Abstract
Broad-range polymerase chain reaction (PCR) is increasingly used in patients with culture-negative infections; however, few studies have assessed the diagnostic utility of this test in this context. We performed a retrospective cohort study of patients who had clinical specimens sent for broad-range PCR, aiming to evaluate performance and determine impact on patient management. Organisms were identified in 21/71 samples. High numbers of polymorphonuclear leukocytes on Gram stain (odds ratio [OR], 4.17; P = .04) and acute inflammation on histopathology (OR, 5.69; P = .02) were significantly associated with a positive result. Management was altered in 18 patients, 11 with positive and 7 with negative results. Overall, broad-range PCR assay had the highest impact in patients with microscopic evidence of inflammation. Physicians ordering this complex, difficult to interpret, and expensive test should carefully consider all available clinical information on an individualized basis to optimize its performance.
Keywords: broad-range PCR, bacterial infection, culture negative
The accurate identification of a specific microbial pathogen is crucial in many infectious disease syndromes as it facilitates a clear diagnosis, allows for targeted therapy, and increases the likelihood of a favorable outcome [1, 2] However, using culture-based diagnostic methods, organisms cannot always be isolated, either due to their fastidious nature or the use of empiric antimicrobials before specimen collection [3–6]. Molecular diagnostics based on nucleic acid target amplification and sequencing technology (also known as “broad-range” polymerase chain reaction [PCR]) are increasingly used for investigation of culture-negative infections. These tests are able to accurately identify microorganisms based on specific regions such as 16S rRNA (for identification of bacteria), rpoB (for mycobacteria), or the internal transcribed spacer region (fungi) and have been used in clinical microbiology for more than 2 decades [7]. These techniques were initially applied to microbial isolates that could not be definitively identified by phenotypic means, and they perform well in this scenario [8]. More recently, they have been increasingly used directly on clinical tissue/fluid samples aiming to identify fastidious organisms that are difficult to culture, or in patients who have been exposed to antimicrobials before specimen collection. In this situation, sensitivity may be lower and methodology more complex due to the tissue extraction process required, potential inhibitors present, and increased likelihood of contamination leading to false-positive results [9]. In 1 large study using 16S rRNA PCR, Rampini et al. demonstrated 91% concordance with bacterial cultures in 394 culture-positive samples and were able to identify 24 bacteria among 184 culture-negative samples [10]. 16S rRNA PCR is also increasingly being used as part of the diagnostic evaluation of prosthetic joint infection [11–13] and culture-negative endocarditis [11, 14–16].
Given the high cost, technical complexity, and time required to perform these tests, attempts have been made in many institutions to try and target their use toward patients most likely to benefit, aiming to optimize test performance and cost-effectiveness. This has included considering factors such as clinical features, serum inflammatory markers, and evidence of infection on microbiologic or histopathologic stains and culture results. Several studies have explored the relationships between these variables and bacteriologic culture results [17–19]. One small study identified a positive association between serum C-reactive protein (CRP) and tissue neutrophil count and level of bacterial DNA measured by cycle threshold [20], but in general the relationships between clinical features and broad-range PCR results are not well studied.
In most prior studies, broad-range PCR has been directly compared with culture-based diagnostic methods, despite the fact that increasingly in modern clinical practice it is being used when cultures are negative [12, 21–25]. Additionally, few studies have examined the overall clinical utility and impact of this test on patient management [21, 26–28]. The aim of this retrospective cohort study was to assess the real-world clinical performance of broad-range PCR at our institution using a “composite clinical gold standard” and determine its impact on antimicrobial decision-making. We also evaluated clinical factors such as pathological findings and presence of polymorphonuclear leukocytes associated with a positive PCR result to guide selection of specimens appropriate for PCR testing.
METHODS
Study Design and Data Collection
The study population consisted of patients at Tufts Medical Center, a 415-bed academic medical center in Boston, Massachusetts, whose tissue and fluid samples were sent for broad-range PCR testing from August 2013 to April 2016, excluding those lost to follow-up (defined as lack of inpatient/outpatient documentation in medical records after the broad-range PCR was performed). The study was approved by the Tufts Medical Center institutional review board; informed consent was not required given the minimal risk and retrospective nature of the study.
Clinical data were collected from medical records. All cases were reviewed by a panel of 3 infectious diseases physicians blinded to PCR results but provided with all other relevant clinical information. The panel determined the presence or absence of infection by using a final gold standard of “composite clinical diagnosis” based on all available data, including medical history, clinical signs and symptoms, operative findings, laboratory testing results including inflammatory markers (C-reactive protein and erythrocyte sedimentation rate), Gram stain results, histopathologic findings, microbiologic, serologic, and radiologic data, and prior antibiotic therapy. Final classification in cases of disagreement was by majority opinion.
Specimen Collection and Laboratory Methods
The majority of samples were collected under sterile conditions in the operating room or interventional radiology suite, divided at the point of collection, and then sent separately for histopathology and microbiology testing. Occasionally, undivided samples were sent, and these were processed first in the microbiology laboratory then sent to histopathology to avoid contamination.
Gram stains (including quantification of bacteria and blood cells) and cultures of tissue specimens were performed in the clinical microbiology laboratory at Tufts Medical Center according to CLSI standards [29]. Tissue specimens were examined for the presence of polymorphonuclear leukocytes (PMNs) and reported in a semiquantitative fashion on a scale of 0–4, corresponding to <1, 1, 2–10, 11–25, and >25 cells per low power field, respectively. For the purposes of analysis, specimens were considered to have a “high” number of leucocytes if they had ≥11 cells per low power field, or ≥3 on this scale. This definition was adapted from prior published studies of prosthetic joint infection and modified to suit our patient population and local diagnostic testing criteria [29–33]. The presence of organisms was recorded based on the number per high power field on a similar scale. Samples were also typically submitted for bacterial, mycobacterial, and fungal cultures. Pathological examination of tissue specimens was performed in the Pathology Laboratory at Tufts Medical Center, and samples were considered to have evidence of infection if there was infiltration with neutrophils, macrophages, or other inflammatory cells, or caseating granulomatous inflammation was seen [34].
PCR testing was performed at the University of Washington Molecular Diagnostic Laboratory, with samples sent for bacterial, fungal, and/or mycobacterial testing using methods previously described [12, 22, 35–40]. Broad-range bacterial PCR targeted the 16S rRNA (forward primer 27F sequence, 5’-AGAGTTTGATCCTGGCTCAG-3’; reverse primer 357-mL sequence, 5’-CTGCTGCCICCCGTAGGAG-3’) [35]. Mycobacterial PCR utilized 3 targets including hsp65 (TB11, 5’-ACCAACGATGGTGTGTCCAT-3’; TB12, 5’-CTTGTCGAACCGCATACCCT-3’) [41], rpoB (MF, 5′-CGACCACTTCGGCAACCG-3′; MR, 5′-TCGATCGGGC ACATCCGG-3′) [42], and 16S (as for bacteria). Fungal PCR also used 3 targets: 28S (NL1, 5′-GCATATCAA TAA GCGGAGGAAAAG-3′; NL4, 5′-GGTCCGTGTTT CAAGACGG-3′), ITS1 (ITS1, 5′-TCCGTAGGTGAACCTGC GG-3′; ITS2, 5′-GCATCGATGAAGAACGCAGC-3′), and ITS2 (ITS3, 5′-GCATCGATGAAGAACGCAGC-3′; ITS4, 5′-GCATATCAATAAGCGGAGGA-3′) [43]. The decision regarding which PCRs to send (bacterial, fungal, and/or mycobacterial) and the timing was determined by the individual clinicians managing each case.
Clinical Diagnosis of Infection, Diagnostic Accuracy, and Outcome Ascertainment
As no uniform criteria have been established for the diagnosis of infection, clinical infection was considered based on the following criteria: (1) presence of clinical manifestations that reflect host damage in the setting of microbial infection such as fever and/or systemic symptoms and/or localizing symptoms of infections and (2) laboratory or radiographic parameters indicative of host damage such as leukocytosis, elevated inflammatory markers, and microbiological, histological, and/or radiological evidence of infection [44]. The presence or absence of infection was defined using a gold standard composite clinical diagnosis based on the assessment of 3 independent infectious diseases physicians, as described above. Following this review, clinical classifications were aligned with PCR results to designate patients as true positive (PCR positive with clinical evidence of infection), false positive (PCR positive without clinical evidence of infection), true negative (PCR negative without clinical infection), or false negative (PCR negative with clinical infection). Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated.
The impact of the test results on clinical decision-making was assessed by examining medical records for documentation regarding antimicrobial changes made after PCR results were available. This included any active optimization of antibiotic therapy such as de-escalation to a narrower-spectrum agent, the transition of patients from intravenous (IV) to oral antibiotics, changing to a different class of antimicrobials, or discontinuation of antimicrobial therapy altogether [45].
Statistical Analysis
Categorical data were reported as percentages, and continuous data were reported as means ± standard deviations if normally distributed and medians with ranges if non-normally distributed. Odds ratios were calculated, and variables were compared across PCR result status using univariate logistic regression. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated by comparing the broad-range PCR result with our composite final clinical diagnosis.
RESULTS
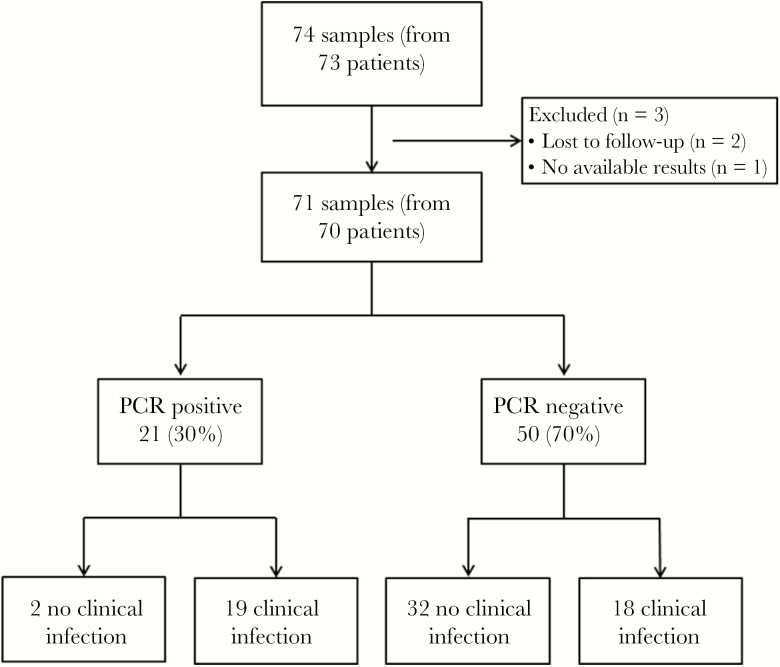
A total of 74 samples from 73 patients were sent for broad-range PCR testing during the study period. Three patients were excluded, 1 for whom no result was available and 2 who were lost to follow-up, leaving 71 samples from 70 patients in our final cohort (Figure 1). Individual sample details are available in the Supplementary Table. Thirty-nine patients (55%) were male, and the mean age was 57.8 ± 15.6 years. There were 13 patients who either did not have samples sent to microbiology at all or did not have sufficient tissue left in microbiology, and as such PCR testing was performed on formalin-fixed, paraffin-embedded (FFPE) tissue. There were 21 tissue samples with positive broad-range PCR results and 50 with negative results, 35 from orthopedic sites and 36 from nonorthopedic sites (Table 1). Two samples were positive for more than 2 organisms. Thirteen bacteria, 3 mycobacteria, and 7 fungi were identified. Organisms were mostly unique, though some were identified more than once, including Propionibacterium acnes (n = 2), Mycobacterium tuberculosis (n = 2), and Aspergillus species (n = 2). Antibiotic use was common (42/71; 59%), with 37 patients actively receiving antibiotics at the time the specimens were obtained, for a median duration (interquartile range) of 15 (6–44) days. There was an array of agents, but they were typically broad spectrum. Four patients were receiving long-term antibiotics for at least several months. An additional 5 patients had received antibiotics within the 4 weeks before sampling.
Figure 1.
Diagram demonstrating number and flow of samples/patients included in the study. Abbreviation: PCR, polymerase chain reaction.
Table 1.
Summary of Specimen Sites and Corresponding PCR Results
| Specimen Site (n = 71) |
No. of Samples | Organisms Identified |
|---|---|---|
| Nonorthopedic sites (n = 36) | ||
| Abdominal abscess | 2 | Aspergillus fumigatus/Ureaplasma urealyticuma |
| Bronchoalveolar lavage | 1 | Cryptococcus neoformans b |
| Brain | 1 | |
| Cerebrospinal fluid | 3 | Fusobacterium nucleatum b |
| Epidural abscess | 1 | |
| Eye | 2 | |
| Heart valve | 5 | Bartonella henselae,bCunninghamella,bStreptococcus mitisb |
| Liver | 1 | |
| Lung | 2 | Pneumocystis jirovecii b |
| Lymph node | 2 | |
| Muscle | 1 | |
| Myocardium | 3 | Aspergillus species b |
| Pleural fluid | 1 | |
| Psoas abscess | 1 | Mycobacterium tuberculosis b |
| Sinus | 2 | Rhizopus oryzae b |
| Spine | 7 | Malassezia restricta,bMycobacterium tuberculosis,bPropionibacterium acnesb |
| Testis | 1 | |
| Orthopedic sites (n = 35) | ||
| Ankle | 2 | Streptococcus agalactiae b |
| Hip | 11 | Streptococcus pneumoniae,bMycobacterium avium complexb |
| Knee | 15 | Staphylococcus epidermidis,bStreptococcus mitisb |
| Phalanx | 1 | |
| Tibia | 4 | Propionibacterium acnes,bPseudomonas aeruginosa,bStaphylococcus pettenkoferi/Staphylococcus pseudolugdunensisa |
| Wrist | 2 |
Abbreviation: PCR, polymerase chain reaction.
aBoth organisms identified in the same sample.
bIsolated from a single specimen alone, with no other organisms.
In deciding presence or absence of clinical infection, there was complete agreement in 58% of cases, and the remainders were by majority opinion. Thirty-seven patients (52%) were thought to have likely infection according to our composite clinical gold standard diagnosis. Of these, 19 had positive PCR results (true positives), and 18 had negative PCR results (false negatives). Of the 34 without clinical infection, 2 patients had positive PCR results (false positives), and 32 patients had negative PCR results (true negatives). This correlated to a sensitivity of 51%, specificity of 94%, PPV of 91%, and NPV of 64% (Table 2). Patients with false positives were also examined in more detail. One sample classified as false positive was Malassezia restricta, obtained from a computed tomography–guided biopsy of the spine and considered to represent a sample retrieval contaminant given the nature of the organism. The second false positive was an Aspergillus species from a routine myocardial biopsy of a heart transplant recipient. The patient was asymptomatic, but the sample was sent for PCR due to the presence of micro-abscesses seen on pathologic examination. After discussion with a fungal expert from the reference laboratory, it was considered an environmental or laboratory contaminant as the Aspergillus PCR was positive in only 1 out of the 2 runs, and the result was inconsistent with the clinical context.
Table 2.
Sensitivity, Specificity, Positive and Negative Predictive Values of Universal PCR as Compared With a Gold Standard of Composite Clinical Diagnosis Determined by a Panel of Infectious Diseases Physicians
| Infection Present | Infection Absent | ||
|---|---|---|---|
| PCR positive | 19 (true positive) | 2 (false positive) | PPV = 91% |
| PCR negative | 18 (false negative) | 32 (true negative) | NPV = 64% |
| Sensitivity = 51% | Specificity = 94% |
Abbreviations: NPV, negative predictive value; PCR, polymerase chain reaction; PPV, positive predictive value.
Although the standard approach in our institution was to only send samples with negative cultures for broad-range PCR, on review 11 samples were actually culture positive. The growth in these cultures usually occurred after at least several days of incubation, by which time samples had already been sent for PCR testing. Of 19 patients with true-positive PCR results, 6 patients also had positive cultures; 4 of these were concordant with the PCR (2 M. tuberculosis isolates from a psoas abscess and spinal tissue, Streptococcus pneumoniae from hip tissue, and Pseudomonas aeruginosa from a tibial sample) and 2 were discordant (Cryptococcus neoformans from bronchoalveolar lavage and Rhizopus oryzae from sinus tissue, both identified by PCR but not cultures). The isolates identified in the 5 culture-positive, PCR-negative specimens included 1 Mycobacterium avium complex, 2 Propionibacterium acnes, 1 P. aeruginosa, and 1 coagulase-negative Staphylococcus.
A comparison of patient characteristics stratified by PCR results is shown in Table 3. The only factors significantly associated with a positive PCR result were high PMN count and signs of inflammation on histopathologic examination. No significant differences were detected between PCR result and specimen site (orthopedic vs nonorthopedic) or nature of the specimen (fresh vs paraffin-embedded), macroscopic operative findings, or inflammatory markers. Antibiotic therapy was significantly more likely to be modified in patients with positive PCR results compared with those with negative results (52% vs 14%; OR, 6.44; P = .002). Details of the specific cases where the result clearly affected management are shown in Table 4. In total, 18/71 patients (25%) had management alterations due to PCR results.
Table 3.
Patient Characteristics Stratified by Universal PCR Result
| Characteristic | PCR Negative (n = 50) | PCR Positive (n = 21) | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|
| Age, mean ± SD, y | 57.5 ± 15.5 | 58.7 ± 16.1 | 1.005 (0.97–1.04) | .77 |
| Orthopedic site, No. (%) | 27 (54) | 8 (38) | 0.52 (0.18–1.46) | .22 |
| ≥3+ PMNs on gram stain (n = 60), No. (%) | 5/43 (12) | 6/17 (35) | 4.17 (1.06–16.67) | .04 |
| CRP | 48 ± 50 | 73 ± 62 | 1.008 (0.997–1.019) | .13 |
| ESR | 58 ± 33 | 67 ± 36 | 1.0079 (0.9888–1.028) | .42 |
| Pathologic signs infection, No. (%) | 16/37 (43) | 13/16 (81) | 5.69 (1.52–27.96) | .02 |
| Operative signs of infection, No. (%) | 18/41 (44) | 7/13 (54) | 1.49 (0.42–5.39) | .53 |
| Receiving antibiotics at time of specimen collection, No. (%) | 29 (58) | 13 (62) | 1.18 (0.42–3.45) | .76 |
| Antibiotic change following result, No. (%) | 7 (14) | 11 (52) | 6.44 (2.05–21.89) | .002 |
| FFPE sample (vs fresh), No. (%) | 8 (16) | 5 (24) | 0.61 (0.18–2.27) | .44 |
Abbreviations: CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FFPE, formalin-fixed, paraffin-embedded; PCR, polymerase chain reaction; PMN, polymorphonuclear leukocyte.
Table 4.
Description of the 18 Patients in Whom PCR Results Had Significant Impact on Antibiotic Therapy
| Diagnosis | Initial Antibiotics | Culture Result | PCR Result | Antibiotic Change |
|---|---|---|---|---|
| Nonunion following open tibial fracture | Vancomycin, cefepime | No growth | Staphylococcus pettenkoferi, Staphylococcus pseudolugdunensis | Vancomycin/cefepime stopped, linezolid started |
| Knee swelling in the setting of PTLD | Vancomycin, cefepime | No growth | Negative | Antibiotics stopped |
| Disseminated fungal infection | Linezolid, micafungin, voriconazole | No growth | Cunninghamella | Patient expired before change could be madea |
| Right prosthetic knee joint infection | Vancomycin, ampicillin | No growth | Streptococcus mitis | Changed to ceftriaxoneb |
| Left hip pain (native joint) | Vancomycin | No growth | Negative | Antibiotic stopped |
| Imaging evidence of 1-cm right frontal lobe mass, with question of granuloma | None | No growth | Negative | No antibiotic was started, and patient was discharged from ID clinic after PCR result |
| Brain abscess | Vancomycin, ceftriaxone, metronidazole, levofloxacin | No growth | Fusobacterium nucleatum | Vancomycin/levofloxacin stopped, ceftriaxone/metronidazole continued |
| Pelvic abscess | Vancomycin, cefepime | Streptococcus pneumoniae | Streptococcus pneumoniae | Cefepime stopped, vancomycin continued |
| Treated left MSSA septic knee, tested before revision | Cefazolin | No growth | Negative | Antibiotic stopped |
| Destructive cervical spine lesion by imaging | Vancomycin, ceftriaxone | No growth | Negative | Antibiotics stopped |
| Pelvic abscess | Ciprofloxacin, meropenem, daptomycin, micafungin | No growth | Ureaplasma urealyticum, Aspergillus fumigatus | Changed to moxifloxacin, fluconazolec |
| Right ankle infection with hardware in situ | Vancomycin | No growth | Propionibacterium acnes | Changed to penicillin |
| HIV with pneumonia | None | Haemophilus influenzae, lactobacillus, Candida glabrata | Cryptococcus neoformans | Fluconazole started following PCR result |
| Left hip prosthetic infection | Vancomycin, ertapenem | No growth | Mycobacteria avium complex | Rifampin and azithromycin started |
| Rheumatoid arthritis with failed multiple therapies to rule out infection | None | No growth | Negative | ID cause was ruled out |
| T10-T11 osteomyelitis with hardware in situ | Vancomycin, ertapenem | No growth | Propionibacterium acnes | Changed to penicillin |
| Question of right prosthetic joint infection | Daptomycin, meropenem | No growth | Negative | De-escalated to daptomycin and ciprofloxacin |
| Left ankle septic joint with exposed hardware | Cipofloxacin, bactrim | No growth | Streptococcus agalactiae | Changed to amoxicillin |
Abbreviations: ID, infectious diseases; PCR, polymerase chain reaction; PTLD, post-transplant lymphoproliferative disorder; T10, 10th thoracic vertebra; T11, 11th thoracic vertebra.
aIncluded in table due to actionable result despite death before change could be made.
bChanged to ceftriaxone for ease of administration, not due to suspicion of ampicillin resistance.
c Aspergillus was thought to be a contaminant given the clinical picture; no treatment for Aspergillus was initiated, but fluconazole was added instead of micafungin for possible intra-abdominal candidasis.
DISCUSSION
Obtaining an accurate microbiologic diagnosis is 1 of the key factors informing choice of antimicrobial therapy in patients with serious infections. Though it is costlier than culture-based methods, our findings suggest that broad-range PCR is a valuable addition to the diagnostic workup of patients with culture-negative infections. Although some studies have suggested that PCR is not superior to culture [21, 26], in our cohort, use of the broad-range PCR assay led to identification of infecting pathogens in approximately half of the patients who were clinically suspected of having infection. Had the assay not been performed, it is possible that 1 patient would have been inappropriately untreated, 4 patients would have been overtreated based on suspected infection, and 11 patients would have been treated with a less effective or inappropriate antibiotic regimen. Given the serious consequences of untreated infections, the broad-range PCR assay was of considerable value in our patient population. It also demonstrated significant value from an antibiotic stewardship perspective, especially in PCR-positive cases, leading to de-escalation in 8 cases, change from intravenous to oral in 3 cases, starting appropriate therapy in 1 case, and stopping antibiotics in 4 cases.
This study is 1 of the first to examine the performance and clinical impact of broad-range PCR in real-world clinical practice, in a setting where most patients had negative cultures [10, 11, 28]. Previous studies have demonstrated the ability of broad-range PCR to identify organisms from heart valve tissue [11, 14–16], joint tissue [12], and other sterile sites, with sensitivity and specificity ranging from 43% to 96% and 72% to 95%, respectively. However, these studies typically compared PCR results with standard culture results as a gold standard. In this study, we reviewed each case in detail and used a “composite clinical diagnosis” as the gold standard rather than only culture results, allowing us to calculate a more realistic sensitivity and specificity than prior studies, which likely explains why our values were somewhat lower than what has been previously described.
Our findings suggest that a positive result is more clinically impactful than a negative result and plays a bigger role in antimicrobial decision-making. In our cohort, the likelihood of obtaining a positive result was significantly higher in patients with signs of inflammation on microbiologic or histological examination. These findings are consistent with prior studies [17, 20, 46]. Factors such as these could be taken into consideration by ordering clinicians aiming to increase the diagnostic yield. Although there have been some studies that reviewed the effect of the PCR result on antibiotic management [27, 28, 47], none of these assessed if there was any difference in antimicrobial management between patients testing PCR positive vs negative.
There are some important limitations that should be kept in mind when interpreting these results. Statistical power was limited by a relatively small sample size, and given the retrospective nature of the study, all available tests were not performed on all samples. Selection bias may influence interpretation of our results and generalizability, as many of our patients were complex with multiple comorbidities, prolonged hospitalization, and antimicrobial exposure. Although we attempted to approximate a true gold standard, incorporating all available clinical information, in some cases it was difficult to truly know if infection was present or absent. Despite the fact that this test was supposed to be performed only in culture-negative patients, there were 11 cases with positive cultures; although it may have been clinically justified to order the broad-range PCR despite this in some cases, it is important for clinicians to remember to wait sufficient time for initial cultures to be finalized in order to avoid performing PCR unnecessarily. Although antimicrobial changes were temporally associated with PCR results’ availability, it is possible that other unmeasured factors influenced this decision-making. Finally, we were unable to assess other clinical outcomes beyond antibiotic selection.
In summary, our findings suggest that broad-range PCR is a clinically useful test that has an important role in the diagnostic evaluation of patients with culture-negative infections. Optimizing specimen selection by considering the full clinical scenario including microbiological and histopathological data can increase the likelihood of a positive result, which in our population had the biggest impact on antimicrobial decision-making. Infectious diseases physicians should carefully consider these advantages and limitations on an individualized basis before requesting this complex, difficult to interpret, and expensive test.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors would like to acknowledge Winston Edwards for help with specimen identification, Dhruba J. SenGupta, PhD, for PCR method description, and Jacob Garrell for assistance with manuscript editing.
Financial support. This work was supported by the Tufts Medical Center Division of Geographic Medicine and Infectious Disease Francis P. Tally MD Fellowship.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Prior presentation: This work was previously presented at ID Week, and the abstract was published by Open Forum Infectious Diseases (Clinical utility of universal PCR and its real-world impact on patient management. Open Forum Infect Dis 2017; 4(Suppl 1):S627).
References
- 1. Harbarth S, Nobre V, Pittet D. Healthcare epidemiology: does antibiotic selection impact patient outcome?Clin Infect Dis 2007; 44:87–93. [DOI] [PubMed] [Google Scholar]
- 2. Kollef MH. Inadequate antimicrobial treatment: an important determinant of outcome for hospitalized patients. Clin Infect Dis 2000; 31(Suppl 4):S131–8. [DOI] [PubMed] [Google Scholar]
- 3. Tunkel AR, Hartman BJ, Kaplan SL, et al. Practice guidelines for the management of bacterial meningitis. Clin Infect Dis 2004; 39:1267–84. [DOI] [PubMed] [Google Scholar]
- 4. Tunkel AR, Glaser CA, Bloch KC, et al. ; Infectious Diseases Society of America The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 2008; 47:303–27. [DOI] [PubMed] [Google Scholar]
- 5. Osmon DR, Berbari EF, Berendt AR, et al. ; Infectious Diseases Society of America Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 2013; 56:e1–e25. [DOI] [PubMed] [Google Scholar]
- 6. Baddour LM, Wilson WR, Bayer AS, et al. ; American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals from the American Heart Association. Circulation 2015; 132:1435–86. [DOI] [PubMed] [Google Scholar]
- 7. Kolbert CP, Persing DH. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr Opin Microbiol 1999; 2:299–305. [DOI] [PubMed] [Google Scholar]
- 8. Clarridge JE., III Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004; 17:840–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petti CA. Detection and identification of microorganisms by gene amplification and sequencing. Clin Infect Dis 2007; 44:1108–14. [DOI] [PubMed] [Google Scholar]
- 10. Rampini SK, Bloemberg GV, Keller PM, et al. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis 2011; 53:1245–51. [DOI] [PubMed] [Google Scholar]
- 11. Marín M, Muñoz P, Sánchez M, et al. Group for the Management of Infective Endocarditis of the Gregorio Marañón Hospital Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 2007; 86:195–202. [DOI] [PubMed] [Google Scholar]
- 12. Bémer P, Plouzeau C, Tande D, et al. Centre de Référence des Infections Ostéo-articulaires du Grand Ouest (CRIOGO) Study Team Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J Clin Microbiol 2014; 52:3583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kobayashi N, Procop GW, Krebs V, et al. Molecular identification of bacteria from aseptically loose implants. Clin Orthop Relat Res 2008; 466:1716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Faraji R, Behjati-Ardakani M, Moshtaghioun SM, et al. The diagnosis of microorganism involved in infective endocarditis (IE) by polymerase chain reaction (PCR) and real-time PCR: a systematic review. Kaohsiung J Med Sci 2018; 34:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bosshard PP, Kronenberg A, Zbinden R, et al. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin Infect Dis 2003; 37:167–72. [DOI] [PubMed] [Google Scholar]
- 16. Fournier PE, Thuny F, Richet H, et al. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 2010; 51:131–40. [DOI] [PubMed] [Google Scholar]
- 17. Bori G, Soriano A, García S, et al. Neutrophils in frozen section and type of microorganism isolated at the time of resection arthroplasty for the treatment of infection. Arch Orthop Trauma Surg 2009; 129:591–5. [DOI] [PubMed] [Google Scholar]
- 18. Del Pozo JL, Patel R. Clinical practice. Infection associated with prosthetic joints. N Engl J Med 2009; 361:787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Francés Borrego A, Martínez FM, Cebrian Parra JL, et al. Diagnosis of infection in hip and knee revision surgery: intraoperative frozen section analysis. Int Orthop 2007; 31:33–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyamae Y, Inaba Y, Kobayashi N, et al. Quantitative evaluation of periprosthetic infection by real-time polymerase chain reaction: a comparison with conventional methods. Diagn Microbiol Infect Dis 2012; 74:125–30. [DOI] [PubMed] [Google Scholar]
- 21. Borde JP, Häcker GA, Guschl S, et al. Diagnosis of prosthetic joint infections using UMD-Universal Kit and the automated multiplex-PCR Unyvero i60 ITI(®) cartridge system: a pilot study. Infection 2015; 43:551–60. [DOI] [PubMed] [Google Scholar]
- 22. Grif K, Heller I, Prodinger WM, et al. Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis. J Clin Microbiol 2012; 50:2250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haag H, Locher F, Nolte O. Molecular diagnosis of microbial aetiologies using SepsiTest™ in the daily routine of a diagnostic laboratory. Diagn Microbiol Infect Dis 2013; 76:413–8. [DOI] [PubMed] [Google Scholar]
- 24. Rothman R, Ramachandran P, Yang S, et al. Use of quantitative broad-based polymerase chain reaction for detection and identification of common bacterial pathogens in cerebrospinal fluid. Acad Emerg Med 2010; 17:741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marín M, Garcia-Lechuz JM, Alonso P, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol 2012; 50:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson KE, Kiyatkin DE, An AT, et al. PCR offers no advantage over culture for microbiologic diagnosis in cellulitis. Infection 2012; 40:537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akram A, Maley M, Gosbell I, et al. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis 2017; 57:144–9. [DOI] [PubMed] [Google Scholar]
- 28. Alraddadi B, Al-Azri S, Forward K. Influence of 16S ribosomal RNA gene polymerase chain reaction and sequencing on antibiotic management of bone and joint infections. Can J Infect Dis Med Microbiol 2013; 24:85–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garcia LS, Isenberg HD.. Clinical Microbiology Procedures Handbook. Washington, DC: ASM Press; 2010. [Google Scholar]
- 30. Lonner JH, Desai P, Dicesare PE, et al. The reliability of analysis of intraoperative frozen sections for identifying active infection during revision hip or knee arthroplasty. J Bone Joint Surg Am 1996; 78:1553–8. [DOI] [PubMed] [Google Scholar]
- 31. Banit DM, Kaufer H, Hartford JM. Intraoperative frozen section analysis in revision total joint arthroplasty. Clin Orthop 2002; 401:230–8. [DOI] [PubMed] [Google Scholar]
- 32. Wong YC, Lee QJ, Wai YL, Ng WF. Intraoperative frozen section for detecting active infection in failed hip and knee arthroplasties. J Arthroplasty 2005; 20:1015–20. [DOI] [PubMed] [Google Scholar]
- 33. Zhao X, Guo C, Zhao G-S, et al. Ten versus five polymorphonuclear leukocytes as threshold in frozen section tests for periprosthetic infection: a meta-analysis - ScienceDirect. J Arthroplasty 2013; 28:913–7. [DOI] [PubMed] [Google Scholar]
- 34. Guarner J. Incorporating pathology in the practice of infectious disease: myths and reality. Clin Infect Dis 2014; 59:1133–41. [DOI] [PubMed] [Google Scholar]
- 35. Lee SA, Plett SK, Luetkemeyer AF, et al. Bartonella quintana aortitis in a man with AIDS, diagnosed by needle biopsy and 16S rRNA gene amplification. J Clin Microbiol 2015; 53:2773–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rakeman JL, Bui U, Lafe K, et al. Multilocus DNA sequence comparisons rapidly identify pathogenic molds. J Clin Microbiol 2005; 43:3324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen YC, Eisner JD, Kattar MM, et al. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J Clin Microbiol 2001; 39:4042–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harrington AT, Creutzfeldt CJ, Sengupta DJ, et al. Diagnosis of neurocysticercosis by detection of Taenia solium DNA using a global DNA screening platform. Clin Infect Dis 2009; 48:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stockinger DE, Roellich KM, Vogel KW, et al. Primary hepatic Mycobacterium tuberculosis complex infection with terminal dissemination in a pig-tailed macaque (Macaca nemestrina). J Am Assoc Lab Anim Sci 2011; 50:258–62. [PMC free article] [PubMed] [Google Scholar]
- 40. Adékambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 2003; 41:5699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brunello F, Ligozzi M, Cristelli E, et al. Identification of 54 mycobacterial species by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J Clin Microbiol 2001; 39:2799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itoh S, Kazumi Y, Abe C, Takahashi M. Heterogeneity of RNA polymerase gene (rpoB) sequences of Mycobacterium gordonae clinical isolates identified with a DNA probe kit and by conventional methods. J Clin Microbiol 2003; 41:1656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leaw SN, Chang HC, Sun HF, et al. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microbiol 2006; 44:693–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pirofski LA, Casadevall A. The meaning of microbial exposure, infection, colonisation, and disease in clinical practice. Lancet Infect Dis 2002; 2:628–35. [DOI] [PubMed] [Google Scholar]
- 45. Barlam TF, Cosgrove SE, Abbo LM, et al. IDSA: implementing an antibiotic stewardship program. Clin Infect Dis 2016; 62:e51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyamae Y, Inaba Y, Kobayashi N, et al. Different diagnostic properties of C-reactive protein, real-time PCR, and histopathology of frozen and permanent sections in diagnosis of periprosthetic joint infection. Acta Orthop 2013; 84:524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gilbert EM, Yucebay F, Malczynski M, et al. Use of organism identification by 16S ribosomal RNA polymerase chain reaction to shorten antimicrobial length of therapy. Diagn Microbiol Infect Dis 2017; 88:163–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.