Abstract
Background
Glioblastoma (GBM) is the most frequent and malignant primary brain tumor in adults and despite the progress in surgical procedures and therapy options, the overall survival remains very poor. Glutamate and α-KG are fundamental elements necessary to support the growth and proliferation of GBM cells. Glutamate oxidative deamination, catalyzed by GLUD2, is the predominant pathway for the production of α-KG.
Methods
GLUD2 emerged from the RNA-seq analysis of 13 GBM patients, performed in our laboratory and a microarray analysis of 77 high-grade gliomas available on the Geo database. Thereafter, we investigated GLUD2 relevance in cancer cell behavior by GLUD2 overexpression and silencing in two different human GBM cell lines. Finally, we overexpressed GLUD2 in-vivo by using zebrafish embryos and monitored the developing central nervous system.
Findings
GLUD2 expression was found associated to the histopathological classification, prognosis and survival of GBM patients. Moreover, through in-vitro functional studies, we showed that differences in GLUD2 expression level affected cell proliferation, migration, invasion, colony formation abilities, cell cycle phases, mitochondrial function and ROS production. In support of these findings, we also demonstrated, with in-vivo studies, that GLUD2 overexpression affects glial cell proliferation without affecting neuronal development in zebrafish embryos.
Interpretation
We concluded that GLUD2 overexpression inhibited GBM cell growth suggesting a novel potential drug target for control of GBM progression. The possibility to enhance GLUD2 activity in GBM could result in a blocked/reduced proliferation of GBM cells without affecting the survival of the surrounding neurons.
Keywords: Glioblastoma, GLUD2, Glutamate, Mitochondrial metabolism, Tumor progression, Zebrafish
Research in context.
Evidence before this study
The involvement of GLUD2 in glioma metabolism and growth has already been suggested, but only in the presence of IDH1 mutations (Cancer Res, 2018 and Proc Natl Acad Sci U S A, 2014). It has been described that GLUD1 and GLUD2 are overexpressed in IDH1 mutant tumors. In this context, the expression of GLUD2 makes cells resistant to the inhibitory effects of growth of the IDH1R132H mutation, by providing a-KG to feed the citric acid cycle and support the synthesis of lipids. In other studies, instead, it was investigated the inhibition of glutamate dehydrogenase activity in glioma, without distinguishing the two existing isoforms GLUD1 and GLUD2 (Cancer Res, 2009). To the best of our knowledge, this is the first work where GLUD2 is investigated as a key player in GBM progression. The research in PubMed database, according to the terms “GLUD2” or “GDH2” and “glioblastoma”, does not provide any other results except for the work, previously mentioned, that was published on Proc Natl Acad Sci U S A in 2014.
Added value of this study
In this study we found an association of GLUD2 mRNA expression levels to the prognosis and survival of patients with GBM. Thereafter, through in-vitro functional studies using human GBM cell lines and in-vivo studies in zebrafish model, we investigated the importance of GLUD2 regulation in cell behavior, metabolism and development. GLUD2 expression was related to the histopathological classification, prognosis and survival of patients with GBM. Moreover, differences in GLUD2 expression level affected cell proliferation, migration, invasion, colony formation abilities, cell cycle phases, mitochondrial function and ROS production. In support of these findings, we also demonstrated that GLUD2 overexpression decreases glial cell proliferation without affecting neurons development in zebrafish embryos.
Implications of all the available evidence
The possibility to enhance GLUD2 activity in GBM could result in a blocked/reduced proliferation of GBM cells without affecting the survival of the surrounding neurons. To the best of our knowledge, this is the first work where GLUD2 is considered the key player in GBM progression. These observations may provide a new target for therapeutic interventions in GBM to reduce tumor progression and aggressiveness.
Alt-text: Unlabelled Box
1. Introduction
Glioblastoma (GBM, World Health Organization grade IV) is the most common malignant primary brain tumor, characterized by an extremely aggressive clinical phenotype due to inter- and intra-patient genomic and histopathological diversity, diffuse infiltrative growth, intense vascularization and innate treatment resistance [[1], [2], [3], [4], [5], [6]]. Despite significant advances in both neurosurgical techniques and medical therapy, GBM treatment remains difficult, as present-days therapies are not curative and the latest improvements in radio-chemotherapy have been reported by Stupp et al. [7] >10 years ago [2,8].
GBM prognosis is extremely poor with a median overall survival between 12 and 15 months and a 5-year survival rate of <5%, making this survival rate one of the worst observed in modern-day oncology [1,2,5]. Almost all patients, under current standard therapy, including maximal safe surgical resection followed by radiotherapy and temozolomide chemotherapy, will develop recurrent diseases with progressive neurological deficits and inevitable death. Second surgery is an applicable therapy option in limited cases; however, an increased risk of neurological morbidity often limits this secondary resection [9].
Unlike in other tumors, where in the past 50 years, the prognosis and life expectancy have strongly increased due to intensive research efforts into tumor cell biology, in GBM, such investigations, have raised more doubts than they have solved [3]. Therefore, there is still an urgent need for novel and effective therapeutic strategies for treating these tumors and ultimately improve GBM patients' chances of survival.
One of the emerging hallmarks of cancer is the deregulation of cellular energetics and the most significant reprogramming occurs in the metabolic machinery in GBM [1,10,11]. Thus, cancer cell metabolism is now a field of intensive investigation to discover new valuable therapeutic targets and biomarkers [10]. In particular, it is well known that glutamine metabolism plays key roles in cellular growth and invasion, supporting tumor progression and poor patient outcomes [12]. The initial step in glutamine degradation involves its conversion into glutamate catalyzed by glutaminase and the conversion of glutamate into α-ketoglutarate (α-KG), by glutamate dehydrogenase. Especially in tumor hypoxia condition, glutamate and α-KG are fundamental elements in glutaminolysis and reductive carboxylation, needed to sustain cancer cell growth and proliferation. Moreover, glutamate release can affect nearby cells, since high glutamate levels induce astrocyte swelling and apoptosis, favoring tumor expansion [13]. Furthermore, an efficient excitatory amino acids clearance from astrocytes is essential in order to maintain low extracellular glutamate concentrations and guarantee an adequate regulation of synaptic transmission and prevents glutamate neurotoxicity [14].
Glutamate dehydrogenase 2 (GLUD2) catalyzes the reversible interconversion of glutamate to α-KG and ammonia while reducing NAD(P) + to NAD(P)H as cofactors. This enzyme plays a key role in cellular homeostasis, being at the interface between amino acid and carbohydrate metabolism. GLUD2 is linked to important cellular processes including Krebs cycle, ammonia control and energy generation [15,16].
In this study, we investigated the correlation of GLUD2 expression to patients' prognosis and survival. We investigated, through in-vitro and in-vivo functional studies, how an alteration of GLUD2 expression could affect the behavior of human glioblastoma cells. Furthermore, we evaluated the effect of GLUD2 expression on the behavior of neurons and glial cells in the zebrafish developing brain.
2. Materials and methods
2.1. Transcriptome analysis
NGS and microarray analysis data were obtained from our previous paper [17] and the Gene Expression Omnibus (GEO) GSE4271 dataset [18] respectively, together with molecular and clinical information. In particular, NGS analysis was performed on FFPE tumor tissues from 13 primary human GBM subjects selected from the archives of the Anatomy Pathology Institute of the University of Pisa, Italy. Subjects were chosen by the same pathologist, they have same histology, similar condition and treatment and were grouped depending on time of recurrence free survival (RFS) after first surgery: 6 Short (S) <6 months, 3 Medium (M) between 16 and 23 months and 4 Long (L) over 25 months. NGS data were analyzed and visualized with Partek Flow software (Partek, Inc., St. Louis, MO, USA). Microarray GSE4271 profile was downloaded from GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4271). GSE4271 contains the mRNA profile of 77 primary grade III and IV astrocytomas characterized by molecular class (proneural, proliferative and mesenchymal) [18] and overall survival.
2.2. Immunohistochemistry
2.2.1. Immunohistochemistry performed on FFPE GBM tissues
Sections of 5 μm thickness were deparaffinized in xylene and rehydrated in graded alcohols. Immunohistochemistry was performed using the Mouse specific HRP/DAB (ABC) Detection IHC Kit (Abcam, Cambridge, UK) according to manufacturer's protocol. The antigen unmasking was achieved with MS-unmasker solution (DIAPATH, Martinengo, BG, Italy) in microwave. GLUD2 primary antibody (cat. number SAB1400112, Sigma Aldrich, St Louis, MO) was used at 1:150 dilution for 1 h at room temperature. Slides were developed with diaminobenzidine chromogen (DAB) (DAKO, Glostrup, DK) and counterstained with hematoxylin. Negative controls included the omission of the primary antibody. Slides were analyzed using the inverted microscope CARL ZEISS Axio Observer Z1FLMot, and images were taken with CARL ZEISS AXIOCAM Icc1 camera (Zeiss, Oberkochen, Germany).
2.2.2. Immunohistochemistry performed on embryos
Immunohistochemistry was performed following standard procedures. Embryos were blocked in 20% lamb serum and incubated with mouse GLUD2 antibody (1:250 dilution, cat. number SAB1400112, Sigma Aldrich), mouse HuC/D antibody (1:500, cat. number A-21271, Invitrogen), or rabbit phospho-histone H3 antibody (1:500, cat. number 06–570, Millipore). Horseradish peroxidase-conjugated secondary antibodies goat anti-mouse (1:500, cat. number G21040, Invitrogen), or goat anti-rabbit (1:500, cat. number G21234, Invitrogen) were used to detect primary antibodies, and DAB (Roche) was used as a substrate for peroxidase. Images of embryos were acquired using a stereomicroscope (SMZ1500, Nikon) equipped with digital camera with LAS Leica Imaging software (Leica, Wetzlar, Germany). Images were processed using Adobe Photoshop software (Adobe System Incorporated, San Josè, CA, USA). The same magnification was always maintained within each control and GLUD2-injected image pair.
2.3. Cell lines and transfection
T98G and U118 GBM cell lines were obtained from American Type Culture Collection (ATCC, Rockville, MD). To ensure the quality and integrity of human cell lines, cells from the initial thawed vials were used for up to a maximum of 10 passages in all the experiments, as recommended by the supplier. T98G and U118 were grown as monolayers in Dulbecco's Modified Eagle Medium (DMEM) low glucose and high glucose respectively, supplemented with 10% FBS and 1% Penicillin-Streptomycin. Cells were tested for the presence of mycoplasma (EZ-PCR Mycoplasma Test Kit; Biological Industries, Beth Haemek, Israel) with negative results. Amplification of GLUD2 gene was performed by PCR with the 5′ end primer (5′-TAActcgagGACCCTTCCTTCCTAGTCGC-3′) containing XhoI-site and with the 3′ end primer (5′-CGggatccTCAGCCATGATCCATCTATGTGA-3′) containing BamHI-site. PCR product was subcloned into pIRES2-AcGFP vector (Clontech Laboratories, Mountain View, CA, USA). GLUD2 sequence was confirmed by Sanger sequencing. Plasmid transfection was performed with Lipofectamine 3000 Reagent, following manufacturer's instructions. GLUD2 was silenced using GLUD2 siRNA Silencer Select (Thermo Fisher Scientific) and the Silencer Select Negative Control No. 1 siRNA was used as non-targeting negative control siRNA (Thermo Fisher Scientific). SiRNA transfection was performed with Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific) following manufacturer's instructions. Cells were incubated for 48 h after GLUD2 overexpression/silencing prior to characterization and functional experiments.
2.4. Quantitative real-time PCR
2.4.1. Cell lines mRNA expression analysis
Total cellular RNA was extracted from GBM cells using the Maxwell 16 LEV simplyRNA kit (Madison, WI) according to the manufacturer's instructions, and quantitated using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA). Total RNA was reverse transcribed into cDNA using the RT-NanoScript kit (PrimerDesign, Southampton, UK). Real Time PCR was performed following the manufacturer's instruction of the SsoAdvanced SYBR Green Supermix kit (Bio-Rad, Hercules, CA) on CFX96 instrument (Bio-Rad). TBP expression values were used for normalization. The following primers were used: GLUD2, 5′-CACTCTGCCTTGGCATACAC-3′ and 5′-CTCAGGTCCAATCCCAGGTT-3′, TBP, 5′-AGTTCTGGGATTGTACCGCA-3′, 5′-TTATATTCGGCGTTTCGGGC-3′, Cyclin E, 5′-TTCTTGAGCAACACCCTCTTCTGCAGCC-3′ and 5′-TCGCCATATACCGGTCAAAGAAATCTTGTGCC-3′, Cyclin D1, 5′-ACAAACAGATCATCCGCAAACAC-3′, 5′-TGTTGGGGCTCCTCAGGTTC-3′. Gene expression analysis was performed using CFX Manager Software (Bio-Rad). All expression experiments were performed in triplicate.
2.4.2. Embryos mRNA expression analysis
Total RNA was extracted from zebrafish embryos (30 per experimental group) using Nucleospin® RNA (Macherey-Nagel) according to manufacturer's instructions. cDNA was synthesized from total RNA using iScript™ cDNA Synthesis Kit (Bio-Rad) and quantitative real-time PCR was performed following the manufacturer's protocol of GoTaq® qPCR master mix (Promega). Ct values were obtained for each gene and normalized to β-actin. Fold change was calculated relative to control embryos expression level using the 2-ΔΔCt method. The following primers were used: pcna (F: 5′-TCGGGTGAGTTTGCCCGCATC-3′; R: 5′-GCCCAGCTCTCCGCTGGCAGA-3′), cyclin D1 (F: 5′-CTGCGCAAACACGCCCAGAC-3′; R: 5′-TACCGCTGCAGCAACACTGCC-3′), gfap (F: 5′-GCAGACAGGTGGATGGACTCA-3′; R: 5′-GGCCAAGTTGTCTCTCTCGATC-3′), β-actin (F: 5′-CGAGCAGGAGATGGGAACC-3′; R: 5′-CAACGGAAACGCTCATTGCC-3′).
2.5. Western blot
For each samples 40 μg of proteins were loaded on the 10% Mini-PROTEAN TGX Gel (Bio-Rad). Proteins were transferred from gels to membrane with Trans-Blot Turbo transfer system (Bio-Rad). GLUD2 and β-tubulin primary antibodies (HPA043640, Sigma Aldrich) were used at 1:250 and 1:200 dilution respectively. Secondary antibodies Goat Anti-Mouse IgG H&L (HRP) (ab6789, Abcam, Cambridge, UK) and Goat Anti-Rabbit IgG H&L (HRP) (ab6721, Abcam) were used at a dilution of 1:2000. Protein detection was performed using the Bio-Rad Clarity western ECL substrate (Bio-Rad). The ChemiDoc MP imager and Image Lab software (Bio-Rad) were used to validate western blotting data via total protein normalization in conjunction with housekeeping proteins (β-tubulin).
2.6. Immunofluorescence
Cells were grown on cell culture chamber slides, and fixed in 1.5% paraformaldehyde for 15 min. Cells were permeabilized with 0.1% Triton X-100 for 15 min and blocked with 2% BSA for 45 min. GLUD2 primary antibody was diluted 1:250 and incubated for 60 min at RT. Phycoerythrin conjugated secondary antibody (P9287, Sigma Aldrich) was diluted 1:20 and incubated for 30 min. Cells were counterstained with Hoechst (Thermo Fisher Scientific) and visualized using the inverted microscope CARL ZEISS Axio Observer 3 Z1FLMot (Zeiss).
2.7. Glutamate dehydrogenase activity
Intracellular glutamate dehydrogenase activity was measured by using the GDH Activity Assay Kit (Sigma-Aldrich) according to the manufacturer's protocol. The activity of glutamate dehydrogenase was assayed photometrically (absorbance at 450 nm) following glutamate consumption by GDH generating NADH, which reacts with a probe generating a colorimetric product proportional to the GDH activity present. After the addition of the Master Reaction Mix the plate was incubated for 3 min at 37 °C degrees and the first measurement was taken. Then, every 10 min the absorbance at 450 nm was measured up to 60 min. The results were visualized as generated NADH nanomoles per minutes, normalized on the total protein concentration of each sample (Bradford Reagent assay, Sigma-Aldrich). The experiment was performed in triplicate.
2.8. Cell viability assay
Cell viability was determined using the WST1 assay (Clontech Laboratories, Mountain View, CA, USA). A total of 5000 cells per well were seeded in a 96-well plate format. At the time of seeding (T0) and after 24 h (T1), 48 h (T2) and 72 h (T3), the WST1 reagent was added and incubated for a further 60 min before reading the plate. Each assay was conducted in triplicate. The quantity of formazan dye is directly related to the number of metabolically active cells, and was quantified by measuring the absorbance at 450 nm in a multiwell plate reader (Tecan, Mannedorf, Switzerland). OD values at 24 h (T1), 48 h (T2) and 72 h (T3) were normalized to T0.
2.9. Clonogenic survival assay
Cells were seeded at 500 cells/well in 6-well plates and incubated for 2 weeks. Cells were fixed with 70% ethanol and stained with 0.01% crystal violet for 30 min. The mean ± SD number of colonies with >50 μm in diameter was counted under a microscope in five non-overlapping fields in three independent experiments.
2.10. Wound healing assay
Cells were plated in Culture-Insert 2 Well in μ-Dish 35 mm (IBIDI, Martinsried, Germany) until cells were confluent or nearly confluent (>90%). After the removal of the insert, cell migration in the wound area was observed and digitally photographed. Wound healing was measured on the images by using the free, open-source software ImageJ [36] and the % of closure was calculated at each time (T0-T3, 0–72 h) as the area to be healed divided the area of the original wound * 100. Relative invasion ability of pIRES-GLUD2 and pIRES Vector transfected cells was measured by counting GFP signal (transfected cells) into the wound. Experiments were performed in triplicate.
2.11. Transwell assay
Cell invasion was assessed using 24-well inserts (Sarstedt, Nuembrecht, Germany) with 5-μm pores according to manufacturer's instructions. In brief, 1 × 105 cells were seeded into the upper chamber with 1% FBS medium and were allowed to invade the lower reservoir, containing 10% FBS, at 37 °C for 24 h. Non-invading cells in the upper surface of the filters were removed using a cotton swab. The remaining cells were fixed in 70% ethanol and stained with 0.01% crystal violet for 30 min. Cells that passed through the membrane were counted in five visual fields as migrated cells. The experiment was performed in triplicate.
2.12. Cell-cycle analysis
Approximately 1 × 106 cells were fixed in 70% ethanol for 30 min at 4 °C. The cells were then labeled with 25 μg/ml of propidium iodide (Sigma Aldrich), 1 mg/ml RNase A (Sigma Aldrich), 0,1% v/v of Triton X-100 (Sigma Aldrich) and incubated 30 min in the dark at 4 °C. The percentage of cells in different phases of the cell cycle was measured by flow cytometry using CyFlow1 Cube 8 Sorter Flow Cytometer (Sysmex Partec, Gorlitz, Germany). Data analysis was performed using FCS express 4 software (BD Bioscience San Jose, CA). The experiment was performed in triplicate.
2.13. Mitochondrial respiration and glycolysis analysis
Cell mitochondrial function was evaluated by using the Seahorse XFp Cell Mito Stress Test Kit on the Seahorse XFp Analyzer (Agilent Technologies, Santa Clara, CA). Cells were seeded at 20,000 cells per well into XFp well cell culture plates and incubated overnight at 37 °C in a 5% CO2 humidified atmosphere in Seahorse XF Base Medium (Agilent Technologies) with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose. Cartridge compounds were loaded in order to have as final concentration 1 μM Oligomycin, 1 μM FCCP and 0.5 μM Rotenone/ antimycin A. Data were analyzed and visualized using Wave 2.3.0 software (Agilent Technologies) and values of OCR and ECAR were normalized to the total protein levels (Bradford Reagent assay, Sigma-Aldrich) in each well. The experiment was performed in three replicates.
2.14. Oxidative stress
To assess oxidative stress/reactive oxygen species (ROS), cells were transfected with GLUD2 overexpression vector and silencing siRNA and relative controls for two days in a 96 well plate. CellROX Green reagent was added at final concentration of 5 uM to the cells and incubate for 30 min at 37 °C. Medium was removed and cells were washed three times with PBS. The quantity of oxidative stress was quantified by measuring the fluorescence at an excitation/emission wavelength of 485/535 nm in a multiwell plate reader (Tecan). The experiment was performed in three replicates and fluorescence in each well was measured in four multiple reads.
2.15. Zebrafish husbandry
Danio rerio (AB strain) was raised and bred at a temperature of 28 °C with a photoperiod of 14 h light/10 h dark. Animal care was performed in strict accordance with protocols approved by the Italian Ministry of Public Health and the University of Pisa Ethical Committee (authorization 99/2012-A, 19.04.2012), in compliance with EU legislation (Directive 2010/63/EU). Zebrafish embryos were obtained by natural spawning, staged according to the hours post fertilization [37] and raised at 28 °C in 1× E3 medium (5.0 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 0.1% methylene blue) in Petri dishes.
2.16. Zebrafish embryos experimental plan
To determine the zebrafish embryos number to analyze in each experimental replicate we followed the indication reported in Busquet et al., 2014 [38]. The authors reported zebrafish embryo experiments results replicated in at least 3 independent laboratories suggesting that the use of 20 embryos per treatment should be maintained to ensure the accuracy of the experimental test. In our experiments, we processed 20–30 embryos per experimental group. Only properly developing embryos between the 4- and 128-cell stages with an intact chorion were used in our experiments as suggested in Busquet et al., 2014 [38].
2.17. Constructs generation and RNA microinjection
The open reading frame of GLUD2 was subcloned into PCS2+ vector. Capped mRNA encoding the full coding sequence of GLUD2 was synthesized using mMESSAGE mMACHINE™ SP6 transcription kit (Thermo Fisher Scientific), following manufacturer's instructions. DNAse treatment to remove template DNA was followed by phenol/chloroform extraction and isopropanol precipitation, according to the kit procedures.
A capped mRNA encoding the full coding sequence of an enhanced green fluorescent protein (eGFP) reporter was also produced as described above.
To perform gene gain of function experiments, GLUD2 capped mRNA was injected into the yolk sac of one-cell stage embryos (~200 pg per embryo). 200 pg of eGFP capped RNA were co-injected to verify successful injections using a fluorescence stereomicroscope.
For the selection of the correctly injected embryos we performed a double check procedure with two different operators in order to minimize any possible human bias in the embryo selection.
In all experiments, GLUD2-injected embryos were compared with embryos injected with the only eGFP capped mRNA at the same developmental stage and cultured in the same conditions (same medium, temperature, same incubator etc.) as a control.
Microinjections were performed using a FemtoJet microinjector (Eppendorf).
2.18. Whole-Mount in situ hybridization
Zebrafish embryos were manually dechorionated, fixed in 4% paraformaldehyde at the desired developmental stages and stored in methanol at −20 °C. In situ hybridization was then performed as previously described [39]. Digoxigenin-UTP labeled antisense RNA probes to detect pcna, gfap, slc1a3a, slc1a2b and cyclin D1 transcripts were generated via in-vitro transcription according to the manufacturer's instructions (Roche). The enzymes used for plasmids linearization and polymerases for probes transcription are indicated: pcna (NotI, T7), gfap (SalI, SP6), slc1a3a (BamHI, SP6), slc1a2b (EcoRV, T7), cyclin D1 (SpeI, T7). The color reaction was carried out using the BM Purple substrate (Roche). After color development, embryos were post-fixed and bleached under light to remove the pigment.
The constructs were kindly provided by Prof. Wolfgang Driever (cyclin D1), Prof. Gerald B. Downes (slc1a2b), Prof. Yi-Chuan Cheng (gfap, slc1a3a). Pcna construct was generated as described in Baumgart et al., 201439.
2.19. Statistical analysis
All results are presented as mean ± SD of at least three independent experiments. For qRT-PCR experiments, graphs are representative of three independent experiments with three technical replicates each. Data were statistically analyzed applying student's t-test and visualized using GraphPad Prism software (GraphPad Software, La Jolla, CA). Differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
3. Results
3.1. GLUD2 expression is associated with GBM prognosis and histopathological classification
3.1.1. RNA-Seq
We previously used whole-transcriptome RNA sequencing [17] to analyze 13 newly diagnosed cases of primary FFPE (formalin-fixed and paraffin-embedded) GBM, specifically selected for different length of recurrence-free survival time (RFS). We defined three groups: the short-term group (S) with RFS < 6 months (n = 6), the medium term group (M) with 16 < RFS < 23 months (n = 3) and the long-term group (L) with RFS > 25 months (n = 4). Here, we performed a functional enrichment analysis of the identified transcripts, finding statistically significant differences in glutamate metabolism genes (Supplementary Table S1). In particular, GLUD2 mRNA expression means in the two extreme groups, S and L RFS GBM patients, were respectively 10.49 and 143.05 with a fold change of 13.64, and a p-value of 0.05 (Fig. 1a). Moreover, immunohistochemical analysis revealed that the protein expression of GLUD2 detected in long RFS GBM tissues was higher than that in short GBM (Fig. 1b).
Fig. 1.
GLUD2 mRNA expression is related to recurrence free survival, prognosis, and histopathological classification in high-grade glioma patients. (a) GLUD2 mRNA expression in GBM patients with short RFS (<6 months) and long RFS (>25 months) from NGS analysis. (b) Immunohistochemical stain of GLUD2 protein in GBM patients with short and long RFS. (c) GLUD2 mRNA expression in GBM patients with long survival (>104 weeks) and short survival (<103 weeks) from GSE4271 dataset. (d) GLUD2 mRNA expression in WHO grade III and WHO grade IV gliomas from GSE4271 dataset. (e) GLUD2 mRNA expression association with molecular sub classification in high-grade gliomas and relative prognosis from GSE4271 dataset. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
3.1.2. Geo dataset analysis
From the GEO dataset we retrieved seventy-seven samples from newly diagnosed cases of high-grade gliomas, which were profiled via microarray analysis, to identify changes in GLUD2 mRNA expression that relates to both survival and disease progression (Gene Expression Omnibus dataset accession number: GSE4271, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4271).
We divided patients' cohort into two groups, depending on their clinical outcome: overall survival (OS) <2 years (OS < 103 weeks) and OS > 2 years (OS > 104 weeks). The longer survival group is defined by higher expression levels of GLUD2 (Fig. 1c; p = 0.0016). Then we associated GLUD2 expression levels to the World Health Organization (WHO) classification system of glioma grading. GLUD2 resulted more expressed in grade III glioma tumors, compared to the most aggressive form of glioma, GBM grade IV (Fig. 1d; p < 0.0001). Finally, based on the molecular sub-classification of high-grade astrocytoma, reported on the literature [18], we evaluated GLUD2 expression levels into the proneural, proliferative and mesenchymal classes. Among these three groups, the proneural group, associated with the most favorable outcome, was characterized by higher levels of GLUD2 (Fig. 1e; p < 0.0001).
3.2. Two human GBM cell lines with different GLUD2 expression levels reveal distinct proliferation rate and colony formation capacity
3.2.1. GLUD2 molecular status in human Gbm cell lines
We selected two GBM human cell lines, T98G and U118, to investigate GLUD2 expression level and enzyme activity. U118 cells had higher levels of both GLUD2 mRNA (Supplementary Fig. S1a; p = 0.0009) and protein (Supplementary Fig. S1b–c) than T98G cells. Moreover, glutamate dehydrogenase activity, in the direction of oxidative deamination of glutamate, showed higher enzymatic activity in U118 than T98G cells (Supplementary Fig. S1d; p < 0.0001). The whole gene GLUD2 (including promoter region) of both cell lines were sequenced and no mutations were identified. Moreover, mutations in codon 132 of IDH1 and codons 140 or 172 of IDH2 have not been identified.
3.2.2. U118 and T98G cell behavior
Once established statistically significant differences in GLUD2 expression level and enzyme activity, we performed in-vitro cell proliferation and tumorigenic assays to investigate U118 and T98G cell behavior dissimilarities.
Cell proliferation rate in T98G at four different time points, T0, T1 (24 h), T2 (48 h) and T3 (72 h), as shown in Supplementary Fig. S1e, was higher than in U118, (p = 0.0128 T1, 0.0003 T2 and 0.0015 T3).
Colony-forming assay revealed a higher colonies number in T98G cells compared to U118 cells (Supplementary Fig. S1f; p < 0.0001). Therefore, GLUD2 lower expression was associated with enhancing cell proliferation signal and higher colony formation ability.
3.3. GLUD2 overexpression decreases proliferation, migration, invasion and colony formation abilities of GBM Cells
3.3.1. GLUD2 overexpression in human GBM cell lines
We evaluated the effects of GLUD2 overexpression on T98G cells, selected due to lower GLUD2 expression. T98G cells were transfected with GLUD2-IRES-GFP plasmid system (pIRES-GLUD2). As a control, we transfected the same cells with the empty vector (pIRES Vector). After transfection, GLUD2 mRNA expression (Supplementary Fig. S2a; p < 0.0001), GLUD2 protein expression (Supplementary Fig. S2b–c) and GLUD2 enzymatic activity (Supplementary Fig. S2d; p < 0.0001) were increased in pIRES-GLUD2 cells compared to cells with empty vector.
3.3.2. Cell functional in-vitro studies on GLUD2 overexpressing T98G cells
We used a WST-1 cell proliferation assay to determine the effect of GLUD2 overexpression in T98G cells at four different time points, T0, T1 (24 h), T2 (48 h) and T3 (72 h). We found that overexpression of GLUD2 inhibits cell proliferation (Fig. 2a; p = 0.0018 T1, 0.0006 T2, 0.0146 T3).
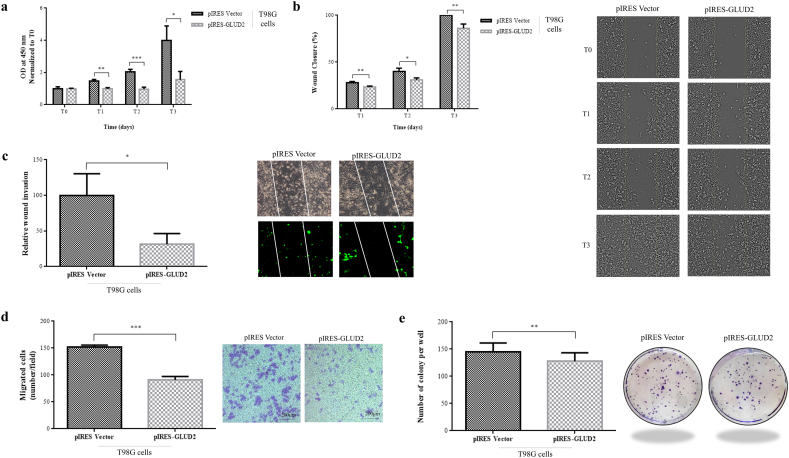
Fig. 2.
GLUD2 overexpression decreases GBM cells proliferation, migration, invasion and colony formation abilities. (a) Cell viability of T98G cells after GLUD2 overexpression (pIRES-GLUD2) and control (pIRES Vector) at the time of seeding (T0) and after 24 h (T1), 48 h (T2) and 72 h (T3). (b) Wound healing assay of T98G cells after GLUD2 overexpression (pIRES-GLUD2) and control (pIRES Vector) and (c) their relative wound invasion ability. (d) Transwell migration assay of T98G cells after GLUD2 overexpression (pIRES-GLUD2) and control (pIRES Vector). Cells that passed through the membrane were counted in five visual fields as migrated cells. (e) Colony formation assay of T98G cells after GLUD2 overexpression (pIRES-GLUD2) and control (pIRES Vector). Colonies were counted in five non-overlapping fields. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
We investigated, by wound healing assay, the effect of GLUD2 overexpression on migration in T98G cells at three different time points, T0, T1 (24 h), T2 (48 h) and T3 (72 h). The wound healing assay results showed that GLUD2 overexpression suppressed the wound healing ability compared with empty vector transfected cells (Fig. 2b; p = 0.0087 T1, 0.0213 T2, 0.0058 T3). We then analyzed wound healing assay only looking at the GFP signal, to compare migration capacity of transfected cells in relation to their different GLUD2 expression levels. We observed that pIRES-GLUD2 cells were almost unable to move into the wound whereas control cells transfected with vector alone (pIRES Vector) invaded the wound area (Fig. 2c; p = 0.0250). To determine the effect of GLUD2 overexpression on T98G cell invasion, we performed a transwell invasion assay. The number of migrating cells was distinctly lower in pIRES-GLUD2 cells than in control cells (Fig. 2d; p < 0.0001).
Colony formation assays were performed in order to evaluate GLUD2 overexpression effect on the clonogenic survival. GLUD2 overexpression reduced the T98G colony formation ability (Fig. 2e; p = 0.0045).
3.4. GLUD2 silencing increases proliferation, migration, invasion and colony formation abilities of GBM cells
3.4.1. GLUD2 silencing in human GBM cell lines
We examined the effects of GLUD2 silencing selecting U118 cells due to their higher GLUD2 expression. U118 cells were transfected with GLUD2 siRNA system (siRNA GLUD2). As control, we transfected the same cells with a non-targeting negative control siRNA (siRNA C+). As shown in Supplementary Fig. S1, after transfection with siRNA GLUD2, GLUD2 mRNA expression (Supplementary Fig. S1a; p < 0.0001), GLUD2 protein expression (Supplementary Fig. S1b and c) and GLUD2 enzymatic activity (Supplementary Fig. S1d; p < 0.0001) were decreased compared to control cells.
3.4.2. Cell functional in-vitro studies on GLUD2 silenced U118 cells
We used a WST-1 cell proliferation assay to determine the effect of GLUD2 silencing in U118 cells. We found that silencing of GLUD2 enhances cell proliferation (Fig. 3a; p < 0.0001 T1, p < 0.0001 T2, p = 0.0002 T3).
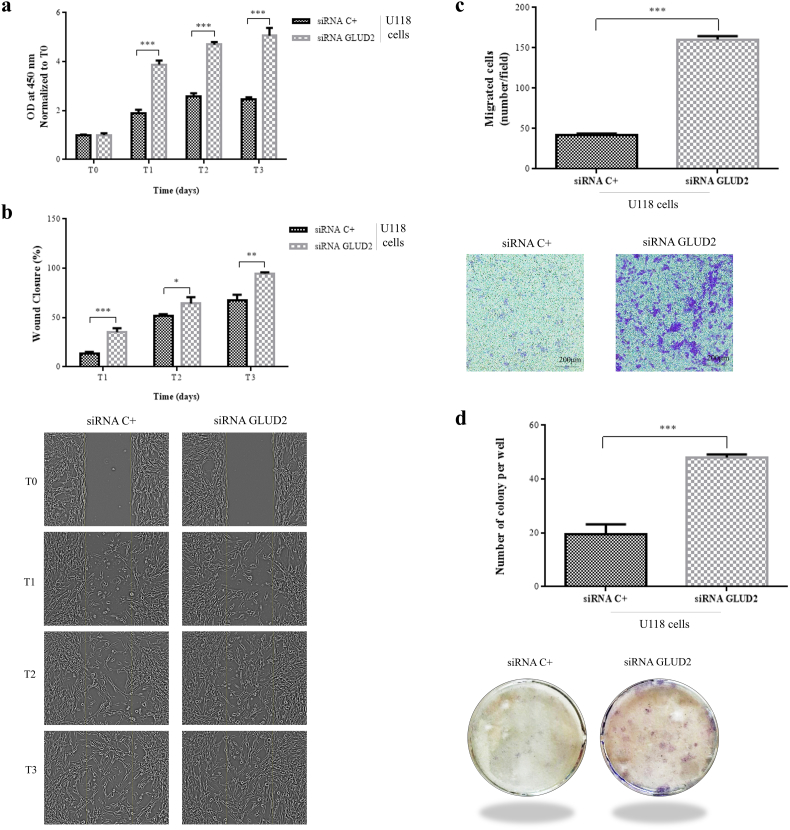
Fig. 3.
GLUD2 silencing increases GBM cells proliferation, migration, invasion and colony formation abilities. (a) Cell viability of U118 cells after GLUD2 silencing (siRNA GLUD2) and control (siRNA C+) at the time of seeding (T0) and after 24 h (T1), 48 h (T2) and 72 h (T3). (b) Wound healing assay of U118 cells after GLUD2 silencing (siRNA GLUD2) and control (siRNA C+). (c) Transwell migration assay of U118 cells after GLUD2 silencing (siRNA GLUD2) and control (siRNA C+). Cells that passed through the membrane were counted in five visual fields as migrated cells. (d) Colony formation assay of U118 cells after GLUD2 silencing (siRNA GLUD2) and control (siRNA C+). Colonies were counted in five non-overlapping fields. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
We investigated the effect of GLUD2 silencing on migration in U118 cells by wound healing at three different time points, T0, T1 (24 h), T2 (48 h) and T3 (72 h). The wound healing assay results showed that GLUD2 silencing improved the wound healing ability compared to control siRNA transfected cells (Fig. 3b; p = 0.0010 T1, 0.0272 T2, 0.0015 T3). To determine the effect of GLUD2 silencing on U118 cell invasion, we performed a transwell invasion assay. The number of migrating cells was distinctly higher in siRNA GLUD2 cells than in control cells (Fig. 3c; p < 0.0001).
Colony formation assays were performed in order to evaluate GLUD2 silencing effect on the clonogenic survival. GLUD2 silencing increased the U118 colony formation ability (Fig. 3d; p < 0.0001).
3.5. GLUD2 expression levels influence G1/S transition regulating cyclins D1 and E in human GBM cells
We studied by flow cytometry the effect of both GLUD2 overexpression and silencing on T98G and U118 cell cycle. GLUD2 overexpression in T98G cells showed a significant increase in G0/G1 phase and a statistically significant decrease in S and G2/M phases when compared to control cells (Fig. 4a; p < 0.0001 G0/G1 and p = 0.0008 S). GLUD2 silencing in U118 cells with siRNA did not lead to a statistically significant change in the cell cycle (Fig. 4a).
Fig. 4.
GLUD2 overexpression and silencing in GBM cells modulate the expression of cyclin D1 and cyclin E, influencing cell cycle phases and affect mitochondrial function and ROS production. (a) Cell cycle distribution in T98G GLUD2 overexpressed cells (pIRES-GLUD2) and U118 GLUD2 silenced cells (siRNA GLUD2) compared to relative control cells. (b) Cyclin D1 and Cyclin E mRNA expression in GLUD2 overexpressed cells (pIRES-GLUD2) and GLUD2 silenced cells (siRNA GLUD2) compared to relative control cells. (c) Oxygen consumption rate (OCR) of T98G control cells (pIRES Vector), T98G GLUD2 overexpressed cells (pIRES-GLUD2), U118 control cells (siRNA C+) and U118 GLUD2 silenced cells (siRNA GLUD2) in Seahorse XFp Cell Mito Stress Test. (d) Basal and maximal respiration, non-mitochondrial respiration, H+ (Proton) leak, ATP-linked respiration, spare respiratory capacity and coupling using modulators of cellular respiration. (e) ROS levels of T98G GLUD2 overexpressed cells (pIRES-GLUD2), U118 GLUD2 silenced cells (siRNA GLUD2) and relative controls quantified by measuring the fluorescence at an excitation/emission wavelength of 485/535 after CellROX Green probe incubation. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
We then evaluated the mRNA expression of G1 phase cell cycle regulators, cyclin D1 and cyclin E. GLUD2 overexpression in T98G cells is characterized by cyclin D1 and cyclin E downregulation (Fig. 4b; p = 0.0083 D1 and p = 0.0003 E). GLUD2 silencing in U118 cells showed an overexpression of both cyclin D1 and cyclin E (Fig. 4b; p = 0.0124 D1 and p = 0.0336 E).
3.6. Changes in GLUD2 expression levels determine alterations of mitochondrial functions in human GBM cells
To obtain insight into mitochondria functional differences between GLUD2 overexpressed and silenced cells, we analyzed mitochondrial function and metabolic phenotype by quantifying oxygen consumption rate (OCR) with Seahorse XFp extracellular flux analyzer.
We examined GLUD2 overexpression and silencing effects on basal respiration, ATP-linked respiration, H+ (Proton) leak, maximal respiration, spare respiratory capacity, and non-mitochondrial respiration, using the Seahorse XFp Cell Mito Stress Test Kit (Supplementary Fig. S3). GLUD2 overexpression in T98G cells led to a statistically significant increase in the basal levels of mitochondrial respiration (p = 0.0015) and maximal respiration (p < 0.0001) compared to empty vector control (Fig. 4c–d). Instead, GLUD2 silencing in U118 caused a statistically significant reduction of basal mitochondrial respiration (p = 0.0416), whereas no statistically significant differences were observed in the maximal respiration compared to the respective control (Fig. 4c–d). Non-mitochondrial respiration was also statistically increased in T98G cells with GLUD2 overexpression (p = 0.0004) and decreased in U118 cells with GLUD2 silencing (p = 0.0057) compared to respective controls (Fig. 4c–d). Despite proton leak and ATP production were increased in T98G cells with GLUD2 overexpression and decreased in U118 cells with GLUD2 silencing, compared to the respective controls, differences were not statistically significant (Fig. 4c–d). Spare respiratory capacity was significantly increased in GLUD2 overexpressed T98G cells (p = 0.0006) whereas no significant differences were observed in the spare respiratory capacity in GLUD2 silenced U118 cells. No statistically significant differences, in couple efficiency, were observed in both GLUD2 overexpressed T98G cells and GLUD2 silenced U118 cells compared to their respective controls (Fig. 4c–d).
To evaluate reactive oxygen species (ROS) levels, GLUD2 overexpressed cells, GLUD2 silenced cells and relative controls were stained with CellROX Green reagent, followed by fluorescence quantification of oxidative stress. We found higher levels of ROS in the GLUD2 overexpressed cells (p < 0.0001) and lower levels in the GLUD2 silenced cells (p < 0.0001) compared to controls (Fig. 4e).
3.7. GLUD2 overexpression in zebrafish embryos
The GLUD2 overexpression ability to reduce the proliferation rate of GBM cell line T98G well correlates with the GLUD2 higher expression in the long RFS group of patients. Nevertheless, the tissue culture condition could not entirely recapitulate the in-vivo condition in which multiple signaling factors and cell-cell interactions could influence cell behavior and metabolism. We therefore decided to test the function of GLUD2 in an in-vivo system to verify the GLUD2 overexpression influence on cell proliferation in the central nervous system. We chose Danio rerio (zebrafish) as experimental model as it is a well accepted tool in biomedical research, including brain cancer research [19,20], allowing us to overexpress the human GLUD2 and to evaluate its effect on neurons and glial cells behavior in the developing brain.
Embryos were injected with GLUD2 and eGFP mRNAs at one-cell stage (Fig. 5a). As a control, a group of embryos was injected with only eGFP mRNA. Effectively injected embryos were selected detecting eGFP expression by fluorescence stereomicroscope (Fig. 5b).
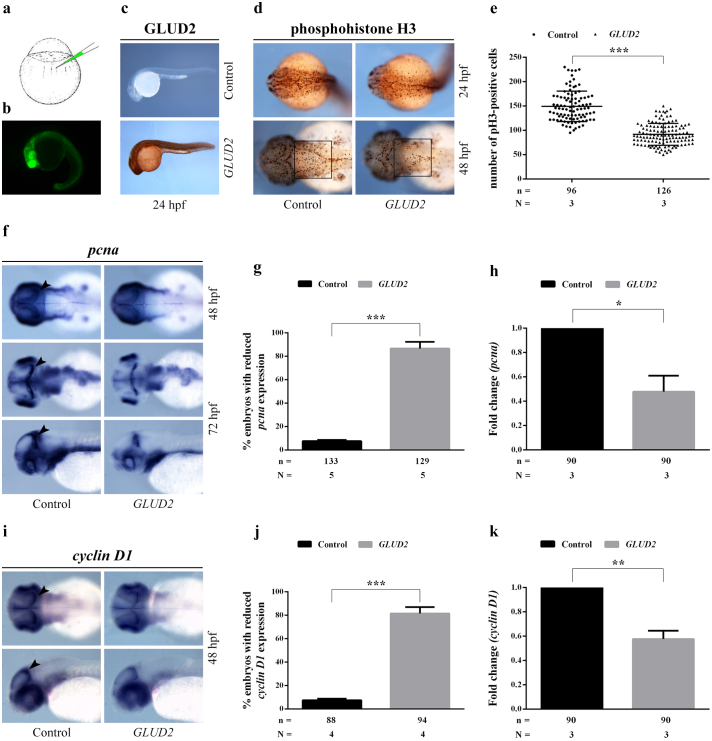
Fig. 5.
GLUD2 overexpression affects cell proliferation in zebrafish embryos. (a) Schematic mRNA microinjection procedure into the yolk sac of a one-cell stage zebrafish embryo. (b) Zebrafish embryo injected with eGFP mRNA at 24 hpf. (c) Control immunohistochemical staining of GLUD2 protein on 24 hpf zebrafish embryos injected with eGFP mRNA (control) or GLUD2 and eGFP mRNAs (GLUD2), showing GLUD2 expression in GLUD2-injected embryos. (d) Immunohistochemical staining of pH 3 (phosphohistone H3) on 24 and 48 hpf embryos injected with only eGFP or eGFP and GLUD2 mRNAs. (e) Number of pH 3-positive cells was counted in the hindbrain area (black square in d) of 48 hpf embryos; n, total number of analyzed embryos, N, number of independent experiments. (f) Whole mount in situ hybridization on both GLUD2-injected and control embryos at 48 and 72 hpf, showing a reduction of pcna expression in GLUD2-injected embryos; arrowheads indicate the proliferative region of the optic tectum. 72 hpf embryos are shown in dorsal and lateral view. (g) Percentage of GLUD2-injected and control embryos with reduced pcna expression; n, total number of analyzed embryos, N, number of independent experiments. (h) qRT-PCR analysis of pcna expression in GLUD2-injected and control embryos at 48 hpf; n, total number of analyzed embryos, N, number of independent experiments. (i) Whole mount in situ hybridization on 48 hpf embryos, showing a reduced cyclin D1 expression in GLUD2-injected embryos compared to controls; arrowheads indicate the proliferative region of the optic tectum. 48 hpf embryos are shown in dorsal and lateral view. (j) Percentage of GLUD2-injected and control embryos with reduced cyclin D1 expression; n, total number of analyzed embryos, N, number of independent experiments. (k) qRT-PCR analysis of cyclin D1 expression level in control and GLUD2-injected embryos; n, total number of analyzed embryos, N, number of independent experiments. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
We first assessed the ability of zebrafish embryos to express the human GLUD2 protein; GLUD2 expression was observed in GLUD2-injected embryos by immunohistochemistry, but not in the control group (Fig. 5c).
3.7.1. GLUD2 overexpression affects cell proliferation in zebrafish embryos
We used a phosphohistone H3 antibody to analyze cell proliferation in the central nervous system and, in particular, to visualize cells during M phase of the cell cycle in zebrafish embryos at two different developmental stages, 24 hpf (hours post fertilization) and 48 hpf (Fig. 5d). The result revealed a decreased number of proliferating cells in GLUD2-injected embryos compared to controls. This observation was confirmed by counting phosphohistone H3-positive cells in the hindbrain area of embryos at 48 hpf (Fig. 5e).
We then performed a whole mount in situ hybridization (WISH) using an antisense RNA probe to detect pcna (proliferating cell nuclear antigen) mRNA, which encodes a non-histone nuclear protein used as a marker for cells in S phase. At 48 hpf, GLUD2-injected embryos showed reduced pcna transcript expression level compared to controls, that was confirmed by quantitative real-time RT-PCR (qRT-PCR) (Fig. 5f–h). At later stages, 72 hpf, the effect became more pronounced highlighting a severely reduced pcna gene expression clearly detectable in proliferative regions of the optic tectum of GLUD2-injected embryos (Fig. 5f, g). These results suggested that GLUD2 overexpression could cause a reduction of cells in both S and M phases confirming the results obtained in T98G cells.
As in GBM cell line T98G overexpressing GLUD2 the reduced proliferation correlates with a reduction of cyclin D1 expression, we evaluated cyclin D1 mRNA level in GLUD2 overexpressing embryos both by in situ hybridization and qRT-PCR (Fig. 5i–k). Cyclin D1 WISH signal intensity was strongly decreased in the tectal proliferative region of 48 hpf GLUD2-injected embryos in comparison to controls (Fig. 5i, j) and was found to be significantly downregulated in GLUD2-injected embryos also by qRT-PCR analysis (Fig. 5k).
3.7.2. GLUD2 overexpression impairs glial cells formation without affecting neurons development
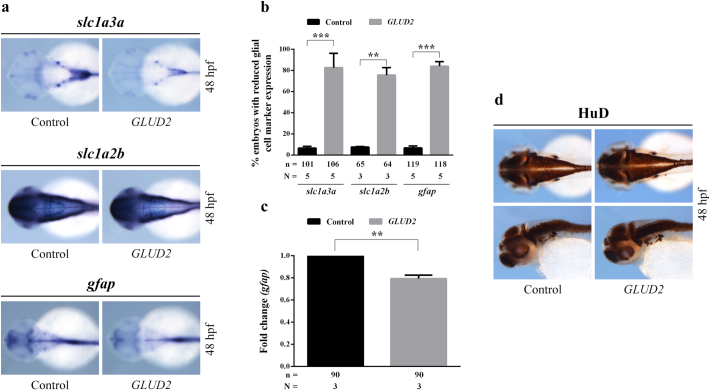
An altered glutamate metabolism could affect not only glial cells expressing GLUD2 but also the nearby neurons survival. Zebrafish model allowed us to further investigate the effects of GLUD2 overexpression on neural and glial development. We first questioned whether this enzyme might have an influence on zebrafish gliogenesis. We therefore assessed the expression pattern of different glial markers by WISH on 48 hpf embryos. Overexpression of GLUD2 mRNA caused a WISH signal reduction of slc1a3a (Fig. 6a, b), an early glial marker encoding an excitatory amino acid transporter (EAAT) expressed at glutamatergic synapses [21]. In addition, we analyzed the expression of slc1a2b (Fig. 6a, b), an EAAT mainly present in a subset of glial cells [22], which was found decreased in GLUD2-injected embryos compared to controls. To further evaluate whether the reduction of slc1a3a was due to increased glial differentiation or not, we examined the expression of a mature glial cells marker, gfap, also used as an astrocytes marker [21]. Gfap downregulation after GLUD2 mRNA injection was detected by WISH (Fig. 6a, b) and confirmed by qRT-PCR (Fig. 6c), suggesting an impairment of gliogenesis caused by GLUD2 overexpression.
Fig. 6.
GLUD2 overexpression impairs glial cells formation, without affecting neurons development in zebrafish embryos. (a) Whole mount in situ hybridization on 48 hpf embryos, indicating a reduction of the expression of three different glial cell markers (slc1a3a, slc1a2b and gfap) in GLUD2-injected embryos compared to controls. (b) Percentage of GLUD2-injected and control embryos with decreased glial cell marker expression; n, total number of analyzed embryos, N, number of independent experiments. (c) qRT-PCR analysis confirming the reduced transcript level of gfap in GLUD2-injected embryos relative to control embryos; n, total number of analyzed embryos, N, number of independent experiments. (d) Immunohistochemical staining of HuD on 48 hpf embryos shown in dorsal and lateral view (n = 84 for control embryos and n = 92 for GLUD2-injected embryos subdivided in three independent experiments) revealing no visible differences in the expression of this postmitotic neuronal marker between GLUD2-injected and control embryos. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: *p < 0.05, **p < 0.01 and ***p < 0.001.
With the aim of investigating a potential effect of GLUD2 overexpression on neurogenesis, we performed an immunohistochemical analysis visualizing the postmitotic neuronal marker HuD distribution (Fig. 6d). Compared to controls, GLUD2-injected embryos didn't display visible alterations of HuD expression pattern at 48 hpf, indicating that GLUD2 overexpression could affect glial cell development without impairing neuronal differentiation and survival.
4. Discussion
Glutamate dehydrogenase (GDH) is a mitochondrial enzyme that catalyzes the reversible inter-conversion of glutamate to α-KG by using NADP and/or NAD as cofactors. GDH in human exists in GLUD1 and GLUD2 gene-encoded isoforms (hGDH1 and hGDH2, respectively). GLUD1 gene is located on 10q and is expressed widely, whereas GLUD2 is an intronless gene located on the X chromosome with expression specificity for nervous and testicular tissues [23,24]. In the brain, GLUD2 expression is mainly associated with astrocytes [24,25] with a substantial mitochondrial localization, representing up to 10% of the matrix protein [10,25,26]. GLUD2 contributes to important cellular processes, such as the Krebs cycle, ammonia homeostasis and energy production [10,15,24,26,27]; however, its role in cell biology is still incompletely understood [26]. GLUD2 has the ability to act on both directions of the reaction depending on the availability of the substrate, but in the cancer it is thought that it works mainly in the direction of the oxidative deamination of glutamate and α-KG production [28].
GLUD2 initially emerged from our previous NGS analysis [17] conducted on 13 GBM samples from patients with different recurrence time (RFS). GLUD2 was overexpressed in the tumors of patients with long RFS. We then interrogated the Gene Expression Omnibus dataset in which 77 samples from newly diagnosed cases of high-grade gliomas were profiled via microarray analysis. We found that GLUD2 overexpression was significantly associated with increased overall survival and lower glioma grading.
We then performed in-vitro functional studies on human GBM cells evaluating cellular effect due to changes in GLUD2 expression levels. In particular, we overexpressed and silenced GLUD2 in two different cell lines, T98G and U118, respectively. GLUD2 overexpression led to a significant decrease in cell proliferation, invasion and migration capacity and colony formation ability; at the same time, GLUD2 silencing caused a statistically significant increase in tumor aggressiveness and tumorigenicity.
There is increasing evidence that glutamate oxidative deamination is the predominant pathway for α-KG production and therefore its abundance strictly depends on GLUD2 activity [16,29]. This also suggests another possible explanation of decrease of proliferation following an increase in the enzymatic activity of GLUD2 and vice versa. In fact, when there is higher consumption of glutamate as a substrate, there is higher attraction of glutamate from the outside towards the inside of the cell and as a consequence, the extracellular glutamate decreases with less effect on the promotion of cell proliferation through the activation of its receptors [30]. Other studies have observed that the increase of glutamate uptake within the glioma cell, through the functional increase of its transporters, decreases tumor proliferation [30,31].
Afterward, we investigated how GLUD2 expression levels could affect the cell cycle phases and their regulation by checkpoints. Both cyclin D1 and cyclin E expression levels are reduced after GLUD2 overexpression and this is reflected on the G0/G1 phase increase and S and G2/M phase reduction. However, a variation of cell cycle phases is not appreciable in GLUD2 silenced cells, although there is a significant increase in both cyclin D1 and cyclin E expression levels,. This is probably due to the small difference in the increase of cyclin E in GLUD2 silenced cells compared to control cells. The role of cyclin D1, in fact, is to advance the G1 phase and prepare cells for the S phase; whereas cyclin E is responsible for G1-to-S-phase transition [32].
We also evaluated the consequences of GLUD2 expression levels variation on mitochondrial function and metabolic phenotype, by measuring oxygen consumption rate (OCR) after GLUD2 overexpression and silencing. Statistically significant effects, in both overexpression and silencing of GLUD2 with opposite trends, were observed in the baseline oxygen consumption and non-mitochondrial respiration. GLUD2 overexpression seems, therefore, correlated with an increase in oxygen consumption, which however does not translate into an increase in oxidative phosphorylation, as there are no significant differences in the production of ATP. The fact that there are differences in non-mitochondrial respiration has led us to investigate the production of reactive oxygen species (ROS). We have indeed observed that GLUD2 overexpression increased the production of ROS and on the other hand, after GLUD2 silencing, the production of ROS was decreased, as expected. The effect of GLUD2 on ROS production, depending on its expression level, helped clarifying the mechanisms by which altered levels of GLUD2 expression can modify tumor progression and development.
The correlation between GLUD2 expression levels and ROS production has already been described in the literature [27]. In particular, it has been demonstrated that increased glutamate oxidation by an enhanced GLUD2 activation by the Ala445 variant in the regulatory domain, may improperly boost mitochondrial oxidative metabolism with consequent increased ROS production [25]. This suggests that a significant gain in GLUD2 activity can cause severe mitochondrial dysfunction, as mitochondria fail to manage the large amounts of GLUD2 produced α-KG. Moreover, it has also been described that GLUD2 NADPH generation affects ROS homeostasis trough NADPH oxidase activation and ROS production [28]. Furthermore, an increase in ROS generation following GLUD2 overexpression may also explain the resulting cell cycle block in G0 / G1, since it has been shown that increased levels of ROS lead to a decrease in cyclin D1/E expression and promote cell-cycle arrest [33,34].
All these metabolic variations observed in-vitro could be differentially regulated in-vivo by extracellular stimuli and cell–cell interactions that give rise to a complex and integrated modulation of cell behavior. In order to test the possible effect of GLUD2 overexpression in-vivo we took advantage of zebrafish embryos in which we can easily alter GLUD2 expression and monitor cell cycle progression in the developing central nervous system. Over the past years, zebrafish has become an effective and alternative tool to the classical mouse model for studying many human diseases including CNS pediatric to adult tumors [19,20]. However obvious evolutive divergences between fishes and humans have always to be taken into account. In this work we decided to use the zebrafish embryo just to evaluate GLUD2 gene function in a cellular context in which all the metabolic pathways are active and the interaction and exchanges between glia, neurons, hormones, neurotransmitters and metabolites are orchestrated in a complex and dynamic environment that is not reproducible in-vitro.
In GLUD2-overexpressing embryos we observed a reduced number of mitotic cells in the developing brain that correlated with a decreased level of cyclin D1 expression mirroring the data obtained in the T98G human GBM cell line. These observations strongly corroborated the in-vitro data adding important hints on the possible side effects of the GLUD2 modulation. The in-vivo system, in fact, allowed us to verify how glial cells overexpressing GLUD2 proceed in their developmental program and how they can influence the behavior of the surrounding neurons. In GLUD2 overexpressing embryos we observed a failure in the generation of differentiated glial cells, probably due to a block of proliferation of gliogenic precursors, but interestingly neurogenic precursors seem not to be affected as post mitotic neurons developed normally. This aspect could reflect the different metabolic activity of glial cells and neurons and could be of great interest in a therapeutic perspective. The possibility to enhance GLUD2 activity in GBM could result in a blocked/reduced proliferation of glial cells without affecting the survival of the surrounding neurons.
To the best of our knowledge, this is the first work where GLUD2 is considered as a key player in GBM progression. These observations may provide a new target for therapeutic interventions in GBM to reduce tumor progression and aggressiveness. On this, however, there is still much to learn about when and how to manipulate glutamate metabolic system as a target and to further clarify the intracellular signaling pathways associated with pathological states. In particular, preclinical studies on more complex animal models than zebrafish, such as murine, will be necessary. This work could represent a starting point to deepen the role of GLUD2 in GBM in-vivo and to design and test possible pharmacological treatments. GLUD2 enzymatic activity could be increased through the induction of an activating mutation, for example the Ala445 variant in the regulatory domain [25], with a highly advanced genome editing system such as CRISPR/Cas9. Alternatively, activation of GLUD2 could be achieved by allosteric effectors, such as leucine [35], although in this case the assessment of specificity is extremely important.
Author contributions
SF, DC, MO and CMM designed the project and experimental work. SF and PA analyzed the data from RNA-seq and microarray analysis. SF performed the cell culture experiments with additional help from FL, ET, CS and PC. MM supervised histological analysis. FP and AGN gave clinical support. DC performed the zebrafish experiments. SF and DC wrote the manuscript. MO and CMM contributed to data review and interpretation. All authors commented and reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Acknowledgements/funding
None. No funding to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.008.
Contributor Information
Sara Franceschi, Email: s.franceschi@fpscience.it.
Michela Ori, Email: michela.ori@unipi.it.
Supplementary data
Multimedia Component 1 Fig. S1. GBM cell lines with different GLUD2 expression levels reveal distinct cell functional behaviors. (a) GLUD2 mRNA expression in U118 and T98G human GBM cell lines. (b) Immunofluorescence stain of GLUD2 protein in U118 and T98G cells. (c) Western blot analysis of GLUD2 in U118 and T98G cells. GLUD2 protein was quantified by ImageJ software and normalized to the quantified value of B-Tubulin protein. The normalized values were further normalized to T98G cell value. (d) Glutamate dehydrogenase (GDH) activity of U118 and T98G cells. (e) U118 and T98G cells proliferation rate. (f) Colony formation ability in U118 and T98G. Colonies were counted in five non-overlapping fields. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: * p < 0.05, ** p < 0.01 and *** p < 0.001. Fig. S2. GLUD2 expression and activity in T98G cells after GLUD2 overexpression and silencing. (a) GLUD2 mRNA expression in GLUD2 overexpressed T98G cells (pIRES-GLUD2), GLUD2 silenced U118 cells (siRNA GLUD2) and relative controls. (b) Western blot analysis of pIRES-GLUD2, siRNA GLUD2 and control cells. GLUD2 protein was quantified by ImageJ software and normalized to the quantified value of β-Tubulin protein. Normalized values were further normalized to control cells values. (c) Immunofluorescence stain of GLUD2 protein in pIRES-GLUD2 cells, siRNA GLUD2 cells and relative controls. (d) Glutamate dehydrogenase (GDH) activity of pIRES-GLUD2 cells and siRNA GLUD2 cells compared to relative controls. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: * p < 0.05, ** p < 0.01 and *** p < 0.001. Fig. S3. Parameter calculations performed in the Seahorse XF Cell Mito Stress Test. (a) The Seahorse assay. Oxygen consumption rate is measured before and after adding pharmacological agents to respiring cells. (b) Complexes of the ETC and the target of action of all of the compounds in the Seahorse XF Cell Mito Stress Test Kit. Oligomycin inhibits ATP synthase (complex V), and the decrease in OCR following injection of oligomycin correlates to the mitochondrial respiration associated with cellular ATP production. Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) is an uncoupling agent that collapses the proton gradient and disrupts the mitochondrial membrane potential. As a result, electron flow through the ETC is uninhibited, and oxygen is maximally consumed by complex IV. (c) Seahorse XF Cell Mito Stress Test parameters glossary.
RNA-seq data analysis using Partek Flow software. Differential gene expression between the short-term group (S) with recurrence free survival (RFS) < 6 months (n = 6), medium group (M) with 16 < RFS < 23 months (n = 3) and the long group (L) with RFS > 25 months (n = 4).
References
- 1.Alfardus H., McIntyre A., Smith S. MicroRNA regulation of glycolytic metabolism in glioblastoma. Biomed. Res. Int. 2017;2017 doi: 10.1155/2017/9157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbruzzese C., Matteoni S., Signore M. Drug repurposing for the treatment of glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2017;36(1):169. doi: 10.1186/s13046-017-0642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoyanov G.S., Dzhenkov D., Ghenev P., Iliev B., Enchev Y., Tonchev A.B. Cell biology of glioblastoma multiforme: from basic science to diagnosis and treatment. Med. Oncol. 2018;35(3):27. doi: 10.1007/s12032-018-1083-x. [DOI] [PubMed] [Google Scholar]
- 4.Bayin N.S., Frenster J.D., Sen R. Notch signaling regulates metabolic heterogeneity in glioblastoma stem cells. Oncotarget. 2017;8(39):64932–64953. doi: 10.18632/oncotarget.18117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szopa W., Burley T.A., Kramer-Marek G., Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. Biomed. Res. Int. 2017;2017 doi: 10.1155/2017/8013575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H., Romero-Lopez M., Benitez L.I. 3D mathematical modeling of glioblastoma suggests that transdifferentiated vascular endothelial cells mediate resistance to current standard-of-care therapy. Cancer Res. 2017;77(15):4171–4184. doi: 10.1158/0008-5472.CAN-16-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stupp R., Mason W.P., van den Bent M.J. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 8.Anjum K., Shagufta B.I., Abbas S.Q. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: a review. Biomed. Pharmacother. 2017;92:681–689. doi: 10.1016/j.biopha.2017.05.125. [DOI] [PubMed] [Google Scholar]
- 9.Torok J.A., Wegner R.E., Mintz A.H., Heron D.E., Burton S.A. Re-irradiation with radiosurgery for recurrent glioblastoma multiforme. Technol. Cancer Res. Treat. 2011;10(3):253–258. doi: 10.7785/tcrt.2012.500200. [DOI] [PubMed] [Google Scholar]
- 10.Comelli M., Pretis I., Buso A., Mavelli I. Mitochondrial energy metabolism and signalling in human glioblastoma cell lines with different PTEN gene status. J. Bioenerg. Biomembr. 2017;50(1):33–52. doi: 10.1007/s10863-017-9737-5. [DOI] [PubMed] [Google Scholar]
- 11.Libby C.J., Tran A.N., Scott S.E., Griguer C., Hjelmeland A.B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim. Biophys. Acta. 2018;1869(2):175–188. doi: 10.1016/j.bbcan.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G., Zhu J., Yu M. Glutamate dehydrogenase is a novel prognostic marker and predicts metastases in colorectal cancer patients. J. Transl. Med. 2015;13:144. doi: 10.1186/s12967-015-0500-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maus A., Peters G.J. Glutamate and alpha-ketoglutarate: key players in glioma metabolism. Amino Acids. 2017;49(1):21–32. doi: 10.1007/s00726-016-2342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danbolt N.C., Furness D.N., Zhou Y. Neuronal vs glial glutamate uptake: resolving the conundrum. Neurochem. Int. 2016;98:29–45. doi: 10.1016/j.neuint.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Plaitakis A., Latsoudis H., Spanaki C. The human GLUD2 glutamate dehydrogenase and its regulation in health and disease. Neurochem. Int. 2011;59(4):495–509. doi: 10.1016/j.neuint.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Plaitakis A., Kalef-Ezra E., Kotzamani D., Zaganas I., Spanaki C. The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology (Basel) 2017;6(1) doi: 10.3390/biology6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschi S., Lessi F., Aretini P. Cancer astrocytes have a more conserved molecular status in long recurrence free survival (RFS) IDH1 wild-type glioblastoma patients: new emerging cancer players. Oncotarget. 2018;9(35):24014–24027. doi: 10.18632/oncotarget.25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips H.S., Kharbanda S., Chen R. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 19.Corallo D., Candiani S., Ori M., Aveic S., Tonini G.P. The zebrafish as a model for studying neuroblastoma. Cancer Cell Int. 2016;16:82. doi: 10.1186/s12935-016-0360-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idilli A.I., Precazzini F., Mione M.C., Anelli V. Zebrafish in translational cancer research: insight into leukemia, melanoma, glioma and endocrine tumor biology. Genes (Basel) 2017;8(9) doi: 10.3390/genes8090236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y.C., Chiang M.C., Shih H.Y. The transcription factor hairy/E(spl)-related 2 induces proliferation of neural progenitors and regulates neurogenesis and gliogenesis. Dev. Biol. 2015;397(1):116–128. doi: 10.1016/j.ydbio.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 22.McKeown K.A., Moreno R., Hall V.L., Ribera A.B., Downes G.B. Disruption of Eaat2b, a glutamate transporter, results in abnormal motor behaviors in developing zebrafish. Dev. Biol. 2012;362(2):162–171. doi: 10.1016/j.ydbio.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanavouras K., Mastorodemos V., Borompokas N., Spanaki C., Plaitakis A. Properties and molecular evolution of human GLUD2 (neural and testicular tissue-specific) glutamate dehydrogenase. J. Neurosci. Res. 2007;85(15):3398–3406. doi: 10.1002/jnr.21576. [DOI] [PubMed] [Google Scholar]
- 24.Spanaki C., Kotzamani D., Plaitakis A. Widening spectrum of cellular and subcellular expression of human GLUD1 and GLUD2 glutamate dehydrogenases suggests novel functions. Neurochem. Res. 2017;42(1):92–107. doi: 10.1007/s11064-016-1986-x. [DOI] [PubMed] [Google Scholar]
- 25.Shashidharan P., Plaitakis A. The discovery of human of GLUD2 glutamate dehydrogenase and its implications for cell function in health and disease. Neurochem. Res. 2014;39(3):460–470. doi: 10.1007/s11064-013-1227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaganas I., Kanavouras K., Mastorodemos V., Latsoudis H., Spanaki C., Plaitakis A. The human GLUD2 glutamate dehydrogenase: localization and functional aspects. Neurochem. Int. 2009;55(1–3):52–63. doi: 10.1016/j.neuint.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Tarasenko V.I., Garnik E.Y., Shmakov V.N., Konstantinov Y.M. Induction of Arabidopsis gdh2 gene expression during changes in redox state of the mitochondrial respiratory chain. Biochemistry (Mosc) 2009;74(1):47–53. doi: 10.1134/s0006297909010076. [DOI] [PubMed] [Google Scholar]
- 28.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16(11):749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 29.Schousboe A., Scafidi S., Bak L.K., Waagepetersen H.S., McKenna M.C. Glutamate metabolism in the brain focusing on astrocytes. Adv. Neurobiol. 2014;11:13–30. doi: 10.1007/978-3-319-08894-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhoutte N., Hermans E. Glutamate-induced glioma cell proliferation is prevented by functional expression of the glutamate transporter GLT-1. FEBS Lett. 2008;582(13):1847–1852. doi: 10.1016/j.febslet.2008.04.053. [DOI] [PubMed] [Google Scholar]
- 31.de Groot J., Sontheimer H. Glutamate and the biology of gliomas. Glia. 2011;59(8):1181–1189. doi: 10.1002/glia.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pucci B., Kasten M., Giordano A. Cell cycle and apoptosis. Neoplasia. 2000;2(4):291–299. doi: 10.1038/sj.neo.7900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li C., Peng W., Song X., Wang Q., Wang W. Anticancer effect of icaritin inhibits cell growth of colon cancer through reactive oxygen species, Bcl-2 and cyclin D1/E signaling. Oncol. Lett. 2016;12(5):3537–3542. doi: 10.3892/ol.2016.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X., Liu J., Jiang L. Bach1 induces endothelial cell apoptosis and cell-cycle arrest through ROS generation. Oxidative Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/6234043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anno T., Uehara S., Katagiri H. Overexpression of constitutively activated glutamate dehydrogenase induces insulin secretion through enhanced glutamate oxidation. Am. J. Physiol. Endocrinol. Metab. 2004;286(2):E280–E285. doi: 10.1152/ajpendo.00380.2003. [DOI] [PubMed] [Google Scholar]
- 36.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 38.Busquet F., Strecker R., Rawlings J.M. OECD validation study to assess intra- and inter-laboratory reproducibility of the zebrafish embryo toxicity test for acute aquatic toxicity testing. Regul. Toxicol. Pharmacol. 2014;69(3):496–511. doi: 10.1016/j.yrtph.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 39.Baumgart M., Groth M., Priebe S. RNA-seq of the aging brain in the short-lived fish N. furzeri – conserved pathways and novel genes associated with neurogenesis. Aging Cell. 2014;13(6):965–974. doi: 10.1111/acel.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia Component 1 Fig. S1. GBM cell lines with different GLUD2 expression levels reveal distinct cell functional behaviors. (a) GLUD2 mRNA expression in U118 and T98G human GBM cell lines. (b) Immunofluorescence stain of GLUD2 protein in U118 and T98G cells. (c) Western blot analysis of GLUD2 in U118 and T98G cells. GLUD2 protein was quantified by ImageJ software and normalized to the quantified value of B-Tubulin protein. The normalized values were further normalized to T98G cell value. (d) Glutamate dehydrogenase (GDH) activity of U118 and T98G cells. (e) U118 and T98G cells proliferation rate. (f) Colony formation ability in U118 and T98G. Colonies were counted in five non-overlapping fields. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: * p < 0.05, ** p < 0.01 and *** p < 0.001. Fig. S2. GLUD2 expression and activity in T98G cells after GLUD2 overexpression and silencing. (a) GLUD2 mRNA expression in GLUD2 overexpressed T98G cells (pIRES-GLUD2), GLUD2 silenced U118 cells (siRNA GLUD2) and relative controls. (b) Western blot analysis of pIRES-GLUD2, siRNA GLUD2 and control cells. GLUD2 protein was quantified by ImageJ software and normalized to the quantified value of β-Tubulin protein. Normalized values were further normalized to control cells values. (c) Immunofluorescence stain of GLUD2 protein in pIRES-GLUD2 cells, siRNA GLUD2 cells and relative controls. (d) Glutamate dehydrogenase (GDH) activity of pIRES-GLUD2 cells and siRNA GLUD2 cells compared to relative controls. Data are presented as mean ± SD and differences were considered statistically significant when p < 0.05 and represented as: * p < 0.05, ** p < 0.01 and *** p < 0.001. Fig. S3. Parameter calculations performed in the Seahorse XF Cell Mito Stress Test. (a) The Seahorse assay. Oxygen consumption rate is measured before and after adding pharmacological agents to respiring cells. (b) Complexes of the ETC and the target of action of all of the compounds in the Seahorse XF Cell Mito Stress Test Kit. Oligomycin inhibits ATP synthase (complex V), and the decrease in OCR following injection of oligomycin correlates to the mitochondrial respiration associated with cellular ATP production. Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP) is an uncoupling agent that collapses the proton gradient and disrupts the mitochondrial membrane potential. As a result, electron flow through the ETC is uninhibited, and oxygen is maximally consumed by complex IV. (c) Seahorse XF Cell Mito Stress Test parameters glossary.
RNA-seq data analysis using Partek Flow software. Differential gene expression between the short-term group (S) with recurrence free survival (RFS) < 6 months (n = 6), medium group (M) with 16 < RFS < 23 months (n = 3) and the long group (L) with RFS > 25 months (n = 4).