Google Scholar]
- Cho EA, Patterson LT, Brookhiser WT, Mah S, Kintner C, and Dressler GR (1998). Differential expression and function of cadherin-6 during renal epithelium development. Development 125, 803–812. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Huerta I, Kim IJ, Voinescu PE, and Sanes JR (2012). Direction-selective retinal ganglion cells arise from molecularly specified multipotential progenitors. Proc Natl Acad Sci U S A 109, 17663–17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A (2016). Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci 17, 22–35. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, De la Huerta I, and Sanes JR (2014). Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 158, 793–807. [DOI] [PubMed] [Google Scholar]
- Duan X, Qiao M, Bei F, Kim IJ, He Z, and Sanes JR (2015). Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron 85, 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Mizoguchi A and Takai Y (2016). Cooperative roles of nectins with cadherins in physiological and pathological processes In The Cadherin Superfamily: Key Regulators of Animal Development and Physiology, Suzuki SHST, ed. (Tokyo: Springer; ), pp. pp. 115–156. [Google Scholar]
- Greene MJ, Kim JS, Seung HS, and EyeWirers (2016). Analogous Convergence of Sustained and Transient Inputs in Parallel On and Off Pathways for Retinal Motion Computation. Cell Rep 14, 1892–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, and Takeichi M (2012). Cadherins in brain morphogenesis and wiring. Physiol Rev 92, 597–634. [DOI] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, and Wong RO (2014). Functional architecture of the retina: development and disease. Progress in retinal and eye research 42, 44–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, and Barres BA (2009). Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron 62, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulpiau PG, I.S.; van Roy F (2016). Evolution of cadherins and associated catenins In The Cadherin Superfamily: Key Regulators of Animal Development and Physiology, Suzuki SHST, ed. (Tokyo: Springer; ), pp. pp. 13–37. . [Google Scholar]
- Kay JN, Chu MW, and Sanes JR (2012). MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature 483, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, De la Huerta I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, and Sanes JR (2011). Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci 31, 7753–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Meister M, and Sanes JR (2010). Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci 30, 1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, and Tessier-Lavigne M (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropouli E, and Kolodkin AL (2014). Semaphorins and the dynamic regulation of synapse assembly, refinement, and function. Curr Opin Neurobiol 27, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostadinov D, and Sanes JR (2015). Protocadherin-dependent dendritic self-avoidance regulates neural connectivity and circuit function. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A, Yamagata M, Duan X, Hong YK, and Sanes JR (2015). Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature 524, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwako K, Nishimoto Y, Kawase S, Okano HJ, and Okano H (2014). Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep 9, 311–323. [DOI] [PubMed] [Google Scholar]
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, and Sanes JR (2012). Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 488, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Reggiani JDS, Laboulaye MA, Pandey S, Chen B, Rubenstein JLR, Krishnaswamy A, and Sanes JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nat Neurosci 21, 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, and Cepko CL (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A 101, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KK, Liu K, Hu Y, Smith PD, Wang C, Cai B, Xu B, Connolly L, Kramvis I, Sahin M, et al. (2008). Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, Tran NM, Krishnaswamy A, Kostadinov D, Martersteck EM, and Sanes JR (2017). Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron 95, 869–883 e866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray TA, Roy S, Kozlowski C, Wang J, Cafaro J, Hulbert SW, Wright CV, Field GD, and Kay JN (2018). Formation of retinal direction-selective circuitry initiated by starburst amacrine cell homotypic contact. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Balthasar N, Olson D, Scott M, Berglund E, Lee CE, Choi MJ, Lauzon D, Lowell BB, and Elmquist JK (2011). Melanocortin-4 receptors expressed by cholinergic neurons regulate energy balance and glucose homeostasis. Cell Metab 13, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Masland RH (2015). The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci 38, 221–246. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Yamagata M (2009). Many paths to synaptic specificity. Annu Rev Cell Dev Biol 25, 161–195. [DOI] [PubMed] [Google Scholar]
- Sanes JR, and Zipursky SL (2010). Design principles of insect and vertebrate visual systems. Neuron 66, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama Y, Tsujimoto G, Kitajima M, and Natori M (2000). Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J 349, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LO, Jiang Z, Rivlin-Etzion M, Hand R, Brady CM, Matsuoka RL, Yau KW, Feller MB, and Kolodkin AL (2013). On and off retinal circuit assembly by divergent molecular mechanisms. Science 342, 1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholm S, Johnson K, Li X, Smith RG, and Awatramani GB (2011). Parallel mechanisms encode direction in the retina. Neuron 71, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney DI, Sivyer B, and Taylor WR (2012). Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat Rev Neurosci 13, 194–208. [DOI] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S Jr., and Lowell BB (2011). Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, and Feller MB (2011). Organization and development of direction-selective circuits in the retina. Trends Neurosci 34, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Duan X, and Sanes JR (2018). Cadherins Interact With Synaptic Organizers to Promote Synaptic Differentiation. Frontiers in molecular neuroscience 11, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev S, and Shen K (2014). Cellular and molecular mechanisms of synaptic specificity. Annu Rev Cell Dev Biol 30, 417–437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.