Abstract
Background
Cryptococcal meningitis (CM) is a significant source of mortality, the pathogenesis of which has not been fully understood, especially in non-HIV infected populations. We aimed to explore the potential genetic influence of Toll-like receptor (TLR) on non-HIV CM.
Methods
This observational cohort study was done in two stages: a discovery stage and a validation stage. A case-control genetic association study was conducted between 159 non-HIV CM patients and 468 healthy controls. TLR SNPs significantly related to susceptibility went further validation in a second cohort of 583 subjects from a certain district. Associations among TLR SNPs, cerebrospinal fluid (CSF) cytokine concentrations, and clinical severity were explored in a third cohort of 99 previously untreated non-HIV CM patients. Logistic regression model was used to determine the independent predictors for disease severity.
Findings
In the discovery stage, eight TLR SNPs exhibited significant genetic susceptibility to non-HIV CM, one of which was validated in a population validation of HIV-infected cases while none survived in non-HIV cases. CSF cytokine detections showed that 18 cytokines were significantly over-expressed in severely ill patients. Two of the 8 SNPs (rs5743604 and rs3804099) were also significantly associated with disease severity. Specifically, the rs3804099 C/T genotype was further found to be correlated to 12 of the 18 up-regulated cytokines in severe patients. In addition, high levels of interleukin (IL)-10 in CSF (OR 2·97, 95% CI 1·49–5·90; p = 0·002) was suggested as an independent predictor for severity after adjusted for possible confounders.
Interpretation
TLR participates in both the occurrence and the pathogenesis of non-HIV CM. The in situ immune responses of CM were under genetic influence of TLR and contributed to disease severity.
Fund
National Natural Science Foundation of China and National Key Basic Research Program of China (973 Program).
Keywords: Toll-like receptor, Non-HIV cryptococcal meningitis, Genetic susceptibility, Disease severity, CSF cytokine
Research in context.
Evidence before this study
Possible deficiencies in innate immune system of non-HIV population may contribute to cryptococcal meningitis (CM). Toll-like receptors (TLRs) are well characterized in central nervous system infections other than CM. We searched PubMed for articles published before Jan, 2018, without any language restrictions using the terms “Toll-like receptor” or “TLR”, combined with “Cryptococcus”, “cryptococcosis”, or “cryptococcal meningitis”. We identified 38 publications, 14 of which highlighted the role of TLR in the host defence against Cryptococcus, using TLR knockout mice or specific antibodies. TLR2 and TLR4 have gained much attention, but yielding no consistent results. We found 3 in vitro studies focused TLR on CM, however, data on human studies are lacking, as the genetic influence of TLR on non-HIV CM remains unclear.
Added value of this study
To our knowledge, this observational study is the first to examine the genetic correlation between TLR and CM in non-HIV cohorts, which combines a discovery stage and a validation stage, using separate patient cohorts and immunological approaches. TLR SNPs was found to predict not only CM susceptibility but also the clinical severity of patients. Moreover, cytokine concentrations in cerebrospinal fluid (CSF) were found under TLR genetic influence and associated with clinical severity. The pairwise correlations we observed among TLR SNPs, CSF cytokines, and clinical severity strongly implied a causal role for TLR in the pathogenesis of non-HIV CM.
Implications of all the available evidence
The identification of genetic polymorphisms in TLR as predictors of susceptibility and disease severity of non-HIV CM provides new avenues towards early evaluation and interventions for patients. Further exploration may target candidate TLR SNPs (eg, cytological research or animal model) to clarify the specific causative mechanisms, which may also inform of possible host-based therapeutics for CM.
Alt-text: Unlabelled Box
1. Introduction
Cryptococcus neoformans and C. gatti are important causes of central nervous system (CNS) infections with significant mortality, remaining a great public health challenge worldwide. Commonly seen as an opportunistic infection in adults with HIV/AIDS, cryptococcal meningitis (CM) accounts for 15% of HIV-related mortality globally [1]. In addition, a growing number of non-HIV CM patients have been witnessed in recent years with fatality approaching 30% in some areas [2,3]. It occurs in both those with natural or iatrogenic immunosuppression, as well as the apparently immunocompetent individuals. Approximately 65–70% of non-HIV CM patients were without any predisposing factors, particularly in the East Asia [4,5]. It remains unsolved whether these “healthy hosts” are carrying underlying immune deficiencies. One previous genetic association study has indicated that the deficient genotypes of mannose-binding lectin (MBL), a member of the C-type lectin receptors of germline-encoded pattern-recognition receptors (PRRs), were correlated with increased susceptibility to non-HIV CM [6]. However, this accounted for only 16·5% of all CM and 19·2% of apparently immunocompetent patients, raising the possibility that other genetic deficiencies in host immune system may also contribute to CM.
Toll-like receptors (TLRs) are the first identified and the most well characterized PRRs that detect both exogenous pathogen-associated molecular patterns (PAMPs) and endogenous damage-associated molecular patterns (DAMPs). TLRs could recognize almost every pathogen as soon as they contact the host via any route of exposure, and they initiate an effective innate immune response at the early stage of infection. Expressed on various immune cells, TLR uses its extracellular leucine-rich repeat domain to mediate PAMPs and its cytosolic Toll–interleukin (IL)-1 receptor domain to recruit adaptor proteins. The subsequent cascades of signaling events result in the expression of proinflammatory cytokines and chemokines, which may further activate the adaptive immunity.
We aimed to explore the potential genetic influence of TLR by comparing TLR single nucleotide polymorphisms (SNPs) of non-HIV CM patients with those of healthy controls and assessing the links among TLR SNPs, cytokine concentrations in cerebrospinal fluid (CSF), and clinical severity.
2. Methods
2.1. Study design and participants
We performed this prospective observational cohort study in two stages: a discovery stage and a validation stage (appendix). Participants were of Chinese Han ethnicity, and were recruited at Huashan Hospital (Shanghai, China), Mengchao Hepatobiliary Hospital (Fuzhou, Fujian, China), Fujian HIV/AIDS Diagnosis and Treatment Center (Fuzhou, Fujian, China), and No. 476 Hospital of Fuzhou General Hospital (Fuzhou, Fujian, China). No restriction in terms of age or sex was applied. In the discovery cohort, a case-control genetic association study was conducted between non-HIV patients with proven diagnosed CM and healthy controls. The identified TLR SNPs related to susceptibility then underwent further validations. A second cohort was used for population validation, consisting of subjects divided into four groups according to their HIV serological status and CM diagnosis. All subjects are permanent residents in Fujian, a province on China's southeastern coast that has a year-round warm and humid climate. To further explore the role of TLR in the pathogenesis of non-HIV CM, cytokine expressions in CSF were detected and correlations among TLR SNPs, CSF cytokines, and clinical severity of disease were examined in a third cohort consisted of previously untreated patients with proven non-HIV CM.
Oral consent for storage of surplus sample and clinical data was obtained from both control participants and patients, or close relatives of patients who were unconscious. Ethical approval was obtained from the medical ethics committee of Huashan Hospital, Mengchao Hepatobiliary Hospital, Fujian HIV/AIDS Diagnosis and Treatment Center, and No. 476 Hospital of Fuzhou General Hospital.
2.2. Procedures
We used a salting out method to extract genomic DNA from peripheral blood obtained from each participant. Genotyping was performed in both stages by multiplex SNaPshot technology using an ABI fluorescence-based assay allelic discrimination method (Applied Biosystems, Foster City, CA, USA). SNPs in TLR1, TLR2, TLR4, TLR6 and TLR9 were selected from NCBI and HapMap databases with the criteria of a minor allele frequency (MAF) >0·1 and r2 >0·8 in the Chinese Han population. The 5-kb regions upstream and 2-kb regions downstream of the gene were included. As supplements, we also selected SNPs within the candidate genes that have been reported in previous studies to be associated with infectious diseases. A total of 40 SNPs were finally selected in the discovery stage. Genotyped SNPs and primers are described in detail in the appendix.
CSF was collected from each patient hospitalized in Huashan Hospital. Initial lumbar puncture was done before the antifungal treatment. CSF was tested for Cryptococcus with microscopy and culture and centrifuged and stored at −80 °C until analysis using a sandwich enzyme-linked immunosorbent assay for 27 cytokines (Luminex, Bio-Rad, Hercules, CA, USA). Phenotypic characterization of clinical isolates were identified by chemotyping on canavanine-glycine-bromothymol blue (CGB) medium. Multilocus sequence typing (MLST) was also performed using the International Society of Human and Animal Mycology (ISHAM) consensus MLST scheme for C. neoformans and C. gatti, which includes seven genetic loci. Cryptococcal antigens in CSF and blood were determined by diluted CSF with use of the Latex-Cryptococcus antigen detection system (IMMY, Norman, OK, USA) before Jan, 2013, followed by the use of rapid lateral flow assay (IMMY, Norman, OK, USA), and a titre of ≥1:10 was considered to be positive.
Detailed clinical data were collected from the previously untreated cohort. We obtained demographic data, predisposing factors, and clinical records on admission, including clinical manifestation, CSF opening pressure and laboratory examination, enhanced cranial magnetic resonance imaging (MRI), initial therapy, and time from symptom onset to treatment. Severe cases were defined as those who developed mental status changes (Glasgow Coma Scale score, [GCS] <15) or died within two weeks after admission.
2.3. Statistical analysis
The χ2 analyses were used for each individual SNP to test for deviation from the Hardy-Weinberg equilibrium. Differences in allele frequencies and genotype distributions were analysed with the use of SNPstats, an online software. Linkage disequilibrium and haplotype analysis were performed with Haploview (version 4·2).
We tested categorical data by χ2 or Fisher exact tests, and continuous data without normal distribution were tested with Mann-Whitney tests. Multivariable logistic regression models were constructed using stepwise regression with the objective of determining the factors at baseline associated independently with disease severity. All variables with a value of p < 0·05 in the univariate analysis were included. The results of the multivariate analysis were expressed as odds ratio (OR) and the corresponding 95% confidence intervals (CIs).
Data were analysed with the use of SPSS statistical package (version 17·0). All tests were two-sided, and a value of p < 0·05 denoted statistical significance.
3. Results
3.1. Genetic susceptibility of TLR in non-HIV CM
In the discovery stage, we enrolled 159 cases of non-HIV CM and 468 healthy controls between Jan 1, 2001, and Dec 31, 2012. The median age was 45 years (range 14–78) in patients and 65·4% were men. Among healthy subjects, the median age was 37 years (range 17–75) and 41·9% were men. Seventy-four (46·5%) patients had one or more predisposing factors, of which corticosteroid use was the most common (40 [25·2%] of 74), followed by autoimmune diseases (26 [16·4%] of 74).
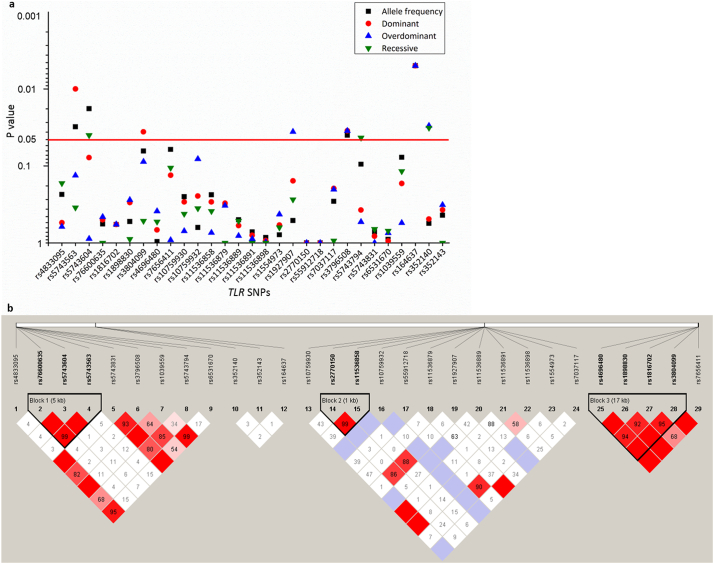
Four samples from the control group were excluded because they failed in genotyping assay. Eleven of the 40 genotyped TLR SNPs showed no polymorphism in all the subjects, so that only 29 SNPs were eligible for the subsequent analysis. All these 29 SNPs were in Hardy-Weinberg equilibrium in 464 controls (data not shown), with MAFs ranged from 0·11% to 49·12%. Association analysis suggested that allele frequencies of four SNPs were significantly correlated with CM. The rs164637 C allele was the most significant (OR 0·07, 95% CI 0·01–0·58; p = 0·005) (Table 1; Fig. 1a). Nine of the 29 SNPs were defined into three haplotype blocks (Fig. 1b). Haplotypes with frequencies >0·01 were analysed; however, none of them were associated with CM (appendix). In addition, comparisons of genotype distributions indicated that eight SNPs showed significant differences, including six SNPs with increased risks of CM (Table 1; Fig. 1a): rs5743563 T/T (OR 1·66, 95% CI 1·13–2·46; p = 0·010), rs5743604 T/T (OR 1·53, 95% CI 1·02–2·29; p = 0·040), rs3804099 T/T (OR 1·47, 95% CI 1·02–2·11; p = 0·036), rs3796508 G/A (OR 1·79, 95% CI 1·04–3·10; p = 0·035), rs164637 C/T (OR 15·03, 95% CI 1·74–129·67; p = 0·005), and rs352140 T/T (OR 1·69, 95% CI 1·04–2·75; p = 0·032). We further made subgroup analyses, comparing allele and genotype distributions between immunocompetent patients and controls (Table 1). Similar to results from the overall patient group, associations were found in rs3804099 T/T, rs352140 C/T, and rs164637 C/C and C/T genotypes (Table 1).
Table 1.
Allele and genotype distributions of 8 TLR SNPs associated with non-HIV CM in the discovery stage.
| SNP | Position | Gene | Genotype | Control |
All patients |
Immunocompetent patients |
All patients vs control |
Immunocompetent patients vs control |
||
|---|---|---|---|---|---|---|---|---|---|---|
| (n = 464) | (n = 159) | (n = 85) | OR (95% CI) | p value | OR (95% CI) | p value | ||||
| rs5743563 | 4:38804398 | TLR1 | T/T (Dominant) | 112 (24·1) | 55 (34·6) | 25 (29·4) | 1·66 (1·13–2·46) | 0·010 | 1·31 (0·78–2·19) | 0·302 |
| C/T (Over-dominant) | 248 (53·5) | 74 (46·5) | 43 (50·6) | 0·76 (0·53–1·09) | 0·133 | 0·89 (0·56–1·42) | 0·627 | |||
| C/C (Recessive) | 104 (22·4) | 30 (18·9) | 17 (20·0) | 0·81 (0·51–1·27) | 0·348 | 0·87 (0·49–1·54) | 0·622 | |||
| T (Allelic) | 472 (50·9) | 184 (57·9) | 93 (54·7) | 1·33 (1·03–1·72) | 0·031 | 1·17 (0·84–1·62) | 0·357 | |||
| rs5743604 | 4:38799664 | TLR1 | C/C (Dominant) | 142 (30·6) | 37 (23·3) | 19 (22·4) | 0·69 (0·45–1·04) | 0·078 | 0·65 (0·38–1·13) | 0·125 |
| C/T (Over-dominant) | 222 (47·8) | 75 (47·2) | 42 (49·4) | 0·97 (0·68–1·40) | 0·883 | 1·06 (0·67–1·69) | 0·790 | |||
| T/T (Recessive) | 100 (21·6) | 47 (29·6) | 24 (28·2) | 1·53 (1·02–2·29) | 0·040 | 1·43 (0·85–2·41) | 0·175 | |||
| C (Allelic) | 506 (54·5) | 149 (46·9) | 80 (47·1) | 0·74 (0·57–0·95) | 0·018 | 0·74 (0·53–1·03) | 0·073 | |||
| rs3804099 | 4:153703504 | TLR2 | T/T (Dominant) | 192 (41·4) | 81 (50·9) | 46 (54·1) | 1·47 (1·02–2·11) | 0·036 | 1·67 (1·05–2·66) | 0·029 |
| C/T (Over-dominant) | 226 (48·7) | 65 (40·9) | 32 (37·6) | 0·73 (0·51–1·05) | 0·088 | 0·64 (0·40–1·02) | 0·060 | |||
| C/C (Recessive) | 46 (9·9) | 13 (8·2) | 7 (8·2) | 0·81 (0·43–1·54) | 0·518 | 0·82 (0·36–1·87) | 0·630 | |||
| T (Allelic) | 610 (65·7) | 227 (71·4) | 124 (72·9) | 1·30 (0·98–1·72) | 0·064 | 1·41 (0·98–2·02) | 0·066 | |||
| rs1927907 | 9:117710486 | TLR4 | G/G (Dominant) | 259 (55·8) | 99 (62·3) | 53 (62·4) | 1·31 (0·90–1·89) | 0·156 | 1·31 (0·82–2·11) | 0·264 |
| G/A (Over-dominant) | 171 (36·9) | 44 (27·7) | 23 (27·1) | 0·66 (0·44–0·97) | 0·036 | 0·64 (0·38–1·06) | 0·082 | |||
| A/A (Recessive) | 34 (7·3) | 16 (10·1) | 9 (10·6) | 1·42 (0·76–2·64) | 0·273 | 1·50 (0·69–3·25) | 0·304 | |||
| G (Allelic) | 689 (74·2) | 242 (76·1) | 129 (75·9) | 1·11 (0·82–1·49) | 0·511 | 1·09 (0·75–1·60) | 0·653 | |||
| rs3796508 | 4:38828495 | TLR6 | G/G (Dominant) | 424 (91·4) | 136 (85·5) | 72 (84·7) | 0·56 (0·32–0·97) | 0·035 | 0·52 (0·27–1·03) | 0·055 |
| G/A (Over-dominant) | 40 (8·6) | 23 (14·5) | 13 (15·3) | 1·79 (1·04–3·10) | 0·035 | 1·91 (0·98–3·75) | 0·055 | |||
| G (Allelic) | 888 (95·7) | 295 (92·8) | 157 (92·4) | 0·58 (0·34–0·98) | 0·040 | 0·54 (0·28–1·04) | 0·062 | |||
| rs5743794 | 4:38831106 | TLR6 | G/G (Dominant) | 157 (33·8) | 60 (37·7) | 30 (35·3) | 1·19 (0·82–1·72) | 0·373 | 1·07 (0·66–1·73) | 0·794 |
| G/A (Over-dominant) | 226 (48·7) | 82 (51·6) | 44 (51·8) | 1·12 (0·78–1·61) | 0·533 | 1·13 (0·71–1·80) | 0·604 | |||
| A/A (Recessive) | 81 (17·5) | 17 (10·7) | 11 (12·9) | 0·57 (0·32–0·99) | 0·043 | 0·70 (0·36–1·38) | 0·305 | |||
| G (Allelic) | 540 (58·2) | 202 (63·5) | 104 (61·2) | 1·25 (0·96–1·63) | 0·095 | 1·13 (0·81–1·58) | 0·467 | |||
| rs164637 | 3:52231199 | TLR9 | C/C (Dominant) | 463 (99·8) | 154 (96·9) | 82 (96·5) | 0·07 (0·01–0·57) | 0·005 | 0·06 (0·01–0·57) | 0·013 |
| C/T (Over-dominant) | 1 (0·2) | 5 (3·1) | 3 (3·5) | 15·03 (1·74–129·67) | 0·005 | 16·94 (1·74–164·83) | 0·013 | |||
| C (Allelic) | 927 (99·9) | 313 (98·4) | 167 (98·2) | 0·07 (0·01–0·58) | 0·005 | 0·06 (0·01–0·58) | 0·013 | |||
| rs352140 | 3:52222681 | TLR9 | C/C (Dominant) | 184 (39·7) | 68 (42·8) | 44 (51·8) | 1·13 (0·79–1·64) | 0·490 | 1·63 (1·03–2·60) | 0·037 |
| C/T (Over-dominant) | 224 (48·3) | 61 (38·4) | 28 (32·9) | 0·67 (0·46–0·96) | 0·030 | 0·53 (0·32–0·86) | 0·009 | |||
| T/T (Recessive) | 56 (12·1) | 30 (18·9) | 13 (15·3) | 1·69 (1·04–2·75) | 0·032 | 1·32 (0·68–2·53) | 0·410 | |||
| C (Allelic) | 592 (63·2) | 197 (61·9) | 116 (68·2) | 0·95 (0·73–1·23) | 0·679 | 1·22 (0·86–1·73) | 0·266 | |||
Data are n (%). TLR = Toll-like receptor. SNP = single nucleotide polymorphism. HIV = human immunodeficiency virus. CM = cryptococcal meningitis.
Fig. 1.
Genetic analyses between patients and controls in the discovery stage.
(a) Distribution of p values for allele and genotype comparisons between patients and controls in the discovery stage. Dashed line indicates the p value correspond with p = 0·05. (b) Haplotype blocks among SNPs of the TLR1, TLR2, TLR4, TLR6 and TLR9 genes in 623 genotyped subjects. TLR = Toll-like receptor. SNP = single nucleotide polymorphism.
3.2. Population validation of TLR SNPs
Population validation was conducted in the validation stage to replicate the genetic association study. We put the discovered 8 TLR SNPs significantly related to susceptibility in a larger cohort from Fujian province (recruited between Apr 1, 2014, and Sep 30, 2016), including 53 HIV cases with CM, 368 HIV cases without CM, 59 non-HIV cases with CM, and 103 healthy controls. Comparisons were conducted in HIV and non-HIV cases, respectively, which indicated that HIV patients carrying 1 rs3796508 A/A genotype could exhibited a 14·39-fold (95% CI 1·28–161·57; p = 0·013) increased risk of CM (appendix). No significant differences were found between non-HIV CM patients and healthy controls (appendix).
3.3. Associations among TLR SNPs, CSF cytokines, and severity of non-HIV CM
Associations among TLR SNPs, CSF cytokine concentrations, and clinical severity were explored in another independent cohort. We prospectively recruited 99 previously untreated patients with proven non-HIV CM from Jan 1, 2014, to Dec 31, 2017. The median age was 45 years (range, 19–86) and 69·7% were men. Ninety-seven (98·0%) patients were from central and eastern regions of China. Fifty (50·5%) patients had one or more predisposing factors. A total of 76 (76·8%) Cryptococcus strains were obtained, of which 71 (71·7%) were identified as C. neoformans and 5 (5·1%) were C. gatti. The genotype VN1/ST5 (64 [90·1%] of 99) was the most frequently isolated (appendix). The median time from symptom onset to diagnosis was 30 days (range, 3–398 days). Amphotericin B deoxycholate ([AmB] 0.5–0.7 mg/kg/day) induction therapy was used in 85 (85·9%) patients and 5 (5·1%) underwent surgical interventions during the treatment. Thirty-one (31·3%) patients with GCS <15 or died within two weeks were classified as a severe group, and the rest fell into a mild group.
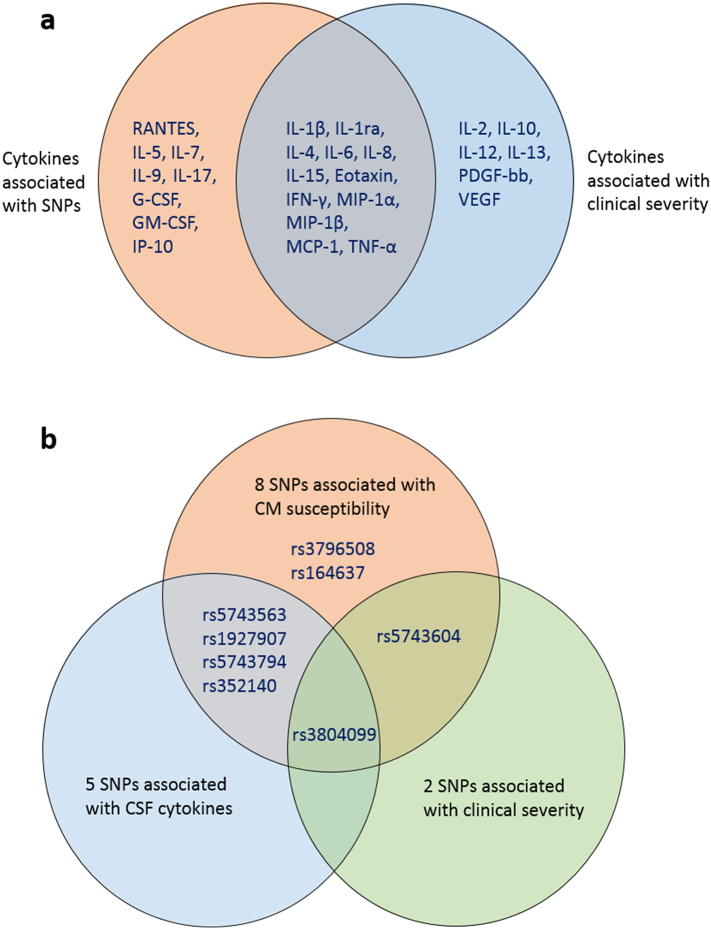
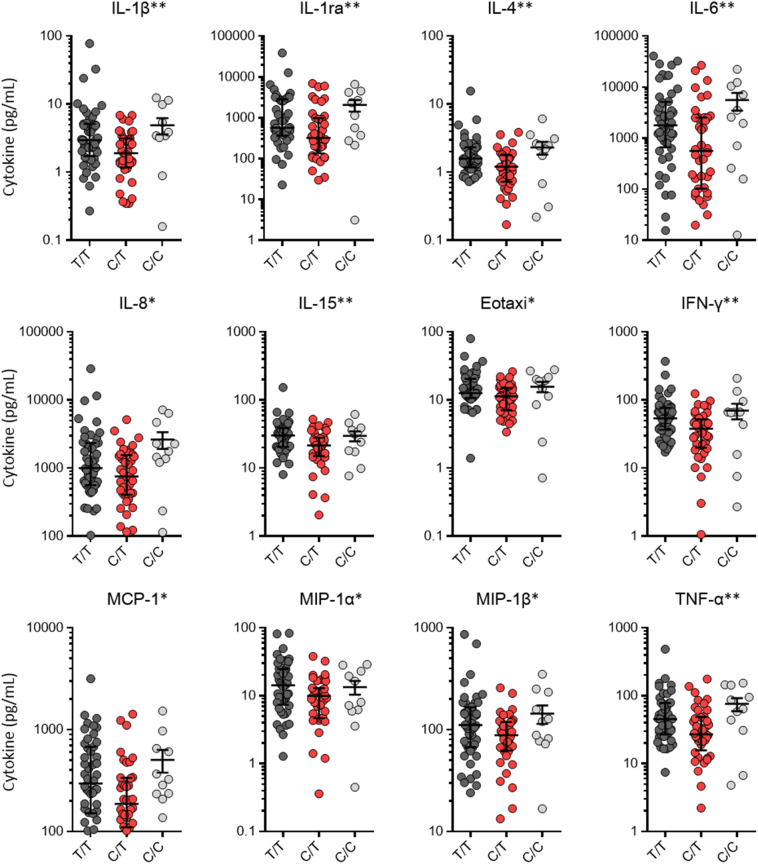
Univariate analysis found that 18 cytokines were significantly over-expressed in severely ill patients (Fig. 2a). Meanwhile, eight SNPs significantly related to CM susceptibility were included in the analysis, of which two SNPs were also notably associated with disease severity (Fig. 2b). The rs5743604 C/C genotype was over-presented (OR 2·54, 95% CI 1·06–6·07; p = 0·034) while the rs3804099 C/T genotype was less frequently detected (OR 0·39, 95% CI 0·15–1·00; p = 0·046) in severe patients (Table 2). In addition, five SNPs (rs5743563, rs3804099, rs1927907, rs5743794, and rs352140) were significantly associated with 20 CSF cytokine expressions after infection (Fig. 2a). Specifically, the rs3804099 C/T genotype was further found to be associated with lower expressions of 12 of the 18 cytokines significantly related to disease severity (Fig. 3). Therefore, pairwise correlations were verified among rs3804099 C/T genotype, CSF cytokine levels, and clinical severity.
Fig. 2.
Summary of interactions among TLR SNPs, CSF cytokine concentrations and clinical severity.
(a) Two cycles represent CSF cytokines associated with TLR SNPs and severity, respectively. The intersection indicates that 12 cytokines are related to both. (b) Three cycles represent TLR SNPs associated with CM susceptibility, CSF cytokine concentrations and clinical severity, respectively. Their intersections show that rs3804099 is correlated to all. CM = cryptococcal meningitis. TLR = Toll-like receptor. SNP = single nucleotide polymorphism. CSF = cerebrospinal fluid. IL = interleukin. IFN = interferon. MIP = macrophage inflammatory protein. MCP = monocyte chemo attractant protein. TNF = tumor necrosis factor. G-CSF = granulocyte-colony stimulating factor. GM-CSF = granulocyte-macrophage colony-stimulating factor. IP = interferon-induced protein. PDGF = platelet-derived growth factor. VEGF = vascular endothelial growth factor.
Table 2.
Univariate and multivariate analysis of factors associated with clinical severity of 99 non-HIV CM patients.
| Variable | Univariate analysisa |
Multivariate analysis |
|||
|---|---|---|---|---|---|
| Mild group |
Severe group |
p value | OR (95% CI) | p value | |
| (n = 68) | (n = 31) | ||||
| Symptoms at presentation | |||||
| Vomiting | 28/68 (41·2) | 21/31 (67·7) | 0·014 | ||
| Epilepsy | 10/68 (14·7) | 11/31 (35·5) | 0·019 | ||
| CSF | |||||
| Cryptococcal antigen (≥1:1280) | 32/68 (47·1) | 22/31 (71·0) | 0·027 | ||
| Cytokine (pg/mL) | |||||
| IL-1β | 2·06 (1·26, 4·06) | 3·22 (2·33, 6·23) | 0·008 | ||
| IL-1ra | 395·04 (188·90, 1236·57) | 931·71 (449·10, 3008·76) | 0·003 | ||
| IL-2 | 4·32 (2·10, 6·48) | 7·33 (3·79, 12·38) | 0·003 | ||
| IL-4 | 1·28 (0·84, 1·93) | 1·76 (1·33, 2·48) | 0·017 | ||
| IL-6 | 837·81 (168·22, 2796·41) | 2673·00 (1233·49, 9831·26) | 0·003 | ||
| IL-8 | 708·11 (429·33, 1500·46) | 1905·49 (988·96, 3079·69) | 0·000 | ||
| IL-10 | 38·84 (25·12, 66·00) | 81·16 (43·91, 128·84) | 0·000 | 2·97 (1·49–5·90) | 0·002 |
| IL-12 | 3·95 (1·58, 6·69) | 5·72 (3·26, 9·11) | 0·010 | ||
| IL-13 | 4·31 (2·35, 12·25) | 9·11 (3·56, 21·69) | 0·029 | ||
| IL-15 | 22·18 (16·24, 32·23) | 31·16 (23·03, 41·14) | 0·001 | ||
| Eotaxin | 11·62 (7·97, 16·54) | 14·68 (11·04, 22·60) | 0·020 | ||
| IFN | 40·89 (22·34, 64·92) | 65·48 (43·80, 95·57) | 0·002 | ||
| MCP-1 | 205·16 (121·07, 431·35) | 367·25 (234·56, 726·76) | 0·003 | ||
| MIP-1α | 9·41 (4·68, 16·28) | 19·25 (8·97, 28·97) | 0·000 | ||
| PDGF-bb | 6·09 (3·71, 10·29) | 7·38 (5·68, 12·29) | 0·049 | ||
| MIP-1β | 83·51 (55·30, 119·20) | 151·82 (105·73, 227·85) | 0·000 | ||
| TNF-α | 31·68 (19·11, 65·08) | 52·34 (37·42, 79·49) | 0·005 | ||
| VEGF | 16·75 (13·68, 25·09) | 22·15 (19·51, 34·45) | 0·008 | ||
| Blood | |||||
| Positive culture of Cryptococcus | 11/67 (16·4) | 12/29 (41·4) | 0·009 | ||
| CRP (>8·20 mg/L) | 26/65 (40·0) | 19/28 (67·9) | 0·014 | 6·24 (1·61–24·18) | 0·008 |
| Pretreatment cranial MRI | |||||
| Ventriculomegaly | 5/60 (8·3) | 13/30 (43·3) | 0·000 | 11·23 (2·58–48·96) | 0·001 |
| TLR SNP | |||||
| rs5743604 C/C | 22/68 (32·4) | 17/31 (54·8) | 0·034 | ||
| rs3804099 C/T | 32/68 (47·1) | 8/31 (25·8) | 0·046 | ||
Data are n (%) or median (IQR). Missing data not provided by the sites are indicated by the denominators in each variable. HIV = human immunodeficiency virus. CM = cryptococcal meningitis. CSF = cerebrospinal fluid. MRI = magnetic resonance imaging. CRP=C-reactive protein. IL = interleukin. IFN = interferon. MCP = monocyte chemo attractant protein. MIP = macrophage inflammatory protein. PDGF = platelet-derived growth factor. TNF = tumor necrosis factor. VEGF = vascular endothelial growth factor. TLR = Toll-like receptor. SNP = single nucleotide polymorphism. Odds ratio for all numerical variables are per quartile increase, and for binary variables represent presence versus absence.
Factors included in the initial univariate analysis not shown in the table: [1] clinical factors: age, sex, predisposing factors, vital signs at presentation (fever, headache, and cranial nerve palsy), CSF examinations (CSF pressure, white blood cell count, lymphocyte count, glucose level, protein level, positive Indian ink smear, and positive culture for Cryptococcus), blood tests (cryptococcal antigen ≥1:1280, erythrocyte sedimentation rate, procalcitonin, D-dimmer, and serum calcium), Cryptococcus strain (Cryptococcus gatti), pretreatment cranial MRI (meningeal enhancement and single/multiple parenchymal lesions) and treatment (time to diagnosis >90 days, Amphotericin B-based treatment, and surgical intervention); [2] CSF cytokines: IL-5, IL-7, IL-9, IL-17, fibroblast growth factor-basic, granulocyte-colony stimulating factor, granulocyte-macrophage colony-stimulating factor, interferon-induced protein-10, and RANTES; [3] TLR SNPs: rs5743563, rs1927907, rs3796508, rs5743794, rs164637, and rs352140.
Fig. 3.
CSF cytokine concentrations in non-HIV CM patients carrying different rs3804099 genotypes.
Comparisons were made based on overdominant models (C/T vs T/T + C/C). *p < 0·05. **p < 0·01. CSF = cerebrospinal fluid. HIV = human immunodeficiency virus. CM = cryptococcal meningitis. TLR = Toll-like receptor. IL = interleukin. IFN = interferon. MCP = monocyte chemo attractant protein. MIP = macrophage inflammatory protein. TNF = tumor necrosis factor.
3.4. Associations of clinical factors with severity of non-HIV CM
We also determined associations of clinical factors including demographic characteristics, symptoms, and laboratory and imaging features at baseline with severity of non-HIV CM (Table 2). Patients presented with severe disease, often with vomiting (p = 0·014) or epilepsy (p = 0·019). Laboratory-tested biomarkers including abnormal C-reactive protein level (>58·20 mg/L), higher CSF cryptococcal capsular polysaccharide antigen titre (≥1:1280), and positive blood culture for Cryptococcus were at p < 0·05. Pre-treatment enhanced cranial MRI had greater association with severity. Interpretations of the MRIs were based on the uses of contrast and FLAIR imaging. Patients with ventriculomegaly, indicating impaired drainage of CSF or intracranial hypertension, were five times more likely to be severely ill (p = 0·000).
3.5. Multivariate analysis of severity of non-HIV CM
We further developed a logistic regression model for severity that includes the following factors: vomiting, epilepsy, CSF cryptococcal antigen titre, C-reactive protein level, blood culture, ventriculomegaly, 18 CSF cytokines, and two TLR SNPs. Increased C-reactive protein level (OR 6·24, 95% CI 1·61–24·18; p = 0·008), ventriculomegaly (OR 11·23, 95% CI 2·58–48·96; p = 0·001) and high IL-10 levels in CSF (OR 2·97, 95% CI 1·49–5·90; p = 0·002) were independently predictive of severity (Table 2).
4. Discussion
Our study firstly identified TLR SNPs as predictors of non-HIV CM susceptibility, and we further conducted a population validation. Additionally, TLR SNPs were found in relation to CSF cytokine concentrations, and pairwise correlations were found among TLR SNPs, CSF cytokine concentrations, and clinical severity. Collectively, our data suggested that TLR is crucial for both the occurrence and the pathogenesis of non-HIV CM.
Although earlier in vitro and animal studies had explored the role of TLR in cryptococcal infection, evidence remains controversial. TLR2 and TLR4 were intensively investigated. TLR4 but not TLR2 was reported to confer responsiveness of cryptococcal glucuronoxylomannan on CHO/CD14 cells [7]. However, recent study on human peripheral blood mononuclear cells suggested the involvement of impaired expression of TLR2 in defective host defence [8]. Additionally, in intraperitoneal C. neoformans–challenged mice, TLR2 knockout mice showed decreased survival, increased fungal load and decreased inflammatory cytokine expression compared with TLR4-deficient and wild-type mice [9]. While for intratracheally infected model, no significant difference existed, and this finding was confirmed in bone marrow–derived dendritic cells (BM-DCs) [10]. Interestingly, a TLR9-dependent manner was later discovered in the process of BM-DC activation caused by cryptococcal DNA, but the role of TLR9 in the pathogenesis of C. neoformans was still obscure [11]. In our study, one SNP in TLR2 gene and one in TLR4, and two SNPs in TLR1, TLR6, and TLR9, respectively, were all shown genetic correlations with non-HIV CM susceptibility. Three SNPs in TLR2 and TLR9 remained statistically significant among patients without predisposing factors. To our knowledge, this is the first to confirm a genetic association between TLR and non-HIV CM susceptibility in human.
Of CNS infections, TLR has been reported to participate in pathogen clearing and inflammation, taking an important part in disease progression. In a murine Streptococcus pneumonia meningitis model, TLR2 knockout mice had an increase in disease severity and a higher activity of tumor necrosis factor (TNF)-α in the cortex [12]. When infected with Staphylococcus aureus, TLR2 knockout mice showed a significantly reduced expression of proinflammatory mediators in brain abscesses during the acute phase, but the mortality was comparable to that of the wild-type [13]. For tuberculous meningitis, TLR2 and TLR9 cooperation was indicated in mediating resistance to Mycobacterium tuberculosis in mice [14]. A case-control study demonstrated that TLR2 SNPs could influence M. tuberculosis dissemination [15]. The direct link between SNPs and tuberculous meningitis prognosis was also validated, but the mechanism is still under investigation [16]. However, data on CNS cryptococcal infection are lacking. To our knowledge, this study is the first to determine TLR SNPs as predictors for the clinical severity of non-HIV CM. Of note, host-pathogen interactions at the site of infection are crucial for disease progression, but present understandings on CNS infections are limited. Recently, CNS-resident cells, mainly microglia and astrocytes, are known to participate in immune responses and lead to a growing appreciation of the dynamic immune kinetics within CNS. It used to be hypothesized that peripheral cells that infiltrated the CNS in response to local chemokines after cryptococcal infection may further stimulate the expression of proinflammatory cytokines by both CNS resident and trafficked cells [17,18]. Together with the existence of the blood–brain barrier, brain histopathology and CSF immunological examinations are seen as direct approaches for detecting the in situ immune response of CNS. Of HIV-associated CM, in consistent with in vitro studies showing that interferon (IFN)-γ/TNF-α predominant response was correlated with survival [18], higher levels of Th1 cytokines in CSF, especially IFN-γ, contributed to a protective immune response [19]. In addition, lower levels of CSF IFN-γ, closely associated with slower rate of Cryptococcus clearance, were seen as a risk factor for acute death as well [20]. Different from the HIV/AIDS population, paradoxical immune responses were observed in severely ill non-HIV CM patients. Reduced macrophage proportions were coexisted with a highly activated immune status in CSF [21]. Both the Th1 and Th2 cytokines in CSF were up-regulated in comparison with healthy individuals [21]. Our present study found that 18 CSF cytokines including Th1 and Th2 cytokines, and chemokines were remarkably elevated in severe non-HIV CM patients compared with those with mild illness. The up-regulation of CSF cytokines in response to cryptococcal infection may influence the balance between pathogen clearance and immunopathology in CM.
We further built a logistic regression model by using demographic, clinical, and laboratory data, and we identified three independent predictors for the severity of non-HIV CM. IL-10 was still significant after adjustment. Strongly associated with TLR signaling cascades, IL-10 production can inhibit the synthesis of proinflammatory cytokines such as TNF-α, which causes the disruption of Th1/Th2 balance and the abnormal activation of macrophages, resulting in pathogen escape [22]. Cryptococcal glucuronoxylomannan was found to induce IL-10 secretion in vitro [23]. Moreover, one recent study in Aspergillus demonstrated that a SNP in IL-10 along with the enhanced production of IL-10 are risk factors for invasive aspergillosis [24]. The dichotomy between IL-10 and TNF-α production according to genotypes of the SNP was confirmed in human macrophages [24]. In addition, IL-10 induced during M. tuberculosis infection has also been shown to block phagosome maturation [25]. However, we failed to find the direct correlations between TLR SNPs and IL-10. The regulatory pathways or the activated cell-specific transcription factors are needed to clarify and may be shared across these infections.
No TLR SNP albeit survived in logistic regression model, we were surprised to find TLR genetic correlations in CSF cytokine concentrations in univariant analysis, suggesting that the in situ immune response of CNS after cryptococcal infection was under TLR genetic influence. Interestingly, patients carrying rs3804099 C/T genotype presented significant lower levels of 12 CSF cytokines, predominantly proinflammatory cytokines (IL-1β, IL-1α, IL-6, IL-8, IL-15, TNF-α, and IFN-γ) and chemokines (eotaxin, monocyte chemo attractant protein [MCP]-1, macrophage inflammatory protein [MIP]-1α, and MIP-1β), all of which were significantly up-regulated in severe patients. Notably, the correlation was also determined between rs3804099 C/T genotype and decreased risk of disease severity. Pairwise correlations among rs3804099 C/T genotype, CSF cytokine concentrations, and clinical severity strongly implied the causal relationships, suggesting that rs3804099 C/T genotype may contribute to the improvement of non-HIV CM prognosis by down-regulating CNS immune responses. The rs3804099 is a synonymous SNP of TLR2. Several possible explanations existed for how such synonymous mutations exert their impact on gene functions are the perturbations of mRNA splicing [26], the stability and structure of mRNA [27,28], or their influence on protein folding [29]. These changes are reported to have a significant effect on the function of proteins, change in cellular response to therapeutic targets, and often explain the different responses of individual patients to a certain medication [27].
Strengths of our study include the enrolment of patients from a total of 16 provinces and areas in China, the prospective and comprehensive description of illness, the reasonable validation of findings in another two separate groups, and the combination of genetics, proteins, and clinical severity. On the other hand, this study has noteworthy limitations. First, the number of cases recruited in cohorts was limited. Some genotypes of SNPs have low frequencies, which may restrict statistical power. Second, the detected TLR SNPs were selected from TLR1, TLR2, TLR4, TLR6, and TLR9, all of which except for TLR9 encode cell-surface TLRs. Several other TLRs expressed in intracellular endosomes or the interactions in between were not assessed but may also be the potential targets. Third, population validation was conducted in patients with relatively same genetic backgrounds. Only 1 SNP related to susceptibility remained significant in HIV-positive patients, which may due to the small sample size and the composition of population. Interestingly, this suggested that HIV-infected patients with such genetic variants might be more susceptible to CM. Moreover, the pairwise correlations we observed strongly implied the causal relationships. Further investigation by cytological research or animal model is therefore needed to clarify the causative mechanism on how TLR SNPs exert their influence on immunopathology and clinical severity after cryptococcal infection.
In summary, TLR SNPs could predict not only CM susceptibility in non-HIV patients but also clinical severity. The in situ immune responses reflected by CSF cytokine expressions were under TLR genetic influence and correlated with the clinical severity of patients. Overall, our data suggested the critical role of TLR in the occurrence and the pathogenesis of non-HIV CM. Our finding is of significance in evaluating non-HIV CM patients and providing early interventions. Further investigation of the causative mechanisms may be helpful to target TLR on which to base host-directed immunotherapy.
Contributors
Y-KJ, J-QW, H-ZZ, and L-PZ conceived, designed, and managed the study. L-PZ, XW, R-YW, Y-HC, and HL supervised patient recruitment. Y-KJ, J-QW, H-ZZ, L-HZ, J-HC, Y-HC, and HL contributed to sample collections. Y-KJ, J-QW, XW, R-YW, and C-WY conducted the experiment for genotyping. Y-KJ, H-ZZ, L-HZ, and L-PH contributed the microbiological data. C-WY, L-PH, and J-HC contributed to the epidemiological investigations. Y-KJ, J-QW, H-ZZ, and L-PZ did the genomic, epidemiological, and statistical analyses, and drafted the manuscript. All authors contributed to and reviewed the final article.
Declaration of interests
We declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 81271803, Grant No. 81571968, and Grant No. 81071333) and the National Key Basic Research Program of China (973 Program) (Grant No. 2013CB531600). The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
We thank all the health workers in the Huashan Hospital, Mengchao Hepatobiliary Hospital, Fujian HIV/AIDS Diagnosis and Treatment Center, and No. 476 Hospital of Fuzhou General Hospital for their support. We thank Shanghai Genesky Bio-Tech Genetic Core Lab for their assistance in genotyping techniques.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.045.
Appendix A. Supplementary data
Supplementary material
References
- 1.Rajasingham R., Smith R.M., Park B.J. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brizendine K.D., Baddley J.W., Pappas P.G. Predictors of mortality and differences in clinical features among patients with Cryptococcosis according to immune status. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratton E.W., El Husseini N., Chastain C.A. Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L.P., Wu J.Q., Xu B., Ou X.T., Zhang Q.Q., Weng X.H. Cryptococcal meningitis in non-HIV-infected patients in a Chinese tertiary care hospital, 1997–2007. Med Mycol. 2010;48:570–579. doi: 10.3109/13693780903437876. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Varma A., Diaz M.R., Litvintseva A.P., Wollenberg K.K., Kwon-Chung K.J. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008;14:755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ou X.T., Wu J.Q., Zhu L.P. Genotypes coding for mannose-binding lectin deficiency correlated with cryptococcal meningitis in HIV-uninfected Chinese patients. J Infect Dis. 2011;203:1686–1691. doi: 10.1093/infdis/jir152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoham S., Huang C., Chen J.M., Golenbock D.T., Levitz S.M. Toll-like receptor 4 mediates intracellular signaling without TNF-alpha release in response to Cryptococcus neoformans polysaccharide capsule. J Immunol. 2001;166:4620–4626. doi: 10.4049/jimmunol.166.7.4620. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L., Liu T., Kong W. Decreased TLR2 signal expression in peripheral blood mononuclear cell from patients with cryptococcal meningitis. Microbiol Immunol. 2015;59:357–364. doi: 10.1111/1348-0421.12264. [DOI] [PubMed] [Google Scholar]
- 9.Biondo C., Midiri A., Messina L. MyD88 and TLR2, but not TLR4, are required for host defense against Cryptococcus neoformans. Eur J Immunol. 2005;35:870–878. doi: 10.1002/eji.200425799. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura K., Miyagi K., Koguchi Y. Limited contribution of Toll-like receptor 2 and 4 to the host response to a fungal infectious pathogen, Cryptococcus neoformans. FEMS Immunol Med Microbiol. 2006;47:148–154. doi: 10.1111/j.1574-695X.2006.00078.x. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura K., Miyazato A., Xiao G. Deoxynucleic acids from Cryptococcus neoformans activate myeloid dendritic cells via a TLR9-dependent pathway. J Immunol. 2008;180:4067–4074. doi: 10.4049/jimmunol.180.6.4067. [DOI] [PubMed] [Google Scholar]
- 12.Bohland M., Kress E., Stope M.B., Pufe T., Tauber S.C., Brandenburg L.O. Lack of Toll-like receptor 2 results in higher mortality of bacterial meningitis by impaired host resistance. J Neuroimmunol. 2016;299:90–97. doi: 10.1016/j.jneuroim.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Kielian T., Haney A., Mayes P.M., Garg S., Esen N. Toll-like receptor 2 modulates the proinflammatory milieu in Staphylococcus aureus-induced brain abscess. Infect Immun. 2005;73:7428–7435. doi: 10.1128/IAI.73.11.7428-7435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bafica A., Scanga C.A., Feng C.G., Leifer C., Cheever A., Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thuong N.T., Hawn T.R., Thwaites G.E. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 2007;8:422–428. doi: 10.1038/sj.gene.6364405. [DOI] [PubMed] [Google Scholar]
- 16.van Laarhoven A., Dian S., Aguirre-Gamboa R. Cerebral tryptophan metabolism and outcome of tuberculous meningitis: An observational cohort study. Lancet Infect Dis. 2018;18:526–535. doi: 10.1016/S1473-3099(18)30053-7. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis J.N., Meintjes G., Bicanic T. Cerebrospinal fluid cytokine profiles predict risk of early mortality and immune reconstitution inflammatory syndrome in HIV-associated cryptococcal meningitis. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis J.N., Casazza J.P., Stone H.H. The phenotype of the Cryptococcus-specific CD4+ memory T-cell response is associated with disease severity and outcome in HIV-associated cryptococcal meningitis. J Infect Dis. 2013;207:1817–1828. doi: 10.1093/infdis/jit099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bicanic T., Muzoora C., Brouwer A.E. Independent association between rate of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: analysis of a combined cohort of 262 patients. Clin Infect Dis. 2009;49:702–709. doi: 10.1086/604716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvis J.N., Bicanic T., Loyse A. Determinants of mortality in a combined cohort of 501 patients with HIV-associated Cryptococcal meningitis: implications for improving outcomes. Clin Infect Dis. 2014;58:736–745. doi: 10.1093/cid/cit794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panackal A.A., Wuest S.C., Lin Y.C. Paradoxical immune responses in non-HIV Cryptococcal meningitis. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Retini C., Kozel T.R., Pietrella D., Monari C., Bistoni F., Vecchiarelli A. Interdependency of interleukin-10 and interleukin-12 in regulation of T-cell differentiation and effector function of monocytes in response to stimulation with Cryptococcus neoformans. Infect Immun. 2001;69:6064–6073. doi: 10.1128/IAI.69.10.6064-6073.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vecchiarelli A. Fungal capsular polysaccharide and T-cell suppression: the hidden nature of poor immunogenicity. Crit Rev Immunol. 2007;27:547–557. doi: 10.1615/critrevimmunol.v27.i6.50. [DOI] [PubMed] [Google Scholar]
- 24.Cunha C., Gonçalves S.M., Duarte-Oliveira C. IL-10 overexpression predisposes to invasive aspergillosis by suppressing antifungal immunity. J Allergy Clin Immunol. 2017;140:867–870. doi: 10.1016/j.jaci.2017.02.034. [DOI] [PubMed] [Google Scholar]
- 25.O'Leary S., O'Sullivan M.P., Keane J. IL-10 blocks phagosome maturation in mycobacterium tuberculosis-infected human macrophages. Am J Respir Cell Mol Biol. 2011;45:172–180. doi: 10.1165/rcmb.2010-0319OC. [DOI] [PubMed] [Google Scholar]
- 26.Egan M.F., Straub R.E., Goldberg T.E. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci U S A. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt R., Sauna Z.E., Ambudkar S.V., Gottesman M.M., Kimchi-Sarfaty C. Silent (synonymous) SNPs: Should we care about them? Methods Mol Biol. 2009;578:23–39. doi: 10.1007/978-1-60327-411-1_2. [DOI] [PubMed] [Google Scholar]
- 28.Shen L.X., Basilion J.P., Stanton V.P., Jr. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci U S A. 1999;96:7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material