Abstract
Background
Dysregulation of the cell cycle has been implicated in esophageal squamous cell carcinoma (ESCC) progression. This study aimed to evaluate the role of miR-424 in cell cycle regulation and ESCC proliferation.
Methods
The role of miR-424 in cell proliferation was evaluated in vitro and in vivo. Transcriptional activation of miR-424 was determined using chromatin immunoprecipitation, and binding of miR-424 to targets was verified using miRNA ribonucleoprotein complex immunoprecipitation.
Findings
miR-424 was upregulated and correlated with poor survival in ESCC patients. Repression or overexpression of miR-424 respectively decreased or increased ESCC cell proliferation in vitro and in vivo. miR-424 expression is transcriptionally regulated by E2F1 and increased during G1/S transition. Knockdown or overexpression of miR-424 respectively inhibited or promoted both G1/S and G2/M cell cycle transitions in ESCC cells, and these effects were mediated by two newly identified miR-424 targets, PRKCD and WEE1, respectively. Consequently, elevation of PRKCD by miR-424 knockdown led to enhanced stability of the p21Cip1 protein via increased activation of PRKCD and downstream p38 MAPK and JNK signaling to block CDK2 activation and G1/S transition, while elevated WEE1 maintained CDC2 in an inactive state to block G2/M transition. However, circLARP4 could sponge the binding of miR-424 to PRKCD, thus compromising the regulation of G1/S progression by miR-424.
Interpretation
miR-424 coordinates a previously unknown, multilayered regulation of ESCC cell cycle progression to promote ESCC proliferation, and may be used as a novel prognostic marker and an effective therapeutic target for ESCCs.
Fund
National Natural Science Foundation of China.
Keywords: miR-424, Esophageal squamous cell carcinoma, Cell cycle, Cell proliferation
Research in context.
Evidence before this study
miR-424 was previously reported to exert a primarily tumor suppressive role in most types of cancers, such as breast cancer, cervical cancer, hepatocellular carcinoma, colon cancer, and leukemia. However, our preliminary microarray studies identified miR-424 as one of the most significantly upregulated miRNAs in esophageal squamous cell carcinoma (ESCC) tissues compared with normal esophageal tissues, suggesting a tumor-promoting role of it in ESCCs.
Added value of this study
Here, we proved that miR-424 is upregulated in ESCC and correlates with poor survival among ESCC patients. The expression of miR-424 is elevated along with cell cycle progression from G0/G1 to S-phase, which is transcriptionally regulated by G1/S transcription factor E2F1. In ESCCs, miR-424 exerts its tumor-promoting role by cell-cycle-phase specifically targeting PRKCD and WEE1, which induces subsequent changes in the expression levels or activities of downstream cyclin-dependent kinases (CDKs) or CDK inhibitory proteins (CKIs), to regulate G1/S and G2/M cell cycle transition, respectively. Furthermore, circLARP4, a natural sponge of miR-424 during G1/S transition, decreases miR-424 binding to PRKCD and modulates the effect of miR-424 on G1/S transition.
Implications of all the available evidence
Our study highlights an important tumor-promoting role for miR-424 in regulating ESCC cell cycle progression; miR-424 may possibly be used as a novel prognostic marker and an effective therapeutic target for ESCCs.
Alt-text: Unlabelled Box
1. Introduction
Esophageal cancer is one of the most aggressive malignancies of the gastrointestinal tract. Esophageal squamous cell carcinoma (ESCC) is the globally predominant pathological type of esophageal cancer [1]. In China, ESCC, which accounts for most malignant esophageal tumors, ranks as the third most common malignancy and the fourth most common cause of cancer-related death [2]. The risk factors for ESCC are thought to be related to dietary and lifestyle habits, as well as genetic polymorphisms [3]. However, the complicated molecular mechanisms underlying ESCC development and progression are not yet fully understood.
The cell cycle, the process by which cell division occurs, is a series of highly regulated steps that are orchestrated at the molecular level by the sequential activation or inactivation of cyclin-dependent kinases (CDKs); the activities of CDKs depend upon physical interactions with positive regulatory subunits cyclins or negative regulatory subunits known as CDK-inhibitory proteins (CKIs) [4]. Impaired function of critical gatekeepers of cell cycle progression caused by the accumulation of alterations involving the cell-cycle regulatory machinery will allow unscheduled persistent cell proliferation, which is a hallmark of cancer [5]. Dysregulation of the cell cycle by genomic perturbations, genetic mutations, and (or) altered expression of key molecules has been implicated in ESCC development [3,6].
MicroRNAs (miRNAs) are single-stranded non-coding small RNA segments that operate via sequence-specific interactions with the 3′ untranslated regions (3′UTRs) of mRNA targets to suppress translation and mRNA decay to regulate gene expression post-transcriptionally [7]. These molecules have been reported to be dysregulated in virtually all human cancer types, including ESCC, and function as either tumor suppressors or oncogenes [8]. To identify miRNAs that are potentially involved in ESCC development, we evaluated the miRNA profiles of ESCC and esophageal normal epithelia (NEs) tissues and identified miR-424 as one of the most significantly upregulated miRNAs in ESCC tissues compared with NE tissues, suggesting a tumor-promoting role of miR-424 in ESCCs. A role of miR-424 in inhibiting epithelial-mesenchymal transition and decreasing invasion and migration of ESCC cells has been previously shown [9]. Moreover, miR-424 exerts a primarily tumor-suppressing role in most types of cancers, such as breast cancer [10], cervical cancer [11,12], hepatocellular carcinoma [13], and leukemia [14]. miR-424 has also been demonstrated to target the cell cycle regulators cyclin E1, cyclin D1 [15], CHK1 [12] and CDC25A [16] to impair G1/S or G2/M cell cycle transition and delay cell proliferation.
The specific role of miR-424 in ESCC proliferation and the underlying mechanisms remain unknown. In this study, we characterized the tumor-promoting role of miR-424 in ESCC proliferation. Further in-depth studies led to the identification of previously unrecognized transcriptional regulators of miR-424 expression and the elucidation of the direct targets and capacities of miR-424 in coordinating ESCC cell cycle progression.
2. Materials and methods
2.1. Clinical specimens
This study was approved by the Research Ethics Committee of Sun Yat-sen University Cancer Center. The 30 ESCC samples and ten NE samples used for miRNA profiling analysis were collected as described in our previous studies [17,18]. Freshly frozen tissues from 60 paired NE and ESCC tissues and 190 ESCC tissues used for quantitative real-time polymerase chain reaction (qRT-PCR) analysis of miR-424 expression were obtained from ESCC patients undergoing complete surgical esophagectomy with no neoadjuvant or adjuvant treatment from March 2002 to October 2008 in the Department of Thoracic Oncology. The fresh tumor samples were taken from macroscopically judged neoplastic regions, and NE samples were from macroscopically judged normal regions at the surgical resection margin at least 3 cm distant from tumor regions. For each sample, an adjacent tissue sample was stained with hematoxylin and eosin and assessed for the presence or absence of ESCC or NE cells by pathologists. In addition, 110 formalin-fixed and paraffin-embedded (FFPE) ESCC tissue and 60 paired NE tissue samples that were randomly selected from these above 190 ESCC cases were used for immunohistochemistry (IHC) analyses of PRKCD and WEE1 protein expression. Tumor stage was evaluated according to the seventh edition of the American Joint Committee on Cancer tumor-node-metastasis (TNM) staging system.
2.2. Reagents and antibodies
Puromycin (Cat# P9620), thymidine (Cat# T18S95), nocodazole (Cat# M1404), propidium iodide (Cat# P4170), cycloheximide (Cat# C7698), and crystal violet (Cat# C6158) were purchased from Sigma-Aldrich Corporation (St. Louis, MO, USA). Rabbit anti-pHH3 (Ser10, Cat# 3377) and a goat anti-rabbit IgG secondary antibody conjugated with Alexa Fluor 647 (Cat# A-21244) used for flow cytometry were purchased from Cell Signaling Technology (Danvers, MA, USA) and Life Technologies/Thermo Fisher Scientific Technology (Carlsbad, CA, USA), respectively.
For western blotting, the primary antibodies anti-PRKCD (Cat# 9616), anti-p-PRKCD (Thr505, Cat# 9374), anti-WEE1 (Cat# 4936), anti-p21Cip1 (Cat# 2947), anti-p27Kip1 (3686), anti-p-CDK2 (Thr160, Cat# 2561), anti-CDK2 (Cat# 2546), anti-p-CDC2 (Tyr15, Cat# 4539), anti-LATS2 (Cat# 13646), anti-PDCD4 (Cat# 9535), anti-CDC25A (Cat# 3652), anti-cyclin D1 (Cat# 2922), anti-cyclin E1 (Cat# 4129), anti-cyclin B1(Cat# 12231), anti-p-JNK (Thr183/Tyr185, Cat# 4668), anti-JNK (Cat# 9252), anti-p38 MAPK (Cat# 8690), anti-p-p38 MAPK (Thr180/Tyr182, Cat# 4511), and anti-E2F1 (Cat# 3742) were purchased from Cell Signaling Technology, anti-CHK1 (Cat# sc-8408) and anti-CDC2 (Cat# sc-954) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA), and anti-GAPDH (Cat# KC-5G4) was purchased from (KangChen Bio-tech, Shanghai, China). The horseradish peroxidase-conjugated secondary antibodies anti-rabbit (Cat# NA934) and anti-mouse (Cat# NA931) IgG were purchased from GE Healthcare (Little Chalfont, UK). An anti-AGO2 antibody (Cat# RN003M) used for miRNA ribonucleoprotein complex immunoprecipitation (miRNA-IP) assays was purchased from MBL (Nagoya, Aichi, Japan). An anti-E2F1 antibody (Cat# 17-10061) used for chromatin immunoprecipitation (ChIP) assays was purchased from Merck Millipore (Burlington, MA, USA). Anti-PRKCD (Cat# sc-8402) and anti-WEE1 (Cat# 13084) antibodies used for IHC were purchased from Santa Cruz and Cell Signaling Technology, respectively.
2.3. Cell lines
The ESCC cell lines KYSE-410 and KYSE-510 were obtained from DSMZ, the German Resource Center for Biological Materials [19], and cultured in Dulbecco's modified Eagle's medium (Life Technologies/Thermo Fisher Scientific; Cat# C11995500BT) supplemented with 10% fetal bovine serum (Life Technologies/Thermo Fisher Scientific; Cat# 10270). The immortalized esophageal epithelial cell line NE1 was provided by Professor GS Tsao (The University of Hong Kong) and cultured in EpiLife medium with 60 μM calcium (Life Technologies/Thermo Fisher Scientific; Cat# MEPI500CA) mixed with defined keratinocyte-SFM (Life Technologies/Thermo Fisher Scientific; Cat# 10744019). All cell lines were cultured in a humidified incubator containing 5% CO2 at 37 °C.
2.4. Vectors, lentiviral transduction, and transfection
Lentiviral constructs expressing miRZip-424 (System Biosciences, Palo Alto, CA, USA: Cat# MZIP424-PP-1), which knocks down miR-424 functionally, lenti-miR-424 (System Biosciences; Cat# PMIRH424PP-1), which expresses the miR-424 precursor, or the corresponding controls (System Biosciences; Cat# MZIP000-PP-1 and PMIRH000PP-1) were packaged using the ViraPower Lentiviral Packaging Mix (Life Technologies/Thermo Fisher Scientific; Cat# K497500) in 293FT cells, respectively. Stable cell lines expressing miRZip-424, lenti-miR-424, or the corresponding controls were generated via lentiviral transduction and selected with puromycin or by flow cytometry sorting for GFP positive populations. Small interfering RNAs (siRNAs) targeting E2F1 and a scrambled control were purchased from RiboBio Co., Ltd. (Guangzhou, China; Cat# stQ0001999-1). Transfection of siRNA or plasmids was conducted using Lipofectamine 2000 reagent (Life Technologies/Thermo Fisher Scientific; Cat# 11668019) according to the manufacturer's instructions. The shRNA and siRNA sequences are presented in Supplementary Table S1.
2.5. RNA isolation, miRNA microarray, and qRT-PCR analysis
For the 30 ESCC and ten NE tissue samples used for miRNA profiling analysis, total RNA was isolated using the mirVana miRNA Isolation Kit (Life Technologies/Thermo Fisher Scientific; Cat# AM1561). Human miRNA microarrays based on miRBase Release 18.0 (Agilent Technologies, Santa Clara, CA, USA; Cat# G4872A) were adopted for miRNA expression profiling analysis. Genespring 12.0 (Agilent Technologies) software was used to analyze the microarray data. Student's t-test for unpaired samples was used to identify differentially expressed miRNAs between ESCC and NE samples. Hierarchical clustering was performed using the Pearson centered correlation as a distance metric and the average linkage algorithm based on significantly different genes to combine cluster branches.
For the tissue and cell samples used for qRT-PCR analysis, total RNA was extracted using TRIzol reagent (Life Technologies/Thermo Fisher Scientific; Cat# 15596018). TaqMan miR-424 (Assay ID: 000604, Life Technologies/Thermo Fisher Scientific; Cat# 4427975) qRT-PCR analysis was performed as described previously [18] with RNU6B (Assay ID: 001093, Life Technologies/Thermo Fisher Scientific; Cat# 4427975) as an internal control. For qRT-PCR analyses of pri-miR-424, pre-miR-424, and mRNAs, total RNA was reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis Kit (Life Technologies/Thermo Fisher Scientific; Cat# K1621). PCR amplification of pri-miR-424 was then performed using the TaqMan pri-miRNA assay kit (Assay ID: Hs03303697_pri, Life Technologies/Thermo Fisher Scientific; Cat# 4427012) and the TaqMan Universal PCR Master Mix (Life Technologies/Thermo Fisher Scientific; Cat# 4304437), with GAPDH (Assay ID: 4333764, Life Technologies/Thermo Fisher Scientific; Cat# 4333764T) as an internal control. PCR amplification of pre-miR-424 and mRNAs was performed using Power SYBR Green PCR Master Mix (Life Technologies/Thermo Fisher Scientific; Cat# 4367660) with GAPDH as an internal control. The specific primers used for qRT-PCR are presented in Supplementary Table S1. All PCR analyses were performed in a Light Cycler 480 thermocycler (Roche Diagnostics, Indianapolis, IN, USA). qRT-PCR for each gene was performed in triplicate. Relative gene expression was analyzed using the 2-ΔCp method [20].
2.6. Cell proliferation in vitro
For growth curve measurements, KYSE-410 and KYSE-510 cells stably expressing lenti-miR-424, miRZip-424, or the corresponding control vectors and NE1 cells transduced with lenti-miR-424 or the control vectors for 3 days were prepared as single-cell suspensions and seeded in 96-well plates. The cell proliferation rate was detected using a CCK-8 cell proliferation kit (Dojindo, Minato-ku, Tokyo, Japan; Cat# CK04). For the colony formation assay, 200 cells were seeded into 6-well plates. After ten days of culture, the surviving colonies (>50 cells/colony) were counted with 1% crystal violet staining. Three independent experiments were performed.
2.7. Tumor formation in nude mice
All animal experiments were performed according to the guidelines of the Council on Animal Care and approved by Sun Yat-sen University. Female BALB/c nude mice, aged 4–5 weeks, were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. A total of 6 × 106 KYSE-410 and KYSE-510 cells stably expressing miRZip-424 or the control vectors were injected subcutaneously into the dorsal flanks of nude mice. Each group contained five mice. Tumor size was measured every 3 days. After 5–6 weeks, the mice were killed, and the tumors were dissected. Tumor volumes were calculated as follows: volume = (D × d2)/2, where D is the longest diameter and d is the shortest diameter.
2.8. Cell synchronization and cell cycle assay by flow cytometry
ESCC cells stably expressing lenti-miR-424, miRZip-424, or the corresponding control vectors were synchronized in G0/G1-phase by serum starvation, at the onset of S-phase using a double-thymidine block (2 mM), or in mitosis with 100 ng/ml nocodazole treatment for 20 h as described previously [21]. The cells were collected at the indicated time points after release from synchronization, fixed in 70% ethanol, and stained with 50 μg/ml propidium iodide. If further discrimination between G2-phase and M-phase was required, the fixed cells were stained with rabbit anti-pHH3 (Ser10) antibody and a secondary antibody conjugated with Alexa Fluor 647, followed by propidium iodide staining. The cell cycle distribution was monitored with a Gallios flow cytometer (Beckman Coulter, Brea, CA, USA) and analyzed with FlowJo v10.
2.9. Western blotting
Cells were lysed in ice-cold lysis buffer with 1× protease inhibitor cocktail (Roche Diagnostics; Cat# 4693116001) and 1× phosphatase inhibitor cocktail 2 (Sigma-Aldrich; Cat#P5726) and 3 (Sigma-Aldrich; Cat# P0044). Protein concentrations were determined using the Protein Assay Reagent (Bio-Rad Laboratories, Cat# 5000002). Cell lysates were subjected to electrophoresis on acrylamide gels with a proper concentration. Separated proteins were transferred onto PVDF membranes (Merck Millipore; Cat# IPVH00010) and probed with primary antibodies after blocking with 5% dried milk and secondary antibodies. The antigen-antibody complexes were visualized with ECL Western Blotting Substrate (Life Technologies/Thermo Fisher Scientific; Cat#32106) on a ChemiDoc Touch (Bio-Rad Laboratories, Hercules, CA, USA). The specific intensity of each protein band was measured by Image Lab (Bio-Rad Laboratories) software and expressed as the ratio of the optical density of the band for each protein to that of GAPDH.
2.10. 3′UTR cloning and luciferase assay
Oligonucleotide pairs of PRKCD and WEE1 3′UTRs containing predicted binding sites for miR-424 (Supplementary Table S1) were synthesized, annealed, and ligated into the pmirGLO Dual-Luciferase miRNA target expression vector (Promega, Madison, WI, USA; Cat# E1330). HEK293 cells were transfected with the luciferase reporter plasmid plus lenti-miR-424 or miRZip-424. After 48 h, luciferase activities were assessed using the Dual-Luciferase 1000 Assay System (Promega; Cat# E1980) by measuring the luminescence signal in a GLOMAX luminometer (Promega). The experiments were performed in triplicate and repeated three times with negative controls.
2.11. miRNA-IP assay
KYSE-410 and KYSE-510 cells stably expressing miR-424 precursor or control vectors were collected after release from G0/G1-phase for 12 h or from S-phase onset for 9 h. miRNA-IP assays of the collected cells were performed using the RiboCluster Profile RIP-Assay kit for microRNA (MBL; Cat# RN1005) following the manufacturer's protocol with Protein A Plus Agarose (Life Technologies/Thermo Fisher Scientific; Cat# 22180) and an anti-AGO2 antibody. RNA isolated from the immune complex of miRNA-IP was used for cDNA synthesis. qRT- PCR was performed to determine PRKCD, WEE1 and circLARP4 enrichment in the RNA-induced silencing complex (RISC); the average values of RISC-associated GAPDH, B2M, and GUSB were used for normalization [22]. The primers used for qRT-PCR of miRNA-IP product are presented in Supplementary Table S1. Data are presented as the 2-ΔΔCp, where ΔCp = CpRIP – CpInput, and ΔΔCp = ΔCp – ΔCpcontrol.
2.12. ChIP assay
KYSE-410 cells were collected after release from G0/G1 for 8 h. ChIP assay of the collected cells were performed using the EZ-Magna ChIP HiSens Kit (Merck Millipore, Burlington, MA, USA; Cat# 17-10461) according to the manufacturer's instructions with an anti-E2F1 antibody. ChIP DNA enrichment was analyzed by qRT-PCR using primers presented in Supplementary Table S1. Data are presented as 2-ΔΔCp, where ΔCp = CpChIP – Cpinput, and ΔΔCp = ΔCp – ΔCpnegative. The quantification of DNA enrichment in the CDC2 gene promoter was used as a positive control, and GAPDH was used as a negative control.
2.13. miR-424 promoter cloning and luciferase assay
A 1750-bp putative promoter sequence that contained a portion of pri-miR-424 and was therefore located upstream of miR-424 was synthesized and cloned into the pEZX-LvPG04 lentiviral reporter vector (GeneCopoeia, Rockville, MD, USA). A QuickChange Site-Directed Mutagenesis kit (Agilent Technologies; Cat# 210518) was used to mutagenize the E2F1 binding sites with the primers presented in Supplementary Table S1. KYSE-410 cells were infected with the lentiviral particles and selected with puromycin. The luciferase activities were assessed using a Secrete-Pair Dual Luminescence assay kit (GeneCopoeia, Cat# LF031).
2.14. IHC
IHC was performed using the standard peroxidase anti-peroxidase complex method. Briefly, FFPE sections were deparaffinized and rehydrated. Peroxidase inactivation and antigen retrieval were achieved by incubating samples in 3% H2O2 and EDTA buffer. The primary antibodies used were PRKCD (1:40 dilution) and WEE1 (1:100 dilution). Immunoperoxidase staining was carried out using peroxidase-conjugated polymer solution (ZSGB-Bio, Beijing, China; Cat# PV6000). Then the slides were visualized with 3,3′-diaminobenzidine (ZSGB-Bio; Cat# ZLI-9017) and counterstained with hematoxylin. A pathologist blinded to the clinicopathological information performed the evaluation of PRKCD and WEE1 protein expression by using a semi-quantitative system as described previously [23].
2.15. Statistical analysis
Calculations of the mean value ± standard deviation (SD) were based on three independent experiments. Data analyses, except for microarray analysis, were carried out using the SPSS 22.0 statistics package (IBM, Armonk, NY, USA). Continuous variables were analyzed using Student's t-test. Repeated measures ANOVA was performed to assess the statistical significance of cell growth curves and tumor growth in nude mice. The correlations between miR-424 expression and the expression of target proteins or clinicopathological parameters were analyzed by the Chi-square test. Overall survival was defined as the time from surgery to death, and patients who were still alive at the time of last follow-up were censored. Survival curves were analyzed by the Kaplan-Meier method and the log-rank test. The Cox proportional hazards regression model was used to identify independent prognostic factors. A two-tailed p < 0·05 was considered statistically significant.
2.16. Data sharing
The dataset for the miRNA microarray analysis is available in the Gene Expression Omnibus repository under accession number GSE114110.
3. Results
3.1. miR-424 is upregulated in ESCC and correlates with poor prognosis
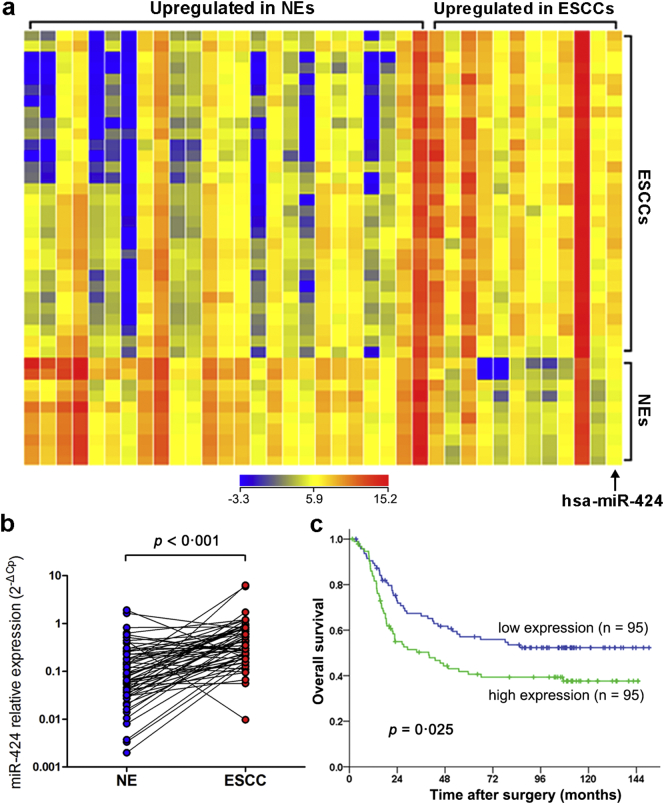
To identify ESCC-related miRNAs, miRNA arrays were used to analyze 30 ESCC and ten NE samples. A total of 37 miRNAs were identified as differentially expressed miRNAs between the two groups (fold change >3, p < 0·001 by Student's t-test with the Benjamin-Hochberg correction, Fig. 1a). miR-424 was one of 12 miRNAs that were upregulated in ESCC samples compared with NE samples (Fig. 1a). The expression level of miR-424 was further evaluated in 60 pairs of ESCC specimens and their matched NE samples by qRT-PCR. The results showed that miR-424 expression was significantly elevated in ESCC specimens compared with that in normal counterparts (p < 0·001 by Student's t-test for paired samples, Fig. 1b).
Fig. 1.
miR-424 is upregulated in ESCC and correlates with poor prognosis. (a) Hierarchical cluster analysis showed differential miRNA expression profiles between 30 ESCC and ten NE samples (fold change >3, p < 0·001 by Student's t-test with the Benjamin-Hochberg correction). Each column represents a miRNA, and each row represents a specimen. Blue and red denote genes that are under- and overexpressed, respectively. (b) qRT-PCR verified the upregulation of miR-424 in 60 ESCC specimens compared with the expression seen in normal counterparts (p < 0·001 by Student's t-test for paired samples). (c) Kaplan-Meier analysis showed that the overall survival (p = 0·025 by log-rank test) of ESCC patients with high miR-424 expression was poorer than that of patients with low miR-424 expression.
To evaluate whether the upregulation of miR-424 correlates with clinical ESCC progression, we examined miR-424 expression by qRT-PCR in a cohort of 190 Stage IB-IIIC ESCC patients who underwent radical surgery with no neoadjuvant or adjuvant treatment. The expression of miR-424 was classified as high or low according to the median miR-424 expression value of the whole cohort. There were no statistically significant associations between miR-424 expression and patients' clinicopathological factors (p > 0·05 by Chi-square test, Table 1). However, both univariate and multivariate survival analyses revealed that high miR-424 expression was significantly associated with poor patient prognosis (p < 0·05 by Kaplan-Meier method with log-rank test for univariate survival analysis and Cox's proportional hazards regression analysis for multivariate survival analysis, Fig. 1c; Table 2, Table 3).
Table 1.
Association between miR-424 expression and clinicopathological characteristics in 190 esophageal squamous cell carcinoma patients.
| Variables |
miR-424 expression in primary tumors |
|||
|---|---|---|---|---|
| Cases | Low (%) | High (%) | p valuea | |
| Gender | 0·095 | |||
| Male | 142 | 76 (53·5) | 66 (46·5) | |
| Female | 48 | 19 (39·6) | 29 (60·4) | |
| Age (years) | 0·384 | |||
| ≤59b | 98 | 52 (53·1) | 43 (46·9) | |
| >59 | 92 | 46 (46·7) | 49 (53·3) | |
| Tumor location | 1·000 | |||
| Upper | 34 | 17 (50·0) | 17 (50·3) | |
| Middle | 102 | 51 (50·0) | 51 (50·0) | |
| Lower | 54 | 27 (50·0) | 27 (50·0) | |
| Histological differentiation | 0·340 | |||
| Well | 48 | 26 (54·2) | 22 (45·8) | |
| Moderate | 96 | 43 (44·8) | 53 (55·2) | |
| Poor | 46 | 26 (56·5) | 20 (43·5) | |
| T stage | 0·401 | |||
| T1–2 | 47 | 26 (55·3) | 21 (44·7) | |
| T3–4 | 143 | 69 (48·3) | 74 (51·7) | |
| N stage | 0·146 | |||
| N0 | 100 | 45 (45·0) | 55 (55·0) | |
| N1–3 | 90 | 50 (55·6) | 40 (44·4) | |
| TNM stage | 0·450 | |||
| Stage I | 15 | 9 (60·0) | 6 (40·0) | |
| Stage II | 92 | 42 (45·7) | 50 (54·3) | |
| Stage III | 83 | 44 (53·0) | 39 (47·0) | |
Chi-square test.
Mean age.
Table 2.
Univariate analysis of miR-424 expression level and clinicopathological factors for overall survival and disease-free survival in 190 surgically resected esophageal squamous cell carcinoma patients.
| Variables | Cases | Overall survival (months) |
|
|---|---|---|---|
| Median | p valuea | ||
| Gender | 0·622 | ||
| Male | 142 | 66·1 | |
| Female | 48 | 40·1 | |
| Age (years) | 0·772 | ||
| ≤59b | 98 | 65·8 | |
| >59 | 92 | 44·5 | |
| Tumor location | 0·578 | ||
| Upper | 34 | NRc | |
| Middle | 102 | 47·2 | |
| Lower | 54 | 66·1 | |
| Histological differentiation | < 0·001 | ||
| Well | 48 | NR | |
| Moderate | 96 | 107·3 | |
| Poor | 46 | 23·4 | |
| T stage | 0·146 | ||
| T1–2 | 47 | NR | |
| T3–4 | 143 | 49·5 | |
| N stage | < 0·001 | ||
| N0 | 100 | NR | |
| N1–3 | 90 | 21·7 | |
| TNM stage | < 0·001 | ||
| Stage I | 15 | NR | |
| Stage II | 92 | NR | |
| Stage III | 83 | 21·5 | |
| miR-424 | 0·025 | ||
| Low expression | 95 | NR | |
| High expression | 95 | 39·8 | |
Kaplan-Meier method, log-rank test.
Median age.
Not reached.
Table 3.
Multivariate analysis for overall survival and disease-free survival in surgically resected esophageal squamous cell carcinoma patients.
| Variable | Overall survival |
||
|---|---|---|---|
| p valuea | HR | 95·0% CI for HR | |
| miR-424 expression | 0·001 | 2·040 | 1·356–3·069 |
| Histological differentiation | 0·008 | 1·533 | 1·120–2·099 |
| N stage | <0·001 | 3·514 | 2·268–5·444 |
Abbreviations: HR hazard ratio, CI confidence internal.
Cox's proportional hazards regression analysis (Forward stepwise).
3.2. miR-424 promotes ESCC cell proliferation
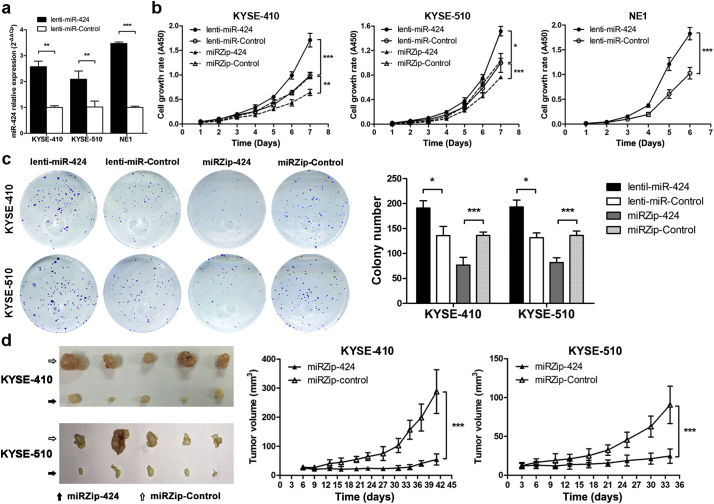
To investigate the biological function of miR-424, the ESCC cell lines KYSE-410 and KYSE-510 were transduced with lentiviruses expressing lenti-miR-424, miRZip-424, or the corresponding control vectors and further selected to generate stable miR-424-overexpressing or miR-424-knockdown ESCC cells. In addition, the immortalized esophageal epithelial cell line NE1 was freshly transduced with lentiviruses expressing lenti-miR-424 and control vectors. Lenti-miR-424 transduction significantly increased miR-424 expression in KYSE-410, KYKSE-510, and NE-1 cells compared with control vectors (Fig. 2a). As miRZip anti-miRNA lentivectors exhibit anti-miRNA activity without directly decreasing miRNA expression [24], we did not determine miR-424 expression after miRZip-424 transduction.
Fig. 2.
miR-424 promotes ESCC cell proliferation in vitro and in vivo. (a) qRT-PCR analyses showed the relative miR-424 expression levels in KYSE-410 and KYSE-510 ESCC cells and NE1 immortalized esophageal epithelial cells expressing the miR-424 precursor (lenti-miR-424) or control vectors. Data are presented as the mean ± SD of three independent experiments. **, p < 0·01 or ***, p < 0·001 by Student's t-test for unpaired samples. (b) Cell growth curves were generated for miR-424-overexpressing (lenti-miR-424) or miR-424-knockdown (miRZip-424) KYSE-410, KYSE-510, and NE1 cells with a CCK-8 cell proliferation kit. Data are presented as the mean ± SD of three independent experiments. *, p < 0·05; **, p < 0·01; or ***, p < 0·001 by repeated measures ANOVA. (c) Representative images and results of colony formation assays of KYSE-410 and KYSE-510 cells expressing miR-424-knockdown (miRZip-424), miR-424-overexpressing, or the corresponding control vectors. Data are presented as the mean ± SD of three independent experiments. ***, p < 0·001 by Student's t-test for unpaired samples. (d) Tumor xenograft experiments with subcutaneous injection of KYSE-410 or KYSE-510 cells stably expressing miRZip-424 or control vectors into nude mice (n = 5 in each group) demonstrated the in vivo tumorigenic ability of miR-424. Images of tumor-bearing mice and tumors from all mice in each group are shown. Tumor volumes were measured on the indicated days, and each data point represents the mean ± SD of five nude mice. ***, p < 0·001 by repeated measures ANOVA.
Cell growth curves and colony formation assays were performed to explore the effect of miR-424 on ESCC cell proliferation in vitro. As shown in Fig. 2b, miR-424 overexpression significantly promoted KYSE-410, KYSE-510, and NE1 cell growth, whereas miR-424 knockdown significantly retarded the growth rate of KYSE-410 and KYSE-510 cells (Fig. 2b). Colony formation assays also showed that miR-424-knockdown in KYSE-410 and KYSE-510 cells results in fewer and smaller colonies than were produced by control cells, whereas the opposite results were observed after overexpressing miR-424 in KYSE-410 and KYSE-510 cells (Fig. 2c).
The in vivo tumorigenic ability of miR-424 was investigated with a tumor xenograft experiment by subcutaneous injection of KYSE-410 and KYSE-510 cells stably expressing miRZip-424 or the control vector into nude mice. As shown in Fig. 2d, the tumors formed by miR-424-knockdown KYSE-410 or KYSE-510 cells were smaller than the tumors formed by control cells.
3.3. Role of miR-424 in ESCC cell cycle progression
The loss of normal cell cycle control is one of the factors that induces sustained cell proliferation [5]. As miR-424 promotes ESCC cell proliferation, we next focused our attention on understanding whether and how miR-424 affects ESCC cell cycle progression.
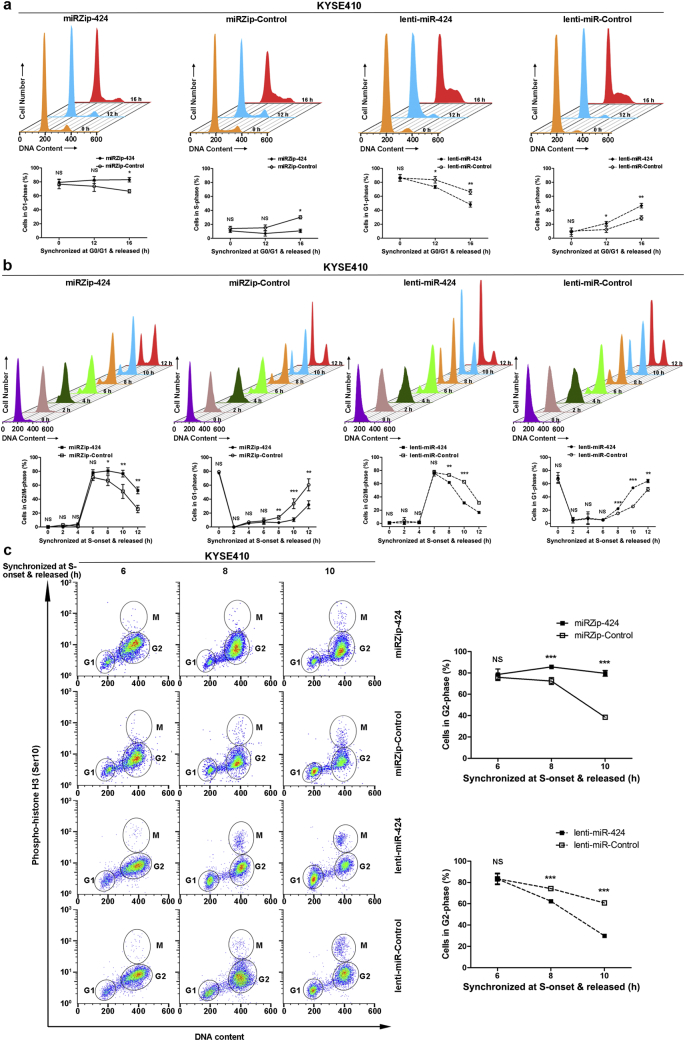
First, KYSE-410 and KYSE-510 cells stably expressing miRZip-424, lenti-miR-424, or the corresponding control vectors were synchronized in G0/G1-phase by serum withdrawal. The cells were then switched to a standard culture medium to allow the cells to re-enter the cell cycle. KYSE-410-miRZip-424 cells displayed an increased percentage of cells in G1-phase and a decreased percentage of cells in S-phase compared with KYSE-410-miRZip-Control cells, whereas KYSE-410-lenti-miR-424 cells exhibited a decreased percentage of cells in G1-phase and an increased percentage of cells in S-phase compared with KYSE-410-lenti-miR-Control cells (Fig. 3a). However, no obvious difference was observed between KYSE-510-miRZip-424 and KYSE-510-miRZip-Control or between KYSE-510-lenti-miR-424 and KYSE-510-lenti-miR-Control cells after release from G0/G1-phase (Supplementary Fig. 1a), suggesting that miR-424 affects the G1/S transition in KYSE-410 cells but not KYSE-510 cells.
Fig. 3.
Effects of miR-424 expression on G1/S and G2/M cell cycle progression in KYSE-410 ESCC cells. (a) Stable miR-424-knockdown (miRZip-424), miR-424-overexpressing (lenti-miR-424), and the corresponding control KYSE-410 cells were synchronized in G0/G1-phase by serum starvation and released, and the cell cycle distribution was monitored by flow cytometry at the indicated time points with propidium iodide staining. Representative images of the cell cycle distribution (upper) and the percentage of cells in G1-phase or S-phase (lower) are shown. (b) Stable miR-424-knockdown (miRZip-424), miR-424-overexpressing (lenti-miR-424), and the corresponding control KYSE-410 cells were synchronized at the onset of S-phase by double-thymidine block and released, and the kinetic transition of the cells through S-phase to G2/M-phase was monitored by flow cytometry at the indicated time points with propidium iodide staining. Representative images of the cell cycle distribution (upper) and the percentage of cells in G2/M- or G1-phase (lower) are shown. (c) Stable miR-424-knockdown (miRZip-424), miR-424-overexpressing (lenti-miR-424), and the corresponding control KYSE-410 cells were synchronized at the onset of S-phase by double-thymidine block and released, and the kinetic transition of the cells through G2-phase to M-phase was monitored by flow cytometry at the indicated time points with propidium iodide and anti-pHH3 and Alexa Fluor 647-conjugated secondary antibody double-staining. Representative images of the cell cycle distribution (left) and the percentage of cells in G2-phase (right) are shown. All the data are presented as the mean ± SD of three independent experiments. *, p < 0·05; **, p < 0·01; ***, p < 0·001; or NS, not significant by Student's t-test for unpaired samples.
Next, the effect of miR-424 on S-phase and G2/M-phase progression in ESCC cells was determined. KYSE-410 and KYSE-510 cells stably expressing miRZip-424, lenti-miR-424, or the corresponding control vectors were synchronized at the onset of S-phase by double-thymidine block, and the cells were then released and harvested at sequential time points for the measurement of S-phase and G2/M-phase events. In KYSE-410 cells, approximately 80% of miRZip-424, lenti-miR-424, and the corresponding control cells were synchronized in G1-phase, and 20% were initially in S-phase. After releasing the cells for 2 h, almost all cells were in S-phase; the cells then gradually entered G2/M-phase, with a peak for G2/M-phase cells at 6 h after release. No significant differences in cell cycle distribution among miRZip-424, lenti-miR-424, and the corresponding control cells were observed during the first 6 h after release from the onset of S-phase (Fig. 3b), indicating that miR-424 did not affect S-phase progression. Later, however, the KYSE-410-miRZip-424 cells displayed an increased percentage of cells in G2/M-phase but a decreased percentage of cells in G1-phase compared with control cells at 8, 10, and 12 h after release from the onset of S-phase, whereas the opposite results were obtained in KYSE-410-lenti-miR-424 cells compared with control cells. These observations suggest that miR-424-knockdown or overexpression respectively blocked or promoted the G2/M cell cycle progression of KYSE-410 cells (Fig. 3b). Similar results were observed in KYSE-510 cells (Supplementary Fig. 1b).
To further discriminate between G2- and M- phase cells, stable KYSE-410-miRZip-424, KYSE-410-lenti-miR-424, and the corresponding control cells were enriched at the onset of S-phase and then released with bivariate analysis of phospho-histone H3 (pHH3, Ser10) expression, a mitosis-specific marker, versus DNA content. As shown in Fig. 3c, after releasing cells from the onset of S-phase for 8–10 h, a greater percentage of miR-424-knockdown but a smaller percentage of miR-424-overexpressing cells remained in G2-phase (pHH3-negative cells) than were observed with control cells, suggesting a role of miR-424 in G2/M transition. However, when mitotic cells were enriched by nocodazole arrest and released, KYSE-410-miRZip-424, KYSE-410-lenti-miR-424, and the corresponding control cells exhibited similar rates of reduction of the mitotic population (pHH3-positive cells) (Supplementary Fig. 2). These results indicated that the expression levels of miR-424 affected G2/M but not mitotic transition in ESCC cells.
Taken together, the cell cycle analyses of ESCC cells after synchronization and release from different cell cycle phases provided evidence that knockdown or overexpression of miR-424 respectively negatively or positively regulates G1/S transition in KYSE-410 cells and G2/M transition in both KYSE-410 and KYSE-510 cells.
3.4. miR-424 expression is transcriptionally regulated by E2F1 during G1/S transition
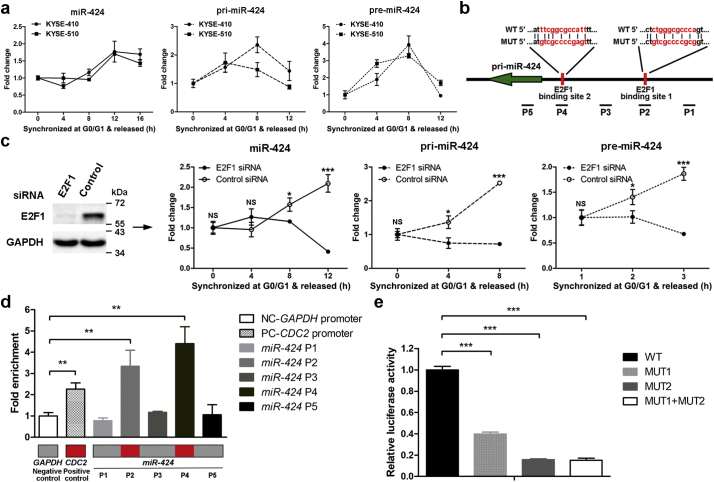
As miR-424 functions in cell cycle progression, we evaluated the expression of miR-424 in various cell cycle phases. qRT-PCR analyses showed that miR-424 expression increased 12 or 16 h after release from G0/G1-phase in both KYSE-410 and KYSE-510 cells (Fig. 4a), suggesting that miR-424 expression increased with cell cycle progression from G0/G1 to S-phase. However, the expression of miR-424 did not show obvious changes during G2/M progression (Supplementary Fig. 3). A small increase in miR-424 expression was observed after release from the onset of S-phase for 10 h (Supplementary Fig. 3), which might be due to increased miR-424 expression during G1/S transition, as some of the cells have already left G2/M-phase and re-entered G1/S-phase as a part of a new division cycle at this time point as shown in Fig. 3b and Supplementary Fig. 1b.
Fig. 4.
Characterization of the miR-424 promoter and transcriptional regulation of miR-424 by E2F1 during G1/S transition. (a) KYSE-410 and KYSE-510 cells were synchronized in G0/G1-phase by serum starvation and released. qRT-PCR analysis shows the expression kinetics of miR-424, pri-miR-424, and pre-miR-424 in KYSE-410 and KYSE-510 cells after release from G0/G1-phase for the indicated time points. Data are presented as the mean ± SD of three independent experiments. (b) A schematic illustration shows two binding sites for E2F1 in the putative promoter region of the miR-424 gene. Specific primers surrounding the promoter were designed. A fragment of the promoter region encompassing the wild-type or mutant E2F1 binding sites was cloned into a reporter vector. (c) Expression of E2F1 was silenced using siRNA (left panel). qRT-PCR showed changes in the expression kinetics of miR-424, pri-miR-424, and pre-miR-424 after knocking down E2F1 in KYSE-410 cells during G1/S transition (right panel). Data are presented as the mean ± SD of three independent experiments. *, p < 0·05; ***, p < 0·001; or NS, not significant by Student's t-test for unpaired samples to compare differences in expression between E2F1-knockdown and control cells at each time point. (d) ChIP was performed with an anti-E2F1 antibody on lysates of KYSE-410 cells after release from G0/G1-phase for 8 h. The quantification of genomic DNA enrichment was performed using specific primers surrounding the promoter, as shown in (b). Putative E2F1-binding sites are indicated by red squares. Quantification of DNA enrichment in the CDC2 gene promoter was used as a positive control, and GAPDH was used as a negative control. Data are presented as the mean ± SD of three independent experiments. **, p < 0·01 by Student's t-test for unpaired samples to compare differences in enrichment between any specific primer pair and the negative control. (e) Luciferase assays showed the different activities of reporters containing either the wild-type or mutated E2F1-binding sites in KYSE-410 cells, which were released from G0/G1-phase for 8 h. Data are presented as the mean ± SD of three independent experiments. ***, p < 0·001 by Student's t-test for unpaired samples to compare differences in relative luciferase activity between reporters with mutant E2F1-binding sites and that with wild-type binding site.
The biogenesis of miRNA is a multistep process. miRNA genes are first transcribed into pri-miRNAs and subjected to a preliminary processing step to form hairpin-loop RNAs (pre-miRNAs), which are then exported to the cytoplasm and cleaved to produce mature miRNAs [25]. In our ESCC cells, qRT-PCR amplification confirmed the upregulation of pri-miR-424 and pre-miR-424 in KYSE-410 and KYSE-510 cells after release from G0/G1-phase (Fig. 4a). Kinetic studies revealed that the increases in pri-miR-424 and pre-miR-424 levels happened before the increase in mature miR-424 levels, suggesting that the elevated miR-424 levels observed after release from G0/G1-phase were primarily due to miR-424 gene transcription. Notably, the generation of mature miR-424 led to reductions of pri-miRNA and pre-miRNA levels 12 h after release from G0/G1-phase. This effect could be due to the cleavage and degradation of the pri-miRNA after the pre-miRNA is excised, the existence of a negative regulatory feedback loop, or a combination of both mechanisms [26].
Llobet-Navas et al. [26] identified the transcription start site (TSS) of the miR-424 gene, and a region located ~2 kb upstream of the TSS that is strongly enriched in active histone promoter marks could be the putative promoter region for its transcription. In the promoter region, two binding sites of E2F1 (−769 to −757 and −145 to −134), a widely studied transcription factor that activates the transcription of genes required for cell cycle entry and DNA synthesis [27], were identified using JASPAR [28] (Fig. 4b). As expected, knockdown of E2F1 decreased the expression of pri-miR-424, pre-miR-424, and mature miR-424 in ESCC KYSE-410 cells (Fig. 4c).
To confirm the direct binding between E2F1 and the miR-424 promoter, a ChIP assay with 5 pairs of primers within the putative promoter region was performed to investigate the presence of E2F1 in the endogenous putative promoter in KYSE-410 cells 8 h after release from G0/G1-phase. The results revealed that the strongest enrichment of E2F1 happened in the two regions (P2 and P4) that contained the predicted E2F1-binding sites, and the levels were higher than those of the bona fide E2F1 target CDC2 (Fig. 4d). In addition, the P4 region of the second E2F1-binding site was enriched more than the P2 region of the first E2F1-binding site.
To evaluate the role of the putative E2F1-binding sites in the upregulation of miR-424 transcription during G1/S progression, a 1750-bp fragment of the putative promoter region encompassing the wild type or mutant E2F1 binding sites was inserted into pEZX-LvPG04, a GLuc-ON transcriptional response element lentiviral clone, using a secreted Gaussia luciferase (GLuc) as the reporter. This reporter was integrated into the genome of KYSE-410 cells by lentiviral delivery to create a pri-miR reporter cell line. After releasing the cells from G0/G1-phase for 8 h, the wild-type miR-424 promoter exhibited significantly higher GLuc activity than the mutant promoters (Fig. 4e). Notably, the mutation of the second E2F1 binding site decreased the luciferase activity more than the mutation of the first binding site.
Taken together, these data demonstrated that E2F1 binds to the miR-424 promoter and directly activates its transcription during the G1/S transition. In addition, the predicted second binding site of E2F1 might play a more important role than the first binding site during E2F1-mediated miR-424 transcriptional activation.
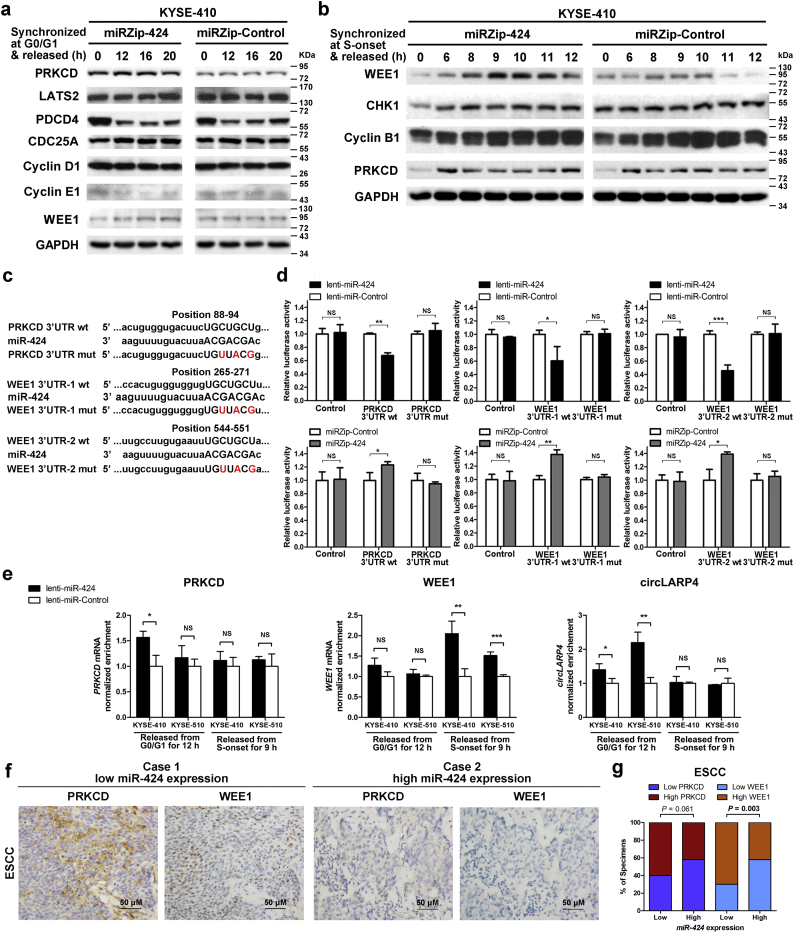
3.5. miR-424 directly targets PRKCD and WEE1 in ESCC cells
The biological functions of miRNAs are mediated by their ability to directly interact with the 3′UTRs of target mRNAs and attenuate their expression. By searching publicly available algorithms (TargetScan and miRanda), we found a series of genes that are involved in cell cycle progression regulation and might be potential targets of miR-424. Among those genes, PRKCD [29], CDC25A [30], PDCD4 [31], LATS2 [32], cyclin D1, and cyclin E1 [33] function primarily in G1/S transition, CHK1 [34] functions primarily in S-phase DNA replication and G2/M transition [34], and WEE1 [35] and cyclin B1 [33] function primarily in G2/M transition.
We analyzed the impact of miR-424 on the levels of the endogenous proteins encoded by the above potential target genes by synchronizing and releasing ESCC cells from G0/G1-phase or the onset of S-phase. During G1/S progression, western blotting revealed that knockdown of miR-424 clearly elevated PRKCD protein in KYSE-410 (Fig. 5a) but not KYSE-510 (Supplementary Fig. 4a) cells. However, no obvious changes in LATS2, PDCD4, CDC25A, cyclin D1, and cyclin E1 expression caused by miR-424 knockdown were observed in either KYSE-410 (Fig. 5a) or KYSE-510 cells (Supplementary Fig. 4a). When the cells were released from the onset of S-phase, WEE1 protein expression increased gradually during S-phase and G2-phase. In addition, this elevation was more pronounced in miR-424-knockdown cells than in control cells (Fig. 5b; Supplementary Fig. 4b). However, expression changes in two additional G2/M progression regulators, CHK1 and cyclin B1, were not observed after knockdown of miR-424 (Fig. 5b; Supplementary Fig. 4b). In addition, knockdown of miR-424 did not induce changes in the expression of either WEE1 protein during the G1/S transition (Fig. 5a) or PRKCD during the G2/M transition (Fig. 5b).
Fig. 5.
miR-424 targets PRKCD and WEE1 in ESCC cells during G1/S and G2/M transitions, respectively. (a) Stable miR-424-knockdown (miRZip-424) and control KYSE-410 cells were synchronized in G0/G1-phase by serum starvation and released for the indicated time points. Western blotting analysis showed the protein expression of potential miR-424 targets during G1/S transition. (b) Stable miR-424-knockdown (miRZip-424) and control KYSE-410 cells were synchronized at the onset of S-phase by double-thymidine block and released for the indicated time points. Western blotting analysis showed the protein expression of potential miR-424 targets during G2/M transition. (c) The illustration shows the predicted miR-424 target sequences in the 3′UTRs of the PRKCD and WEE1 mRNAs. (d) Luciferase assays showed different activities of reporters containing the 3′UTRs or mutated 3′UTRs of PRKCD and WEE1 in miR-424-overexpressing, miR-424-knockdown, and the corresponding control HEK293 cells. Data are presented as the mean ± SD of three independent experiments. *, p < 0·05; **, p < 0·01; ***, p < 0·001; or NS, not significant by Student's t-test for unpaired samples. (e) miRNP-IP was performed with an anti-AGO2 antibody on lysates of stable miR-424-overexpressing or control KYSE-410 and KYSE-510 cells, and the cells were released from G0/G1 for 12 h or from S-phase onset for 9 h. The quantification of RISC enrichment of PRKCD, WEE1, and circLARP4 was determined by qRT-PCR. Data are presented as the mean ± SD of three independent experiments. *, p < 0·05; **, p < 0·01; ***, p < 0·001; or NS, not significant by Student's t-test for unpaired samples. (f) Representative immunohistochemical staining images of PRKCD and WEE1 protein in two ESCC cases with low and high miR-424 expression (Scale bar: 50 μM). (g) The correlation between miR-424 expression and PRKCD or WEE1 protein expression in 110 ESCC samples was analyzed. r = −0·282, p = 0·003 for WEE1, and r = −0·179, p = 0·061 for PRKCD by Chi-square test.
One fragment of the PRKCD 3′UTR and two fragments of the WEE1 3′UTR that contained the miR-424-binding sites predicted by bioinformatics algorithms were subcloned into the pmiRGLO dual-luciferase reporter vector (Fig. 5c). A luciferase assay showed that overexpression of miR-424 attenuated the reporter activities driven by the 3′UTRs of PRKCD and WEE1 transcripts, but knockdown of miR-424 elevated these reporter activities. However, dysregulation of miR-424 did not result in alterations in the reporter activities driven by the mutated 3′UTRs of these transcripts within the miR-424-binding-regions (Fig. 5d).
Next, we demonstrated the direct binding of miR-424 to endogenous PRKCD and WEE1 by immunoprecipitating Ago2, a core component of the RISC. Enrichment of the PRKCD transcript significantly increased in miR-424-overexpressing KYSE-410 cells after release from G0/G1 for 12 h, while WEE1 enrichment increased in both miR-424-overexpressing KYSE-410 and KYSE-510 cells after release from the onset of S-phase for 9 h, compared with the levels in control cells, respectively (Fig. 5e). No enrichment of PRKCD was observed during G2/M progression, and no enrichment of WEE1 was observed during G1/S progression. The results of the miRNP-IP assay were consistent with the above effects of miR-424 on the cell cycle distribution and target gene expression, as shown by flow cytometry and western blotting, indicating that the effects of miR-424 on target genes were cell cycle-phase specific. However, the reason why miR-424 failed to target PRKCD during G1/S progression in KYSE-510 cells was unknown.
Mounting evidence has demonstrated that transcripts of non-coding RNAs (nc-RNAs) that contain miRNA-binding sites can act as natural miRNA sponges to affect the efficacy of miRNA target repression. circLARP4, which is localized in the cytoplasm, was identified previously as a natural sponge of miR-424 and demonstrated to inhibit its activity by directly binding to miR-424 [36]. In ESCC cells, our miRNP-IP assay proved that circLARP4 could be enriched in miR-424-overexpressing KYSE-410 and KYSE-510 cells at G1/S transition, and the effect was more profound in KYSE-510 cells (Fig. 5e). Therefore, we supposed that miR-424 was sponged by circLARP4 in ESCC cells during G1/S transition, which could repress the ability of miR-424 to bind to and target PRKCD. However, knockdown of miR-424 had no influence on the expression of circLARP4 (Supplementary Fig. 5).
Finally, we evaluated the expression levels of PRKCD and WEE1 protein in ESCC and NE specimens by IHC analysis. Positive staining for PRKCD and WEE1 was observed in the cytoplasmic and nuclear regions, respectively (Fig. 5f and Supplementary Fig. 6). The expression of PRKCD or WEE1 protein of each specimen was classified as high or low according to the median protein expression value in the ESCC samples. Correlation studies showed that miR-424 levels determined by qRT-PCR were significantly inversely correlated with the expression of WEE1 (r = −0·282, p = 0·003 by Chi-square test) but not PRKCD (r = −0·179, p = 0·061 by Chi-square test, Fig. 5g) in the ESCC samples. However, no significant correlation between either PRKCD or WEE1 protein expression and miR-424 expression was observed in the NE samples (p > 0.05 by Chi-square test, Supplementary Fig. 6).
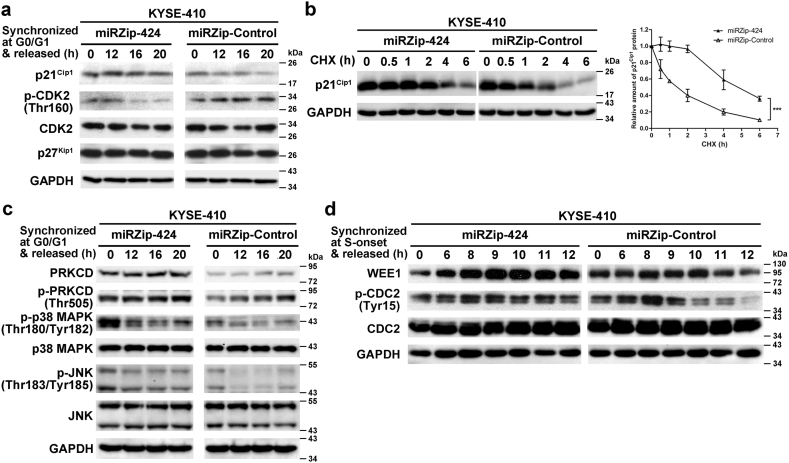
3.6. miR-424 modulates cell cycle regulator expression and activity to regulate ESCC cell cycle progression
The temporal order of cell cycle transition is imposed by the sequential activation or inactivation of CDKs, which is coordinated by interactions with positive regulator cyclins or negative regulator CKIs. Therefore, we examined the active forms and the total protein expression of G1/S and G2/M transition-related cyclins, CDKs, and CKIs in miR-424-knockdown and control ESCC cells.
During G1/S transition, western blotting showed that knocking down miR-424 in KYSE-410 cells increased p21Cip1 protein expression and decreased p-CDK2 (Thr160) levels, which represent the activated state of CDK2 (Fig. 6a), whereas total CDK2 expression levels remained constant. p21Cip1 is a member of the CDK-interacting protein/kinase inhibitory protein (CIP/KIP) family that binds to and inhibits the activity of cyclin E-CDK2 complexes and thus functions as a negative regulator of G1/S progression [37]. However, another member of the CIP/KIP family, p27Kip1, did not exhibit expression changes during G1/S transition in miR-424-knockdown KYSE-410 cells (Fig. 6a). The negative regulation of p21Cip1 by miR-424 did not occur at the transcriptional level, as the mRNA level of p21Cip1 was not altered by miR-424 knockdown in KYSE-410 cells (Supplementary Fig. 7). However, knockdown of miR-424 delayed p21Cip1 protein degradation when KYSE-410 cells were cultured with cycloheximide to inhibit de novo protein synthesis (Fig. 6b), suggesting a possible role of miR-424 in p21Cip1 protein stability. It has been reported that p38 MAPK [38] and JNK [39] activation, downstream signaling of PRKCD expression and activation, can enhance the cellular stability of the p21Cip1 protein without disturbing its mRNA expression [40]. As expected, during G1/S transition, compared with control cells, KYSE-410-miRZip-424 cells exhibited increased levels of total PRKCD, activated PRKCD (p-PRKCD at Thr505), and downstream activated p38 MAPK (p-p38 MAPK at Thr180/Tyr182) and JNK (p-JNK at Thr183/Tyr185) but not total p38 MAPK and JNK (Fig. 6c).
Fig. 6.
Knockdown of miR-424 modulates the expression or activity of CDKs or CKIs to regulate ESCC cell cycle progression. (a) Western blotting analysis showed the expression of p21Cip1, p-CDK2 (Thr160), and total CDK2 in stable miR-424-knockdown or control KYSE-410 cells, which were collected at the indicated time points after release from G0/G1-phase. (b) The stability of the endogenous p21Cip1 protein in stable miR-424-knockdown or control KYSE-410 cells was measured by incubating the cells with 25 μg/ml cycloheximide (CHX). The cells were collected at the indicated time points and analyzed by western blotting (left panel). Quantitation of p21Cip1 protein levels is shown in the right panel. Data are presented as the mean ± SD of three independent experiments. ***, p < 0·001 by repeated measures ANOVA. (c) Western blotting analysis showed the expression changes in PRKCD, p-PRKCD (Thr505), p-p38 MAPK (Thr180/Tyr182), p38 MAPK, p-JNK (Thr183/Tyr185), and JNK in stable miR-424-knockdown or control KYSE-410 cells, which were collected at the indicated time points after release from G0/G1-phase. (d) Western blotting analysis showed the expression changes in WEE1, p-CDC2 (Tyr15), and CDC2 in stable miR-424-knockdown or control KYSE-410 cells, which were collected at the indicated time points after release from the onset of S-phase.
The mitotic kinase CDC2/Cyclin B is critical for G2/M transition and is maintained in an inactive state until the dephosphorylation of Thr-14 and Tyr-15 at the G2/M border activates CDC2. WEE1 is the kinase responsible for phosphorylating Tyr-15 of CDC2 and hence inhibits CDC2 activity [35]. As shown in Fig. 6d, knockdown of miR-424 in KYSE-410 cells increased WEE1 expression and CDC2 phosphorylation at Tyr-15 when the cells were at the G2/M border (6 h after release from the onset of S-phase by double-thymidine treatment) compared with control cells.
Collectively, our results showed that knockdown of miR-424 upregulated PRKCD expression and activated downstream signaling, which further increased p21Cip1 protein stability to delay G1/S transition in ESCC. On the other hand, the WEE1 upregulation induced by knockdown of miR-424 maintained the inactivation of CDC2 to postpone G2/M transition in ESCC.
4. Discussion
Knowledge about the precise molecular mechanisms that underlie ESCC tumorigenesis is crucial for the development of better diagnostic and therapeutic strategies for ESCC patients. Despite the established roles of a variety of extrinsic and intrinsic signals in regulating ESCC development [6], few studies have identified and validated miRNAs with a role in ESCC [41]. Our study is the first to explore the potential tumor promoter roles and regulatory mechanisms of miR-424 during cell cycle progression in ESCC.
Previous studies have reported an association between miR-424 and human cancers. miR-424 is commonly lost in aggressive breast cancers, and the loss of miR-424 promotes breast tumorigenesis and chemoresistance by up-regulating two of its targets: BCL-2 and IGF1R [10]. The expression of miR-424 is lower in cervical cancers than in cervical normal tissues, and enforced expression of miR-424 inhibits cervical cancer cell growth, migration and invasion via targeting CHK1 [12]. In hepatocellular carcinoma, miR-424 inhibits cell proliferation, migration, and invasion by targeting c-Myb [13]. miR-424 directly targets BCR-ABL in chronic myeloid cells, and overexpression of miR-424 suppresses proliferation and induces apoptosis of leukemia cells and sensitizes cells to imatinib treatment [14]. However, miR-424 has been reported to have elevated expression and a tumor-promoting role in tongue squamous cell carcinoma [42] and pancreatic cancer [43]. miR-424 is upregulated in pancreatic cancer and promotes proliferation, migration and invasion while inhibits cell apoptosis in pancreatic cancer cells via targeting SOCS6 to modulate the ERK1/2 signaling pathway [43]. These discrepancies suggest that the role of miR-424 is tumor specific and highly dependent on its targets in different types of cancer cells.
In the present study, we first demonstrated that miR-424 is significantly upregulated in ESCC by microarray analyses, and these results were further validated in paired ESCC and NE tissues. There were some cases in which NE samples exhibited miR-424 expression levels higher than or close to those in the paired ESCC samples. This is probably due to the phenomenon of field cancerization, in which some morphologically normal cells acquire pro-tumorigenic genetic or epigenetic mutations [44], leading to the increased expression of miR-424 in NEs. More importantly, miR-424 expression was identified as an independent predictor of survival in ESCC patients. The poor survival of patients with high miR-424 expression may be attributable to intrinsic characteristics of the tumor cells that are partly determined by miR-424, although no significant correlation was observed between miR-424 expression and patient clinicopathological characteristics, such as tumor pathological stage or differentiation. Consistent with the clinical data, knockdown of miR-424 in ESCC cells remarkably suppressed ESCC proliferation in vitro and tumor formation in a mouse model, while the opposite results were observed after overexpressing miR-424. Additionally, overexpression of miR-424 also promoted the proliferation of the immortalized esophageal epithelial cell line NE1, suggesting that miR-424 might promote the malignant transformation potential of esophageal normal epithelia cells. Therefore, our findings suggest that miR-424 functions as a tumor promoter in ESCCs and may be used as a novel prognostic marker for ESCC patients.
After cell synchronization and subsequent flow cytometry analysis, miR-424 was found to regulate both G1/S and G2/M transitions in ESCC cells. The roles of miR-424 and its targets in cell cycle progression have been partly explored in previous studies. miR-424 has been reported to directly target the cell cycle regulators cyclin E1 [45], CDC25A [16], cyclin D1 [46], and CHK1 [12] to impair G1/S or G2/M cell cycle transition and inhibit cell proliferation in various kinds of cells. However, expression changes in these previously reported genes were not observed in ESCC cells after knockdown of miR-424. When further exploring the underlying mechanisms of miR-424 in the regulation of ESCC cell proliferation, we found that miR-424 suppressed the expression of PRKCD and WEE1 by directly targeting their 3′UTRs during G1/S and G2/M transitions, respectively. The correlation of expression between miR-424 and the PRKCD or WEE1 protein could not be verified in NE samples, suggesting that PRKCD or WEE1 might not be targeted by miR-424 in NEs. Therefore, the promotion of cell proliferation by miR-424 overexpression in the immortalized esophageal cell line NE1 might not be mediated by PRKCD or WEE1. Other previously validated or unidentified miR-424 targets might be functional in NE tissues. The distinct targets of miR-424 in ESCC, NE, and other types of cancers explain the tissue specificity of the role of miR-424 and are consistent with the notion that miRNA-target regulation is context dependent [47,48]. Interestingly, this context-dependent miR-424 function is not only tissue specific, but also biological process specific. For example, biphasic roles of miR-424 in ESCC development and progression have been observed. miR-424 was shown to inhibit ESCC invasion and metastasis by targeting SMAD7 in a previous study [9] but to promote cell cycle progression by targeting PRKCD and WEE1 in the current study. Similarly, biphasic roles of miR-424 have also been identified in breast cancer, where miR-424 facilitates earlier but represses later stage of metastasis [49].
Diverse RNA species, including protein-coding mRNAs and non-coding RNAs, such as long non-coding RNAs, circular RNAs and pseudogenes, can act as natural miRNA sponges. These RNAs communicate with and co-regulate each other by competing specifically for binding to shared miRNAs, thus acting as competing endogenous RNAs (ceRNAs) [50]. In this study, we found that the targeted effects of miR-424 were cell cycle-phase-specific, as PRKCD was targeted during G1/S transition, whereas WEE1 was targeted during G2/M transition in ESCC cells. During G2/M transition, the gradually accumulating WEE1 transcript, which has two binding sites for miR-424, might bind more miR-424 by competing with the PRKCD transcript. Furthermore, circLARP4 was proved to bind to miR-424 in ESCC cells during G1/S transition, especially in KYSE-510 cells, to decrease the binding of miR-424 to its G1/S target PRKCD. It is possible that PRKCD and circLARP4 are ceRNAs during G1/S transition in ESCC cells. However, the reason why circLARP4 completely abrogated miR-424 binding to PRKCD in KYSE-510 but not KYSE-410 cells is unknown. It has been suggested that ceRNA crosstalk applies mainly to a subset of ceRNAs and miRNAs whose cellular concentrations fall within a specific range of values [50]. The quantitation of the absolute levels of miR-424 and its target transcript abundance will allow a more precise determination of the effectiveness of ceRNAs.
PRKCD, a target of miR-424 during G1/S transition in ESCCs, is a member of the protein kinase C (PKC) family and can function as a negative regulator of cell cycle progression to inhibit cancer cell proliferation. PRKCD-dependent induction of p21Cip1 is one of the mechanisms by which PRKCD inhibits G1/S transition [29]. The control of p21Cip1 levels by PRKCD appears to be complex, with involvement of both transcriptional [51] and post-transcriptional [40] regulation. In ESCC KYSE-410 cells, PRKCD induction by miR-424 knockdown enhanced p21Cip1 protein stability by activating downstream p38 MAPK and JAK, which further decreased CDK2 activity to delay G1/S transition. On the other hand, during G2/M transition, miR-424 targeted WEE1 in ESCC cells. WEE1 fulfils a major cell cycle-related function during G2/M transition by directly catalyzing the inhibitory tyrosine 15 phosphorylation of CDC2 and thereby inhibiting the activity of CDC2. Loss of WEE1 activity would lead to increased CDC2 activity, resulting in a loss of control of cell cycle progression and genome integrity, which would contribute to cancer development [35].
The locus of the human miR-424 gene is at Xq26.3, which is not commonly amplified or deleted in ESCCs [52]. We also intended to identify the factors involved in the upregulation of miR-424 in ESCC. Here, we found that expression of miR-424, as well as pri-miR-424 and pre-miR-424, increased during G1/S transition. Interestingly, two binding sites of the G1/S transcriptional activator E2F1 were identified in the promoter of miR-424. Experiments involving silenced expression of E2F1 in ESCC cell lines demonstrated that E2F1 could regulate pri-miR-424 expression. In addition, ChIP and luciferase activity assays indicated that E2F1 directly binds to the promoter region of miR-424 and promotes its transcription and the elevation of miR-424. These results suggest a positive feedback loop in which miR-424 is one of the G1/S genes that are activated by the transcription factor E2F1 and elevation of miR-424 further increases cyclin-CDK activity and decreases CDK-inhibitory proteins, leading to irreversible cell cycle commitment [27]. In addition to E2F1, HIF1α has been reported to transcriptionally activate miR-424 expression under hypoxic conditions, which decreases the sensitivity of colon cancer and melanoma cells to chemotherapy by inhibiting apoptosis [53]. HIF1α is a widely studied transcription factor that is induced by hypoxia to activate downstream target genes associated with angiogenesis, cell survival, chemotherapy and radiation resistance, invasion, and metastasis of tumors, including ESCCs [54]. Moreover, the transcription factor SMAD4, a downstream factor of the activated TGFβ pathway, can activate miR-424 transcription during mammary epithelial involution after pregnancy [26]. TGFβ/SMAD signaling has been reported to play a role in ESCC invasion and migration. It is possible that miR-424 expression is regulated transcriptionally by distinct transcription factors during various biological processes, including but not limited to cell proliferation, invasion, and migration.
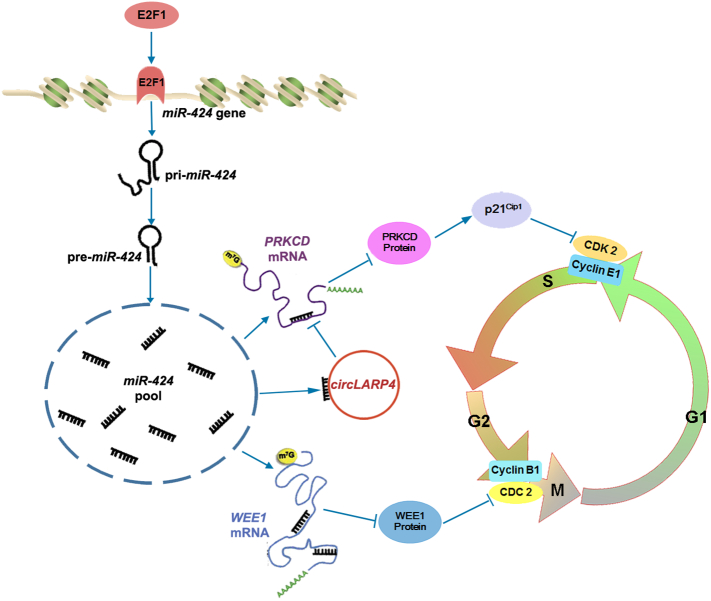
Mechanistically, our data suggest a model (Fig. 7) in which miR-424 regulates ESCC cell proliferation. When the cell cycle progresses to mid- or late-G1-phase, the activation of E2F1 induces the transcription of pri-miR-424. This primary transcript is processed to generate mature miR-424, which in turn regulates the expression of its targets PRKCD and WEE1, which are components of the cell cycle progression regulation network. PRKCD modulates p21Cip1 protein stability to affect CDK2 activity and G1/S progression, while WEE1 affects CDC2 activity directly to regulate G2/M progression. Furthermore, circLARP4 sponges miR-424 during G1/S transition, which decreases miR-424 binding to its target PRKCD and modulates the effect of miR-424 on G1/S transition.
Fig. 7.
Proposed mechanistic model for the role of miR-424 in coordinating ESCC cell cycle progression. During G1/S transition, activation of the transcription factor E2F1 induces the expression of the miR-424 primary transcript. Subsequently, mature miR-424, which is generated from pri-miR-424, targets PRKCD and WEE1 at the G1/S and G2/M borders, respectively, and these proteins further modulate the expression or activity levels of cell cycle regulators such as p21Cip1, CDK2, and CDC2 to regulate ESCC cell cycle progression. However, circLARP4, a natural sponge of miR-424, can diminish the binding of miR-424 to PRKCD and compromise its regulation of G1/S progression.
Sustained cell proliferation induced by the loss of normal cell cycle control is a hallmark of human cancer [5]. In this study, miR-424 was found to be upregulated in ESCC and to correlate with poor survival among ESCC patients. The tumor-promoting role of miR-424 in ESCC was exerted through its regulation of cell cycle progression by directly targeting PRKCD and WEE1 in ESCC cells, which further affected the activity or expression of CDKs or CKIs involved in the G1/S and G2/M cell cycle transitions. miR-424 may possibly be used as a novel prognostic marker and (or) as an effective therapeutic target for ESCCs. Considering the complexity of miRNA roles, which can be tissue specific and biological process specific, future studies are required to fully elucidate the function of miR-424 in distinct ESCC cellular behaviors, such as invasion, migration, survival and apoptosis.
Funding sources
This work was supported by National Natural Science Foundation of China (Grant Nos 81672356 and 81360365), Guangzhou Science Technology and Innovation Commission (Grant No. 201610010127), Guangdong Talents Special Support Program (Grant No. 201629038), and Guangdong Esophageal Cancer Institute Science and Technology Program (Grant Nos Q201404 and M201701).
Declaration of interests
The authors have no financial conflicts to declare.
Author contributions
JW, JF, and HY designed the study and wrote the manuscript. JW, YH, and QL performed the experiments and analyzed the data; YL performed the evaluation of IHC staining of tissues; SZ assisted with miRNA microarray data analyses; KL assisted with statistical analysis; XX assisted with the tumor xenograft experiment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.043.
Appendix A. Supplementary data
Supplementary data
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Dotto G.P., Rustgi A.K. Squamous cell cancers: A unified perspective on biology and genetics. Cancer Cell. 2016;29:622–637. doi: 10.1016/j.ccell.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingham M., Schwartz G.K. Cell-cycle therapeutics come of age. J Clin Oncol. 2017;35:2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Ohashi S., Miyamoto S., Kikuchi O., Goto T., Amanuma Y., Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 7.Shah M.Y., Ferrajoli A., Sood A.K., Lopez-Berestein G., Calin G.A. microRNA therapeutics in cancer - An emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David S., Meltzer S.J. MicroRNA involvement in esophageal carcinogenesis. Curr Opin Pharmacol. 2011;11:612–616. doi: 10.1016/j.coph.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Wang J., Yang X., Chen D., Wang L. MiR-424-5p participates in esophageal squamous cell carcinoma invasion and metastasis via SMAD7 pathway mediated EMT. Diagn Pathol. 2016;11:88. doi: 10.1186/s13000-016-0536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Barrueco R., Nekritz E.A., Bertucci F. miR-424(322)/503 is a breast cancer tumor suppressor whose loss promotes resistance to chemotherapy. Genes Dev. 2017;31:553–566. doi: 10.1101/gad.292318.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian Q., Li Y., Wang F. MicroRNA detection in cervical exfoliated cells as a triage for human papillomavirus-positive women. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J., Li Y., Wang F. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32:976–987. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 13.Yu L., Ding G.F., He C., Sun L., Jiang Y., Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Hershkovitz-Rokah O., Modai S., Pasmanik-Chor M. Restoration of miR-424 suppresses BCR-ABL activity and sensitizes CML cells to imatinib treatment. Cancer Lett. 2015;360:245–256. doi: 10.1016/j.canlet.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q., Fu H., Sun F. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Llobet-Navas D., Rodriguez-Barrueco R., de la Iglesia-Vicente J. The microRNA 424/503 cluster reduces CDC25A expression during cell cycle arrest imposed by transforming growth factor beta in mammary epithelial cells. Mol Cell Biol. 2014;34:4216–4231. doi: 10.1128/MCB.00611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen J., Yang H., Liu M.Z. Gene expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neo-chemoradiotherapy. Ann Oncol. 2014;25:1769–1774. doi: 10.1093/annonc/mdu201. [DOI] [PubMed] [Google Scholar]
- 18.Wen J., Luo K., Liu H. MiRNA expression analysis of pretreatment biopsies predicts the pathological response of esophageal squamous cell carcinomas to neoadjuvant chemoradiotherapy. Ann Surg. 2016;263:942–948. doi: 10.1097/SLA.0000000000001489. [DOI] [PubMed] [Google Scholar]
- 19.Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 20.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Ji P., Smith S.M., Wang Y. Inhibition of gliomagenesis and attenuation of mitotic transition by MIIP. Oncogene. 2010;29:3501–3508. doi: 10.1038/onc.2010.114. [DOI] [PubMed] [Google Scholar]
- 22.Fasanaro P., Greco S., Lorenzi M. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen J., Luo K.J., Hu Y., Yang H., Fu J.H. Metastatic lymph node CHIP expression is a potential prognostic marker for resected esophageal squamous cell carcinoma patients. Ann Surg Oncol. 2013;20:1668–1675. doi: 10.1245/s10434-012-2733-4. [DOI] [PubMed] [Google Scholar]
- 24.Thomson D.W., Bracken C.P., Goodall G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llobet-Navas D., Rodriguez-Barrueco R., Castro V. The miR-424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev. 2014;28:765–782. doi: 10.1101/gad.237404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertoli C., Skotheim J.M., de Bruin R.A. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A., Fornes O., Stigliani A. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx1188. (D260–D6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson D.N., Foster D.A. The enigmatic protein kinase Cdelta: Complex roles in cell proliferation and survival. FASEB J. 2004;18:627–636. doi: 10.1096/fj.03-0979rev. [DOI] [PubMed] [Google Scholar]
- 30.Nilsson I., Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 31.Dorrello N.V., Peschiaroli A., Guardavaccaro D., Colburn N.H., Sherman N.E., Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 32.Li Y., Pei J., Xia H., Ke H., Wang H., Tao W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene. 2003;22:4398–4405. doi: 10.1038/sj.onc.1206603. [DOI] [PubMed] [Google Scholar]
- 33.Hydbring P., Malumbres M., Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nat Rev Mol Cell Biol. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int J Cancer. 2014;134:1013–1023. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorensen C.S., Syljuasen R.G. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Liu H., Hou L. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abukhdeir A.M., Park B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19. doi: 10.1017/S1462399408000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blank V.C., Bertucci L., Furmento V.A., Pena C., Marino V.J., Roguin L.P. A chimeric cyclic interferon-alpha2b peptide induces apoptosis by sequential activation of phosphatidylinositol 3-kinase, protein kinase Cdelta and p38 MAP kinase. Exp Cell Res. 2013;319:1471–1481. doi: 10.1016/j.yexcr.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 39.Yokoyama G., Fujii T., Tayama K., Yamana H., Kuwano M., Shirouzu K. PKCdelta and MAPK mediate G(1) arrest induced by PMA in SKBR-3 breast cancer cells. Biochem Biophys Res Commun. 2005;327:720–726. doi: 10.1016/j.bbrc.2004.12.070. [DOI] [PubMed] [Google Scholar]
- 40.Wakino S., Kintscher U., Liu Z. Peroxisome proliferator-activated receptor gamma ligands inhibit mitogenic induction of p21(Cip1) by modulating the protein kinase Cdelta pathway in vascular smooth muscle cells. J Biol Chem. 2001;276:47650–47657. doi: 10.1074/jbc.M108719200. [DOI] [PubMed] [Google Scholar]
- 41.Harada K., Baba Y., Ishimoto T. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol. 2016;51:520–530. doi: 10.1007/s00535-016-1161-9. [DOI] [PubMed] [Google Scholar]
- 42.Boldrup L., Coates P.J., Laurell G., Wilms T., Fahraeus R., Nylander K. Downregulation of miRNA-424: A sign of field cancerisation in clinically normal tongue adjacent to squamous cell carcinoma. Br J Cancer. 2015;112:1760–1765. doi: 10.1038/bjc.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu K., Hu G., He X. MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19:739–748. doi: 10.1007/s12253-013-9637-x. [DOI] [PubMed] [Google Scholar]
- 44.Curtius K., Wright N.A., Graham T.A. An evolutionary perspective on field cancerization. Nat Rev Cancer. 2018;18:19–32. doi: 10.1038/nrc.2017.102. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima T., Jinnin M., Etoh T. Down-regulation of mir-424 contributes to the abnormal angiogenesis via MEK1 and cyclin E1 in senile hemangioma: its implications to therapy. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merlet E., Atassi F., Motiani R.K. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovasc Res. 2013;98:458–468. doi: 10.1093/cvr/cvt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inui M., Martello G., Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 48.Liu H., Kohane I.S. Tissue and process specific microRNA-mRNA co-expression in mammalian development and malignancy. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drasin D.J., Guarnieri A.L., Neelakantan D. TWIST1-induced miR-424 reversibly drives mesenchymal programming while inhibiting tumor initiation. Cancer Res. 2015;75:1908–1921. doi: 10.1158/0008-5472.CAN-14-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saha K., Eckert R.L. Methylosome protein 50 and PKCdelta/p38delta protein signaling control keratinocyte proliferation via opposing effects on p21Cip1 gene expression. J Biol Chem. 2015;290:13521–13530. doi: 10.1074/jbc.M115.642868. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Zhang L., Zhou Y., Cheng C. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96:597–611. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D., Shi Z., Li M., Mi J. Hypoxia-induced miR-424 decreases tumor sensitivity to chemotherapy by inhibiting apoptosis. Cell Death Dis. 2014;5:e1301. doi: 10.1038/cddis.2014.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Semenza G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data