Abstract
Background
Glioblastoma multiforme (GBM) is a fatal disease without effective therapy. Identification of new biomarkers for prognosis would enable more rational selections of strategies to cure patients with GBM and prevent disease relapse.
Methods
Seven datasets derived from GBM patients using microarray or next generation sequencing in R2 online database (http://r2.amc.nl) were extracted and then analyzed using JMP software. The survival distribution was calculated according to the Kaplan-Meier method and the significance was determined using log-rank statistics. The sensitivity of a panel of GBM cell lines in response to temozolomide (TMZ), salinomycin, celastrol, and triptolide treatments was evaluated using MTS and tumor-sphere formation assay.
Findings
We identified that CD44, ATP binding cassette subfamily C member 3 (ABCC3), and tumor necrosis factor receptor subfamily member 1A (TNFRSF1A) as highly expressed genes in GBMs are associated with patients' poor outcomes and therapy resistance. Furthermore, these three markers combined with MGMT, a conventional GBM marker, can classify GBM patients into five new subtypes with different overall survival time in response to treatment. The four-gene signature and the therapy response of GBMs to a panel of therapeutic compounds were confirmed in a panel of GBM cell lines.
Interpretation
The data indicate that the four-gene panel can be used as a therapy response index for GBM patients and potential therapeutic targets. These results provide important new insights into the early diagnosis and the prognosis for GBM patients and introduce potential targets for GBM therapeutics.
Fund
Baylor Scott & White Health Startup Fund (E.W.); Collaborative Faculty Research Investment Program (CFRIP) of Baylor University, Baylor Scott & White Health, and Baylor College of Medicine (E.W., T.S., J.H.H.); NIH R01 NS067435 (J.H.H.); Scott & White Plummer Foundation Grant (J.H.H.); National Natural Science Foundation of China 816280007 (J.H.H. and Fu.W.).
Keywords: CD44, ABCC3, Cancer stem cell, Glioblastoma multiforme, Brain tumor, Prognosis
Abbreviations: ABCC3, ATP binding cassette subfamily C member 3; ATCC, American Type Culture Collection; ANOVA, Analysis of Variance; CSC, Cancer stem cell; CT, Computed tomography; DMSO, Dimethyl sulfoxide; FDA, Food and Drug Administration; GBM, Glioblastoma multiforme; IDH, Isocitrate dehydrogenases; LOH, Loss of heterozygosity; MGMT, O6-methylguanine DNA methyltransferase; MRI, Magnetic resonance imaging; PDGFR, Platelet derived growth factor receptor; TMZ, Temozolomide; TNFRSF1A, Tumor necrosis factor receptor superfamily member 1A; TTF, Tumor treating fields
Research in context.
Evidence before this study
Glioblastoma multiforme (GBM) is a devastating brain cancer that usually results in death in the first 14.6 months after diagnosis. Currently, diagnosis and prognosis of GBM rely on magnetic resonance imaging (MRI), computed tomography, and biopsy. In addition, therapy resistance is a major problem for GBM treatments. Both intrinsic heterogeneity among GBMs and intratumoral heterogeneity in GBM are considered as the major hurdles in the therapy of GBMs. Moreover, the existence of cancer stem cells (CSCs) in GBMs has been considered as another major reason for therapy resistance.
Added value of this study
We used large datasets derived from GBM patients to analyze the expression of several CSC markers and the CD44 correlated genes ABCC3 and TNFRSF1A. We identified a four-gene signature which could be employed as prognostic markers for GBM patients in the future. We further tested the four-gene model using a panel of GBM cell lines with TMZ and three compounds with the potential to be used as anti-GBM therapy. This study could be a resource in clinical activity for patient stratification.
Implications of all the available evidence
The MGMT methylation, IDH, and EGFR mutation, as well as the identified four-gene index model from this study could aid in the prognosis and target therapy for patients with GBM. The current poor survival of GBM patients may result from delayed diagnosis, which typically occurs after individuals display neurological symptoms and require expensive diagnostic MRI analyses. Therefore, the four-gene signature has a potential role in guiding clinicians towards developing cost-efficient personalized optimal patient care.
Alt-text: Unlabelled Box
1. Introduction
Glioblastoma multiforme (GBM), characterized by the accumulation of multiple genetic alterations in tumor cells, accounts for 50% of central nervous system tumors, and is a deadly disease with no known curable therapy [1]. Even with aggressive treatments, such as surgery followed by chemotherapy and radiotherapy, patients with GBM will eventually die of their disease and the median survival time is approximately 14–16 months [2]. Adjuvant TMZ plus Optune (a tumor treating fields (TTF) device) treatment is currently considered as a standard strategy for maintenance therapy to extend patients' lives after surgery and chemotherapy [2,3]. Currently, diagnosis and prognosis of GBM rely on magnetic resonance imaging (MRI), computed tomography (CT), and biopsy [4]. Though several targeted therapy strategies have been initiated, all these therapies are only suitable for a very small population due to the heterogeneity of the disease [5,6]. Therefore, understanding the complexity of GBM biology in order to identify suitable diagnostic and prognostic biomarkers and therapeutic targets is essential for optimal patient care.
With the aid of the current advanced technologies, GBMs have been classified into several subtypes based on the patients' performances, as well as morphology and gene signature and/or the genetic alterations in the tumors (e.g., isocitrate dehydrogenases (IDH) mutations, platelet derived growth factor receptor (PDGFR), epidermal growth factor receptor (EGFR) mutation or amplification, and 1p/19q loss of heterozygosity (LOH), or changes in epigenetic modifications) [1,[7], [8], [9], [10], [11], [12], [13]]. Clinically relevant subtypes of GBM, classical, neural, proneural, and mesenchymal, with different phenotypes based on their intrinsically genetic alterations and gene expression patterns in the tumor have been also identified [8]. Studies show that both intrinsic heterogeneity among GBMs and intratumoral heterogeneity in each GBM are considered as the major hurdles in management of GBMs [7,14,15]. In addition, the existence of cancer stem cells (CSCs) in GBMs has been considered as another major reason for therapy resistance [16,17]. CD133 is one of the most commonly used markers for selecting GBM CSCs [15,18,19]. Moreover, CD44 expression in GBM cells is critical for GBM invasion and migration [[20], [21], [22], [23]]. Furthermore, Anido et al. reported that GBM cells with high levels of CD44 have notably higher potential for developing tumors in vivo [24]. Recent studies also support the observation that CD44 may be a GBM stem cell marker in vitro and in animal models [15,20,25,26]. However, the molecular mechanism of GBM therapy resistance remains to be determined, and biomarkers to monitor treatment responses are in urgent need.
In this study, we use large microarray datasets derived from GBM patients to analyze the expression of commonly used CSC markers, especially CD44 in GBM patients and CD44 correlated genes in GBMs from patients. GBM patients with a highly methylated O6-methylguanine-methyltransferase (MGMT) promoter or IDH1 mutation in their tumors often exhibit a better response to treatments, while alterations at EGFR result in therapy resistance [1,14]. Here, we determine the contribution of some common genetic modifications/alterations (e.g., MGMT methylation, EGFR amplification, IDH1 mutation, and 1p/19q LOH) on CD44 expression and related genes involved in CD44 signaling and validate these results using a group of GBM cell lines.
2. Materials & methods
2.1. Datasets, data analyses, and statistical testing
2.1.1. Datasets and data analyses
Seven datasets from previously detected microarray with complete information were employed in this study [10,19,[27], [28], [29], [30], [31], [32], [33]]. Using the ‘R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl)’, we analyzed the expression levels of a panel of commonly used CSC markers including CD133, CD44, SOX2, Nestin (NES), and MYC and their correlations in GBM cells, neural stem cells, normal brain tissues, and GBMs, as well as the correlation of gene expression levels with patients' survival outcomes. The levels of commonly used CSC markers and the correlation levels between the CSC markers in GBM cells vs. neural stem cells or normal cortex were determined using the previously generated dataset “Cellline Glioblastoma Stemcells - Pollard – 20” by the GeneChip Human Genome U133 plus 2.0 Array (Affymetrix) [19]. The levels of gene expression in GBM tumors vs. non-brain tumor tissues were determined using the dataset “Mixed Brain Glioma - Sun - 180 - MAS5.0 - u133p2” previously generated using the GeneChip Human Genome U133 plus 2.0 Array [29]. The CD44 regulated target gene signatures (co-expressed gene groups) were determined using three large datasets “Tumor Glioblastoma-TCGA-540-MAS5.0-u133a”, “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331”, and “Tumor Glioma - French - 284 - MAS5.0 - u133p2” [10,28,33,34]. The CD44 signaling related genes were selected based on the degree of positive correlation with CD44 in the GBMs as reported in our previous study [35]. For the correlated genes in GBM with CD44, at least R ≥ 0.65 were selected from each cohort as candidate genes, and among them, the genes with highest correlation defined by R ≥ 0.65 and lowest Log10[pval] in all three cohorts were selected [10,28,33,34]. The levels of selected markers in GBM tissues at different stages and subtypes were determined using the dataset “Tumor Glioma - Lee - 100 - MAS5.0 - u133a” generated by the microarray [31,32]. The levels of selected markers in GBM with or without MGMT methylation in GBM tissues were determined using the dataset “Tumor Glioblastoma - TCGA - 540 - MAS5.0 - u133a” generated by Brennan et al. [33]. The levels of selected markers in GBM with or without IDH1 R132H mutation, EGFR amplification, and 1p/19q LOH in the GBM tissues were determined using microarray data “Tumor Glioma - French - 284 - MAS5.0 - u133p2” generated by Gravendeel et al. [10]. The correlation of gene expression levels with patients' survival outcomes were determined using three datasets with patients' overall survivals in the R2 database [10,28,33,34].
For all analyses using microarray data, we used the probes that could recognize all gene isoforms. We then applied the same probe for the target gene in all datasets. Only consistent results from different cohorts are reported here. For the correlation of gene levels with patient's survival time, we extracted the data from R2 database using the probes selected for gene expression analysis and then analyzed for correlation using JMP software (SAS Institute, Cary, NC). To define high and low expressions, we first evaluated the distribution and mean value for each gene in all samples crossing all datasets. The range of cut-off values for each gene was determined according to the distribution and mean value of each gene in the datasets. The high and low expressions in all samples crossing all datasets employed in this study were determined based on the cut-off values selected for each gene and tested for correlation with patients' survival outcomes using JMP software. The threshold was established based on the test of gene sets using values selected in the distribution range by Kaplan-Meier and log rank methods. Based on the p-values from all tests, the median cut-off value that gave the smallest p-value was selected. The high and low expressions refer to the value above and below the cut-off value selected for the log rank test, respectively. The survival distribution was estimated according to the Kaplan-Meier method and the significance was determined using log-rank statistics.
2.1.2. Statistical testing
All statistical analyses were performed with JMP software. The level of significance for gene expression among different types of tissues was analyzed using student t-test or ANOVA. The log-rank test was used for comparison of patient survival between high and low expression groups for each selected gene. Statistical significance was defined as p < .05.
2.2. Cell culture
GBM cell lines, U87 with TP53 wild type, T98G containing a TP53 Met → Ile mutation at codon 237, LN18 carrying TP53 Cys → Ser mutation at codon 183, and LN229 harboring a TP53 Pro → Leu mutation at codon 98, were obtained from ATCC. Cells were maintained in Minimum Essential Medium (MEM) (Cellgro) supplemented with 4 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 1% sodium pyruvate, 1% nonessential amino acids, and 10% fetal bovine serum (FBS) at 37 °C with 5% CO2.
2.3. Cell proliferation analysis
GBM cells (1 × 105/well) maintained in complete medium were placed in 96-well plates overnight. Salinomycin, celastrol, and triptolide at various concentrations or identical volume of control (DMSO) were added to the appropriate wells. The cells were treated for 48 h before adding MTS solution (Promega) according to the manufacturer's instructions. After 2–4 h incubation, the number of cells in each well was determined by measuring the optical densities at 490 nm. The results were expressed as the percentages of viable cells against the control cultures.
2.4. Tumor-sphere formation assay
Tumor-sphere formation assay was performed as previously described [36]. Tumor-sphere media consisted of a 50:50 mix of F12 and DMEM (Invitrogen), supplemented with 40 ng/ml bFGF (R&D systems), 20 ng/ml EGF (R&D systems), 1% B27 and N2 supplements (Invitrogen), 2 μg/ml heparin (Sigma), 0.1 mM β-mercaptoethanol (Sigma), and 1× antibiotic/antimycotic (Mediatech). 200 GBM cells were seeded in 200 μl medium into each well in Ultra-low attachment 96 well plate (Corning). At day 7, the numbers of tumor-sphere formed in each condition were counted under phase-contrast microscope using the 10× magnification lens.
2.5. Reverse transcription-quantitative PCR (RT-qPCR) analysis
Total RNA was isolated from GBM cell lines with or without treatments using RNeasy (Qiagen). The quantity and quality of extracted RNA were determined using NanoDrop (Thermo Fisher Scientific). 100 ng of total RNA was used to prepare cDNA using iScript Reverse Transcription Supermix (Bio-Rad) by following the instructions provided by the manufacturer. Gene specific primers were designed using Primer-BLAST online tool; and the primer sequences are available upon request. Real time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) and CFX96 real-time system C1000 Touch Thermal Cycler (Bio-Rad). PCR amplification was done at 95 °C for 10 min, denaturation at 95 °C for 15 s followed by annealing/extension at 60 °C for 1 min for 40 cycles. Fold change obtained from Ct values using 2-ΔΔCt methodology [37] was converted into logarithmic base 2 for statistical analysis. P-values < .05 were considered to be statistically significant.
3. Results
3.1. CD44 is an indicator of patients with poor survival
To search for therapy resistant markers in GBM, we determined the expression levels of a panel of commonly used putative CSC markers including CD133, CD44, Nestin (NES), SOX2, and MYC in tumorigenic GBM cell lines, neural stem cell lines, and normal cortex tissues as well as GBM tissues by analyzing microarray datasets [19,33]. As shown in Fig. 1a–b and Supplemental Fig. 1, all CSC markers, especially CD44, are highly expressed in GBM and neural stem cell lines and the levels of these markers are significantly higher than the ones in the normal cortex tissues (p ≤ .01). The expression levels of CD44 in GBM cell lines, neural stem cell lines, and normal cortex tissues correlated with NES (r = 0.851, p < .001) and MYC (r = 0.618, p < .001) extremely well, but not CD133 (r = 0.222, p = .38) (Fig. 1c). The result from the further analysis of the expression and the correlation between CD44 and these CSC markers in GBMs using three large datasets in the R2 database, revealed that the expression level of CD44 is higher than those previously defined CSC markers in the GBM samples (Fig. 1b, p < .001) and a good correlation between CD44 is only detected with NES but not CD133 and MYC in all GBM samples (Fig. 1d). More importantly, higher levels of CD44 and CD133 were associated with significantly shorter survival times (Fig. 1e and Supplemental Figs. 2a and 3). As GBMs are highly heterogeneous, previous studies have identified the existence of at least four subtypes of GBMs: proneural, neural, mesenchymal, and classical subtypes. Among all subtypes, the survival outcome from good to poor are: classical > proneural > mesenchymal > neural [8]. We further analyzed CD44 and CD133 expression in four subtypes of GBM patients and found a relatively low level of CD44 in all classical GBMs compared with mesenchymal and neural GBMs (Fig. 1f–h); CD44 levels classified proneural GBMs into two subtypes, of which, the one with higher levels of CD44 is associated with patients' significantly short survival times (Fig. 1i), while the expression of CD133 can be detected in all four major groups of GBMs (Supplemental Fig. 2b–e). These results indicate that CD44 is a potentially useful marker for evaluating GBM patient survival and treatment resistance.
Fig. 1.
The expression of CD44, CD133, NES, SOX2, and MYC in GBM.
(a) The heat map shows the expression of common CSC markers in GBM cells, neural stem cells, and normal cortex. The data was extracted from the dataset “Cellline Glioblastoma Stemcells - Pollard – 20 in the R2 database”[19] and the clustering was performed based on the nearest distance according to gene expression levels using JMP software. (b) The expression levels of common CSC markers in GBM tissues (ANOVA, p < .001). The data were extracted from "Tumor Glioblastoma-TCGA-540-MAS5.0-u133a" [33]. The boxes represent the interquartile ranges of the analyzed genes' expression levels. The line within each box represents median 2Log expression cut-off value of each gene in the dataset. The minimum and maximum ranges for each gene expression level were shown as whiskers. (c) The correlation of CD44 levels with CD133, NES, and MYC expression in GBM cells and neural stem cells were analyzed using the dataset “Cellline Glioblastoma Stemcells - Pollard – 20” [19]. (d) The correlation of CD44 expression with other CSC markers in GBM tissues using three available complete datasets in the R2 database and analyzed using the R2 software (http://r2.amc.nl) [10,28,33,34]. (e–i) The correlation of CD44 expression with patients' survival time. The correlation of expression of selected markers with patients' overall survivals in three available complete datasets in the R2 database were analyzed using the R2 software [10,28,33,34]. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics. P-values < .05 were considered to be statistically significant.
3.2. Identification of a panel of genes as prognostic markers for GBMs
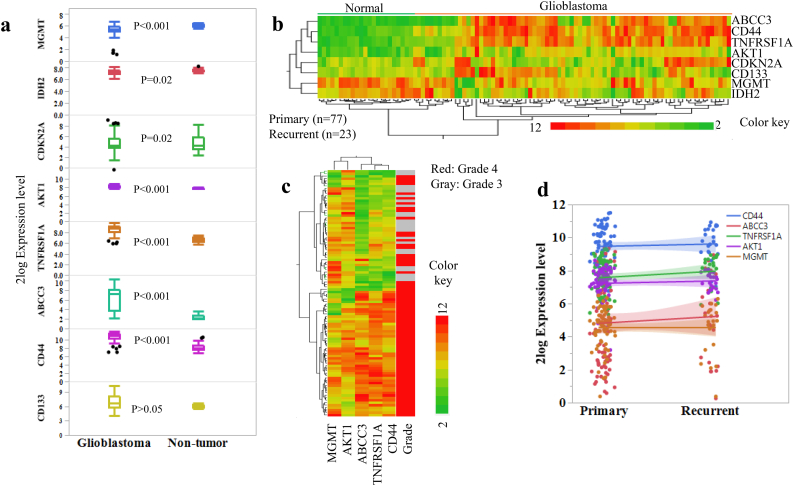
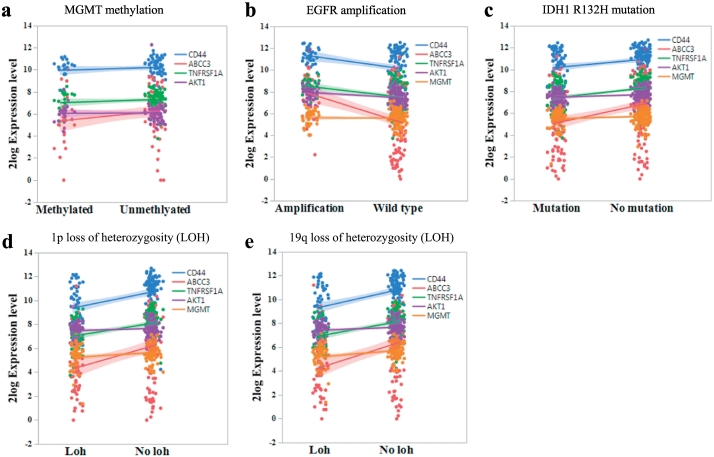
We then searched for CD44 regulated genes defined by its highly positively co-expressed genes (R ≥ 0.65) in GBMs and found that the highly co-expressed genes are mainly regulators involved in DNA repair, apoptosis, cell cycle, and transcription regulators, as well as membrane molecules (Fig. 2a & Supplemental Fig. 4). The expressions of nicotinamide phosphoribosyltransferase (NAMPT), tenascin C (TNC, a glioma associated extracellular matrix antigen), Annexin A1 (ANXA1), tumor necrosis factor receptor superfamily member 1A (TNFRSF1A), complement component 1 subcomponent R (C1R), WW domain containing transcription regulator 1 (WWTR1), ATP binding cassette subfamily C member 3 (ABCC3), complement C1r subcomponent like (C1RL), complement factor I (CFI), epithelial membrane protein 1 (EMP1, a tumor associated membrane protein), and S100 calcium binding protein 10 (S100A10) are highly positively correlated with CD44 expression in GBMs. We then compared the expression levels of CD44, and its highly correlated genes, ABCC3 and TNFRSF1A, in GBM and non-brain tumor cerebellum using the dataset from mixed brain glioma [29]. Among these genes, the expression levels of CD44, ABCC3, and TNFRSF1A are significantly higher in GBMs than in non-brain tumor tissues (Fig. 3a). In parallel, we also compared the expression levels of the most commonly used markers, such as CD133, AKT1, CDKN2A, IDH2, and MGMT in GBM and non-brain tumor tissues (Fig. 3a). We observed that the levels of AKT1 are significantly higher in GBMs than in normal cerebellum, while the levels of MGMT are much higher in the non-brain tumor tissues compared with GBM tissues (Fig. 3a–b). These results indicate that the detection of MGMT levels in GBM may not always be accurate; thus, only utilizing MGMT level as a biomarker to determine the therapy response may not be sufficiently sensitive. Further analysis of these markers in astrocytoma samples from patients at grade III vs. grade IV as well as primary vs. recurrent GBMs using the data generated by microarray [31,32] show that the expression levels of CD44, ABCC3, and TNFRSF1A in GBMs are increased along with disease grades (Fig. 3c), but such correlation is not observed between primary and recurrent GBMs (Fig. 3d). Because MGMT methylation status and IDH1 mutation, as well as abnormal activation of EGFR signaling due to amplification or mutation, are considered as important indicators of chemotherapy resistance [1,14], we also evaluated the effects of MGMT methylation, IDH1 mutation, and EGFR amplification on the expression of all selected markers in GBMs. We did not observe significant differences in expressions of these markers between MGMT-methylated and MGMT-unmethylated GBMs (Fig. 4a). In contrast, we found that the levels of TNFRSF1A and MGMT are higher in GBMs with EGFR amplification than those in wild type (WT) (Fig. 4b). Similarly, significantly elevated levels of CD44, ABCC3, TNFRSF1A, and AKT1 were observed in the GBMs with WT IDH1 than those with the IDH1 R132H mutation (Fig. 4c). The loss of heterozygosity (LOH) of 1p/19q has been previously shown to significantly affect GBM patients' survival outcomes; we thus further analyzed the levels of these markers in tumors with or without 1p/19q LOH. We found that the levels of CD44, ABCC3, and TNFRSF1A were significantly lower in the tumors with 1p/19q LOH (Fig. 4d-e).
Fig. 2.
Identification of CD44 highly correlated genes.
(a) The gene levels highly correlated with CD44 in GBM tissues selected from 540 GBMs "Tumor Glioblastoma-TCGA-540-MAS5.0-u133a" [33]. The genes with a correlation factor R > 0.65 and lowest p-value were highlighted in green as candidate genes. (b) The figure shows the correlation of genes with CD44 “R” and the significance levels of “p” in GBMs.
Fig. 3.
The expression levels of selected markers in GBM tumors and non-brain tumor tissues.
(a) The levels of selected genes in GBM tumors (n = 77) and non-brain tumor tissues (n = 77). The data were extracted from the dataset “Mixed Brain glioma - Sun - 180 - MAS5.0 - u133p2” [29]. The difference between GBM and non-brain tumor was determined using ANOVA test. P-values < .05 were considered to be statistically significant. The boxes represent the interquartile ranges of the analyzed genes' expression levels. The line within each box represents median 2Log expression cut-off value of each gene. The minimum and maximum ranges for each gene expression level were shown as whiskers. Black dots represent outliers. (b) The heat map represents the levels of selected markers in normal and GBM tissues. The clustering was performed based on the nearest distance according to gene expression levels using JMP software. Each row represents a gene and each column represents a sample. The color scale illustrates the relative expression levels of the genes. Colors from green to yellow to red represent the expression levels of each gene from low to intermediate to high. (c) The heat map represents the levels of selected markers in grade three and grade four astrocytoma tissues using the dataset “Tumor Glioma - Lee - 100 - MAS5.0 - u133a” [31]. (d) The expression levels of selected markers in primary (n = 23) and recurrent GBM tissues (n = 77) were determined using the dataset “Tumor Glioma - Lee - 100 - MAS5.0 - u133a” [31] (ANOVA, p < .05).
Fig. 4.
Common genetic modifications or alterations in GBM on CD44 and ABCC3 expression.
(a) The expression levels of selected genes in GBM with or without MGMT methylation (methylated, n = 22; unmethylated, n = 63) were determined using the dataset “Tumor Glioblastoma-TCGA-540-MAS5.0-u133a” [33] (student t-test, p < .05). (b) The expression levels of selected genes in GBM tissues with or without EGFR amplification (WT, n = 108; amplification, n = 43) (student t-test, p < .05) and (c) The expression levels of selected genes in GBM tissues with or without IDH1 mutation (WT, n = 143; mutation, n = 83) were determined using the dataset “Tumor Glioma - French - 284 - MAS5.0 - u133p2” [10] (student t-test, p < .01). (d) The expression levels of selected markers in GBM tissues with or without LOH at 1p (LOH, n = 51; Non LOH, n = 80) (student t-test, p < .01) and (e) with or without LOH at 19q (LOH, n = 50; Non LOH, n = 95) were determined using the dataset “Tumor Glioma-French-284-MAS5.0-u133p2” [10] (student t-test, p < .01).
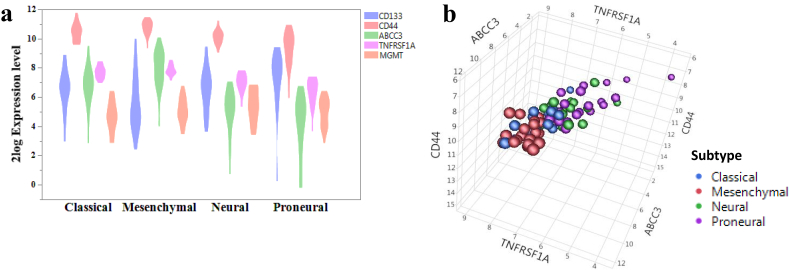
3.3. A four-gene index model for prediction of GBM patients' therapy response
From the further examination of the levels of CD133, CD44, ABCC3, TNFRSF1A, and MGMT in four subtypes of GBMs, we found that CD44, ABCC3, and TNFRSF1A were differentially expressed in all four subtypes and the highest levels were observed in the mesenchymal and classical groups. In contrast, the expression level of CD133 was higher in the proneural group (Fig. 5a). Using a 3D model, we show that expression levels of CD44, ABCC3, and TNFRSF1A could separate GBMs into two groups (classical and mesenchymal group vs. neural and proneural group) (Fig. 5b). As the existence of CSC is one of the major causes of disease relapse and MGMT methylation is a previously demonstrated indicator for GBM, we next analyzed the correlations between the expression levels of four genes (CD44, ABCC3, TNFRSF1A, and MGMT) in a 540 GBM cohort with patients' outcomes in response to radiotherapy and/or chemotherapy, respectively. As shown in Fig. 6a, d, and g, elevated levels of four markers in GBM are associated with those patients' shorter overall survival time in response to therapy. By applying the same standard to other two large cohorts of GBMs, the four-gene index model can classify all GBM patients into five subtypes (4 gene (CD44, ABCC3, TNFRSF1A, and MGMT) expression -low, 1 gene-high, 2 gene-high, 3 gene-high, and 4 gene-high), with different outcomes (Fig. 6b–c) and this index model divides the patients into subtypes in response to both radiation and chemotherapy (Fig. 6d–i). We further evaluated this four-gene index model in the GBMs carrying the 1p/19q LOH or the IDH1 mutation or the EGFR mutation; our results indicate that this four-gene index model could also divide the patients into subtypes with different survival outcomes (Supplemental Fig. 5).
Fig. 5.
Clinical relevance of selected markers.
The pre-normalized data were extracted from the dataset “Tumor Glioblastoma-TCGA-540-MAS5.0-u133a” [33] and 2log ratio values were used. (a) The expression levels of CD133, CD44, ABCC3, TNFRSF1A, and MGMT in four subtypes of GBMs (classical, n = 17; mesenchymal, n = 25; neural, n = 14; proneural, n = 22). (b) The 3D model was built based on the expression levels of three selected markers in four different subtypes of GBMs using JMP software.
Fig. 6.
A four-gene index model for prediction of GBM patients' therapy response.
The pre-normalized data were extracted from three available complete datasets in the R2 database (http://r2.amc.nl) [10,28,33,34] and analyzed using JMP software. The patient survival curves were produced using Kaplan-Meier method and log-rank method. The 540 GBM sample set was used as the training set and the other two sets of data were used for evaluation. 2log ratio values were used for all testing. (a) The correlation of four gene expressions in GBM with patients' survial (training set, p < .002). (b–c) The correlation of four gene expressions in GBM with patients' survial in the 395 and 284 sample sets respectively (validation sets, p < .02 and p < .0001, respectively). (d, e, f) The correlation of four gene expressions in GBM with patients' survial in the 540, 395, and 284 sample sets in response to radiation therapy, respectively (validation, p < .05, p < .01, p < .0001, respectively). (g, h, i) The correlation of four gene expressions in GBM with patients' survial in the 540, 395, and 284 sample sets in response to chemotherapy, respectively (validation, p < .05, p < .05, p < .0001, respectively).
3.4. Anti-GBM drug evaluation for GBM cells with different expression levels of ABCC3, CD44, TNFRSF1A, and MGMT
We also evaluated the expression levels of the abovementioned selected markers in a panel of GBM cell lines and determined the sensitivities of these cell lines in response to TMZ based on previous publications [[38], [39], [40], [41], [42]]. As shown in Table 1, in four GBM cell lines, U87 and LN229 are more sensitive to TMZ treatment than T98G and LN18. In addition, the expression levels of the four genes (ABCC3, CD44, TNFRSF1A, and MGMT) are higher in T98G and LN18 than those in U87 and LN229, though TNFRSF1A shows a slightly different pattern. These results suggest that GBM cell lines (T98G and LN18) which express high levels of the mentioned four genes are less sensitive to TMZ. We then assessed the sensitivity of GBM cells in response to two natural products, celastrol and triptolide, isolated from the root extracts of Tripterygium wilfordii (Thunder god vine) and demonstrated the effectiveness of the latter two compounds against GBM in vitro and in vivo [[43], [44], [45], [46], [47], [48]], as well as the effectiveness of salinomycin, an emerging agent with anti-breast CSC activities [49]. As shown in Fig. 7b–g, in response to celastrol treatments, the EC50 values for T98G and LN18 cells are approximately 5 μM; while the EC50 for LN229 and U87 is 1 μM. In response to triptolide treatments, the EC50 values are 30 nM for LN229, 80 nM for U87, and >80 nM for T98G and LN18. These results show that GBM cells (T98G and LN18) expressing higher levels of all four genes are less sensitive to celastrol and triptolide treatments and also highly resistant to TMZ treatment. However, intriguingly in terms of salinomycin-sensitivity [[50], [51], [52], [53], [54]], T98G and LN229 cells are more sensitive than LN18 and U87 cells accessed by both cell proliferation (the EC50 for T98G and LN18 is approximately 1.25 μM, while the EC50 for LN229 and U87 is about 10 times higher than T98G and LN18) and tumor-sphere formation assays (Fig. 7d, g). Taken together, these results suggest that the cell lines expressing different levels of these four markers are predictive of the efficacies of drugs used in GBM treatment.
Table 1.
The sensitivity of glioblastomas in response to TMZ.
| GBM cell line | TMZ-sensitivity | References |
|---|---|---|
| U87 vs. LN229 | U87 (IC50: 300 μM) > LN229 (IC50: >300 μM) | Cheng YC et al., Oncotarget. 2016 [71] |
| U87 vs. T98G | U87 (IC50:100 μM) > T98G (IC50: >500 μM) | Kanzawa T et al., Br J Cancer. 2003 [39] |
| LN229 vs. T98G | LN229 > T98G | Chen D et al., BMC Cancer. 2014 [41] |
| LN18 vs. LN229 | LN229 > LN18 (IC50: 400 μM) | Harrabi S et al., Int J Radiat Biol. 2013 & Yang N et al., J Transl Med. 2014 [38,42] |
Fig. 7.
The sensitivity of GBM cell lines to treatments.
(a) The expression levels of selected markers in a panel of GBM cell lines determined by real time PCR. (b, c, d) The effects of celastrol, triptolide, and salinomycin on GBM cell proliferation. (e, f, g) The effects of celastrol, triptolide, and salinomycin on GBM cell tumor-sphere formation. The error bars represent the standard error of the mean (SEM) for all graphs in this fig. P-values < .05 were considered to be statistically significant. *p < .05, **p < .01, ***p < .001.
4. Discussion
In this study, through analyzing the expression of the most commonly used conventional putative CSC markers in GBMs and their correlations with patients' outcomes using three large cohorts of patients' data previously generated and stored in the public databases, we have first identified that CD44 is a highly expressed biomarker in GBM. Upon further analysis, we found that the levels of CD44 and its highly correlated genes, ABCC3 and TNFRSF1A, in GBM tissues, are significantly higher in GBMs than those in normal brain tissues. The levels of these three genes are increased with disease grades and elevated levels of CD44, ABCC3, and TNFRSF1A are associated with patients' shorter survival times. We have also demonstrated that these three genes, combined with MGMT, can be used as a four-gene signature for the prediction of GBM patients' response to anti-GBM therapies. In the evaluation of the selected markers in a panel of GBM cell lines in response to TMZ, salinomycin, and two other natural compounds, we demonstrated that GBM cells expressing higher levels of these four selected markers are less sensitive to traditional chemotherapy regimens, but are sensitive to salinomycin.
Given that the existence of CSC in GBM is one of the major causes of therapy resistance and disease relapse, we also compared all commonly used CSC markers in GBMs. Though many publications indicate that CD133, CD44, SOX2, MYC, and NES are putative CSC markers for GBM, good positive correlations in expression of these markers in GBMs were only observed between CD44 vs. NES and CD44 vs. MYC. Among these markers, CD133 is the most commonly used CSC marker in many in vitro and in vivo tumor models and its expression level was associated with GBM patients' poor survival [55]. In this study, we analyzed the correlation of CD133 with GBM patients' outcomes in three cohorts of GBM datasets and our results are in agreement with our previous conclusions, but the correlation is less significant than that between CD44 with patients' outcomes. Thus, we conclude that CD44 is a better prognostic marker for GBM patients than other CSC markers. Overexpression of EGFR could result in enhancing cell growth, migration, and angiogenesis in GBMs and is associated with a poor outcome [56,57]. We observed significantly higher levels of CD44, ABCC3, TNFRSF1A, and AKT1 expression in the GBM with EGFR amplification, which is consistent with a previously published finding [56,57]. Notably, the biological functions of CD44 in different types of cells are variable depending on the cell type and the expression pattern of the particular variants in certain types of cells, as the existence of multiple CD44 isoforms due to alterative splicing (11 splicing variants are reported so far) is linked with its specific function [[58], [59], [60], [61], [62], [63]]. In this study, we used the common CD44 probe in the arrays that detects all CD44 isoforms in the GBMs. It is not clear whether a specific isoform in GBM plays a dominant role, which warrants further investigation.
By combining the levels of CD44, ABCC3, TNFRSF1A, and MGMT, we found these four genes can be used as an index model for glioblastoma patients' therapy response. Initially, we tested the index model using more markers including CD44, ABCC3, TNFRSF1A, MGMT, AKT1, CDKN2A, and CD133. By comparing the results of using three, four, and five gene indexes for the models, we found that utilizing only three genes (not including MGMT), did not provide sufficient patient classification criteria. We also compared the use of four and five genes and found using five genes (including CD133) did not provide additional benefits than using four genes. The four-gene index was first identified using the 540 cohort dataset by analyzing the correlation of these genes in GBM with all patients' overall survival and the results were validated using two additional datasets, which contained 395 and 284 GBM samples, respectively.
To evaluate the four-gene model, we selected four compounds. TMZ was approved by the US FDA in 1999 to treat GBM patients and is now a widely used drug in GBM clinical treatment. For the other three anticancer compounds, salinomycin, celastrol, and triptolide, although they have not been approved by the FDA for use in human subjects at this time, they showed promising effects on CSC elimination and overcoming therapy resistance and some have already been used in clinical trials [[64], [65], [66]]. Salinomycin was identified from a library of 16,000 natural and commercial chemical compounds based on its highly selective inhibitory effect on breast CSCs with >100-fold greater potency than paclitaxel [67]. As we summarized in our recent publication [64], the extraordinary properties and presumed clinical implications of salinomycin evident in this seminal finding laid the foundation for a flurry of studies conducted thereafter examining salinomycin's anti-cancer effects in various cancer types and model systems. Salinomycin has been used for individual clinical therapy [64,68]. Celastrol and triptolide, which are derived from a traditional Chinese medicine, Tripterygium wilfordii, have been reported to exhibit antitumor activity on GBM in vitro [44,48] and minnelide, a pro-drug of triptolide, has been used in a just completed phase 1 trial against GI cancers and is currently awaiting usage in phase 2 trials. As the four-gene signature we identified from this study is related to CSCs and therapy resistance, we tested them in GBM cell lines. We show that the cell lines with higher levels of CD44, ABCC3, TNFRSF1A, and MGMT are much less sensitive to TMZ treatment and to triptolide and celastrol treatments, but not to salinomycin treatment, which indicates that GBM cells expressing elevated levels of these four genes are of CSC characters. The cell line model also confirmed that the expression of four genes in GBM predicted the treatment response well. Thus, we conclude that these markers may be used to identify subtypes of GBM patients with different outcomes and the expression levels of CD44, ABCC3, and TNFRSF1A in GBMs from patients may serve as potential markers for monitoring the efficacy of GBM therapy. In fact, resistance to TMZ was reported to be associated with the expression of ABCC3 in NK but not CD8+ T cells [69,70]. Recently, Pellegatta et al. also showed that hypermethylation of the MGMT promoter was a clinical feature associated with longer progression free survival and overall survival [69]. Therefore, the four-gene signature has a potential role for guiding clinicians for personalized optimal patient care.
Ethics approval and consent to participate
Ethical approval of our experiments was given by the ethics committee of Baylor Scott and White Health.
Consent for publication
The authors of this paper have consented this study for publication.
Data availability
Data and materials are available upon request, contact at Fengfei.Wang@BSWHealth.org.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
F.W. designed the project, analyzed data, and wrote the manuscript; Z.Z., J.G. participated in data analysis and performed experiments; S.Z., X.S. performed some experiments; D.Q., Fu.W., D.W., B.K., T.S. contributed to data interpretation and writing the manuscript; E.F. contributed data interpretation; E.T.W. conceived the project and wrote the manuscript. J.H.H., E.W. conceived the project, wrote the manuscript, and provided financial and administrative supports.
Acknowledgements
We would like to thank the discussions and inputs from other members of the Wang lab, Wu lab, and Huang lab.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.024.
Contributor Information
Fengfei Wang, Email: Fengfei.Wang@BSWHealth.org.
Jason H. Huang, Email: Jason.huang@BSWHealth.org.
Erxi Wu, Email: Erxi.Wu@BSWHealth.org.
Appendix A. Supplementary data
Supplementary figures
Supplemental Fig. 1. The expression of common CSC markers in GBM cells, neural stem cells, and normal cortex. The data were extracted from the dataset “Cellline Glioblastoma Stemcells - Pollard – 20 in the R2 database” [19] and the statistical differences were evaluated with paired student t-test using JMP software. The boxes represent the interquartile ranges of the analyzed genes' expression levels. The line within each box represents median 2Log expression cut-off value of each gene. The minimum and maximum ranges for each gene expression level were shown as whiskers. P-values < .05 were considered to be statistically significant.
Supplemental Fig. 2. The correlation of CD133 levels in GBM with the overall survival of GBM patients. The pre-normalized data were extracted from the dataset “Tumor Glioblastoma-TCGA-540-MAS5.0-u133a” [33] and 2log ratio values were used. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics with Bonferroni based multiple testing corrections. P-values <.05 were considered to be statistically significant. (a) The correlation of CD133 levels in GBM with the overall survival of all GBM patients. (b, c, d, and e) The correlation of CD133 levels in GBM with the overall survival of subtypes of GBM patients.
Supplemental Fig. 3. The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients. The data were extracted from the datasets “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and “French-284-MAS5.0-u133p2” in the R2 database [10,28,34]. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics. P-values < .05 were considered to be statistically significant. (a, b) The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients using “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” dataset. (c, d) The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients using “French-284-MAS5.0-u133p2” dataset.
Supplemental Fig. 4. CD44 highly correlated genes. The gene levels highly correlated with CD44 in GBM tissues selected and extracted from the datasets “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and “French-284-MAS5.0-u133p2” in the R2 database [10,28,34]. The genes with a correlation factor R > 0.60 were used for the final graph. The genes with lowest p-value were selected as candidate genes. (a, b) The highly correlated genes in GBMs of “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and the figure (b) shows the correlation of genes with CD44 “R” and the significance levels of “p” in GBMs. (c, d) The highly correlated genes in GBMs of “French-284-MAS5.0-u133p2” and the figure (d) shows the correlation of genes with CD44 “R” and the significance levels of “p” in GBMs.
Supplemental Fig. 5. The correlation of four gene expressions in GBM with or without 1p/19q LOH, IDH1 mutation or EGFR mutation with patients' survial. The data were extracted from the dataset “Tumor Glioma - French - 284 - MAS5.0 - u133p2” in the R2 database [10]. The correlation of four gene expression in GBM with or without 1p/19q LOH, IDH1 mutation or EGFR mutation with patients' survival was evaluated. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics. P-values <.05 were considered to be statistically significant.
References
- 1.Louis D.N., Perry A., Reifenberger G. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Wong E.T., Lok E., Swanson K.D. Clinical benefit in recurrent glioblastoma from adjuvant NovoTTF-100A and TCCC after temozolomide and bevacizumab failure: a preliminary observation. Cancer Med. 2015;4(3):383–391. doi: 10.1002/cam4.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong E.T., Lok E., Swanson K.D. Response assessment of NovoTTF-100A versus best physician's choice chemotherapy in recurrent glioblastoma. Cancer Med. 2014;3(3):592–602. doi: 10.1002/cam4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryken T.C., Aygun N., Morris J. The role of imaging in the management of progressive glioblastoma: a systematic review and evidence-based clinical practice guideline. J Neurooncol. 2014;118(3):435–460. doi: 10.1007/s11060-013-1330-0. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y.Y., Gao P., Sun Y., Duan Y.R. Development of targeted therapies in treatment of glioblastoma. Cancer Biol Med. 2015;12(3):223–237. doi: 10.7497/j.issn.2095-3941.2015.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polivka J., Jr., Polivka J., Rohan V., Topolcan O., Ferda J. New molecularly targeted therapies for glioblastoma multiforme. Anticancer Res. 2012;32(7):2935–2946. [PubMed] [Google Scholar]
- 7.Aldape K., Zadeh G., Mansouri S., Reifenberger G., von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 8.Verhaak R.G., Hoadley K.A., Purdom E. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colman H., Aldape K. Molecular predictors in glioblastoma: toward personalized therapy. Arch Neurol. 2008;65(7):877–883. doi: 10.1001/archneur.65.7.877. [DOI] [PubMed] [Google Scholar]
- 10.Gravendeel L.A., Kouwenhoven M.C., Gevaert O. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 11.Bender S., Tang Y., Lindroth A.M. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Molinari C., Iorio P., Medri L. Chromosome 1p and 19q evaluation in low-grade oligodendrogliomas: a descriptive study. Int J Mol Med. 2010;25(1):145–151. [PubMed] [Google Scholar]
- 13.Ohgaki H., Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19(4):764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 14.Nathanson D.A., Gini B., Mottahedeh J. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343(6166):72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown D.V., Filiz G., Daniel P.M. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen D. Tumor formation and drug resistance properties of human glioblastoma side population cells. Mol Med Rep. 2015;11(6):4309–4314. doi: 10.3892/mmr.2015.3279. [DOI] [PubMed] [Google Scholar]
- 17.Jackson M., Hassiotou F., Nowak A. Glioblastoma stem-like cells: at the root of tumor recurrence and a therapeutic target. Carcinogenesis. 2015;36(2):177–185. doi: 10.1093/carcin/bgu243. [DOI] [PubMed] [Google Scholar]
- 18.Yan X., Ma L., Yi D. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A. 2011;108(4):1591–1596. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollard S.M., Yoshikawa K., Clarke I.D. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Klank R.L., Decker Grunke S.A., Bangasser B.L. Biphasic dependence of glioma survival and cell migration on CD44 expression level. Cell Rep. 2017;19(3):668. doi: 10.1016/j.celrep.2017.03.074. [DOI] [PubMed] [Google Scholar]
- 21.Harabin-Slowinska M., Slowinski J., Konecki J., Mrowka R. 36(3) Folia neuropathologica/Association of Polish Neuropathologists and Medical Research Centre, Polish Academy of Sciences; 1998. Expression of adhesion molecule CD44 in metastatic brain tumors; pp. 179–184. [PubMed] [Google Scholar]
- 22.Chetty C., Vanamala S.K., Gondi C.S., Dinh D.H., Gujrati M., Rao J.S. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell Signal. 2012;24(2):549–559. doi: 10.1016/j.cellsig.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Murai T., Miyazaki Y., Nishinakamura H. Engagement of CD44 promotes Rac activation and CD44 cleavage during tumor cell migration. J Biol Chem. 2004;279(6):4541–4550. doi: 10.1074/jbc.M307356200. [DOI] [PubMed] [Google Scholar]
- 24.Anido J., Saez-Borderias A., Gonzalez-Junca A. TGF-beta receptor inhibitors target the CD44(high)/Id1(high) glioma-initiating cell population in human glioblastoma. Cancer Cell. 2010;18(6):655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 25.Brown D.V., Daniel P.M., D'Abaco G.M. Coexpression analysis of CD133 and CD44 identifies proneural and mesenchymal subtypes of glioblastoma multiforme. Oncotarget. 2015;6(8):6267–6280. doi: 10.18632/oncotarget.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka S., Nakada M., Yamada D. Strong therapeutic potential of gamma-secretase inhibitor MRK003 for CD44-high and CD133-low glioblastoma initiating cells. J Neurooncol. 2015;121(2):239–250. doi: 10.1007/s11060-014-1630-z. [DOI] [PubMed] [Google Scholar]
- 27.Berchtold N.C., Cribbs D.H., Coleman P.D. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105(40):15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paugh B.S., Qu C., Jones C. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L., Hui A.M., Su Q. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Patel A.P., Tirosh I., Trombetta J.J. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips H.S., Kharbanda S., Chen R. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Lee Y., Scheck A.C., Cloughesy T.F. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brennan C.W., Verhaak R.G., McKenna A. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erdem-Eraslan L., Gravendeel L.A., de Rooi J. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F., Remke M., Bhat K. A microRNA-1280/JAG2 network comprises a novel biological target in high-risk medulloblastoma. Oncotarget. 2015;6(5):2709–2724. doi: 10.18632/oncotarget.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahller Y.Y., Williams J.P., Baird W.H. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One. 2009;4(1) doi: 10.1371/journal.pone.0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Harrabi S., Combs S.E., Brons S., Haberer T., Debus J., Weber K.J. Temozolomide in combination with carbon ion or photon irradiation in glioblastoma multiforme cell lines - does scheduling matter? Int J Radiat Biol. 2013;89(9):692–697. doi: 10.3109/09553002.2013.791406. [DOI] [PubMed] [Google Scholar]
- 39.Kanzawa T., Germano I.M., Kondo Y., Ito H., Kyo S., Kondo S. Inhibition of telomerase activity in malignant glioma cells correlates with their sensitivity to temozolomide. Br J Cancer. 2003;89(5):922–929. doi: 10.1038/sj.bjc.6601193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng Y., Li Y., Ma C. Arsenic trioxide inhibits glioma cell growth through induction of telomerase displacement and telomere dysfunction. Oncotarget. 2016;7(11):12682–12692. doi: 10.18632/oncotarget.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen D., Song M., Mohamad O., Yu S.P. Inhibition of Na+/K+-ATPase induces hybrid cell death and enhanced sensitivity to chemotherapy in human glioblastoma cells. BMC Cancer. 2014;14:716. doi: 10.1186/1471-2407-14-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang N., Yan T., Zhu H. A co-culture model with brain tumor-specific bioluminescence demonstrates astrocyte-induced drug resistance in glioblastoma. J Transl Med. 2014;12:278. doi: 10.1186/s12967-014-0278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maysinger D., Moquin A., Choi J., Kodiha M., Stochaj U. Gold nanourchins and celastrol reorganize the nucleo- and cytoskeleton of glioblastoma cells. Nanoscale. 2018;10(4):1716–1726. doi: 10.1039/c7nr07833a. [DOI] [PubMed] [Google Scholar]
- 44.Boridy S., Le P.U., Petrecca K., Maysinger D. Celastrol targets proteostasis and acts synergistically with a heat-shock protein 90 inhibitor to kill human glioblastoma cells. Cell Death Dis. 2014;5:e1216. doi: 10.1038/cddis.2014.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J., Chen L.Y., Lin Z.X., Zhao M.L. The effect of triptolide on apoptosis of glioblastoma multiforme (GBM) cells. J Int Med Res. 2007;35(5):637–643. doi: 10.1177/147323000703500508. [DOI] [PubMed] [Google Scholar]
- 46.Lin J., Chen L., Lin Z., Zhao M. Inhibitory effect of triptolide on glioblastoma multiforme in vitro. J Int Med Res. 2007;35(4):490–496. doi: 10.1177/147323000703500408. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y.X., Huang Y.L. Antiangiogenic effect of celastrol on the growth of human glioma: an in vitro and in vivo study. Chin Med J (Engl) 2009;122(14):1666–1673. [PubMed] [Google Scholar]
- 48.Zhang H., Zhu W., Su X. Triptolide inhibits proliferation and invasion of malignant glioma cells. J Neurooncol. 2012;109(1):53–62. doi: 10.1007/s11060-012-0885-5. [DOI] [PubMed] [Google Scholar]
- 49.Chen T., Yi L., Li F. Salinomycin inhibits the tumor growth of glioma stem cells by selectively suppressing glioma-initiating cells. Mol Med Rep. 2015;11(4):2407–2412. doi: 10.3892/mmr.2014.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S., Wang F., Zhang Y. Salinomycin suppresses PDGFRbeta, MYC, and notch signaling in human medulloblastoma. Austin J Pharmacol Ther. 2014;2(3) [PMC free article] [PubMed] [Google Scholar]
- 51.Xipell E., Gonzalez-Huarriz M., Martinez De Irujo J.J. Salinomycin induced ROS results in abortive autophagy and leads to regulated necrosis in glioblastoma. Oncotarget. 2016;7(21):30626–30641. doi: 10.18632/oncotarget.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tigli Aydin R.S., Kaynak G., Gumusderelioglu M. Salinomycin encapsulated nanoparticles as a targeting vehicle for glioblastoma cells. J Biomed Mater Res A. 2016;104(2):455–464. doi: 10.1002/jbm.a.35591. [DOI] [PubMed] [Google Scholar]
- 53.Olmez I., Shen W., McDonald H., Ozpolat B. Dedifferentiation of patient-derived glioblastoma multiforme cell lines results in a cancer stem cell-like state with mitogen-independent growth. J Cell Mol Med. 2015;19(6):1262–1272. doi: 10.1111/jcmm.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calzolari A., Saulle E., De Angelis M.L. Salinomycin potentiates the cytotoxic effects of TRAIL on glioblastoma cell lines. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0094438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dahlrot R.H., Hansen S., Jensen S.S., Schroder H.D., Hjelmborg J., Kristensen B.W. Clinical value of CD133 and nestin in patients with glioma: a population-based study. Int J Clin Exp Pathol. 2014;7(7):3739–3751. [PMC free article] [PubMed] [Google Scholar]
- 56.Tortora G., Bianco R., Daniele G. Overcoming resistance to molecularly targeted anticancer therapies: Rational drug combinations based on EGFR and MAPK inhibition for solid tumours and haematologic malignancies. Drug Resist Updates. 2007;10(3):81–100. doi: 10.1016/j.drup.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinojima N., Tada K., Shiraishi S. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63(20):6962–6970. [PubMed] [Google Scholar]
- 58.Friedrichs K., Franke F., Lisboa B.W. CD44 isoforms correlate with cellular differentiation but not with prognosis in human breast cancer. Cancer Res. 1995;55(22):5424–5433. [PubMed] [Google Scholar]
- 59.Gotte M., Yip G.W. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006;66(21):10233–10237. doi: 10.1158/0008-5472.CAN-06-1464. [DOI] [PubMed] [Google Scholar]
- 60.Lopez J.I., Camenisch T.D., Stevens M.V., Sands B.J., McDonald J., Schroeder J.A. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005;65(15):6755–6763. doi: 10.1158/0008-5472.CAN-05-0863. [DOI] [PubMed] [Google Scholar]
- 61.Olsson E., Honeth G., Bendahl P.O. CD44 isoforms are heterogeneously expressed in breast cancer and correlate with tumor subtypes and cancer stem cell markers. BMC Cancer. 2011;11:418. doi: 10.1186/1471-2407-11-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S.J., Wong G., de Heer A.M., Xia W., Bourguignon L.Y. CD44 variant isoforms in head and neck squamous cell carcinoma progression. Laryngoscope. 2009;119(8):1518–1530. doi: 10.1002/lary.20506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Louderbough J.M., Schroeder J.A. Understanding the dual nature of CD44 in breast cancer progression. Mol Cancer Res. 2011;9(12):1573–1586. doi: 10.1158/1541-7786.MCR-11-0156. [DOI] [PubMed] [Google Scholar]
- 64.Zhou S., Wang F., Wong E.T. Salinomycin: a novel anti-cancer agent with known anti-coccidial activities. Curr Med Chem. 2013;20(33):4095–4101. doi: 10.2174/15672050113109990199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu H., Yang W., He L.J. Upregulating Noxa by ER stress, celastrol exerts synergistic anti-cancer activity in combination with ABT-737 in human hepatocellular carcinoma cells. PLoS One. 2012;7(12) doi: 10.1371/journal.pone.0052333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGinn O., Gupta V.K., Dauer P. Inhibition of hypoxic response decreases stemness and reduces tumorigenic signaling due to impaired assembly of HIF1 transcription complex in pancreatic cancer. Sci Rep. 2017;7(1):7872. doi: 10.1038/s41598-017-08447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gupta P.B., Onder T.T., Jiang G. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naujokat C., Steinhart R. Salinomycin as a drug for targeting human cancer stem cells. J Biomed Biotechnol. 2012;2012 doi: 10.1155/2012/950658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pellegatta S., Eoli M., Cuccarini V. Survival gain in glioblastoma patients treated with dendritic cell immunotherapy is associated with increased NK but not CD8(+) T cell activation in the presence of adjuvant temozolomide. Oncoimmunology. 2018;7(4) doi: 10.1080/2162402X.2017.1412901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pessina S., Cantini G., Kapetis D. The multidrug-resistance transporter Abcc3 protects NK cells from chemotherapy in a murine model of malignant glioma. Oncoimmunology. 2016;5(5) doi: 10.1080/2162402X.2015.1108513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Y.C., Hueng D.Y., Huang H.Y., Chen J.Y., Chen Y. Magnolol and honokiol exert a synergistic anti-tumor effect through autophagy and apoptosis in human glioblastomas. Oncotarget. 2016;7(20):29116–29130. doi: 10.18632/oncotarget.8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures
Supplemental Fig. 1. The expression of common CSC markers in GBM cells, neural stem cells, and normal cortex. The data were extracted from the dataset “Cellline Glioblastoma Stemcells - Pollard – 20 in the R2 database” [19] and the statistical differences were evaluated with paired student t-test using JMP software. The boxes represent the interquartile ranges of the analyzed genes' expression levels. The line within each box represents median 2Log expression cut-off value of each gene. The minimum and maximum ranges for each gene expression level were shown as whiskers. P-values < .05 were considered to be statistically significant.
Supplemental Fig. 2. The correlation of CD133 levels in GBM with the overall survival of GBM patients. The pre-normalized data were extracted from the dataset “Tumor Glioblastoma-TCGA-540-MAS5.0-u133a” [33] and 2log ratio values were used. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics with Bonferroni based multiple testing corrections. P-values <.05 were considered to be statistically significant. (a) The correlation of CD133 levels in GBM with the overall survival of all GBM patients. (b, c, d, and e) The correlation of CD133 levels in GBM with the overall survival of subtypes of GBM patients.
Supplemental Fig. 3. The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients. The data were extracted from the datasets “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and “French-284-MAS5.0-u133p2” in the R2 database [10,28,34]. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics. P-values < .05 were considered to be statistically significant. (a, b) The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients using “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” dataset. (c, d) The correlation of CD133 and CD44 levels in GBM with the overall survival of GBM patients using “French-284-MAS5.0-u133p2” dataset.
Supplemental Fig. 4. CD44 highly correlated genes. The gene levels highly correlated with CD44 in GBM tissues selected and extracted from the datasets “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and “French-284-MAS5.0-u133p2” in the R2 database [10,28,34]. The genes with a correlation factor R > 0.60 were used for the final graph. The genes with lowest p-value were selected as candidate genes. (a, b) The highly correlated genes in GBMs of “Tumor Glioblastoma-TCGA-395-MAS5.0-u1331” and the figure (b) shows the correlation of genes with CD44 “R” and the significance levels of “p” in GBMs. (c, d) The highly correlated genes in GBMs of “French-284-MAS5.0-u133p2” and the figure (d) shows the correlation of genes with CD44 “R” and the significance levels of “p” in GBMs.
Supplemental Fig. 5. The correlation of four gene expressions in GBM with or without 1p/19q LOH, IDH1 mutation or EGFR mutation with patients' survial. The data were extracted from the dataset “Tumor Glioma - French - 284 - MAS5.0 - u133p2” in the R2 database [10]. The correlation of four gene expression in GBM with or without 1p/19q LOH, IDH1 mutation or EGFR mutation with patients' survival was evaluated. Survival time was measured from the time of initial diagnosis to the date of death or the date of last follow up. The survival distribution was estimated according to the Kaplan-Meier method using optimal cut-off selection and log-rank statistics. P-values <.05 were considered to be statistically significant.
Data Availability Statement
Data and materials are available upon request, contact at Fengfei.Wang@BSWHealth.org.