Abstract
Background
Rapid tests for HIV testing are essential tools to achieve the 90-90-90 target of the World Health Organization. Many tests are available, some directly from websites. Evaluation of the performance of rapid tests, under close to real-life usage, is therefore needed to ensure accurate diagnosis in the context of the recommendation for their more widespread use.
Method
Nine third- (3G) or fourth-generation (4G) rapid screening tests or self-tests (two bought on websites), were evaluated on an extensive panel of 200 HIV-negative and 312 HIV-positive samples, representative of a wide variety of clinical situations and HIV genetic diversity. A whole blood reconstitution protocol was designed to simulate real-life usage of these tests in community-based and private settings.
Findings
The specificity was high (98.5–100%) and sensitivity excellent (100%) for samples from patients chronically infected with the pandemic strains. The performance for infrequent situations with a major epidemiological and clinical impact, such as infection with divergent viruses or primary infection, was highly variable, depending on the test. One of the two 4G tests allowed detection of additional positive samples from early stages of infection, whereas the second (sold as a 4G test on a website) corresponded in reality to a 3G test.
Interpretation
Our study showed that not all tests are equal for the detection of major HIV variants or early stages of HIV infection; adding the detection of specific p24Ag improved the latter point. This study also showed, for the first time, that buying through web-based vendors can be risky, due to the varying performance of the tests and questionable sales practices. Our results are of particular importance in the context of the increasing use of rapid tests in an “outside laboratory” settings.
Fund
Santé Publique France, COREVIH – Normandie, and Rouen University Hospital.
Research in context.
Evidence before this study
HIV rapid tests are unitary assays used in clinical and non-clinical settings; they facilitate screening of HIV infection due to their technical simplicity and rapid results. The World Health Organisation (WHO) encourages more widespread use of these tests, in particular to extend HIV self-testing. Real-life surveillance of test performance has shown it to be affected by the clinical status of the patient, the genetic nature of the variant responsible for the infection, or the tested population. Many tests are available which may be CE-marked, FDA-approved, WHO prequalified, or not. It is thus essential to continuously control the performance of current and new tests.
Former evidence was identified by searches of MEDLINE/PubMed references from relevant articles using the search terms “rapid diagnostic test”, and “HIV diagnosis”. Abstracts and reports from meetings were included only when they related directly to previously published work. The recommendations and opinions of numerous national and international health organizations and agencies, including the WHO, UNAIDS, ANRS, CDC, ECDC, NIH, and FDA were included. Documents published in English and French between 1983 and 2018 were included.
Added value of this study
We evaluated numerous tests (n = 9) using an extensive panel of 512 samples representative of a wide variety of clinical situations and HIV genetic diversity. This panel allowed us to challenge the various tests, which can be used by anyone and in any region of the world.
This study was conducted on HIV rapid screening tests and self-tests obtained through the regulated market by regional distributors and is the first to also evaluate self-tests available from web-based vendors promoting tests which are not CE-marked, FDA-approved, or WHO prequalified, and for which no published scientific evaluation of performance is available.
Although classical evaluations are performed on plasma/serum samples, this work was performed using fresh whole blood samples or reconstituted whole blood samples to be close to real-life usage. This method could be useful for evaluating other rapid tests, such as those for screening HCV, HBV, or Syphilis infections.
Implications of all the available evidence
This work provides important information on the specific performance and limits of each test. Thus, our data should be particularly useful for helping non-professionals or non-specialists to decide which RSTs/STs to use, according to their HIV-testing context.
The results of our study also showed that the general public should be cautious when buying such medical devices through web-based vendors, due to the varying performance of the tests and questionable sales practices. Tests which are CE-marked, FDA-approved, or WHO prequalified, provided by manufacturers or distributors that respect regional regulations, should be favoured.
Alt-text: Unlabelled Box
1. Introduction
HIV rapid screening tests (RSTs) are unitary immunochromatographic or immunofiltration assays that facilitate screening of HIV infection due to their technical simplicity and rapid results. They have been used for years in high-income countries to provide a rapid indication of HIV status for pregnant women during labour and delivery, patients with AIDS-related symptoms in emergency units, or evaluate the risk of HIV infection in cases of viral exposure of health care workers or sexual exposure between partners [[1], [2], [3], [4]], the HIV status being further controlled and confirmed by supplemental assays according to regional algorithms. Because of their low cost, robustness, and ease of use, they have been promoted for several decades in algorithms for the diagnosis of HIV infection in low- and middle-income countries [5].
RSTs have also used outside of laboratories in high-income countries for many years for community-based testing, increasing the frequency of testing within populations with a high risk of infection [6]. RSTs have also been adapted for HIV self-testing (HIV-ST) for people who do not want to be tested in sexually-transmitted infection (STI) clinics or health or community-based facilities. The person collects his/her own specimen (oral fluid or blood), performs the self-test (ST), and interprets the result himself/herself, with, if necessary, possible phone or internet assistance, according to the details provided by the supplier [7].
RSTs/STs provide an opportunity to achieve the UNAIDS' 90-90-90 objective [8], by easily increasing the frequency of HIV screening, particularly in low-income regions, in which permanent facilities are often limited. The World Health Organisation (WHO) “Guidelines on HIV self-testing and partner notification”, released in December 2016, supports the implementation of HIV-ST in HIV testing services and provide specific guidance on establishing public health policies to improve HIV diagnosis in specific high-risk groups [7]. These recommendations will encourage a growing number of non-specialists to buy RSTs or STs. Given this context of more widespread use, independent evaluations and information concerning the performance of these in vitro diagnostic medical devices are essential, particularly for helping non-professionals to decide which RSTs/STs to use.
For the commercialisation and use of HIV RSTs/STs CE-marking (in Europe), FDA approval (in the US), or WHO prequalification requires performance that matches criteria [[9], [10], [11], [12]] of high sensitivity [100% (CE), ≥99% (FDA-WHO)] and specificity [(≥99% (CE-FDA), ≥98% (WHO)] to avoid a missed diagnosis due to a false-negative result, or an unacceptable false-positive result. Such performance can be guaranteed in the context of these qualifications, but real-life surveillance of test performance has shown it to be affected by the clinical status of the patient, the genetic nature of the variant responsible for the infection, or the tested population, especially in sub-Saharan regions [[13], [14], [15]]. Thus, differences in the sensitivity of 3rd generation (3G) tests, which only detect HIV-1/2 antibodies, have been demonstrated in the early phase of HIV infection [[15], [16], [17]]. The development of 4th generation (4G) RSTs, which detect both HIV-1/2 antibodies and HIV-1 p24 antigen (p24Ag), should improve the screening of such infections [18]. However, the first CE-marked/FDA-approved 4G RST, the Determine HIV-1/2 Ag/Ac Combo assay (Alere/Abbott Chicago Ill), showed limited performance for the specific detection of p24Ag, leading to insufficient added value relative to 3G tests [[19], [20], [21], [22]]. It has also been shown that viral diversity, especially major variants, such as HIV-1 group O or HIV-2, which are circulating in sub-Saharan Africa countries, can affect clinical sensitivity or lead to inefficient discrimination [13,23,24]. Finally, the presence in some regions of numerous undetermined HIV profiles with sticky sera can lead to decreased specificity and false HIV reactivity [13,23]. These issues highlight the need for the continuous evaluation of available and new RSTs/STs.
RSTs/STs are provided by multiple manufacturers, sometimes with limited experience in HIV assays, and a larger number of distributors. Some assays, with no scientific evaluation of their performance, are also sold by web-based vendors (e.g. commercial websites that promote these tests although they are not allowed by regional regulations to sell such medical devices or selling tests that do not respect regional regulations). These points emphasize the need to control the declared performance of the tests.
Moreover, performance evaluations are generally carried out using plasma or serum, although RSTs or STs require finger-stick whole blood. Although some studies have reproduced real-life conditions using whole blood, they did not evaluate performance on a large scale nor in the context of various clinical situations or wide HIV diversity [15].
Taking into account such issues, we designed this work to assess the performance of various 3G and 4G HIV RSTs/STs using an extensive panel of 512 samples representative of varying clinical status and viral diversity. Seven tests were obtained from the regulated market by regional manufacturers or distributors. Given the availability of STs via web-based vendors, we included two supplementary tests randomly bought from such web-based vendors. The nine tests were evaluated using fresh whole blood or reconstituted whole blood samples to be close to the real-life usage of RSTs/STs.
2. Samples and methods
2.1. Rapid screening tests (RSTs) – self-tests (STs)
Six 3G RSTs/STs and one 4G RST (CE-marked or in the process of being CE-marking), allowed in France for clinical, community-based, or self-testing, were provided by regional manufacturers or distributors. The 3G assays were the: EXACTO PRO TEST HIV (Biosynex), Genie Fast HIV1/2 (Bio-Rad), HIVTOP (Biosynex), INSTI (bioLytical), STAT-VIEW HIV 1/2 (Chembio diagnostic systems), and VIKIA HIV 1/2 (bioMérieux). The 4G RST was the HIV Combo (Alere, now Abbott); for the latter, the detection of the p24Ag is performed with a specific line, distinct from the antibody line.
Two supplementary STs were randomly bought from web-based vendors; we searched for websites that directly sell self-tests using the terms “HIV home test”, “HIV self-test”, or “HIV rapid test” with Google or Bing tools. The EZ-TRUST HIV 1 & 2 Rapid Screen Test (CS Innovation Ltd) was sold as a 3G test, whereas the BioTechMed HIV1/2 Rapid-4 (BioTechMed) was sold as a 4G test. No antigen-specific line is present in this 4G test, in contrast to the 4G Alere HIV Combo. No information concerning the CE-marking/FDA-approved/WHO prequalification of these tests was found on the packaging, nor were the tests found on established lists [[25], [26], [27], [28], [29], [30], [31], [32], [33]].
Among these nine tests, two (HIVTOP and HIV1/2 Rapid-4) are designed to discriminate HIV-1 from HIV-2 infections; three lines are present in the test: one for the control, one for the detection of HIV-1 antibodies, and one for the detection of HIV-2 antibodies.
All tests were used following the manufacturers' instructions or indications present on the packaging for the two STs bought on websites. A detailed description of the tests is presented in Table 1.
Table 1.
RST/ST characteristics.
| Name | Manufacturer / distributor | CE approved | FDA approved | WHO prequalification | RST / ST | Detection | Antigen compositiona | Discrimination HIV-1 / HIV-2 | Matrix | Volume | Technology¤ | Control | Early reading time | Final reading time |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EXACTO PRO TEST HIV | Biosynex / id | Yes⁎ | No | No | RST/ST | Ab | ND (gp O mention) | No | Capillary or venous whole blood, serum or plasma | 5 μL | IC | ND (migration) | 10 min | 20 min |
| Genie Fast HIV1/2 | Bio-Rad / id | Yes | No | Yes | RST | Ab | HIV-1 gp120 gp41 & HIV-2 gp36 | No | Capillary or venous whole blood, serum or plasma | 80 μL | IC | ND (migration) | 10 min | 30 min |
| HIV TOP | Biosynex / id | No⁎⁎ | No | No | RST | Ab | ND (gp O mention) | Yes | Capillary or venous whole blood, serum or plasma | 50 μL | IC | ND (migration) | 10 min | 20 min |
| INSTI | bioLytical / Nephrotek | Yes | Yes | Yes | RST/ST | Ab | HIV-1 gp41 & HIV-2 gp36 | No | Capillary or venous whole blood, serum or plasma | 50 μL | IF | protein A | immediatly | NA |
| STAT-VIEW HIV 1/2 | Chembio Diagnostic systems/AAZb | Yes | Yes | Yes | RST/ST | Ab | ND | No | Capillary or venous whole blood, serum or plasma | 2,5 μL | IC | protein A | < (if positive) or = 15 min | 20 min |
| VIKIA HIV 1/2 | bioMérieux / id | Yes | No | Yes | RST | Ab | HIV-1 (gps M & O) gp41 & HIV-2 gp36 | No | Capillary or venous whole blood, serum or plasma | 75 μL | IC | ND (migration) | < (if positive) or = 30 min | 30 min |
| HIV Combo | Alere / Abbott | Yes | No | Yes | RST | Ag/Ab | ND (gp O mention) | No | Capillary or venous whole blood, serum or plasma | 50 μL | IC | ND (migration) | 20 min | 40 min |
| EZ- TRUST HIV 1 & 2 Rapid Screen Test | CS Innovation Ltd. / Web | No | No | No | ST | Ab | ND | No | Capillary whole blood, serum or plasma | 1 drop (with micropipette) | IC | ND (migration) | < (if positive) or = 10 min | 10 min |
| BioTechMed HIV1/2 Rapid-4 | BioTechMed / Web | No | No | No | ST | Ag/Ab | ND | Yes | Capillary whole blood | 1 drop (with micropipette) | IC | ND | < (if positive) or = 15 min | 20 min |
RST: rapid screening test; ST: self-test; Ag: antigen; Ab: antibody; ND: not defined; NA: not applicable; ¤: IC = immunochromatography; IF = immunofiltration.
CE-marking in March 2017; only the version of the test before CE-marking was evaluated.
CE-marking in revision.
From the instruction for use.
Manufactured and distributed as Autotest® VIH by AAZ in France.
2.2. Ethics
Fresh samples were collected from patients after they received written and oral information about the study; no additional blood sample was collected for the study as we used left-over or remnant specimens from our routine analyses or our activities as National Reference Centre on HIV. Reconstituted samples were prepared from samples belonging to registered collections (DC 2015–2533) of our routine and National Reference Centre on HIV activities.
2.3. Whole blood reconstitution
To be close to real-life usage, we reconstituted artificial whole blood by mixing plasma/sera and a globular concentrate of human group O blood cells collected from blood donors. Fifty millilitres globular concentrate was diluted in 50 mL sterile PBS. After centrifugation (3000 rpm, 10 min), the supernatant was removed and the pellet washed once more with an equal volume of sterile PBS. After another round of centrifugation, the red blood cells were suspended in sterile PBS to reach a 45% haematocrit level and then aliquoted into 1.5 mL Eppendorf tubes. Reconstituted whole blood was then obtained by mixing equal volumes of plasma/serum and blood cells. We ensured there were no false results due to the nature of the matrix, by testing 20 paired samples (fresh venous whole blood and reconstituted blood), according to the quality procedure in our laboratory, which conforms to quality regulation norm NF EN ISO 15189. Ten HIV-negative and 10 HIV-positive samples were tested by first performing the test on fresh venous whole blood and then after reconstitution.
The same protocol was performed on the viral supernatants used for the specific detection of p24Ag; supernatants were first mixed with HIV-negative plasma samples, to mimic HIV-positive plasma samples, and then reconstituted as whole blood, as described above. Dilutions of HIV-1 supernatants were performed to obtain a final concentration of approximately 150 pg/mL p24 in whole blood (corresponding to 36·3 UI/mL), measured with the VIDAS® HIV P24 II (P24) (bioMerieux; France) assay, following the manufacturer's instructions.
2.4. Plasma/sera samples
Negative samples (n = 200) were collected from patients attending our hospital-based STI clinic and used as fresh whole blood (n = 39) or reconstituted whole blood (n = 161). The positive samples (n = 300) corresponded to plasma/sera, and all were used as reconstituted whole blood. They were representative of ART-naïve (n = 100) or ART-experienced (n = 125) patients, infected by various subtypes and CRFs of the pandemic group M (HIV-1/M) (characteristics detailed in Table 2 and Fig. 1). Samples representative of divergent variants - HIV-1/O (n = 10) and HIV-2 (n = 15; two were molecularly confirmed to be dually-infected with HIV-1 and HIV-2) - were also tested (Table 2 and Fig. 1). Fifty samples collected during the primary HIV infection (PHI) phase were added to evaluate the sensitivity of detection of early infections; PHI was defined by an incomplete western blot (WB) profile, with the absence of reactivity to Pol proteins, or quantifiable plasma HIV RNA with a negative or weakly reactive EIA, or an interval of less than six months between a negative and positive EIA result, as previously described [34] (viral load, WB, and EIAs results at diagnosis are presented in Table 3).
Table 2.
Patient characteristics.
| Patient characteristics | Negative samples | HIV-1/M ART naive | HIV-1/M ART experienced | HIV-1/O | HIV-2 | PHI |
|---|---|---|---|---|---|---|
| Number | 200 | 100 | 125a | 10 | 15b | 50 |
| Mean Age (±SD) | 28.3 (12.3) | 36.1 (11.9) | 40.5 (11.3) | 42.4 (7.2) | 52.0 (12.8) | ND |
| Sex Ratio (M/F) | 1.0 | 1.7 | 1.3 | 0.1 | 0.4 | 49.0 |
| Mean pVL (log10 cp/mL) (±SD)c | NA | 4.8 (1.06) | 3.7d (0.80) | <1.8 | 3.2e (0.57) | 5.65 (1.17) |
| Min pVL (log10 cp/mL)c | NA | 2.8 | 1.6 | <1.8 | 2.4 | 2.0 |
| Max pVL (log10 cp/mL)c | NA | 6.5 | 6.7 | <1.8 | 3.9 | 7.0 |
| Mean treatment duration (month) (±SD) | NA | NA | 82.7f (66.97) | 72.2 (48.02) | 113.5 (85.98) | NA |
| Mean duration of undetectability (month) (±SD) | NA | NA | 28.2g (29.52) | ND | ND | NA |
| CDC stage | ||||||
| A1 | NA | 11 (11%) | 10 (80%) | ND | 0 (0%) | 50 (100%) |
| A2 | NA | 36 (36%) | 35 (28%) | ND | 5 (33.3%) | 0 (0%) |
| A3 | NA | 19 (19%) | 17 (13.6%) | ND | 0 (0%) | 0 (0%) |
| B1 | NA | 0 (0%) | 2 (1.6%) | ND | 0 (0%) | 0 (0%) |
| B2 | NA | 8 (8%) | 8 (6.4%) | ND | 2 (13.3%) | 0 (0%) |
| B3 | NA | 3 (3%) | 8 (6.4% | ND | 0 (0%) | 0 (0%) |
| C1 | NA | 0 (0%) | 1 (0.8%) | ND | 0 (0%) | 0 (0%) |
| C2 | NA | 2 (2%) | 6 (4.8%) | ND | 0 (0%) | 0 (0%) |
| C3 | NA | 21 (21%) | 30 (24%) | ND | 4 (26.7%) | 0 (0%) |
| ND | NA | 0 (0%) | 8 (6.4%) | ND | 4 (26.7%) | 0 (0%) |
| Total | 200 | 100 | 125 | 10 | 15 | 50 |
| Risk factor | ||||||
| Heterosexual | ND | 56 (56%) | 75 (60%) | 10 (100%) | 14 (93.3%) | ND |
| MSM⁎ | ND | 36 (36%) | 35 (28%) | 0 (0%) | 0 (0%) | ND |
| IDU⁎⁎ | ND | 2 (2%) | 9 (7.2%) | 0 (0%) | 1 (6.7%) | ND |
| MTC⁎⁎⁎ | ND | 0 (0%) | 2 (1.6%) | 0 (0%) | 0 (0%) | ND |
| ND | ND | 6 (6%) | 4 (3.2%) | 0 (0%) | 0 (0%) | ND |
| Total | 200 | 100 | 125 | 10 | 15 | 50 |
ND: not determined; NA: not applicable.
pVL < 1.6 log10 cp/mL (n = 55).
two patients were co-infected with HIV-1/M.
Monitoring pVL: for HIV-1/M, RealTime HIV-1 Abbott Molecular (Rungis, France); for HIV-1/O, group O specific RNA quantification (National Reference Centre on HIV, Rouen, France) [52]; for HIV-2, type 2 specific RNA quantification (National Reference Centre on HIV, Paris, France) [53].
pV L > 1.6 log10 cp/mL (n = 70).
pVL > 1.7 log10 cp/mL (n = 4).
Data available for 108 patients.
Mean calculated from follow-up of 38 patients.
Men who have sex with men.
Intravenous drug user.
Mother-to-child infection.
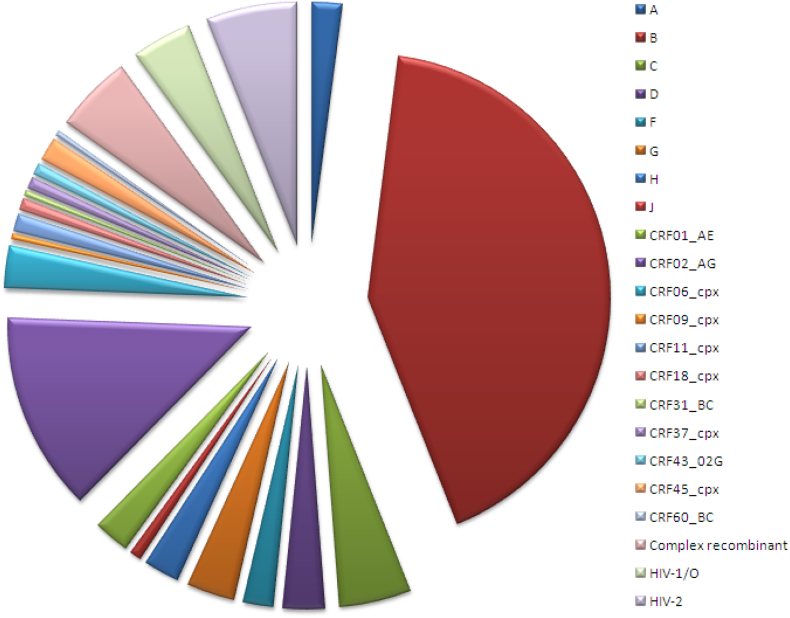
Fig. 1.
Sample genetic diversity.
Diagram representing the genetic diversity of the HIV samples of the clinical sensitivity panel (n = 250). The viruses were characterized for group and subtype by molecular analysis as described previously [[54], [55], [56]].
Table 3.
Virological characteristics of the PHI samples.
| Patient | pVL (log cp/mL) | Western blot |
Architect indexa | BioPlex Ag indexb | BioPlex HIV-1 Ab indexb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gp160 | gp120 | p68 | p55 | gp41 | p40 | p34 | p24 | p18 | |||||
| 1 | 2.0 | − | − | − | − | − | − | − | − | − | 0.14 | 0.14 | 0.04 |
| 2 | 6.1 | − | − | − | − | − | − | − | − | − | 0.11 | 0.12 | 0.06 |
| 3 | 5.1 | − | − | − | − | − | − | − | − | − | 2.68 | 0.26 | 0.06 |
| 4 | 5.1 | − | − | − | − | − | − | − | − | − | 1.08 | 0.59 | 0.06 |
| 5 | 6.2 | − | − | − | − | − | − | − | − | − | 1.25 | 0.43 | 0.08 |
| 6 | 6.7 | − | − | − | − | − | − | − | − | − | 16.12 | 4.47 | 0.06 |
| 7 | 6.3 | − | − | − | − | − | − | − | − | − | 16.41 | ND | ND |
| 8 | 6.9 | − | − | − | − | − | − | − | − | − | 107.02 | 24.46 | 0.03 |
| 9 | 6.6 | − | − | − | − | − | − | − | − | − | 19.95 | ND | ND |
| 10 | 7.0 | − | − | − | − | − | − | − | − | − | 235.33 | ND | ND |
| 11 | 6.7 | − | − | − | − | − | − | − | − | − | 52.21 | 22.80 | 0.11 |
| 12 | 6.8 | − | − | − | − | − | − | − | − | − | 1079.21 | 200.0 | 0.29 |
| 13 | 7.0 | − | − | − | − | − | − | − | − | − | 144.03 | 19.80 | 5.47 |
| 14 | 6.5 | − | − | − | − | − | − | − | − | − | 17.30 | ND | ND |
| 15 | 7.5 | − | − | − | − | − | − | − | − | − | 177.63 | 55.13 | 0.67 |
| 16 | 6.2 | − | − | − | − | − | − | − | − | − | 23.12 | ND | ND |
| 17 | 6.7 | − | − | − | − | − | − | − | − | − | 35.77 | ND | ND |
| 18 | 7.0 | − | − | − | − | − | − | − | − | − | 154.18 | ND | ND |
| 19 | 7.0 | − | − | − | − | − | − | − | − | − | 55.92 | 19.79 | 0.17 |
| 20 | 5.6 | − | − | − | − | − | − | − | + | − | 1.69 | ND | ND |
| 21 | 5.7 | − | − | − | − | − | − | − | + | − | 2.40 | ND | ND |
| 22 | 7.0 | − | − | − | − | − | − | − | + | − | 49.15 | ND | ND |
| 23 | 3.7 | − | − | − | − | − | − | − | + | − | 8.41 | 0.26 | 4.23 |
| 24 | 5.8 | − | − | − | − | − | − | − | + | − | 18.65 | ND | ND |
| 25 | 7.0 | − | − | − | + | − | − | − | + | − | 146.73 | ND | ND |
| 26 | 3.7 | − | − | − | + | − | − | − | + | − | 85.05 | ND | ND |
| 27 | 6.7 | + | − | − | − | − | − | − | + | − | 109.74 | ND | ND |
| 28 | 4.3 | + | − | − | + | − | − | − | + | − | 4.72 | 0.34 | 6.90 |
| 29 | 4.3 | + | − | − | + | − | − | − | + | − | 9.44 | ND | ND |
| 30 | 5.4 | + | − | − | + | − | − | − | + | − | 90.64 | ND | ND |
| 31 | 6.1 | + | − | − | + | − | − | − | + | − | 191.64 | ND | ND |
| 32 | 4.1 | + | − | − | + | − | + | − | + | − | 20.39 | ND | ND |
| 33 | 4.9 | + | − | − | + | − | − | − | + | + | 29.49 | ND | ND |
| 34 | 5.0 | + | − | + | + | − | − | − | + | − | 92.84 | ND | ND |
| 35 | 5.8 | + | + | − | + | − | − | − | + | − | 215.68 | ND | ND |
| 36 | 6.7 | + | − | − | + | − | + | − | + | − | 39.93 | ND | ND |
| 37 | 3.6 | + | + | − | + | − | + | − | + | − | 4.68 | 0.15 | 11.09 |
| 38 | 4.4 | + | + | − | + | − | − | − | + | + | 23.24 | ND | ND |
| 39 | 5.1 | + | + | − | + | − | + | − | + | − | 90.64 | ND | ND |
| 40 | 5.8 | + | − | − | + | − | + | − | + | + | 8.30 | ND | ND |
| 41 | 6.4 | + | − | + | + | − | + | − | + | − | 11.11 | 1.68 | 36.10 |
| 42 | 4.0 | + | + | + | + | − | − | + | + | − | 19.33 | 0.14 | 66.90 |
| 43 | 5.5 | + | + | − | + | + | − | − | + | + | 26.08 | 0.74 | 34.48 |
| 44 | 5.5 | + | + | + | + | − | + | − | + | − | 36.23 | ND | ND |
| 45 | 6.0 | + | + | + | + | − | + | − | + | − | NA | ND | ND |
| 46 | 4.6 | + | + | + | + | + | + | + | + | − | 38.68 | ND | ND |
| 47 | 4.7 | + | + | + | + | + | + | + | + | − | 100.39 | ND | ND |
| 48 | 4.8 | + | + | + | + | − | + | + | + | + | 8.84 | ND | ND |
| 49 | 4.8 | + | + | + | + | + | − | + | + | + | 910.66 | ND | ND |
| 50 | 6.1 | + | + | + | + | + | + | + | + | + | 75.71 | ND | ND |
pVL: plasma HIV RNA viral load; NA: not available; ND: not done.
Architect HIV Ag/Ab Combo Abbott C/O = 1.
BioPlex 2200HIV Ag-Ab Bio-Rad C/O = 1.
Finally, we tested the sensitivity of the 4G RSTs to detect the p24Ag of distinct variants using HIV-culture supernatants (n = 12), representative of HIV-1/M subtypes A/CRF02, B, C, D, H, CRF01_AE, CRF02_AG, and Unique Recombinant Form (n = 1 each) and HIV-1/O (n = 1), HIV-1/N (n = 1), HIV-1/P (n = 1), and HIV-2 (n = 1).
2.5. Data analysis
For each session, one operator blindly prepared the blood samples, performed the tests, and read the results. Another operator performed a second blind read to maintain objectivity. A third operator was solicited for consensus if the results were discordant [35]. Readings were performed according to the time defined by the manufacturer, which generally called for an early and late reading. Specificity, sensitivity, and their corresponding exact binomial 95% confidence interval were estimated using standard methods. Data analysis was performed using Excel and Vassarstats (www.vassarstats.net; 2018).
2.6. Role of the funding source
The study was funded by Santé Publique France, COREVIH Normandie (Regional Coordination against HIV infection), and Rouen University Hospital. None of the funders had any role in the study design; collection, analysis, or interpretation of data; writing of the manuscript; or the decision to submit the paper for publication.
3. Results
3.1. Validation of the reconstitution protocol
There was no difference between the results obtained with the fresh and reconstituted samples from the 20 paired-whole blood samples. No negative sample from the panel was falsely reactive, despite reconstitution, and no positive sample was negative or showed a weaker intensity of the lines after reconstitution.
3.2. Specificity of the results
Results for the specificity obtained for the 200 negative samples are presented in Table 4. For two samples, only one of the three operators observed a very weak signal (with HIVTOP) that was therefore concluded to be negative. The overall specificity ranged from 98.5 to 100%. EXACTO PRO TEST HIV, HIVTOP, and VIKIA HIV 1/2 had the lowest specificities (98·5%, 99·0%, and 99·5%, respectively), as well as Alere HIV Combo (99·5%), due to reactivity on the antigen line for one sample. Complementary analyses - plasma viral load (Abbott) and p24Ag (Vidas, bioMerieux) - were all negative, excluding a true positive p24Ag result for the latter sample (data not shown). All false reactivities were verified and the samples were still reactive upon two retests.
Table 4.
Specificity results with the negative samples (N = 200).
| HST/HT | EXACTO PRO TEST HIV | EZ-TRUST HIV 1 & 2 rapid screen Test | Genie fast HIV1/2 | HIVTOP |
INSTI | STAT-VIEW HIV 1/2 | VIKIA HIV 1/2 | BioTechMed HIV1/2 Rapid-4 a |
HIV combo |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T1 | T2 | Ag | Ac | |||||||
| Nbr of positive samples | 3 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 |
| Specificity | 98,50% | 100% | 100% | 99% | 100% | 100% | 100% | 99,50% | 100% | 100% | 99,50% | 100% |
| 95% CI (%) | 95·3–99·6 | 97·6–100 | 97·6–100 | 96·0–99·8 | 97·6–100 | 97·6–100 | 97·6–100 | 96·8–99·9 | 85·9–100 | 85·9–100 | 96·8–99·9 | 97·6–100 |
Specificity was evaluated on 200 HIV-negative whole blood samples with seven 3G RSTs/STs (left part of the table) and two 4G RSTs/STs (right part of the table). The table reports the false-positive results. For the HIV Combo (Alere) test, the detection of p24Ag is performed with a specific line, distinct from the antibody line. No antigen-specific line was present in the BioTechMed HIV1/2 Rapid-4 test. HIVTOP and BioTechMed HIV1/2 Rapid-4 are designed to discriminate HIV-1 from HIV-2 infections; results for both HIV-1 and HIV-2 are reported.
N = 30 samples.
3.3. Antibody detection performance
Results for the clinical sensitivity of the tests are presented according to the category of samples (Table 5, Table 6). The sensitivity was excellent (100%) for all tests with the 225 samples from patients infected with HIV-1/M, irrespective of subtype or therapeutic status (Table 5). Concerning therapeutic status, there was a weak positive signal in 8/850 tests (0·94%) for ART-naïve patients and 17/1010 tests (1·68%) for ART-experienced patients (p = 0·17). The discrimination between HIV-1 and HIV-2 infection was poor with HIVTOP, as 52% of the HIV-1 samples were falsely reactive for HIV-2; it was more difficult to draw a conclusion for BioTechMed, as only 60 samples could be tested.
Table 5.
Sensitivity results with the positive HIV-1/M samples (N = 225).
| Nbr of positive samples | EXACTO PRO TEST HIV | EZ-TRUST HIV 1 & 2 rapid screen test | Genie fast HIV1/2 | HIVTOP |
INSTI | STAT-VIEW HIV 1/2 | VIKIA HIV 1/2 | BioTechMed HIV1/2 Rapid-4b |
HIV combo |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T1 | T2 | Ag | Ac | |||||||
| N = 100$ | 100 | 100 | 100 | 100 | 54 | 100 | 100 | 100 | 50 | 0 | 6 c | 100 |
| N = 125£ | 125 | 125 | 125 | 125 | 62 | 125 | 125 | 125 | 10 | 0 | 0 | 125 |
| Sensitivity | 100% | 100% | 100% | 100% | NA | 100% | 100% | 100% | 100% | NA | NA | 100% |
| 95% CI (%) | 97·9–100 | 97·9–100 | 97·9–100 | 97·9–100 | NA | 97·9–100 | 97·9–100 | 97·9–100 | 92·5–100 | NA | NA | 97·9–100 |
NA: not applicable.
Sensitivity was evaluated on 225 HIV-positive whole blood samples with seven 3G RSTs/STs (left part of the table) and two 4G RSTs/STs (right part of the table). For the HIV Combo (Alere) test, the detection of p24Ag is performed with a specific line, distinct from the antibody line. No antigen-specific line was present in the BioTechMed HIV1/2 Rapid-4 test. HIVTOP and BioTechMed HIV1/2 Rapid-4 are designed to discriminate HIV-1 from HIV-2 infections; results for both HIV-1 and HIV-2 are reported.
Corresponding to ART naive patients.
Corresponding to ART experienced patients.
N = 60.
Only 2/6 positive at 20 min.
Table 6.
Sensitivity results with the positive HIV-1/O and HIV-2 samples.
| Nbr of positive samples | EXACTO PRO TEST HIV | EZ-TRUST HIV 1 & 2 Rapid Screen Test | Genie fast HIV1/2 | HIVTOP |
INSTI | STAT-VIEW HIV 1/2 | VIKIA HIV 1/2 | BioTechMed HIV1/2 Rapid-4 |
HIV combo |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T1 | T2 | Ag | Ac | |||||||
| HIV-1/O | ||||||||||||
| N = 10 | 8 (7) | 6 | 6 (4) | 8* [7] | 0 | 6 | 9 | 10** | 2 (1) | 0 | 0 | 10 |
| Sensitivity | 80% | 60% | 60% | 80% | NA | 60% | 90% | 100% | 20% | NA | NA | 100% |
| 95% CI (%) | 44·2–96·5% | 27·4–86·3% | 27·4–86·3% | 44·2–96·5% | NA | 27·4–86·3% | 54·1–99·5% | 65·5–100% | 3·5–55·8% | NA | NA | 65·5–100% |
| HIV-2 | ||||||||||||
| N = 13 | 13 | 13 | 13(12) | 4(3) | 13 | 13 | 13 | 13 | 1 | 7 | 0 | 13 |
| Sensitivity | 100% | 100% | 100% | NA | 100% | 100% | 100% | 100% | NA | 54% | NA | 100% |
| 95% CI (%) | 71·7–100% | 71·7–100% | 71·7–100% | NA | 71·7–100% | 71·7–100% | 71·7–100% | 71·7–100% | NA | 26·1–79·6% | NA | 71·7–100% |
| HIV-1 + 2 | ||||||||||||
| N = 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 0 | 2 |
| Sensitivity | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC |
| 95% CI (%) | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC | NC |
(): early reading time; *6/8 weakly positive; **one sample was retested twice and considered to be a weak positive result; NA: not applicable; NC: not calculable.
Sensitivity towards divergent viruses was evaluated on 10 HIV-1/O, 13 HIV-2, and two HIV-1 + 2 coinfected positive whole blood samples with seven 3G RSTs/STs (left part of the table) and two 4G RSTs/STs (right part of the table). For the HIV Combo (Alere) test, the detection of p24Ag is performed with a specific line, distinct from the antibody line. No antigen-specific line was present in the BioTechMed HIV1/2 Rapid-4 test. HIVTOP and BioTechMed HIV1/2 Rapid-4 are designed to discriminate HIV-1 from HIV-2 infections; results for both HIV-1 and HIV-2 are reported.
The sensitivity for samples from patients infected with the major variant HIV-1/O (n = 10) was variable and generally poor; only VIKIA HIV 1/2 and Alere HIV Combo detected 100%, with the others detecting the presence of HIV in 20 to 90% of the samples, the worst being the BioTechMed test (Table 6). Moreover, we only observed positivity at the late recommended reading time, which was very weak, for 5/90 assays (corresponding to five patients). The sensitivity was excellent (100%) for the patients infected with HIV-2 or co-infected with HIV-1 and HIV-2 (n = 15), except for the BioTechMed test, which detected only 54% of the mono-infected HIV-2 patients and one co-infected patient. However, we observed numerous cross-reactivities for the HIV-1 line with HIVTOP (4/13) and BioTechMed (1/13), leading to false HIV-1 + HIV-2 results.
3.4. Performance for samples from primary HIV infection and specific p24Ag detection
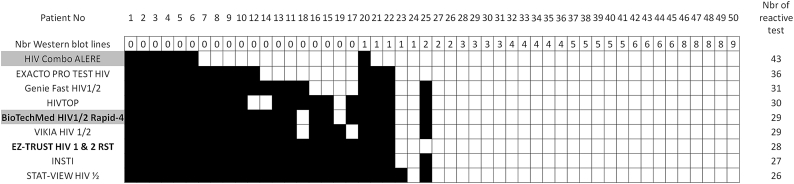
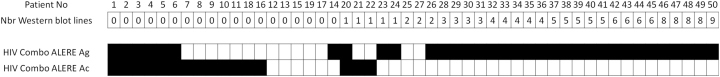
Results for samples collected from patients during PHI were classified according to the WB profiles (Fig. 2, Fig. 3). The first six samples for which no test was reactive corresponded to patients at a very early stage of infection, as confirmed by the absence of or weak reactivity of the 4G EIAs used at screening (Table 3). The number of reactive samples varied from 26 to 43 among the tests (Fig. 2); the 4G test Alere HIV Combo was the test that detected the earliest phase, detecting seven more samples [samples n° [7], [8], [9], [10],12,21,22] than the earliest detecting 3G test, EXACTO PRO. The second 4G test, BioTechMed, detected only 29 samples, similar to the results of the other 3G tests. Of note, five samples [samples n° [20], [21], [22], [23],25] presenting at least one reactive line by WB (corresponding to three subtype B, one CRF02_AG, and one subtype F), were non-reactive with at least one test. Analysis of the specific data obtained with Alere HIV Combo, which allows independent detection of Ag and Ab reactivities, showed overall detection of 43 (86%) of 50 samples, with nine only reactive on the Ag line (Fig. 3).
Fig. 2.
Performance of the nine RSTs/STs on the panel of PHI samples, classified according to the WB profiles.
The performance of seven 3G RSTs/STs and two 4G RSTs/STs (underlined in grey) was evaluated on a panel of 50 reconstituted whole blood samples from 50 patients during the PHI stage. The number of western blot lines are reported. The tests bought on websites are shown in bold. For the HIV Combo (Alere) test, the detection of p24Ag is performed with a specific line, distinct from the antibody line; we report the combined (Ag + Ab) result. A positive result was assigned to the sample if either the HIV-1 or HIV-2 line was positive for the HIVTOP and BioTechMed HIV1/2 Rapid-4 tests. The positive (in white) or negative result (in black) is represented for each sample.
Fig. 3.
Difference in the detection of antigens and antibodies with the HIV Combo (ALERE) test on the panel of PHI samples, classified according to the WB profiles.The positive (in white) or negative result (in black) is represented for each sample. An HIV-positive result is given if at least either Ag or Ab detection is reactive.
The results of specific p24Ag detection by the two 4G tests in reconstituted whole blood containing viral supernatants showed that all but three strains (HIV-1/M URF, HIV-1/P and HIV-2) were detected with Alere test and none with BioTechMed test (Table 7). Among the three samples non-reactive with Alere, the HIV-1/M URF and HIV-1/P strains gave a positive signal at higher concentrations (894 and 5992 pg/mL, respectively), but the HIV-2 sample remained non-reactive even at the highest concentration. The results with BioTechMed remained negative, even at the highest concentration.
Table 7.
Detection of p24 antigen by the 4th generation RSTs/STs.
| Group | Subtype | Final concentration (pg/mL)a | BioTechMed HIV1/2 Rapid-4 |
HIV combo |
||
|---|---|---|---|---|---|---|
| T1 | T2 | Ag | Ab | |||
| HIV-1/M | B | 145 | − | − | + | − |
| HIV-1/M | CRF01_AE | 147 | − | − | + | − |
| HIV-1/M | CRF02_AG | 145.5 | − | − | + | − |
| HIV-1/M | A | 144 | − | − | + | − |
| HIV-1/M | C | 134.5 | − | − | + | − |
| HIV-1/M | D | 137.5 | − | − | + | − |
| HIV-1/M | H | 130 | − | − | + | − |
| HIV-1/M | cpx | 149 | − | − | − | − |
| HIV-1/O | H | 153.6 | − | − | + | − |
| HIV-1/N | NA | 100 | − | − | + | − |
| HIV-1/P | NA | 149.8 | − | − | − | − |
| HIV-2 | B | ND | − | − | − | − |
The performance of the two 4G tests to detect p24Ag only was evaluated on a panel of 12 reconstituted whole blood samples loaded with HIV culture supernatant. The p24 concentrations mimic the physiopathological in vivo concentration during PHI.
After reconstitution of whole blood.
3.5. Retesting rate
Finally, among the >4000 assays performed, only eight retests (0·19%) were required, because the result could not be read/interpreted (two with STAT-VIEW, three with HIVTOP, two with VIKIA, and one with Genie Fast).
4. Discussion
HIV RSTs and STs are useful, easy-to-use devices to scale-up HIV testing in a large number of contexts. The WHO has urged more HIV testing outside clinical settings and now promotes HIV rapid tests to more accurately target those who need it the most [36]. This further favours the growing use of RSTs/STs, thus requiring that they be of high quality and perform well.
Currently, evaluations of such tests are mainly performed on plasma or sera, owing to their use in laboratory settings where whole blood is decanted. This is also because it is almost impossible to obtain fresh whole blood from hundreds of patients corresponding to various clinical situations (such as PHI or those who are ART-naïve or ART-experienced) or infection with strains representative of pandemic and divergent viruses, such as HIV-2 or HIV-1/O. Thus, prospective studies with finger-stick blood were mainly conducted to evaluate feasibility, acceptability, and effectiveness [[36], [37], [38]]. Here, we used a reconstituted whole blood panel, which is a good substitute for fresh whole blood, allowing extensive evaluation of the performance of rapid tests under close to real-life usage. This should be useful for testing other RSTs for the screening of HCV, HBV, or syphilis infection, even if fresh whole blood (especially finger-stick) is suitable for studies.
This method allowed us to test an extensive panel of 512 samples representative of most situations encountered, such as a negative status in at-risk populations, primary HIV infection, and infection with pandemic strains (HIV-1 group M), as well as infections with divergent HIV-2 and HIV-1/O strains, which challenge HIV diagnosis in regions where they circulate. Specificity ranged from 98.5 to 100%, comparable to that of most of EIAs [39], and sensitivity for the detection of various HIV-1/M infections was excellent, irrespective of the subtype. Although our panel contained various pandemic strains, further evaluations are needed in specific regional contexts, especially in sub-Saharan countries, in which numerous HIV variants circulate. Previous reports have highlighted failures to detect HIV-positive patients effectively treated for years or those treated at the primary infection stage; this may be explained by a low titre of circulating antibodies, especially for tests based on oral fluids [15,40]. Here, there was no difference in the results between the 100 naïve patients and the 125 ART-experienced HIV-1/M infected patients (of whom 44% had undetectable viral loads), highlighting no major impact of ART. We cannot exclude that failures in antibody detection may occur on rare occasions in patients with undetectable plasma viral load for years; however, this may not represent an important public health issue, because such individuals do not correspond to those targeted by these tests.
We report a frequent failure to diagnose patients infected with divergent variants. For HIV-2, the BioTechMed web test detected only 54% of mono-infections. Another challenge for rapid tests is to discriminate between HIV-1 and HIV-2 infections, as confirmed here using HIVTOP and BioTechMed. Although this point has been previously reported [24,41], such tests are still largely used in diagnosis and typing algorithms [42,43]. Previous data and those here highlight the need of manufacturers to improve these tests and that type-specific molecular tools are the only way to accurately diagnose a mono- or dual infection. The problem was more acute for HIV-1/O, as among the nine tests, only VIKIA HIV 1/2 and HIV Combo were able to detect all positive samples, with BioTechMed detecting only two. These samples, mainly from treated patients, revealed clear differences between the tests. These results may reflect a validation step of the tests in which a limited number of HIV-1/O samples, not representative of the high intra-group genetic diversity, is probably used [44]. Even if the global prevalence of HIV-1 group O and HIV-2 infections is relatively low, such poor clinical sensitivity is still problematic, as up to two million patients are infected by these major variants and live in countries where rapid tests are already extensively used. It is therefore essential to have tools that can distinguish between true HIV-2 mono-infection and dual HIV-1 + 2 infections, as well as correctly detect group O infections, owing to their specific management [45,46].
Results from samples collected during primary infection showed that 3G tests can be highly sensitive, despite the absence of p24Ag detection. The most sensitive RST, EXACTO PRO, could detect 36/50 samples versus the 26/50 detected with the less sensitive STAT-VIEW. The results were better with the new 4G Alere test, HIV Combo, which could detect 43/50 samples, allowing detection of seven additional samples (of which six were Ag reactive only) relative to the earliest-detecting 3G test. Specific analysis of p24Ag detection showed that this RST appears to be well adapted to various strains (except HIV-2, in accordance with the manufacturer's instructions), although we did not evaluate analytical sensitivity. Our data are in accordance with recent results showing that this test can shorten serological windows [[47], [48], [49]]. Nonetheless, this needs to be confirmed on a larger scale and compared to other 4G RSTs, such as the WHO prequalified SD Bioline test, recently evaluated using a serum matrix [49,50]. Here, we performed a comparison with the BioTechMed non-CE-marked/FDA-approved/WHO prequalified test, sold as a 4G SF by a web-based vendor; finally, this test detected fewer PHI samples than three 3G tests (29 versus 30, 31, and 36) and none of the p24Ag samples reconstituted from viral supernatants.
This last observation highlights the concerns of STs available via commercial websites. As no scientific evaluation was provided by these websites or found in literature, we wished to determine the performance of such tests, which are not CE-marked, FDA-approved, or WHO prequalified, on our extensive panel. Our results showed varying performance depending on the test; indeed, if the EZ Trust HIV 1&2 Rapid Screen Test did not provide globally bad results, except for the detection of HIV-1/O, the second one, BioTechMed HIV1/2 Rapid-4, clearly performed poorly for the detection of HIV-1/O and HIV-2. Another point is the sales practices. First, although the sale of HIV STs on websites in France is forbidden, except on allowed pharmacy websites, these vendors were clearly able to override it, since there was no restriction nor information concerning a restriction when we ordered from France. Another concern is the discrepancy between ordered and received devices; indeed, data obtained with BioTechMed HIV1/2 Rapid-4 on the PHI panel, associated with the absence of a specific Ag reading line, clearly demonstrate that it was a 3G test sold as a 4G test. Thus, this did not correspond to the order, and is of concern for people who believe that they are buying a test that can detect the early stages of the HIV infection. The concern was different for the other test; during the evaluation, we received two tests with different packaging, the first entitled EZ Trust HIV 1&2 Rapid Screen Test and the second iCare HIV 1&2 Rapid Screen Test, without a reason for this switch provided by the vendor. These two tests are, in fact, the same device provided by twin companies [CS Innovation Ltd. and JAL Innovation Pte Ltd., respectively]. Altogether, these data show that buying STs through certain web-based vendors is clearly risky, consistent with a recent investigation in Switzerland, highlighting that buying such “susceptible” devices on such websites is not recommended [51].
In conclusion, our data, based on an extensive panel of samples representative of varying clinical status and HIV genetic diversity, show that care must still be taken for the detection of major HIV variants and that adding specific p24Ag detection allows identification during early stages of HIV infection. This study also shows, for the first time, that buying via web-based vendors can be risky, due to the varying performance of non-qualified tests and questionable sales practices; CE-marked, FDA-approved assays, or at least those prequalified by the WHO, provided by manufacturers or distributors that respect regional regulations, should therefore be favoured. Our results are of particular importance in the context of the increasing use of rapid tests in an “outside laboratory” setting.
Author's contributions
TM, VL, MLC, and JCP conceived and designed the study. VD, CD, ME, and MLC contributed to the sampling. TM and VL performed the experiments. TM, VL, and JCP analysed the data. TM, VL, and JCP wrote the first draft of the manuscript. VD, CD, EAG, ME, FS, and MLC revised the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
We declare no competing interests.
HIV Combo (Alere) and EXACTO PRO TEST HIV (Biosynex) were provided by the manufacturers. Genie Fast HIV1/2 (Bio-Rad), HIVTOP (Biosynex), INSTI (bioLytical), STAT-VIEW HIV 1/2 (Chembio diagnostic systems), and VIKIA HIV 1/2 (bioMérieux) were bought directly from French distributors or manufacturers. EZ-TRUST HIV 1 & 2 Rapid Screen Test (CS Innovation Ltd) and BioTechMed HIV1/2 Rapid-4 (BioTechMed) were bought directly from websites.
Acknowledgments
We want to thank our funders Santé Publique France, COREVIH Normandie (Regional Coordination against HIV infection), and Rouen University Hospital. We also want to thank Dr. Nadia Mahjoub, Saint Louis Hospital, and all the technicians from Rouen and Saint Louis laboratories.
References
- 1.Jamieson D.J., Cohen M.H., Maupin R., Nesheim S., Danner S.P., Lampe M.A. Rapid human immunodeficiency virus-1 testing on labor and delivery in 17 US hospitals: the MIRIAD experience. Am J Obstet Gynecol. 2007;197(3 Suppl):S72–S82. doi: 10.1016/j.ajog.2007.03.067. [DOI] [PubMed] [Google Scholar]
- 2.Landrum M.L., Wilson C.H., Perri L.P., Hannibal S.L., O'Connell R.J. Usefulness of a rapid human immunodeficiency virus-1 antibody test for the management of occupational exposure to blood and body fluid. Infect Control Hosp Epidemiol. 2005;26(9):768–774. doi: 10.1086/502615. [DOI] [PubMed] [Google Scholar]
- 3.Puro V., Francisci D., Sighinolfi L., Civljak R., Belfiori B., Deparis P. Benefits of a rapid HIV test for evaluation of the source patient after occupational exposure of healthcare workers. J Hosp Infect. 2004;57(2):179–182. doi: 10.1016/j.jhin.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Lubelchek R., Kroc K., Hota B., Sharief R., Muppudi U., Pulvirenti J. The role of rapid vs conventional human immunodeficiency virus testing for inpatients: effects on quality of care. Arch Intern Med. 2005;165(17):1956–1960. doi: 10.1001/archinte.165.17.1956. [DOI] [PubMed] [Google Scholar]
- 5.Njouom R., Ngono L., Mekinda-Gometi D.D., Nde C.K., Sadeuh-Mba S.A., Vernet M.A. Evaluation of the performances of twelve rapid diagnostic tests for diagnosis of HIV infection in Yaounde, Cameroon. J Virol Methods. 2017;243:158–163. doi: 10.1016/j.jviromet.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Suthar A.B., Ford N., Bachanas P.J., Wong V.J., Rajan J.S., Saltzman A.K. Towards universal voluntary HIV testing and counselling: a systematic review and meta-analysis of community-based approaches. PLoS Med. 2013;10(8) doi: 10.1371/journal.pmed.1001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO. Guidelines on HIV self-testing and partner notification: Supplement to consolidated guidelines on HIV testing services. [Guidelines] December 2016 [cited 27 September 2018]; Available from: http://www.who.int/hiv/pub/vct/hiv-self-testing-guidelines/en/ [PubMed]
- 8.UNAIDS 90-90-90 An ambitious treatment target to help end the AIDS epidemic. 2014 October. http://www.unaids.org/en/resources/909090 2014 [cited 27 september 2018; Available from:
- 9.EC Commission decision of 27 November 2009 amending Decision 2002/364/EC on common technical specifications for in vitro diagnostic medical devices, 2009/886/EC. 2009. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=uriserv:OJ.L_.2009.039.01.0034.01.ENG [cited 27 September 2018; Available from:
- 10.FDA BPAC Reclassification of HIV Point of Care and Laboratory-based serological and NAT diagnostic devices from Class III (PMA) to Class II 510(k) July 19 2018. https://www.fda.gov/downloads/AdvisoryCommittees/CommitteesMeetingMaterials/BloodVaccinesandOtherBiologics/BloodProductsAdvisoryCommittee/UCM611839.pdf [cited; 12 September 2018:[Available from:
- 11.WHO Selecting and purchasing HIV, HBsAg and HCV in vitro diagnostics. 2018. http://www.who.int/diagnostics_laboratory/procurement/purchase/en/ [cited 12 september 2018]; Available from:
- 12.WHO. EXPERT COMMITTEE ON BIOLOGICAL STANDARDIZATION Human immunodeficiency virus (HIV) rapid diagnostic tests for professional use and/or self-testing Technical Guidance Series for WHO prequalification of in vitro diagnostic medical devices. 2017 [cited 27 September 2018; Available from: http://www.who.int/biologicals/expert_committee/ECBS_2017_BS2305_Human_immunodeficiency_virus_(HIV)_rapid_diagnostic_tests.pdf.
- 13.Aghokeng A.F., Mpoudi-Ngole E., Dimodi H., Atem-Tambe A., Tongo M., Butel C. Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PLoS One. 2009;4(11) doi: 10.1371/journal.pone.0007702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gautheret-Dejean A., Mesmin-Poho S., Birguel J., Lemee V., Huraux J.M., Plantier J.C. Unequal detection of HIV type 1 group O infection by simple rapid tests. Clin Infect Dis. 2008;46(12):1936–1937. doi: 10.1086/588561. [DOI] [PubMed] [Google Scholar]
- 15.Pavie J., Rachline A., Loze B., Niedbalski L., Delaugerre C., Laforgerie E. Sensitivity of five rapid HIV tests on oral fluid or finger-stick whole blood: A real-time comparison in a healthcare setting. PLoS One. 2010;5(7) doi: 10.1371/journal.pone.0011581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayaphi S.H., Martin D.J., Quinn T.C., Laeyendecker O., Olorunju S.A., Tintinger G.R. Detection of acute and early HIV-1 infections in an HIV hyper-endemic area with limited resources. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guanira J.V., Leigler T., Kallas E., Schechter M., Sharma U., Glidden D. Streamlining HIV testing for HIV preexposure prophylaxis. J Clin Microbiol. 2015;53(1):179–183. doi: 10.1128/JCM.01540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ly T.D., Laperche S., Courouce A.M. Early detection of human immunodeficiency virus infection using third- and fourth-generation screening assays. Eur J Clin Microbiol Infect Dis. 2001;20(2):104–110. doi: 10.1007/s100960000430. [DOI] [PubMed] [Google Scholar]
- 19.Conway D.P., Holt M., McNulty A., Couldwell D.L., Smith D.E., Davies S.C. Multi-centre evaluation of the Determine HIV Combo assay when used for point of care testing in a high risk clinic-based population. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0094062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duong Y.T., Mavengere Y., Patel H., Moore C., Manjengwa J., Sibandze D. Poor performance of the determine HIV-1/2 Ag/Ab combo fourth-generation rapid test for detection of acute infections in a National Household Survey in Swaziland. J Clin Microbiol. 2014;52(10):3743–3748. doi: 10.1128/JCM.01989-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laperche S., Leballais L., Ly T.D., Plantier J.C. Failures in the detection of HIV p24 antigen with the determine HIV-1/2 Ag/Ab combo rapid test. J Infect Dis. 2012;206(12):1946–1947. doi: 10.1093/infdis/jis616. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg N.E., Kamanga G., Phiri S., Nsona D., Pettifor A., Rutstein S.E. Detection of acute HIV infection: A field evaluation of the determine(R) HIV-1/2 Ag/Ab combo test. J Infect Dis. 2012;205(4):528–534. doi: 10.1093/infdis/jir789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray R.H., Makumbi F., Serwadda D., Lutalo T., Nalugoda F., Opendi P. Limitations of rapid HIV-1 tests during screening for trials in Uganda: Diagnostic test accuracy study. BMJ. 2007;335(7612):188. doi: 10.1136/bmj.39210.582801.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gautheret-Dejean A., Bocobza J., Brunet S., Damond F., Plantier J.C., Barin F. Performance of rapid tests for discrimination between HIV-1 and/or HIV-2 infections. J Med Virol. 2015;87(12):2061–2066. doi: 10.1002/jmv.24282. [DOI] [PubMed] [Google Scholar]
- 25.WHO WHO list of prequalified in vitro diagnostic products. 2018 12 September. http://www.who.int/diagnostics_laboratory/evaluations/PQ_list/en/ [cited 27 September 2018]; Available from:
- 26.WHO WHO list of prequalified in vitro diagnostic products. 2018. http://www.who.int/diagnostics_laboratory/evaluations/180912_prequalified_product_list.pdf cited 27 September 2018; Available from:
- 27.WHO HIV assays: laboratory performance and other operational characteristics: rapid diagnostic tests (combined detection of HIV-1/2 antibodies and discriminatory detection of HIV-1 and HIV-2 antibodies): report 18. 2018. http://www.who.int/diagnostics_laboratory/publications/evaluations/en/ [cited 27 September 2018; Available from:
- 28.The_Global_Fund List of HIV Diagnostic test kits and equipments classified according to the Global Fund Quality Assurance Policy. 2018. https://www.theglobalfund.org/en/search/?q=List+of+HIV+Diagnostic+test+kits [cited 2 October 2018]; Available from:
- 29.The_Global_Fund List of HIV Diagnostic test kits and equipments classified according to the Global Fund Quality Assurance Policy. 2018. https://www.theglobalfund.org/media/5878/psm_productshiv-who_list_en.pdf [cited 2 October 2018]; Available from:
- 30.FDA Complete list of donor screening assays for infectious agents and HIV diagnostic assays. 2018. https://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm080466.htm cited 27 September 2018; Available from:
- 31.CDC HIV/AIDS HIV testing laboratory tests. 2018. https://www.cdc.gov/hiv/testing/laboratorytests.html [cited 27 September 2018]; Available from:
- 32.CDC Rapid HIV tests suitable for use in clinical settings. 2016. https://www.cdc.gov/hiv/pdf/testing/rapid-hiv-tests-clinical-moderate-complexity.pdf 08/10/2016 [cited 27 September 2018; Available from:
- 33.CDC Rapid HIV tests suitable for use in non-clinical settings. 2018. https://www.cdc.gov/hiv/pdf/testing/rapid-hiv-tests-non-clinical.pdf 06/11/2018 [cited 27 September 2018; Available from:
- 34.Troude P., Chaix M.L., Tran L., Deveau C., Seng R., Delfraissy J.F. No evidence of a change in HIV-1 virulence since 1996 in France. AIDS. 2009;23(10):1261–1267. doi: 10.1097/QAD.0b013e32832b51ef. [DOI] [PubMed] [Google Scholar]
- 35.WHO WHOROfAC, APHL AoPHL Guidelines for appropriate evaluations of HIV testing technologies in Africa. 2003. http://apps.who.int/medicinedocs/en/m/abstract/Js15211e/ cited 27 September 2018; Available from:
- 36.Baggaley R. 8th International AIDS Society Conference on HIV Pathogenesis, Treatment, and Prevention; 2015 July 19–22 2015; Vancouver. 2015. Launch of the WHO consolidated HIV testing services guidelines: Overview of the guidelines. [Google Scholar]
- 37.Prazuck T., Karon S., Gubavu C., Andre J., Legall J.M., Bouvet E. a finger-stick whole-blood HIV self-test as an HIV screening tool adapted to the general public. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0146755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornton A.C., Delpech V., Kall M.M., Nardone A. HIV testing in community settings in resource-rich countries: a systematic review of the evidence. HIV Med. 2012;13(7):416–426. doi: 10.1111/j.1468-1293.2012.00992.x. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell E.O., Stewart G., Bajzik O., Ferret M., Bentsen C., Shriver M.K. Performance comparison of the 4th generation Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA on the EVOLIS automated system versus Abbott ARCHITECT HIV Ag/Ab Combo, Ortho Anti-HIV 1+2 EIA on Vitros ECi and Siemens HIV-1/O/2 enhanced on Advia Centaur. J Clin Virol. 2013;58(Suppl. 1):e79–e84. doi: 10.1016/j.jcv.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Stefic K., Novelli S., Mahjoub N., Seng R., Molina J.M., Cheneau C. IAS conference on HIV Science; 2017 23–26 July 2017; Paris. 2017. Non-reactive HIV-1 self-tests after sustained viral suppression following early antiretroviral therapy. [Google Scholar]
- 41.Tchounga B.K., Inwoley A.A., Coffie P.A., Minta D.D., Messou E.E., Bado G.G. Re-testing and misclassification of HIV-2 and HIV-1&2 dually reactive patients among the HIV-2 cohort of the West African Database to evaluate AIDS collaboration. J Int AIDS Soc. 2014;17:19064. doi: 10.7448/IAS.17.1.19064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.CDC 2018 Quick reference guide: Recommended laboratory HIV testing algorithm for serum or plasma specimens. 2018. https://stacks.cdc.gov/view/cdc/50872 [cited 9 september 2018]; Available from:
- 43.Ekouevi D.K., Balestre E., Coffie P.A., Minta D., Messou E., Sawadogo A. Characteristics of HIV-2 and HIV-1/HIV-2 dually seropositive adults in West Africa presenting for care and antiretroviral therapy: the IeDEA-West Africa HIV-2 Cohort Study. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0066135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plantier J.C., Djemai M., Lemée V., Reggiani A., Leoz M., Burl L., Vessière A., Rousset D., Poveda J.D., Henguell C., Gautheret-Dejean A., Barin F. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J Clin Microbiol. 2009;47(9):2906–2911. doi: 10.1128/JCM.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mourez T., Simon F., Plantier J.C. Non-m variants of human immunodeficiency virus type 1. Clin Microbiol Rev. 2013;26(3):448–461. doi: 10.1128/CMR.00012-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visseaux B., Damond F., Matheron S., Descamps D., Charpentier C. Hiv-2 molecular epidemiology. Infect Genet Evol. 2016;46:233–240. doi: 10.1016/j.meegid.2016.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Delaugerre C., Antoni G., Mahjoub N., Pialoux G., Cua E., Pasquet A. Assessment of HIV screening tests for use in preexposure prophylaxis programs. J Infect Dis. 2017;216(3):382–386. doi: 10.1093/infdis/jix297. [DOI] [PubMed] [Google Scholar]
- 48.Livant E., Heaps A., Kelly C., Maharaj R., Samsunder N., Nhlangulela L. The fourth generation AlereTM HIV Combo rapid test improves detection of acute infection in MTN-003 (VOICE) samples. J Clin Virol. 2017;94:15–21. doi: 10.1016/j.jcv.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fransen K., de Baetselier I., Rammutla E., Ahmed K., Owino F., Agingu W. Performance of serological and molecular tests within acute HIV infection. J Clin Virol. 2017;93:81–84. doi: 10.1016/j.jcv.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Stafylis C., Klausner J.D. Evaluation of two 4th generation point-of-care assays for the detection of Human Immunodeficiency Virus infection. PLoS One. 2017;12(8) doi: 10.1371/journal.pone.0183944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim E. 2013. Test VIH à domicile trop facile d'accès et dangereux. [Google Scholar]
- 52.Gueudin M., Leoz M., Lemee V., De Oliveira F., Vessiere A., Kfutwah A. A new real-time quantitative PCR for diagnosis and monitoring of HIV-1 group O infection. J Clin Microbiol. 2012;50(3):831–836. doi: 10.1128/JCM.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Damond F., Gueudin M., Pueyo S., Farfara I., Robertson D.L., Descamps D. Plasma RNA viral load in human immunodeficiency virus type 2 subtype A and subtype B infections. J Clin Microbiol. 2002;40(10):3654–3659. doi: 10.1128/JCM.40.10.3654-3659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plantier J.C., Dachraoui R., Lemee V., Gueudin M., Borsa-Lebas F., Caron F. HIV-1 resistance genotyping on dried serum spots. AIDS. 2005;19(4):391–397. doi: 10.1097/01.aids.0000161768.98534.e7. [DOI] [PubMed] [Google Scholar]
- 55.Vessiere A., Rousset D., Kfutwah A., Leoz M., Depatureaux A., Simon F. Diagnosis and monitoring of HIV-1 group O-infected patients in Cameroun. J Acquir Immune Defic Syndr. 2010;53(1):107–110. doi: 10.1097/QAI.0b013e3181b97ec1. [DOI] [PubMed] [Google Scholar]
- 56.Charpentier C., Eholie S., Anglaret X., Bertine M., Rouzioux C., Avettand-Fenoel V. Genotypic resistance profiles of HIV-2-treated patients in West Africa. AIDS. 2014;28(8):1161–1169. doi: 10.1097/QAD.0000000000000244. [DOI] [PMC free article] [PubMed] [Google Scholar]