Abstract
Intervertebral disc (IVD) degeneration and trauma is a major socio‐economic burden and the focus of cell‐based regenerative medicine approaches. Despite numerous ongoing clinical trials attempting to replace ailing IVD cells with mesenchymal stem cells, a solid understanding of the identity and nature of cells in a healthy mature IVD is still in need of refinement. Although anatomically simple, the IVD is comprised of heterogeneous cell populations. Therefore, methods involving cell pooling for RNA profiling could be misleading. Here, by using RNA in situ hybridization and z proportion test, we have identified potential novel biomarkers through single cell assessment. We quantified the proportion of RNA transcribing cells for 50 genetic loci in the outer annulus fibrosus (AF) and nucleus pulposus (NP) in coccygeal bovine discs isolated from tails of four skeletally mature animals. Our data reconfirm existing data and suggest 10 novel markers such as Lam1 and Thy1 in the outer AF and Gli1, Gli3, Noto, Scx, Ptprc, Sox2, Zscan10 and LOC101904175 in the NP, including pluripotency markers, that indicate stemness potential of IVD cells. These markers could be added to existing biomarker panels for cell type characterization. Furthermore, our data once more demonstrate heterogeneity in cells of the AF and NP, indicating the need for single cell assessment by methods such as RNA in situ hybridization. Our work refines the molecular identity of outer AF and NP cells, which can benefit future regenerative medicine and tissue engineering strategies in humans.
Keywords: adult, annulus fibrosus, bovine, heterogeneity, IVD, nucleus pulposus, regenerative medicine, RNA in situ hybridization
Introduction
Degeneration of the intervertebral disc (IVD) is frequently associated with severe and chronic low back pain (LBP), one of today's most prevalent musculoskeletal problems (Cheung et al. 2009; Waterman et al. 2012). Annual expenditures related to medical healthcare and lost workdays due to severe and chronic LBP in the US typically exceed the combined costs of common ailments such as coronary artery disease or stroke, therefore imposing an enormous socio‐economic burden (Katz, 2002).
The mature healthy IVD is situated between the vertebrate bodies of the vertebral column and is composed of anatomically distinct areas of different composition: A hydrogel‐like central nucleus pulposus (NP) is encapsulated in the outer ligamentous annulus fibrosus (AF) and sandwiched between the cartilaginous endplates (Eyre, 1979; Bayliss et al. 1988; Humzah & Soames, 1988; Oegema, 1993; Bedore et al. 2014; Erwin & Hood, 2014). In the human and bovine IVD, cells are of thin and elongated nature in the outer AF and round in the NP (Errington et al. 1998; Kraus et al. 2017; Fig. 1). Both longitudinal and round cells were present in the bovine inner AF (TZ) using Mallory's tetrachrome staining (Kraus et al. 2017; and data not shown) with round cells being more prevalent (Errington et al. 1998; Kraus et al. 2017). Despite a relatively simple anatomy, the IVD is a unique and challenging organ in many ways: hypoxic, slightly acidic and nutrient‐deficient (Urban et al. 1977, 2004; Antoniou et al. 1996; Wuertz et al. 2008; Liang et al. 2012), where cells are sparse in a vast amount of extracellular matrix (ECM) (Errington et al. 1998; Kraus et al. 2017; Lama et al. 2018). Regenerative medicine aims to restore the function of compromised tissues or entire organs via cell‐based approaches, and clinical trials employing mesenchymal stem cells (MSC) to treat disc degeneration are on their way (Sivakamasundari & Lufkin, 2013; Sakai & Andersson, 2015; Pennicooke et al. 2016; Kraus & Lufkin, 2017). However, the harsh environment in the avascular mature IVD could limit the density of viable cells and impact on the ability of NP cells to produce sufficient glycosaminoglycans (GAG) (Urban et al. 1977; Bibby & Urban, 2004; Wuertz et al. 2008; Grunhagen et al. 2011; Liang et al. 2012), so is unclear whether introduced MSC for therapeutic purposes can initiate sufficient de novo ECM production to restore proper function of degenerated IVDs (Sakai & Schol, 2017). A gradual change in ECM synthesis and composition from outer AF to central NP was demonstrated through immunohistochemistry in human IVDs for abundant structural macromolecules such as Collagen I, II and GAG (Eyre & Muir, 1976; Bushell et al. 1977; Antoniou et al. 1996). Type I collagen is prominent in the AF, whereas type II collagen is mostly associated with the NP (Antoniou et al. 1996). Proteoglycans, particularly those of the long and modular type, are important components of the ECM in general, but in particular the negatively charged, large, aggregating bottlebrush proteoglycan Aggrecan is considered a key player in providing the swelling capacity that pulls water into the disc against compressive loads (Bibby et al. 2001; Singh et al. 2009). Currently, a solid understanding of the molecular identity of mature IVD cells is lacking and a heterogeneous cell population in vivo is suspected (Chelberg et al. 1995; Errington et al. 1998; Alini et al. 2008; Gilson et al. 2010; Pattappa et al. 2012; Lee et al. 2015; Molinos et al. 2015; Risbud et al. 2015; Sakai & Andersson, 2015; Moriguchi et al. 2016; Thorpe et al. 2016; Turner et al. 2016; Kraus et al. 2017). Identifying NP and AF biomarkers is an ongoing quest in the field and crucial to assure quality control measures for cultured cells destined for regenerative treatment (Risbud et al. 2015; Thorpe et al. 2016; van den Akker et al. 2017; Kraus et al. 2017). Active transcription of genes encoding ECM molecules could point to differences between AF and NP cells. Other genes encoding structural molecules investigated in this context are summarized in Table 1. Of those, three members of the Keratin family: Krt8, Krt18, and Krt19, although typically associated with epithelial cells, are frequently discussed as IVD biomarkers (Minogue et al. 2010b; Rodrigues‐Pinto et al. 2016; Richardson et al. 2017). We also investigated whether the expression of crucial signalling and transcription factors during early patterning of the axial skeleton and IVD persists in the mature disc for the purpose of tissue maintenance (for details see Table 1). As a progenitor cell potential of IVD cells has been suggested previously (Risbud et al. 2007, 2015; Henriksson et al. 2009; Tekari et al. 2016; Thorpe et al. 2016; Kraus et al. 2017; Liu et al. 2017) we included several pluripotency and stemness markers in our analysis of outer AF and NP cells (for details see Table 1). Given the hypoxic environment cells encounter in the avascular mature IVD and that anaerobic lactic acid formation creates acidic conditions in the IVD niche (Wuertz et al. 2008; Grunhagen et al. 2011; Liang et al. 2012), in a broader metabolic context we also investigated the expression of genes encoding catabolic enzymes and those involved in pH balance, along with the expression of genes encoding proteins otherwise considered relevant to the IVD or vertebral column development. Cell proliferation potential was assessed through Ki67 transcripts. Also analyzed was the novel lncRNA LOC101904175, the homologue of murine Klhl14as that was recently identified in the developing IVD through transcriptome profiling of cells in the axial skeleton of Pax1/Pax9 mutant mouse embryos (Sivakamasundari et al. 2017; Kraus et al. 2018a; see Table 1).
Figure 1.
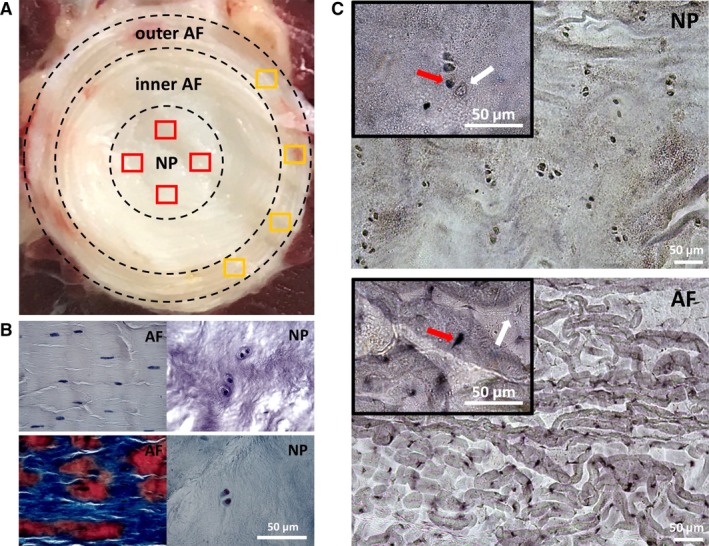
Illustration of the principle of data collection (a) indicating the random selection of 20 non‐overlapping fields in the outer AF area (oAF) as indicated by four examples of yellow frames and the NP area as indicated by four examples of red frames on a mature bovine coccygeal IVD. Haematoxylin staining (b, top) and Mallory's tetrachrome staining (b, bottom) was performed to identify cells and tissue types. (c) AP‐RISH identification of cells transcribing (red arrow) or not transcribing (white arrow) a particular genetic locus, as shown here for the example of Col1a1 expression, can be observed within the same tissue on a section. Images are shown at 10× and 40× (insert) magnification. Scale bar: 50 μm.
Table 1.
List of genetic loci investigated using AP‐RISH for transcription in outer AF and NP cells in skeletally mature bovine IVDs
| Functional category | Genetic locus | Gene symbol | Function | References |
|---|---|---|---|---|
| Structural proteins | Aggrecan | Acan |
Major component of cartilage and IVD ECM Provides shock absorbing function of the NP |
Watanabe et al. (1994), Day et al. (2004), Le Maitre et al. (2007) |
| Biglycan | Bgn |
Small leucine‐rich proteoglycan (SLRP) Role in ECM assembly |
Schonherr et al. (1995), Fisher et al. (1989), Wilda et al. (2000), Marfia et al. (2014) | |
| Chondromodulin‐1 | Chm1 | Glycoprotein with anti‐angiogenesis properties | Hiraki & Shukunami (2000), Hiraki et al. (1991) | |
| Collagen Ia1 | Col1a1 | α1(I) chains of Collagen I heterotrimer | Pereira et al. (1993), Khillan et al. (1994), Aszodi et al. (1998) | |
| Collagen Ia2 | Col1a2 | α2(I) chain of Collagen I heterotrimer | Aszodi et al. (1998), Le Maitre et al. (2007) | |
| Collagen IIa1 | Col2a1 | α1(II) chains of Collagen II homotrimer | Vandenberg et al. (1991), Garofalo et al. (1991), Karsenty & Park (1995) | |
| Decorin | Dcn |
Small leucine‐rich proteoglycan (SLRP) Role in ECM assembly |
Iozzo et al. (1999), Wilda et al. (2000) | |
| Fibromodulin | Fmod |
Small leucine‐rich proteoglycan (SLRP) Role in ECM assembly |
Wilda et al. (2000), Jan et al. (2016) | |
| Heparan sulphate proteoglycan 2 | Hspg2 |
Role in IVD development Structural similarity to Laminin α |
Noonan et al. (1991), Sasaki et al. (1988), Melrose et al. (2001) | |
| Keratin 8 | Krt8 | Intermediate filament proteins | Bader et al. (1988), Moll et al. (2008) | |
| Keratin 18 | Krt18 | Intermediate filament proteins | ||
| Keratin 19 | Krt19 | Intermediate filament proteins | ||
| Laminin1 | Lam1 |
Glycoprotein in basal lamina ECM Interacts with collagens, integrins and proteoglycans |
Ekblom et al. (1998) | |
| Talin1 | Tln1 | Connects cells to the ECM | Critchley & Gingras (2008) | |
| Tenomodulin | Tnmd |
ChM1 related transmembrane glycoprotein Tendon and tendon progenitor cell marker |
Shukunami et al. (2001) | |
| Tenascin XB | Tnxb |
Glycoprotein with anti‐adhesive properties Mutations associated with Ehlers Danlos Syndrome |
Chiquet‐Ehrismann & Tucker (2011), Burch et al. (1997), Mao et al. (2002) | |
| Transcription factors | Forkhead box F1 | Foxf1 |
Required for the differentiation of the lateral plate mesoderm in mouse Proposed as NP specific in humans |
Mahlapuu et al. (2001), Minogue et al. (2010b), Thorpe et al. (2016) |
| Glioma‐associated oncogene 1 | Gli1 | Downstream mediator of Shh and Ihh signaling | Ahn & Joyner (2005), Buttitta et al. (2003) | |
| Glioma‐associated oncogene 3 | Gli3 |
Downstream mediator of Shh and Ihh signaling Impact on Pax1, Pax9, and Sox9 expression |
Buttitta et al. (2003), Shin et al. (1999) | |
| Myoblast determination protein 1 | MyoD |
Early differentiation marker for myogenic commitment Serves as marker of the non‐chondrogenic lineage |
Rudnicki et al. (1993) | |
| Notochord | Noto |
Involved in early notochord development Acts downstream of brachyury |
Abdelkhalek et al. (2004), McCann et al. (2012) | |
| Paired box protein 1 | Pax1 | Synergistically regulate the development of the vertebral column in mice | Peters et al. (1999), Sivakamasundari et al. (2017) | |
| Paired box protein 9 | Pax9 | |||
| Scleraxis | Scx |
In connective tissues like tendons and ligaments Involved in regulating Tnmd expression Implicated in skeletogenesis during mouse embryonic development |
Cserjesi et al. (1995), Shukunami et al. (2006, 2018), Schweitzer et al. (2001) | |
| Sex determining region Y‐box 5 | Sox5 |
Crucial roles in organogenesis Key regulators of the chondrogenic pathway |
Lefebvre et al. (2001), Lee et al. (2017), Smits et al. (2004), Barrionuevo et al. (2006), Ikeda et al. (2004), Chatterjee et al. (2014), Zhang et al. (2006) | |
| Sex determining region Y‐box 6 | Sox6 | |||
| Sex determining region Y‐box 9 | Sox9 | |||
| Brachyury | T |
Conserved function in bilateral animals Regulates notochord formation Biomarker for spine tumors (chordomas) |
Tang et al. (2012), Nibu et al. (2013), Herrmann et al. (1990), Vujovic et al. (2006) | |
| Signalling factors | Bone morphogenetic protein 4 | Bmp4 |
Involved in bone and cartilage development Belongs to TGF‐beta superfamily |
Nifuji et al. (1997), Wijgerde et al. (2005), Zhang et al. (2006) |
| Growth differentiation factor 5 | Gdf5 | Related to BMP and TGF‐beta superfamily | Storm et al. (1994), Francis‐West et al. (1999) | |
| Indian hedgehog | Ihh | Involved in axial and appendicular skeleton development | Vortkamp et al. (1996), St‐Jacques et al. (1999), Ingham & McMahon (2001), Maeda et al. (2007) | |
| Sonic hedgehog | Shh |
Linked to Bmp4 signaling Crucial in axial and appendicular skeleton development Absence results in aberrant vertebral column and NP development |
DiPaola et al. (2005), Dahia et al. (2012), Chiang et al. (1996), Kraus et al. (2001), Ahn & Joyner (2005), Ingham & McMahon (2001), Choi et al. (2012) | |
| Pluripotency and stem cell markers | Endoglin | Eng |
Cell surface marker Part of a marker panel defining multipotent mesenchymal stromal cells |
Dominici et al. (2006) |
| Estrogen‐related receptor beta | Esrrb |
Direct Nanog target Fibroblasts reprogramming factor |
Feng et al. (2009), Festuccia et al. (2012), Doege et al. (2012) | |
| Nanog | Nanog |
Guardian of pluripotency Levels correlate with the self‐renewal potential of stem cells |
Mitsui et al. (2003), Chambers et al. (2007) | |
| Octamer‐binding transcription factor 4 | Oct4 |
Essential for the pluripotency self‐renewal capacity of stem cells Fibroblasts reprogramming factor |
Nichols et al. (1998), Niwa et al. (2009) | |
| Tyrosine phosphate receptor type C | Ptprc |
Cell surface marker Part of a marker panel defining multipotent mesenchymal stromal cells |
Dominici et al. (2006) | |
| Sex determining region Y‐box 2 | Sox2 |
Essential for the pluripotency and self‐renewal capacity of stem cells Fibroblasts reprogramming factor |
Niwa et al. (2009), Takahashi & Yamanaka (2006) | |
| Thymocyte differentiation antigen 1 | Thy1 |
Cell surface marker Part of a marker panel defining multipotent mesenchymal stromal cells |
Dominici et al. (2006) | |
| Zinc finger and SCAN domain containing 10 | Zscan10 | Associated with progenitor cell subpopulations or impact on their fate choice decisions in mouse | Wang et al. (2007), Kraus et al. (2014) | |
| Metabolic context | Carbonic anhydrase 12 | Ca12 |
Hypoxia induced enzyme Involved in pH balance Suggested as NP specific |
Chiche et al. (2009), Power et al. (2011), Minogue et al. (2010b) |
| Glyceraldehyde 3‐phosphate dehydrogenase | Gapdh | Catabolic enzyme in glycolysis | Seidler (2013), Lopa et al. (2016) | |
| Hypoxia‐inducible factor 1‐alpha | Hif1α |
Hypoxia induced transcription factor Loss of function in mouse resulted in morphological abnormalities of the NP |
Wuertz et al. (2008), Merceron et al. (2014) | |
| Lactate dehydrogenase A | LdhA | Catabolic enzyme involved in anaerobic energy production | Sudo et al. (1992a) | |
| Lactate dehydrogenase B | LdhB | Catabolic enzyme involved in anaerobic energy production | Sudo et al. (1992b) | |
| Malate dehydrogenase 2 | Mdh2 | Catabolic enzyme in the citric acid cycle | Bell et al. (2001) | |
| Others | Annexin A4 | Anxa4 |
Regulates ion channel activity Modulates the mobility of membrane proteins |
Piljic & Schultz (2006) |
| Ki67 | Ki67 |
Proliferation marker found throughout the active cell cycle Immune positive cell clusters in degenerated disc |
Johnson et al. (2001), Li et al. (2015) | |
| lnc RNA LOC101904175 | LOC101904175 |
Long non‐coding RNA Orthologue of murine Klhl14as Downregulated in axial skeleton of Pax1/Pax9 mutant mouse embryos |
Sivakamasundari et al. (2017), Kraus et al. (2018a) | |
| Synaptosomal‐associated protein 25 | Snap25 |
Neuron‐specific in mouse hippocampus Functions in docking and membrane fusion of synaptic vesicles Suggested as NP marker |
Zhao et al. (1994), Minogue et al. (2010a) |
We provide quantitative values for the proportion of cells expressing respective mRNAs in the bovine outer AF and NP. Our data clearly demonstrate cellular heterogeneity in the IVD, a finding obscured in quantitative expression profiling such as microarray analysis or qRT‐PCR that relies on cell pooling for mRNA extraction. It is of concern when cell pooling is applied, that in a heterogeneous cell population, such as the AF and NP, non‐transcribing cells could be masked by a few individual cells with high expression levels. This indicates the need for including single cell assessment by methods such as RISH. As such our work aids in refining AF and NP biomarkers in the adult bovine IVD with possible implications for future regenerative medicine and tissue engineering studies in humans.
Materials and methods
Tissue collection and processing
Four tails from skeletally mature adult cows were collected fresh on ice from local abattoirs and immediately transported for dissection. Skin and most skeletal muscle was removed (Fig. 1a). For RISH, typically coccygeal discs three to seven were isolated leaving the endplates behind and immediately fixed in > 5× volume of fresh cold 4% (w/v) paraformaldehyde (PFA) for 24 h. Intervertebral discs were then slowly dehydrated in graded ethanol (EtOH) baths of 30%, 70%, 90% EtOH in nuclease‐free water, followed by 2× 100% EtOH, 1× equal volume EtOH:HistoChoice™ and 3× 100% HistoChoice™ prior to paraffin embedding (Wang et al. 2000; Kraus et al. 2017). For RISH and histological analysis, 7‐μm cross‐sections were cut on a rotary microtome and up to three consecutive sections were mounted on VistaVision HistoBond (VWR) glass slides (Kraus & Lufkin, 1999; Kraus et al. 2017). All procedures were performed according to the ethical standards of Clarkson University. No live animals or human material was included in this study.
Histological tissue assessment
The 7 μm paraffin sections were de‐waxed in 3× 100% HistoChoice™ and slowly rehydrated in graded EtOH baths, essentially reversing the steps above (Robledo & Lufkin, 2006). Haematoxylin stain (VWR; Fig. 1b, top) or Mallory's Tetrachrome stain (Fig. 1b, bottom) containing Groat's haematoxylin, Acid Fuchsin, Aniline Blue and Orange G (Kraus et al. 2017) was adapted from Lufkin et al. (1992) and used to differentiate outer AF from inner AF [or transition zone (TZ); Kraus et al. 2017] and NP tissue as indicated (Fig. 1a). In the IVDs used for RISH, vasculature was only observed in the outer periphery of the outer AF.
RNA in situ hybridization on paraffin sections (RISH)
Fifty RNA probe templates were PCR‐amplified from bovine genomic DNA isolated from the skeletal muscle of the collected tails. Gene specific primers were designed with NCBI software (Supporting Information Table S1). As sense and antisense probes, modified T3 (5′‐ CCGAATTC_T3‐3′) or T7 (5′‐ CCAAGCTT_T7 ‐3′) promoter sequences were added to the 5′ end of the forward or reverse base primer, respectively (Kraus et al. 2018b). For cell counts, probes were labelled with digoxygenin (DIG)‐UTP and detected with Sheep anti‐DIG‐AP Fab fragments (Roche; Kraus et al. 2018b). Hybridization was carried out 62 °C and washes were performed in slide mailing jars with buffers as described in Kraus et al. (2017). Chromogenic signal detection was performed with NBT/BCIP (Roche; Kraus et al. 2018b). The colour was developed by adding nitro blue tetrazolium (NBT) and 5‐bromo‐4‐chloro‐3‐indolylphosphate (BCIP) (Roche) substrate to the sections. Stained (red arrows in Fig. 1C) and unstained cells (white arrows in Fig. 1C) within each section as well as adjacent tissues served as positive/negative controls for each probe. This approach is hereafter referred to as AP‐RISH. To validate our AP‐RISH approach of gene expression analysis through cell count and z proportion test, RNA expression was further quantified through the acquisition of single‐cell fluorophore‐labelled expression intensities and confocal microscopy for two genes: Col2a1, a widely accepted NP marker, and LOC101904175, a novel lnc RNA, both present in the mature IVD according to our AP‐RISH data. Here mouse anti‐digoxin (1 : 100, Jackson IR) followed by Alexa Fluor 488‐conjugated AffiniPure goat anti‐mouse (1 : 1000, Jackson IR) antibodies were used instead of chromogenic signal development, and To‐Pro‐3 (1 : 1000, Thermo Fisher) marked the nucleus. This approach is hereafter referred to as FL‐RISH.
Data collection and statistical analysis
We provide AP‐RISH in situ transcription data for 50 genomic loci (Table 1), focusing on cells in the NP and outer AF (Fig. 1a), omitting the inner AF (or TZ) (Fig. 1a) and see also (Kraus et al. 2017) to allow for a clear distinction between the two tissue types analyzed. To compare the number of cells transcribing a gene (thereafter denoted as positive cells) and the number of cells without noticeable transcription (thereafter denoted as negative cells), for each of the 50 analyzed genes, 20 non‐overlapping frames (n = 20) from three independent IVD sections (n = 3) were selected for cell counts in the outer AF and NP area post AP‐RISH under 40× magnification using a Motic BA310 compound scope (Fig. 1a). These IVD areas should be similar to the outer AF and NP tissue subjected to a study by van den Akker et al. (2017: fig. 1) and close to the study by Minogue et al. (2010b) where the tissue was described as discs macroscopically dissected into AF and NP, removing any transition zone. All counting was performed by the same individual to avoid inter‐rater variability. The percentage of positive cells was calculated and graphed with GraphPad prism 5. To avoid inflating the statistical significance of our results, we used averaging techniques to aggregate across the three replicates. For each gene and tissue type, an average proportion value was estimated from the data collected across the frames analyzed for each gene. This average was computed as the ratio of average number of positive cells to the average number of cells counted for each gene and tissue type (% positive cells). The z proportion test for differences between two population proportions was applied to determine significant difference in the proportion of positive cells between outer AF and NP for each gene when the normality assumption held. Fisher's exact test was used otherwise. A confidence level of α = 0.05 was selected for all test conducted. Differences were considered significant for P < 0.05. The statistical power indicates the probability of a statistically significant effect (power of 1 = 100%). The NP/AF ratio was generated and compared with previously reported IVD biomarkers (Table 2). For assay validation by FL‐RISH, fluorescent signal intensities representing gene transcription were captured for cells in the outer AF and NP with a Leica DMi8 confocal microscope. The established NP biomarker Col2a1 and a novel locus LOC10190417 were chosen as examples. The data were averaged per cell in the respective tissue using imagej and graphed with GraphPad prism 5. Student's t‐test was applied to assess the significant difference of the mean intensities.
Table 2.
RISH data of all investigated genetic loci organized by functional categories including mean values (standard errors), statistical power, P‐values and relative gene expression ratios of NP/AF (fold changes)
| Functional Category | Genetic locus | Symbol | RISH data | Gene expression ratio (NP/AF) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total NP/AF cell count | positive cells in the NP in % (Mean SE) | positive cells in the AF in % (Mean SE) | P‐value | Power | RNA SISH | qPCR | Micro array | |||
| Structural proteins | Aggrecan | Acan | 151/143 | 89.6 (4.3) | 31.4 (6.7) | 3.8E‐09 | 1.0 | 2.9 |
3.2** n.s.* |
>10* |
| Biglycan | Bgn | 140/121 | 32.0 (6.8) | 23.2 (6.6) | 0.35 | 0.15 | 1.4 | |||
| Chondromodulin‐1 | Chm1 | 136/159 | 36.1 (7.1) | 33.6 (6.5) | 0.82 | 0.042 | 1.1 | |||
| Collagen Ia1 | Col1a1 | 150/204 | 41.6 (7.0) | 84.8 (4.4) | 3.3E‐07 | 1.0 | 0.5 | <0.1** | <0.1* | |
| Collagen Ia2 | Col1a2 | 148/148 | 66.3 (6.7) | 5.1 (3.1) | 1.6E‐10 | 1.0 | 13.0 | <0.1* | ||
| Collagen IIa1 | Col2a1 | 226/181 | 88.7 (3.7) | 21.8 (5.3) | 7.6E‐16 | 1.0 | 4.1 |
7.1** n.s.* |
0.3* | |
| Decorin | Dcn | 223/224 | 1.9 (2.0) | 0 | – | 0.030 | – | |||
| Fibromodulin | Fmod | 125/127 | 0 | 11.0 (4.9) | – | 0.38 | 0 | |||
| Heparan sulphate proteoglycan 2 | Hspg2 | 120/144 | 38.9 (7.8) | 35.1 (6.9) | 0.82 | 0.042 | 1.1 | |||
| Keratin 8 | Krt8 | 151/154 | 30.9 (6.5) | 19.1 (5.5) | 0.12 | 0.34 | 1.6 | >100* | >100* | |
| Keratin 18 | Krt18 | 175/168 | 54.2 (6.5) | 25.2 (5.8) | 0.0011 | 0.91 | 2.2 | >10* | >100* | |
| Keratin 19 | Krt19 | 171/145 | 87.3 (4.4) | 14.4 (5.0) | 3.8E‐14 | 1.0 | 6.1 |
5.8** <10* |
>50* | |
| Laminin1 | Lam1 | 211/322 | 10.3 (3.6) | 37.8 (4.7) | 9.4E‐05 | 1.0 | 0.3 | |||
| Talin1 | Tln1 | 118/136 | 2.4 (2.4) | 4.4 (3.1) | 1.0 | 0.010 | 0.5 | |||
| Tenomodulin | Tnmd | 125/143 | 26.6 (6.8) | 23.9 (6.3) | 1.0 | 0.031 | 1.1 | |||
| Tenascin XB | Tnxb | 118/129 | 5.2 (3.5) | 15.6 (5.6) | 0.16 | 0.28 | 0.3 | |||
| Transcription factors | Forkhead box F1 | Foxf1 | 221/270 | 48.5 (5.8) | 47.3 (5.3) | 0.91 | 0.032 | 1.0 |
0.7* n.s.** |
|
| Glioma‐associated oncogene 1 | Gli1 | 300/284 | 75.3 (4.3) | 33.9 (4.9) | 9.3E‐09 | 1.0 | 2.2 | |||
| Glioma‐associated oncogene 3 | Gli3 | 279/407 | 83.8 (3.8) | 57.0 (4.3) | 2.0E‐05 | 1.0 | 1.5 | |||
| Myoblast determination protein 1 | MyoD | 144/137 | 38.7 (7.0) | 29.3 (6.7) | 0.37 | 0.15 | 1.3 | |||
| Notochord | Noto | 207/216 | 64.8 (5.7) | 14.9 (4.2) | 4.8E‐10 | 1.0 | 4.3 | |||
| Paired box protein 1 | Pax1 | 310/298 | 51.8 (4.9) | 47.7 (5.0) | 0.53 | 0.092 | 1.1 | n.s.** | ||
| Paired box protein 9 | Pax9 | 177/144 | 32.8 (6.1) | 25.8 (6.3) | 0.20 | 0.25 | 1.3 | |||
| Scleraxis | Scx | 286/298 | 63.6 (4.9) | 28.9 (4.5) | 1.2E‐07 | 1.0 | 2.2 | |||
| Sex determining region Y‐box 5 | Sox5 | 126/144 | 22.3 (6.4) | 19.2 (5.7) | 0.75 | 0.051 | 1.2 | |||
| Sex determining region Y‐box 6 | Sox6 | 130/134 | 28.9 (6.9) | 14.1 (5.2) | 0.10 | 0.37 | 2.0 | |||
| Sex determining region Y‐box 9 | Sox9 | 138/144 | 60.1 (7.2) | 41.3 (7.1) | 0.068 | 0.50 | 1.5 | 7* | ||
| Brachyury | T | 145/149 | 46.3 (7.2) | 37.1 (6.9) | 0.27 | 0.20 | 1.2 | >100** | ||
| Signaling factors | Bone morphogenetic protein 4 | Bmp4 | 135/152 | 51.2 (7.5) | 43.1 (7.0) | 0.43 | 0.12 | 1.2 | ||
| Growth differentiation factor 5 | Gdf5 | 113/153 | 12.9 (5.5) | 7.2 (3.6) | 0.49 | 0.11 | 1.8 | |||
| Indian hedgehog | Ihh | 148/144 | 22.2 (5.9) | 21.1 (5.9) | 0.68 | 0.061 | 1.1 | |||
| Sonic hedgehog | Shh | 151/149 | 49.8 (7.0) | 25.1 (6.1) | 0.013 | 0.71 | 2.0 | |||
| Pluripotency and stem cell markers | Endoglin | Eng | 139/138 | 48.1 (7.3) | 30.7 (6.8) | 0.094 | 0.40 | 1.6 | ||
| Estrogen related receptor beta | Esrrb | 202/257 | 30.2 (5.6) | 31.7 (5.0) | 0.80 | 0.044 | 1.0 | |||
| Nanog | Nanog | 135/123 | 21.9 (6.1) | 18.1 (6.0) | 0.73 | 0.047 | 1.2 | |||
| Octamer‐binding transcription factor 4 | Oct4 | 140/147 | 21.2 (6.0) | 24.1 (6.1) | 0.71 | 0.027 | 0.9 | |||
| Tyrosine phosphate receptor type C | Ptprc | 240/294 | 73.7 (4.9) | 32.0 (4.7) | 7.6E‐08 | 1.0 | 2.3 | |||
| Sex determining region Y‐box 2 | Sox2 | 253/239 | 79.5 (4.4) | 24.6 (4.8) | 1.4E‐12 | 1.0 | 3.2 | |||
| Thymocyte differentiation antigen 1 | Thy1 | 201/248 | 6.2 (3.2) | 27.2 (4.9) | 0.0020 | 0.88 | 0.2 | |||
| Zinc finger and SCAN domain containing 10 | Zscan10 | 254/348 | 82.7 (4.1) | 50.2 (4.6) | 2.5E‐06 | 1.0 | 1.6 | |||
| Metabolic context | Carbonic anhydrase 12 | Ca12 | 153/163 | 64.2 (6.8) | 21.3 (5.6) | 9.7E‐06 | 1.0 | 3.0 | ||
| Glyceraldehyde 3‐phosphate dehydrogenase | Gapdh | 225/246 | 16.2 (5.5) | 12.4 (4.7) | 0.46 | 0.11 | 1.3 | |||
| Hypoxia‐inducible factor 1‐alpha | Hif1α | 206/254 | 12.5 (5.2) | 13.8 (4.8) | 0.80 | 0.044 | 1.4 | 0.3* | ||
| Lactate dehydrogenase A | LdhA | 126/155 | 5.4 (3.5) | 12.3 (4.8) | 0.29 | 0.13 | 0.4 | |||
| Lactate dehydrogenase B | LdhB | 125/146 | 4.8 (3.3) | 17.4 (5.4) | 0.10 | 0.32 | 0.3 | |||
| Malate dehydrogenase 2 | Mdh2 | 140/155 | 12.3 (7.2) | 16.1 (5.1) | 0.66 | 0.064 | 0.8 | |||
| Others | Annexin A4 | Anxa4 | 106/119 | 2.8 (2.8) | 4.0 (3.7) | 1.0 | 0.010 | 0.7 | 0.56* | |
| Ki67 | Ki67 | 141/151 | 4.0 (2.8) | 6.7 (3.5) | 1.0 | 0.020 | 0.6 | |||
| lnc RNA LOC101904175 | LOC101904175 | 255/195 | 77.7 (4.6) | 15.3 (4.5) | 6.1E‐14 | 1.0 | 5.1 | |||
| Synaptosomal‐associated protein 25 | Snap25 | 132/162 | 30.0 (7.0) | 17.3 (5.1) | 0.11 | 0.36 | 1.7 | >100* | ||
Results and Discussion
Evaluation of techniques
Identifying AF‐ or NP‐specific biomarkers has proven challenging and remains a hot topic in the field (Lv et al. 2014; Thorpe et al. 2016). On the transcriptional level, this is often achieved by microarray analysis. Technical challenges related to microarray transcriptomics with non‐standard or outbred organisms could further bias results (Kraus et al. 2012). Validation of microarray expression profiling data is recommended and is typically achieved by qRT‐PCR (Minogue et al. 2010b); however, both technologies rely on cell pooling (Minogue et al. 2010b; van den Akker et al. 2017), which does not take potential cellular heterogeneity into account. RISH allows assessment of every cell within a population for the active transcription of a gene and highlights cellular heterogeneity. Hybridization of complementary nucleic acid sequences is highly specific and allows the study of non‐protein coding markers, such as lncRNAs. Proteome analysis through immunohistochemistry can be misleading if the exact epitope for antibody recognition is unknown and there is cross‐reactivity with closely related proteins (Craig et al. 1998). More importantly, secreted proteins might not remain cell associated and those with a long half‐life might no longer be actively transcribed and therefore may not accurately reflect mature cell identity.
Evaluation of IVD sources
Despite increasing requests for data from human IVD sources, there are shortfalls in their use, particularly for transcriptome‐based analysis. Surgically removed human IVDs are typically degenerated and the avascular nature of the IVDs would preclude sufficient RNA fixation through perfusion of the body. We and others therefore chose to use the adult coccygeal bovine IVD as a research model because it appears anatomically, histologically and biochemically similar to a human lumbar disc of a healthy young adult between 15 and 40 years of age (Oshima et al. 1993; Demers et al. 2004; Kraus et al. 2017) and can be harvested fresh and sufficiently fixed in 4% PFA through diffusion. Here, we focus on mRNA expression in cells of mature bovine coccygeal IVDs, representative of a human IVD from a healthy young adult. However, conclusions made regarding disc degeneration based on results obtained from non‐human sources need to consider that degenerated human IVDs exhibit reduced cell density, increased concentration of Collagen I, along with increased collagen cross‐linking, reduced ECM turn‐over as well as reduced proteoglycan and water content; the result is likely to be cells with reduced replicative potential (Antoniou et al. 1996; Sakai & Andersson, 2015; Lama et al. 2018).
Structural proteins
Given the vast amount of ECM and low cell count in the mature IVD, most studies focus on key components of the ECM. Microarray data proposed a NP/AF ratio of < 0.1 for Col1a1 (Minogue et al. 2010b), and qPCR data showed a 56.8‐fold higher expression of Col1a1 in the outer AF over NP tissue (van den Akker et al. 2017). AP‐RISH identified Col1a1‐expressing cells in both the outer AF and NP, although with different prevalence. The AP‐RISH NP/AF ratio of Col1a1‐positive cells was 0.5, therefore higher in the outer AF (P < 0.001). The NP/AF ratio of Col1a2‐positive cells was 13.0 and significantly higher in the NP (P < 0.001; Table 2; Fig. 2), yet the fold changes were reported as < 0.1 by microarray analysis (Minogue et al. 2010b). Unlike the common assumption that the NP tissue is rich in Collagen II, microarray data on bovine IVDs suggested an approximately three‐fold higher Col2a1 expression in the AF than in the NP (Minogue et al. 2010b; Lv et al. 2014). AP‐RISH indicated 4.1× more Col2a1‐positive cells in the NP (P < 0.001) (Table 2, Fig. 2, Supporting Information Fig. S1), similar to a 7.1× upregulated expression level in the bovine NP over the outer AF described by qPCR (van den Akker et al. 2017). Quantitative validation by FL‐RISH of Col2a1 mRNA expression indicated a 2.5× higher expression level in NP cells over those in the outer AF (P = 0.0073; Fig. 3). While not exclusive to AF or NP tissue or the IVD in general, the expression of Col1a1 should serve in a panel of potential AF markers just as Col2a1 is widely accepted as a NP marker in the IVD or cells derived thereof.
Figure 2.
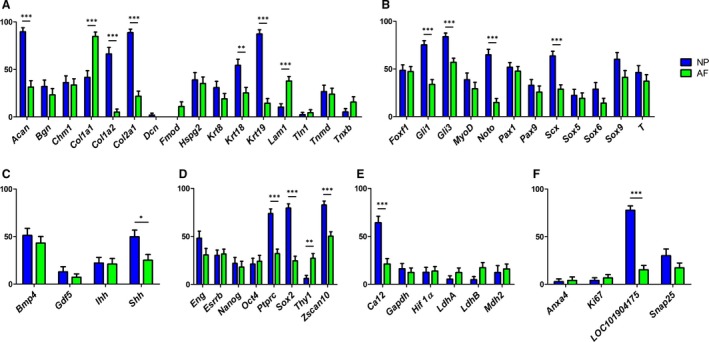
Identification of AF and NP biomarkers of the mature bovine intervertebral disc through z proportion test analysis. Investigated genes encoding structural proteins (a), transcription factors (b), signaling factors (c), pluripotency and stem cell marker (d), markers in a broader metabolic context (e), and other proteins related to the IVD (f) are displayed on the x‐axis. The percentage of cells transcribing a gene is represented on the y‐axis, Blue bars represent NP and green bars represent AF cells. Statistical significance: *P < 0.05, **P < 0.01, ***P < 0.001.
Figure 3.
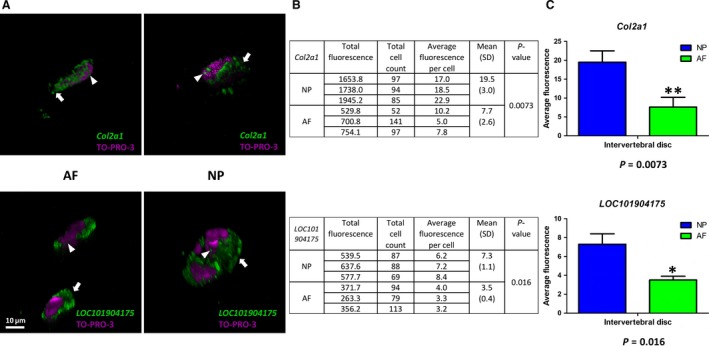
Transcription of the established NP marker Col2a1 and the novel marker LOC101904175 was analyzed in cells of the outer AF and NP via FL‐RISH and confocal microscopy to validate our findings from AP‐RISH. (a) Gene expression is indicated by Alexa‐488 (green) and the nucleus is visualized through To‐Pro3 (magenta). (b) Raw data to determine average fluorescence intensities per cell for Col2a1 and LOC101904175 was generated in imagej. (c) Average fluorescence per cell is represented in graph form using GraphPad prism 5. Student t‐test indicates significantly higher transcription in the NP over the AF for both Col2a1 and LOC101904175 (*P < 0.05, **P < 0.01).
Laminins as basal membrane proteins are important ECM components interacting with larger structural ECM molecules such as collagens. Increased Laminin 1 and 3 was described in the immature rat and pig AF (Chen et al. 2009). We describe a significantly higher proportion of Lam 1‐transcribing cells in the outer AF of bovine IVDs (P < 0.001) through AP‐RISH with a NP/AF ratio of 0.3 (Table 2, Fig. 2, Supporting Information Fig. S7), suggesting that Lam1 should be added to a panel of AF markers.
An increased Aggrecan/Collagen II ratio was proposed as NP‐specific (Risbud et al. 2015). While a ~15× increase of Acan in the NP over AF tissue was noted in microarray expression profiling (Minogue et al. 2010b), no significant increase in Acan or Col2a1 expression was reported in bovine NP over AF samples by qRT‐PCR in the same study. AP‐RISH indicated Acan as a NP biomarker with a significantly higher proportion of Acan‐expressing cells in the NP and an NP/AF ratio of 2.9, comparable to previous qPCR data with a ratio of 3.2 (van den Akker et al. 2017). None of the other glycoproteins examined here by AP‐RISH showed a significant difference in the proportion of positive cells between the outer AF and NP (Table 2, Figs 2 and S1). Although analyzed by AP‐RISH, Dcn was only detected in NP cells and Fmod was only found in AF cells; however, the small number of positives cells precluded statistical analysis (Table 2, Figs 2 and S1),
The presence of the Keratin family, especially Krt8, Krt18 and Krt19, has been described before in different species including human and bovine IVDs and considered a marker for remnant notochord cells (Minogue et al. 2010b; Rodrigues‐Pinto et al. 2016; Richardson et al. 2017). A consistently high NP/AF ratio for Krt19 was described and microarray analysis identified Krt8, Krt18, and Krt19 as bovine NP markers (Minogue et al. 2010a,b; Rodrigues‐Pinto et al. 2013; Lv et al. 2014). AP‐RISH on bovine IVDs only confirmed Krt18 and Krt19 as NP markers with an NP/AF ratio of 2.2 (Krt18, P = 0.0011) and 6.1 (Krt19, P < 0.0001) for the proportion of positive cells, which is supported by recent qPCR data; however, the qPCR NP/AF ratio for Krt18 was nearly 5× higher (van den Akker et al. 2017; Table 2, Figs 2 and S1).
Transcription and signaling factors
Many transcription factors act as molecular switches in cellular fate determination early in development and might have a function later in live for tissue maintenance. We investigated transcripts of key transcription factors during axial skeleton development and differentiation of the chondrocyte lineage Sox5, Sox6 and Sox9, but did not observe a significant difference in the number of positive cells between the outer AF and NP. Of the notochord lineage associated transcription factors Noto and T, only Noto showed a significant 4.3× increase in the number of positive cells in the NP (P < 0.001). Brachyury (T), a transcription factor with conserved function that regulates notochord formation and a biomarker for chordomas (Vujovic et al. 2006), was not identified as NP‐specific by AP‐RISH and z proportion test (Table 2, Figs 2, S2 and S7); however, it has been reported to be significantly higher by qRT‐PCR in the NP over the outer AF in a bovine study (van den Akker et al. 2017). Our AP‐RISH data support findings that only a few, if any, bovine notochordal cells remain present at birth (Demers et al. 2004). As Noto is acting downstream of Brachyury (Abdelkhalek et al. 2004) and AP‐RISH identified a significantly higher proportion of Noto but not T‐transcribing cells in the NP over the outer AF, this could indicate the notochordal lineage origin of these NP cells; however, they no longer exhibit a notochordal phenotype.
The transcription factors FOXF1 and PAX1 are considered NP markers in human (Minogue et al. 2010a; Thorpe et al. 2016; van den Akker et al. 2017). However, a microarray study with bovine tissue identified an increased Foxf1 expression in the AF (Minogue et al. 2010b). Pax1 and Pax9 have a role in AF patterning in mouse (Sivakamasundari et al. 2017) yet are absent in the notochord, the origin of mature murine NP cells (Choi et al. 2012). Our AP‐RISH data did not indicate a significant difference in the proportion of cells expressing Foxf1 or Pax1 between the outer AF and NP, similar to data found through qRT‐PCR on RNA isolated from bovine IVDs reported by others (van den Akker et al. 2017). Also, AP‐RISH did not indicate any significant difference in the proportion of Pax9‐expressing cells (Table 2, Figs 2 and S2).
AP‐RISH identified a significantly (P < 0.001) higher number of cells expressing the transcription factors Gli1, Gli3 and Scx with an NP/AF ratio of 2.2, 1.5 and 2.2, respectively, suggesting that they are potential NP markers that have not previously been reported in the bovine IVD model. Glis are known mediators of hedgehog signalling (Ingham & McMahon, 2001; Buttitta et al. 2003) and we confirmed the signalling factor Shh as an NP biomarker (P = 0.013) by AP‐RISH. However, recognizing Scx as an NP marker in the bovine IVD by AP‐RISH appears to be contrary to data from murine studies, where Scx is expressed in AF tissue but not NP (Pryce et al. 2007; Yoshimoto et al. 2017) and is involved in regulating Tnmd expression, which itself serves as tendon and tendon progenitor cell marker (Shukunami et al. 2006). Whereas Tnmd was reported as significantly increased in bovine AF cells using qRT‐PCR (Minogue et al. 2010b), AP‐RISH data did not indicate a significant difference in cell proportions between the outer AF and NP (Table 2, Figs 2, S2, S3 and S7).The discrepancy might once more reflect the anatomical difference between a mature murine NP, which is entirely notochord‐derived (Choi & Harfe, 2011; Choi et al. 2012), and the adult bovine NP, where only few notochordal cells might remain at birth (Demers et al. 2004). Also, technical differences in trancriptome analyses with qRT‐PCR relying on RNA extraction after cell pooling and AP‐RISH analyzing proportions of cells within a heterogeneous cell population might be contributory. In this context, it is further noteworthy that Scx expression was reported at a higher expression level in passage 2/3 NP cells in monolayer culture compared with the same passage of AF cells (Schulze‐Tanzil et al. 2014), further indicating a difference in cellular composition of the murine NP from that in bovines and humans.
Pluripotency and stem cell markers
As the IVD is of interest in the field of regenerative medicine, the natural presence of progenitor or stem cells might be a key to future therapeutic approaches. In the IVD, AP‐RISH identified a significantly higher number of cells expressing Sox2 (P < 0.001), Ptprc (P < 0.001) and Zscan10 (P < 0.001) in the NP tissue and significantly more Thy1‐expressing cells in the AF (P = 0.002), whereas the proportion of cells expressing Esrrb, Nanog, Oct4 and Eng showed no significant difference (Table 2, Figs 2, S4 and S7). This difference between AF and NP cell populations might be relevant for their therapeutic potential, however, we have previously demonstrated that cells can be isolated from all three tissues, outer AF, inner AF (TZ) and NP, and propagated in vitro in 2D monolayer culture under normal oxygen and zero‐applied pressure (Kraus & Lufkin, 2016; Kraus et al. 2017), which are common culture conditions but are unusual in vivo. This finding further supports the presence of IVD progenitor cells in vivo, as we and others have reported previously (Henriksson et al. 2009; Risbud et al. 2015; Kraus & Lufkin, 2016; Tekari et al. 2016; Thorpe et al. 2016; Kraus et al. 2017; Liu et al. 2017), and might have facilitated the straightforward non‐enzymatic derivation of IVD primary cells (Kraus et al. 2017). Access to oxygen, nutrients and growth factors is limited by diffusion through the dense ECM for cells in the mature IVD (Grunhagen et al. 2011), and ECM stiffness and other chemico/physical properties further impact on cell survival and differentiation in the mature IVD (Guilak et al. 2009; Navaro et al. 2015). Findings by Lama et al. report that high physical pressure and GAG concentrations confine blood vessels to the outer AF in a healthy young human IVD, but vessels reach further into AFs and even NP tissue in severely degenerated or herniated discs when pressure and GAG concentrations drop (Binch et al. 2015; Lama et al. 2018). These vessels could theoretically supply necessary oxygen, nutrients and growth factors to progenitor cells to activate cell metabolism and proliferation; however, no initiation of AF or NP self‐healing in damaged or degenerated discs has been described. Consequently, even if autologous AF or NP progenitor cells are isolated for regenerative purposes from the heterogeneous pool of IVD cells present in vivo, they might encounter similar challenges as MSC upon injection if, in situ chemical and physical conditions are non‐permissive for cell‐mediated damage repair. Ongoing clinical trials need to overcome this hurdle given that only very few trials employing autologous MSC, articular chondrocytes or IVD cells have reported improvements in preliminary studies (Sakai & Andersson, 2015). Clearly, the development of a permissive carrier matrix is as important as the isolation of viable stem cells to successful regenerative approaches. Currently, the NOVOCART™ Disk autologous IVD chondrocyte system is likely the most promising ongoing trial with an estimated Phase I/II completion in August 2021 (Tschugg et al. 2017). Pluripotency markers as identified in our study could serve as a diagnostic tool during quality control measures of such matrices.
Metabolic components and others
Owing to the hypoxic and nutrient‐deprived environment any cell will face in the avascular IVD, cellular adaptation to this environment could point to biomarkers. Of the genes we analyzed by AP‐RISH in this context, only Ca12 encoding the metabolic enzyme carbonic anhydrase XII was expressed in a higher proportion of cells in the NP (P < 0.001) (Table 2, Figs 2 and S5), confirming suggestions of Ca12 as NP marker (Minogue et al. 2010a; Power et al. 2011). Microarray analysis reported a 0.3‐fold change in the NP/AF ratio for Hif1alpha (Minogue et al. 2010b), whereas AP‐RISH indicated no significant difference in the proportion of positive cells between outer AF and NP. Snap25 was proposed as a human and bovine NP marker with significantly higher expression in NP over AF cells by microarray and qRT‐PCR analysis (Minogue et al. 2010b), but AP‐RISH did not identify any significant difference in cell proportions positive for Snap25 between NP and outer AF. There was also no significant difference for any of the genes encoding metabolic enzymes, nor Anxa4 or the cell proliferation marker Ki67. Although no significant difference in the number of cells transcribing the proliferation marker Ki67 was observed, our data indicate that both outer AF and NP tissue harbour cells with a potential to proliferate (Table 2, Figs 2 and S6). Interestingly, the lncRNA LOC101904175 was transcribed in significantly more NP cells (P < 0.001) with a NP/AF ratio of 5.1 (Table 2, Figs 2 and S7). This was further validated through quantification of LOC101904175 transcripts by FL‐RISH, where average fluorescence indicated a 2× increased transcription of LOC101904175 in NP cells than in cells in the outer AF (P = 0.016) (Fig. 3).
In summary, by analyzing the proportion of cells transcribing a gene of interest in a heterogeneous cell population, RISH identified two novel markers in the outer AF, Lam1 and Thy1, and eight novel NP markers, Gli1, Gli3, Noto, Scx, Ptprc, Sox2, Zscan10 and LOC101904175 in the bovine IVD and validated existing biomarkers such as Acan, Col1a1, Col1a2, Clo2a1, Krt18, Krt19, Shh and Ca12, previously identified by others using different methods (Fig. 4). None of these markers is unique to the IVD, but a combination of actively transcribed genes might make it possible to distinguish between outer AF and NP phenotype in cultured cells intended for cell‐based regenerative medicine approaches and provide means of quality control.
Figure 4.
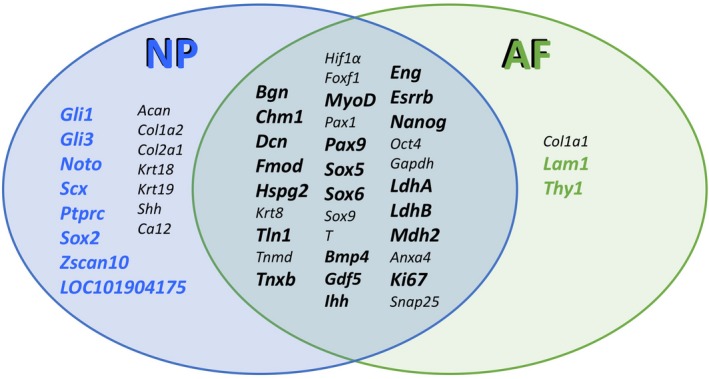
Venn diagram summarizing the genetic loci investigated by AP‐RISH for their biomarker potential in NP (blue) and outer AF (green) tissue. Genes in the centre of the diagram were detected in outer AF and NP tissue without a significant difference in the proportion of positive cells. Genetic loci highlighted in bold indicate novel genes investigated for each group in our study. Ten novel biomarkers were identified by AP‐RISH and z proportion test: two in the AF and eight in the NP.
Conclusions
In a heterogeneous cell population, RISH can provide cell phenotyping with single cell resolution, and distinguish individual cells that remain synthetically active in mature IVDs from others. However, cell pooling‐based transcription analysis such as qRT‐PCR might mask an entire population as positive even if only few cells actively transcribe a gene within a population of negative cells. Identifying synthetically active cells could identify those capable of responding to simulative regenerative treatment, but non‐permissive niche conditions for cell‐initiated tissue healing need to be overcome. Taking the heterogeneous cell population of AF and NP into account, with RISH we were able to identify novel AF (Lam1 and Thy1) and NP (Gli1, Gli3, Noto, Scx, Ptprc, Sox2, Zscan10 and LOC101904175) markers in the bovine IVD which have not been discussed in this context before and should be added to a broader panel of AF and NP biomarkers. Confirmation of several previously identified biomarkers such as Col1a1 in the AF and Col2a1 in the NP further validates our approach, and additional validation is provided through quantification of mRNA expression for Col2a1 and LOC101904175. Unlike methods involving cell pooling for mRNA isolation, RISH allows one to assess the cellular heterogeneity of a tissue on a cell‐by‐cell basis. Ultimately however, only single cell transcriptome analysis, such as single cell RNA sequencing, of cells in their natural environment will definitively clarify the true identity of the cells residing in the AF or NP.
Funding
This work was supported by the Bayard and Virginia Clarkson Endowment Fund granted to Thomas Lufkin. V.K. was supported by the CUPO CStep Program. There are no relevant financial activities outside the submitted work.
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
K.L., P.K. and T.L. designed the study. K.L. acquired the data and performed all cell counts. K.L., D.K. and S.M. performed statistical analysis. B.Y. and V.K. assisted with IVD dissection. K.L., D.K., P.K., S.M. and T.L. drafted and critically revised the manuscript. All authors approved the final version of the manuscript.
Supporting information
Fig. S1 AP‐RISH analysis of genes encoding for structural proteins in the bovine IVD.
Fig. S2 AP‐RISH analysis of genes encoding for relevant transcription factors in the bovine IVD.
Fig. S3 AP‐RISH analysis of genes encoding for signaling factors in the bovine IVD.
Fig. S4 AP‐RISH analysis of genes encoding for stemness and pluripotency markers in the bovine IVD.
Fig. S5 AP‐RISH analysis of genes encoding for metabolic enzymes in the bovine IVD.
Fig. S6 AP‐RISH analysis of other relevant genetic loci in the bovine IVD.
Fig. S7 Ten novel biomarkers identified by AP‐RISH.
Table S1 Gene‐specific sequence of base primer for the amplification of the RISH probe template from bovine genomic DNA.
Acknowledgements
We are grateful to Willard & Sons (Heuvelton, NY, USA), Tritown Meat Packing (Brasher Falls, NY, USA) and Peter Braun of Woodcrest Dairy (Lisbon, NY, USA), for providing us with bovine tails. We greatly appreciate the intellectual input from Darren Sipes and Shantanu Sur and comments by anonymous reviewers.
References
- Abdelkhalek HB, Beckers A, Schuster‐Gossler K, et al. (2004) The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev 18,1725–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S, Joyner AL (2005) In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437,894–897. [DOI] [PubMed] [Google Scholar]
- van den Akker GGH, Koenders MI, van de Loo FAJ, et al. (2017) Transcriptional profiling distinguishes inner and outer annulus fibrosus from nucleus pulposus in the bovine intervertebral disc. Eur Spine J 26,2053–2062. [DOI] [PubMed] [Google Scholar]
- Alini M, Eisenstein SM, Ito K, et al. (2008) Are animal models useful for studying human disc disorders/degeneration? Eur Spine J 17,2–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou J, Steffen T, Nelson F, et al. (1996) The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J Clin Invest 98,996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszodi A, Beier DR, Hiripi L, et al. (1998) Sequence, structure and chromosomal localization of Crtm gene encoding mouse cartilage matrix protein and its exclusion as a candidate for murine achondroplasia. Matrix Biol 16,563–573. [DOI] [PubMed] [Google Scholar]
- Bader BL, Jahn L, Franke WW (1988) Low level expression of cytokeratins 8, 18 and 19 in vascular smooth muscle cells of human umbilical cord and in cultured cells derived therefrom, with an analysis of the chromosomal locus containing the cytokeratin 19 gene. Eur J Cell Biol 47,300–319. [PubMed] [Google Scholar]
- Barrionuevo F, Taketo MM, Scherer G, et al. (2006) Sox9 is required for notochord maintenance in mice. Dev Biol 295,128–140. [DOI] [PubMed] [Google Scholar]
- Bayliss MT, Johnstone B, O'Brien JP (1988) 1988 Volvo award in basic science. Proteoglycan synthesis in the human intervertebral disc. Variation with age, region and pathology. Spine (Phila Pa 1976) 13,972–981. [DOI] [PubMed] [Google Scholar]
- Bedore J, Leask A, Seguin CA (2014) Targeting the extracellular matrix: matricellular proteins regulate cell‐extracellular matrix communication within distinct niches of the intervertebral disc. Matrix Biol 37,124–130. [DOI] [PubMed] [Google Scholar]
- Bell JK, Yennawar HP, Wright SK, et al. (2001) Structural analyses of a malate dehydrogenase with a variable active site. J Biol Chem 276,31156–31162. [DOI] [PubMed] [Google Scholar]
- Bibby SR, Urban JP (2004) Effect of nutrient deprivation on the viability of intervertebral disc cells. Eur Spine J 13,695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby SR, Jones DA, Lee RB, et al. (2001) The pathophysiology of the intervertebral disc. Joint Bone Spine 68,537–542. [DOI] [PubMed] [Google Scholar]
- Binch AL, Cole AA, Breakwell LM, et al. (2015) Nerves are more abundant than blood vessels in the degenerate human intervertebral disc. Arthritis Res Ther 17,370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch GH, Gong Y, Liu W, et al. (1997) Tenascin‐X deficiency is associated with Ehlers‐Danlos syndrome. Nat Genet 17,104–108. [DOI] [PubMed] [Google Scholar]
- Bushell GR, Ghosh P, Taylor TF, et al. (1977) Proteoglycan chemistry of the intervertebral disks. Clin Orthop Relat Res, 129,115–123. [DOI] [PubMed] [Google Scholar]
- Buttitta L, Mo R, Hui CC, et al. (2003) Interplays of Gli2 and Gli3 and their requirement in mediating Shh‐dependent sclerotome induction. Development 130,6233–6243. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, et al. (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450,1230–1234. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Sivakamasundari V, Yap SP, et al. (2014) In vivo genome‐wide analysis of multiple tissues identifies gene regulatory networks, novel functions and downstream regulatory genes for Bapx1 and its co‐regulation with Sox9 in the mammalian vertebral column. BMC Genom 15,1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelberg MK, Banks GM, Geiger DF, et al. (1995) Identification of heterogeneous cell populations in normal human intervertebral disc. J Anat 186(Pt 1),43–53. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Jing L, Gilchrist CL, et al. (2009) Expression of laminin isoforms, receptors, and binding proteins unique to nucleus pulposus cells of immature intervertebral disc. Connect Tissue Res 50,294–306. [PMC free article] [PubMed] [Google Scholar]
- Cheung KM, Karppinen J, Chan D, et al. (2009) Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty‐three individuals. Spine (Phila Pa 1976) 34,934–940. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, et al. (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383,407–413. [DOI] [PubMed] [Google Scholar]
- Chiche J, Ilc K, Laferriere J, et al. (2009) Hypoxia‐inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res 69,358–368. [DOI] [PubMed] [Google Scholar]
- Chiquet‐Ehrismann R, Tucker RP (2011) Tenascins and the importance of adhesion modulation. Cold Spring Harb Perspect Biol, 3,a004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Harfe BD (2011) Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc Natl Acad Sci USA 108,9484–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KS, Lee C, Harfe BD (2012) Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev 129,255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Sanschagrin PC, Rozek A, et al. (1998) The role of structure in antibody cross‐reactivity between peptides and folded proteins. J Mol Biol 281,183–201. [DOI] [PubMed] [Google Scholar]
- Critchley DR, Gingras AR (2008) Talin at a glance. J Cell Sci 121,1345–1347. [DOI] [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Ligon KL, et al. (1995) Scleraxis: a basic helix‐loop‐helix protein that prefigures skeletal formation during mouse embryogenesis. Development 121,1099– 1110. [DOI] [PubMed] [Google Scholar]
- Dahia CL, Mahoney E, Wylie C (2012) Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PLoS One 7,e35944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JM, Olin AI, Murdoch AD, et al. (2004) Alternative splicing in the aggrecan G3 domain influences binding interactions with tenascin‐C and other extracellular matrix proteins. J Biol Chem 279,12511–12518. [DOI] [PubMed] [Google Scholar]
- Demers CN, Antoniou J, Mwale F (2004) Value and limitations of using the bovine tail as a model for the human lumbar spine. Spine (Phila Pa 1976) 29,2793–2799. [DOI] [PubMed] [Google Scholar]
- DiPaola CP, Farmer JC, Manova K, et al. (2005) Molecular signaling in intervertebral disk development. J Orthop Res 23,1112–1119. [DOI] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, et al. (2012) Early‐stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488,652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, et al. (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8,315–317. [DOI] [PubMed] [Google Scholar]
- Ekblom M, Falk M, Salmivirta K, et al. (1998) Laminin isoforms and epithelial development. Ann N Y Acad Sci 857,194–211. [DOI] [PubMed] [Google Scholar]
- Errington RJ, Puustjarvi K, White IR, et al. (1998) Characterisation of cytoplasm‐filled processes in cells of the intervertebral disc. J Anat 192(Pt. 3),369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin WM, Hood KE (2014) The cellular and molecular biology of the intervertebral disc: A clinician's primer. J Can Chiropr Assoc 58,246–257. [PMC free article] [PubMed] [Google Scholar]
- Eyre DR (1979) Biochemistry of the intervertebral disc. Int Rev Connect Tissue Res 8,227–291. [DOI] [PubMed] [Google Scholar]
- Eyre DR, Muir H (1976) Types I and II collagens in intervertebral disc. Interchanging radial distributions in annulus fibrosus. Biochem J 157,267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, et al. (2009) Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 11,197–203. [DOI] [PubMed] [Google Scholar]
- Festuccia N, Osorno R, Halbritter F, et al. (2012) Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11,477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LW, Termine JD, Young MF (1989) Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J Biol Chem 264,4571–4576. [PubMed] [Google Scholar]
- Francis‐West PH, Abdelfattah A, Chen P, et al. (1999) Mechanisms of GDF‐5 action during skeletal development. Development 126,1305–1315. [DOI] [PubMed] [Google Scholar]
- Garofalo S, Vuorio E, Metsaranta M, et al. (1991) Reduced amounts of cartilage collagen fibrils and growth plate anomalies in transgenic mice harboring a glycine‐to‐cysteine mutation in the mouse type II procollagen alpha 1‐chain gene. Proc Natl Acad Sci USA 88,9648–9652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson A, Dreger M, Urban JP (2010) Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther 12,R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunhagen T, Shirazi‐Adl A, Fairbank JC, et al. (2011) Intervertebral disk nutrition: a review of factors influencing concentrations of nutrients and metabolites. Orthop Clin North Am 42,465–477.vii. [DOI] [PubMed] [Google Scholar]
- Guilak F, Cohen DM, Estes BT, et al. (2009) Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5,17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson H, Thornemo M, Karlsson C, et al. (2009) Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine (Phila Pa 1976) 34,2278–2287. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, et al. (1990) Cloning of the T gene required in mesoderm formation in the mouse. Nature 343,617–622. [DOI] [PubMed] [Google Scholar]
- Hiraki Y, Shukunami C (2000) Chondromodulin‐I as a novel cartilage‐specific growth‐modulating factor. Pediatr Nephrol 14,602–605. [DOI] [PubMed] [Google Scholar]
- Hiraki Y, Tanaka H, Inoue H, et al. (1991) Molecular cloning of a new class of cartilage‐specific matrix, chondromodulin‐I, which stimulates growth of cultured chondrocytes. Biochem Biophys Res Commun 175,971–977. [DOI] [PubMed] [Google Scholar]
- Humzah MD, Soames RW (1988) Human intervertebral disc: structure and function. Anat Rec 220,337–356. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Kamekura S, Mabuchi A, et al. (2004) The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum 50,3561–3573. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15,3059–3087. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Moscatello DK, McQuillan DJ, et al. (1999) Decorin is a biological ligand for the epidermal growth factor receptor. J Biol Chem 274,4489–4492. [DOI] [PubMed] [Google Scholar]
- Jan AT, Lee EJ, Choi I (2016) Fibromodulin: A regulatory molecule maintaining cellular architecture for normal cellular function. Int J Biochem Cell Biol 80,66–70. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Eisenstein SM, Roberts S (2001) Cell cluster formation in degenerate lumbar intervertebral discs is associated with increased disc cell proliferation. Connect Tissue Res 42,197–207. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Park RW (1995) Regulation of type I collagen genes expression. Int Rev Immunol 12,177–185. [DOI] [PubMed] [Google Scholar]
- Katz WA (2002) Musculoskeletal pain and its socioeconomic implications. Clin Rheumatol 21(Suppl. 1),S2–S4. [DOI] [PubMed] [Google Scholar]
- Khillan JS, Li SW, Prockop DJ (1994) Partial rescue of a lethal phenotype of fragile bones in transgenic mice with a chimeric antisense gene directed against a mutated collagen gene. Proc Natl Acad Sci USA 91,6298–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Lufkin T (1999) Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J Cell Biochem Suppl 32–33,133–140. [DOI] [PubMed] [Google Scholar]
- Kraus P, Lufkin T (2016) Bovine annulus fibrosus cell lines isolated from intervertebral discs. Genom Data 10,83–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Lufkin T (2017) Implications for a stem cell regenerative medicine based approach to human intervertebral disk degeneration. Front Cell Dev Biol 5,17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Fraidenraich D, Loomis CA (2001) Some distal limb structures develop in mice lacking Sonic hedgehog signaling. Mech Dev 100,45–58. [DOI] [PubMed] [Google Scholar]
- Kraus P, Xing X, Lim SL, et al. (2012) Mouse strain specific gene expression differences for illumina microarray expression profiling in embryos. BMC Res Notes 5,232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Sivakamasundari V, Yu HB, et al. (2014) Pleiotropic functions for transcription factor zscan10. PLoS One 9,e104568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Yerden R, Kocsis V, et al. (2017) RNA in situ hybridization characterization of non‐enzymatic derived bovine intervertebral disc cell lineages suggests progenitor cell potential. Acta Histochem 119,150–160. [DOI] [PubMed] [Google Scholar]
- Kraus P, Sivakamasundari V, Olsen V, et al. (2018a) Klhl14 antisense RNA is a target of key skeletogenic transcription factors in the developing intervertebral disc. Spine (Phila Pa 1976), 10.1097/BRS.0000000000002827. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus P, Yerden R, Sipes D, et al. (2018b) A quantitative and qualitative RNA expression profiling assay for cell culture with single cell resolution. Cytotechnology 70,185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama P, Le Maitre CL, Harding IJ, et al. (2018) Nerves and blood vessels in degenerated intervertebral discs are confined to physically disrupted tissue. J Anat 233,86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Maitre CL, Pockert A, Buttle DJ, et al. (2007) Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans 35,652–655. [DOI] [PubMed] [Google Scholar]
- Lee JT, Cheung KM, Leung VY (2015) Systematic study of cell isolation from bovine nucleus pulposus: Improving cell yield and experiment reliability. J Orthop Res 33,1743–1755. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Chatterjee S, Yap SP, et al. (2017) An integrative developmental genomics and systems biology approach to identify an in vivo sox trio‐mediated gene regulatory network in murine embryos. Biomed Res Int 2017,8932583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Behringer RR, de Crombrugghe B (2001) L‐Sox5, Sox6 and Sox9 control essential steps of the chondrocyte differentiation pathway. Osteoarthritis Cartilage, 9(Suppl. A),S69–S75. [DOI] [PubMed] [Google Scholar]
- Li LT, Jiang G, Chen Q, et al. (2015) Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep 11,1566–1572. [DOI] [PubMed] [Google Scholar]
- Liang CZ, Li H, Tao YQ, et al. (2012) The relationship between low pH in intervertebral discs and low back pain: a systematic review. Arch Med Sci 8,952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liang H, Lee SM, et al. (2017) Isolation and identification of stem cells from degenerated human intervertebral discs and their migration characteristics. Acta Biochim Biophys Sin (Shanghai) 49,101–109. [DOI] [PubMed] [Google Scholar]
- Lopa S, Ceriani C, Cecchinato R, et al. (2016) Stability of housekeeping genes in human intervertebral disc, endplate and articular cartilage cells in multiple conditions for reliable transcriptional analysis. Eur Cell Mater 31,395–406. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Mark M, Hart CP, et al. (1992) Homeotic transformation of the occipital bones of the skull by ectopic expression of a homeobox gene. Nature 359,835–841. [DOI] [PubMed] [Google Scholar]
- Lv F, Leung VY, Huang S, et al. (2014) In search of nucleus pulposus‐specific molecular markers. Rheumatology (Oxford) 53,600–610. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Nakamura E, Nguyen MT, et al. (2007) Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc Natl Acad Sci USA 104,6382–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, et al. (2001) The forkhead transcription factor Foxf1 is required for differentiation of extra‐embryonic and lateral plate mesoderm. Development 128,155–166. [DOI] [PubMed] [Google Scholar]
- Mao JR, Taylor G, Dean WB, et al. (2002) Tenascin‐X deficiency mimics Ehlers‐Danlos syndrome in mice through alteration of collagen deposition. Nat Genet 30,421–425. [DOI] [PubMed] [Google Scholar]
- Marfia G, Campanella R, Navone SE, et al. (2014) Potential use of human adipose mesenchymal stromal cells for intervertebral disc regeneration: a preliminary study on biglycan‐deficient murine model of chronic disc degeneration. Arthritis Res Ther 16,457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MR, Tamplin OJ, Rossant J, et al. (2012) Tracing notochord‐derived cells using a Noto‐cre mouse: implications for intervertebral disc development. Dis Model Mech 5,73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melrose J, Ghosh P, Taylor TK (2001) A comparative analysis of the differential spatial and temporal distributions of the large (aggrecan, versican) and small (decorin, biglycan, fibromodulin) proteoglycans of the intervertebral disc. J Anat 198,3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merceron C, Mangiavini L, Robling A, et al. (2014) Loss of HIF‐1alpha in the notochord results in cell death and complete disappearance of the nucleus pulposus. PLoS One 9,e110768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, et al. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113,631–642. [DOI] [PubMed] [Google Scholar]
- Minogue BM, Richardson SM, Zeef LA, et al. (2010a) Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum 62,3695–3705. [DOI] [PubMed] [Google Scholar]
- Minogue BM, Richardson SM, Zeef LA, et al. (2010b) Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther 12,R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinos M, Almeida CR, Caldeira J, et al. (2015) Inflammation in intervertebral disc degeneration and regeneration. J R Soc Interface 12,20141191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll R, Divo M, Langbein L (2008) The human keratins: biology and pathology. Histochem Cell Biol 129,705–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi Y, Alimi M, Khair T, et al. (2016) Biological treatment approaches for degenerative disk disease: a literature review of in vivo animal and clinical data. Global Spine J 6,497–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaro Y, Bleich‐Kimelman N, Hazanov L, et al. (2015) Matrix stiffness determines the fate of nucleus pulposus‐derived stem cells. Biomaterials 49,68–76. [DOI] [PubMed] [Google Scholar]
- Nibu Y, Jose‐Edwards DS, Di Gregorio A (2013) From notochord formation to hereditary chordoma: the many roles of Brachyury. Biomed Res Int 2013,826435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, et al. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95,379–391. [DOI] [PubMed] [Google Scholar]
- Nifuji A, Kellermann O, Kuboki Y, et al. (1997) Perturbation of BMP signaling in somitogenesis resulted in vertebral and rib malformations in the axial skeletal formation. J Bone Miner Res 12,332–342. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, et al. (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460,118–122. [DOI] [PubMed] [Google Scholar]
- Noonan DM, Fulle A, Valente P, et al. (1991) The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein‐receptor, and the neural cell adhesion molecule. J Biol Chem 266,22939–22947. [PubMed] [Google Scholar]
- Oegema TR Jr (1993) Biochemistry of the intervertebral disc. Clin Sports Med 12,419–439. [PubMed] [Google Scholar]
- Oshima H, Ishihara H, Urban JP, et al. (1993) The use of coccygeal discs to study intervertebral disc metabolism. J Orthop Res 11,332–338. [DOI] [PubMed] [Google Scholar]
- Pattappa G, Li Z, Peroglio M, et al. (2012) Diversity of intervertebral disc cells: phenotype and function. J Anat 221,480–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennicooke B, Moriguchi Y, Hussain I, et al. (2016) Biological treatment approaches for degenerative disc disease: a review of clinical trials and future directions. Cureus 8,e892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R, Khillan JS, Helminen HJ, et al. (1993) Transgenic mice expressing a partially deleted gene for type I procollagen (COL1A1). A breeding line with a phenotype of spontaneous fractures and decreased bone collagen and mineral. J Clin Invest 91,709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Wilm B, Sakai N, et al. (1999) Pax1 and Pax9 synergistically regulate vertebral column development. Development 126,5399–5408. [DOI] [PubMed] [Google Scholar]
- Piljic A, Schultz C (2006) Annexin A4 self‐association modulates general membrane protein mobility in living cells. Mol Biol Cell 17,3318–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power KA, Grad S, Rutges JP, et al. (2011) Identification of cell surface‐specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum 63,3876–3886. [DOI] [PubMed] [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, et al. (2007) Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn 236,1677–1682. [DOI] [PubMed] [Google Scholar]
- Richardson SM, Ludwinski FE, Gnanalingham KK, et al. (2017) Notochordal and nucleus pulposus marker expression is maintained by sub‐populations of adult human nucleus pulposus cells through aging and degeneration. Sci Rep 7,1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud MV, Guttapalli A, Tsai TT, et al. (2007) Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976) 32,2537–2544. [DOI] [PubMed] [Google Scholar]
- Risbud MV, Schoepflin ZR, Mwale F, et al. (2015) Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res 33,283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Lufkin T (2006) Dlx5 and Dlx6 homeobox genes are required for specification of the mammalian vestibular apparatus. Genesis 44,425–437. [DOI] [PubMed] [Google Scholar]
- Rodrigues‐Pinto R, Richardson SM, Hoyland JA (2013) Identification of novel nucleus pulposus markers: Interspecies variations and implications for cell‐based therapiesfor intervertebral disc degeneration. Bone Joint Res 2,169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues‐Pinto R, Berry A, Piper‐Hanley K, et al. (2016) Spatiotemporal analysis of putative notochordal cell markers reveals CD24 and keratins 8, 18, and 19 as notochord‐specific markers during early human intervertebral disc development. J Orthop Res 34,1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, et al. (1993) MyoD or Myf‐5 is required for the formation of skeletal muscle. Cell 75,1351–1359. [DOI] [PubMed] [Google Scholar]
- Sakai D, Andersson GB (2015) Stem cell therapy for intervertebral disc regeneration: obstacles and solutions. Nat Rev Rheumatol 11,243–256. [DOI] [PubMed] [Google Scholar]
- Sakai D, Schol J (2017) Cell therapy for intervertebral disc repair: Clinical perspective. J Orthop Translat 9,8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Kleinman HK, Huber H, et al. (1988) Laminin, a multidomain protein. The A chain has a unique globular domain and homology with the basement membrane proteoglycan and the laminin B chains. J Biol Chem 263,16536–16544. [PubMed] [Google Scholar]
- Schonherr E, Witsch‐Prehm P, Harrach B, et al. (1995) Interaction of biglycan with type I collagen. J Biol Chem 270,2776–2783. [DOI] [PubMed] [Google Scholar]
- Schulze‐Tanzil G, Lemke M, Meier C, et al. (2014) Characterization of human anulus fibrosus– and nucleus pulposus–derived cells during monolayer expansion and in hydrogel cultures. Bone Tissue Regen Insights 5,BTRI.S13604. [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, et al. (2001) Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128,3855–3866. [DOI] [PubMed] [Google Scholar]
- Seidler NW (2013) GAPDH and intermediary metabolism. Adv Exp Med Biol 985,37–59. [DOI] [PubMed] [Google Scholar]
- Shin SH, Kogerman P, Lindstrom E, et al. (1999) GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proc Natl Acad Sci USA 96,2880–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukunami C, Oshima Y, Hiraki Y (2001) Molecular cloning of tenomodulin, a novel chondromodulin‐I related gene. Biochem Biophys Res Commun 280,1323–1327. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Takimoto A, Oro M, et al. (2006) Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev Biol 298,234–247. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Takimoto A, Nishizaki Y, et al. (2018) Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes. Sci Rep 8,3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Masuda K, Thonar EJ, et al. (2009) Age‐related changes in the extracellular matrix of nucleus pulposus and anulus fibrosus of human intervertebral disc. Spine (Phila Pa 1976), 34,10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakamasundari V, Lufkin T (2013) Stemming the degeneration: IVD stem cells and stem cell regenerative therapy for degenerative disc disease. Adv Stem Cells, 2013,724547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakamasundari V, Kraus P, Sun W, et al. (2017) A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol Open 6,187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Dy P, Mitra S, et al. (2004) Sox5 and Sox6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J Cell Biol 164,747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St‐Jacques B, Hammerschmidt M, McMahon AP (1999) Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev 13,2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm EE, Huynh TV, Copeland NG, et al. (1994) Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta‐superfamily. Nature 368,639–643. [DOI] [PubMed] [Google Scholar]
- Sudo K, Maekawa M, Shioya M, et al. (1992a) Molecular analysis of genetic mutation in electrophoretic variant of human lactate dehydrogenase‐A(M) subunit. Biochem Int 27,1051–1057. [PubMed] [Google Scholar]
- Sudo K, Maekawa M, Tomonaga A, et al. (1992b) Molecular characterization of genetic mutations in human lactate dehydrogenase (LDH) B (H) variant. Hum Genet 89,158–162. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126,663–676. [DOI] [PubMed] [Google Scholar]
- Tang X, Jing L, Chen J (2012) Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One 7,e52020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekari A, Chan SC, Sakai D, et al. (2016) Angiopoietin‐1 receptor Tie2 distinguishes multipotent differentiation capability in bovine coccygeal nucleus pulposus cells. Stem Cell Res Ther 7,75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe AA, Binch AL, Creemers LB, et al. (2016) Nucleus pulposus phenotypic markers to determine stem cell differentiation: fact or fiction? Oncotarget 7,2189–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschugg A, Diepers M, Simone S, et al. (2017) A prospective randomized multicenter phase I/II clinical trial to evaluate safety and efficacy of NOVOCART disk plus autologous disk chondrocyte transplantation in the treatment of nucleotomized and degenerative lumbar disks to avoid secondary disease: safety results of Phase I – a short report. Neurosurg Rev 40,155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SA, Wright KT, Jones PN, et al. (2016) Temporal analyses of the response of intervertebral disc cells and mesenchymal stem cells to nutrient deprivation. Stem Cells Int 2016,5415901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JP, Holm S, Maroudas A, et al. (1977) Nutrition of the intervertebral disk. An in vivo study of solute transport. Clin Orthop Relat Res (101),101–114. [PubMed] [Google Scholar]
- Urban JP, Smith S, Fairbank JC (2004) Nutrition of the intervertebral disc. Spine (Phila Pa 1976) 29,2700–2709. [DOI] [PubMed] [Google Scholar]
- Vandenberg P, Khillan JS, Prockop DJ, et al. (1991) Expression of a partially deleted gene of human type II procollagen (COL2A1) in transgenic mice produces a chondrodysplasia. Proc Natl Acad Sci USA 88,7640–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A, Lee K, Lanske B, et al. (1996) Regulation of rate of cartilage differentiation by Indian hedgehog and PTH‐related protein. Science 273,613–622. [DOI] [PubMed] [Google Scholar]
- Vujovic S, Henderson S, Presneau N, et al. (2006) Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol 209,157–165. [DOI] [PubMed] [Google Scholar]
- Wang W, Lo P, Frasch M, et al. (2000) Hmx: an evolutionary conserved homeobox gene family expressed in the developing nervous system in mice and Drosophila . Mech Dev 99,123–137. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Kueh JL, Teh CH, et al. (2007) Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells 25,2173–2182. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Kimata K, Line S, et al. (1994) Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nat Genet 7,154–157. [DOI] [PubMed] [Google Scholar]
- Waterman BR, Belmont PJ Jr, Schoenfeld AJ (2012) Low back pain in the United States: incidence and risk factors for presentation in the emergency setting. Spine J 12,63–70. [DOI] [PubMed] [Google Scholar]
- Wijgerde M, Karp S, McMahon J, et al. (2005) Noggin antagonism of BMP4 signaling controls development of the axial skeleton in the mouse. Dev Biol 286,149–157. [DOI] [PubMed] [Google Scholar]
- Wilda M, Bachner D, Just W, et al. (2000) A comparison of the expression pattern of five genes of the family of small leucine‐rich proteoglycans during mouse development. J Bone Miner Res 15,2187–2196. [DOI] [PubMed] [Google Scholar]
- Wuertz K, Godburn K, Neidlinger‐Wilke C, et al. (2008) Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine (Phila Pa 1976) 33,1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto Y, Takimoto A, Watanabe H, et al. (2017) Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci Rep 7,45010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, An HS, Thonar EJ, et al. (2006) Comparative effects of bone morphogenetic proteins and sox9 overexpression on extracellular matrix metabolism of bovine nucleus pulposus cells. Spine (Phila Pa 1976) 31,2173–2179. [DOI] [PubMed] [Google Scholar]
- Zhao N, Hashida H, Takahashi N, et al. (1994) Cloning and sequence analysis of the human SNAP25 cDNA. Gene 145,313–314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 AP‐RISH analysis of genes encoding for structural proteins in the bovine IVD.
Fig. S2 AP‐RISH analysis of genes encoding for relevant transcription factors in the bovine IVD.
Fig. S3 AP‐RISH analysis of genes encoding for signaling factors in the bovine IVD.
Fig. S4 AP‐RISH analysis of genes encoding for stemness and pluripotency markers in the bovine IVD.
Fig. S5 AP‐RISH analysis of genes encoding for metabolic enzymes in the bovine IVD.
Fig. S6 AP‐RISH analysis of other relevant genetic loci in the bovine IVD.
Fig. S7 Ten novel biomarkers identified by AP‐RISH.
Table S1 Gene‐specific sequence of base primer for the amplification of the RISH probe template from bovine genomic DNA.


