Abstract
Background
Ductal carcinoma in situ (DCIS) is the earliest stage of breast cancer. During DCIS, tumor cells remain inside the mammary duct, growing under a microenvironment characterized by hypoxia, nutrient starvation, and waste product accumulation; this harsh microenvironment promotes genomic instability and eventually cell invasion. However, there is a lack of biomarkers to predict what patients will transition to a more invasive tumor or how DCIS cells manage to survive in this harsh microenvironment.
Methods
In this work, we have developed a microfluidic model that recapitulates the DCIS microenvironment. In the microdevice, a DCIS model cell line was grown inside a luminal mammary duct model, embedded in a 3D hydrogel with mammary fibroblasts. Cell behavior was monitored by confocal microscopy and optical metabolic imaging. Additionally, metabolite profile was studied by NMR whereas gene expression was analyzed by RT-qPCR.
Findings
DCIS cell metabolism led to hypoxia and nutrient starvation; revealing an altered metabolism focused on glycolysis and other hypoxia-associated pathways. In response to this starvation and hypoxia, DCIS cells modified the expression of multiple genes, and a gradient of different metabolic phenotypes was observed across the mammary duct model. These genetic changes observed in the model were in good agreement with patient genomic profiles; identifying multiple compounds targeting the affected pathways. In this context, the hypoxia-activated prodrug tirapazamine selectively destroyed hypoxic DCIS cells.
Interpretation
The results showed the capacity of the microfluidic model to mimic the DCIS structure, identifying multiple cellular adaptations to endure the hypoxia and nutrient starvation generated within the mammary duct. These findings may suggest new potential therapeutic directions to treat DCIS. In summary, given the lack of in vitro models to study DCIS, this microfluidic device holds great potential to find new DCIS predictors and therapies and translate them to the clinic.
Keywords: Breast cancer, DCIS, Microfluidics, Organotypic, 3D model
Graphical abstract
Research in context.
Evidence before this study
During DCIS, the earliest stage of breast cancer, tumor cells grow within the mammary duct under a harsh microenvironment. If left untreated, up to half of DCIS patients will develop an invasive ductal carcinoma, a more aggressive tumor where cancer cells spread to the surrounding tissue and can metastasize to other organs. Some studies have suggested that the DCIS microenvironment plays a critical role during DCIS evolution and progression to an invasive tumor. However, despite the previous research clinicians still lack good predictors to determine what patients will develop an invasive tumor from those that remain as an indolent DCIS.
Added value of this study
In order to understand DCIS evolution, different in vitro models have been developed. However, most of them do not mimic the mammary duct structure with the DCIS trapped inside. Therefore, there is a need for more complex DCIS models that allow the study of cell behavior under a more physiologic microenvironment. In a previous work we developed a DCIS microfluidic model that mimics the mammary duct structure with the DCIS cells trapped inside. In this work, we have used the model to study DCIS evolution and metabolism. The model allowed the co-culture of multiple cell types mimicking the in vivo DCIS and mammary duct architecture. The model revealed that under nutrient starvation, DCIS cells activated multiple survival metabolic and genetic adaptations. These adaptations observed in the model were in good agreement with patient genomic profiles obtained in clinical trials, supporting the relevance of the model to identify new DCIS evolution predictors. Finally, using the model, we also observed how DCIS cells invaded the surrounding tissue, mimicking in vitro the DCIS transition to an invasive ductal carcinoma.
Implications of all the available evidence
To the best of our knowledge, this is the first in vitro model that mimics the DCIS structure and allows the study of DCIS metabolism and survival mechanisms. The good correlation between the in vitro genomic profile with in vivo data shows the potential of this platform to find new DCIS evolution predictors. Finally, the model could also be applied to evaluate new therapies against DCIS.
Alt-text: Unlabelled Box
1. Introduction
Breast cancer is the most common non-cutaneous cancer in women, and it is estimated that one in eight women will develop breast cancer during their lifetime [1]. With increased mammography screening, ductal carcinoma in situ (DCIS), the earliest stage of breast cancer, emerged as a common diagnosis and approximately 53,000 cases of DCIS are diagnosed each year in the US [2]. During DCIS, the tumor cells are non-invasive and remain trapped inside the mammary duct [3]. Thus, patients diagnosed with DCIS have an excellent prognosis, with 5-year breast cancer survival approaching 100% with treatment. Treatment can include breast surgery, radiation therapy and hormonal therapy [4]. However, concerns about the over-treatment of DCIS are increasing. When DCIS is not treated, 25–50% of patients will eventually develop invasive ductal carcinoma (IDC), a more aggressive tumor where tumor cells escape from the duct and may metastasize to other organs (e.g., bone, lung and brain) [5]. Ideally, only the patients with DCIS that are at risk for developing IDC would be treated, and those destined to evade IDC would be monitored. Currently, thousands of women undergo unnecessary treatments as clinicians have limited tools to understand DCIS to guide in these decisions [4]. The mechanisms by which these tumor cells survive within the mammary duct and eventually invade the surrounding tissue remain unclear. Recently, it has been suggested that the DCIS tumor microenvironment (TME) plays a critical role during DCIS development. The DCIS TME is characterized by hypoxia, nutrient starvation, and waste product accumulation inside the mammary duct [6].
Consequently, tumor cells may activate multiple survival responses such as hypoxia-inducible factor (HIF) expression, altered metabolism (i.e., changes in the redox ratio, glucose/amino acid/fatty acid metabolism) and autophagy activation [6,7]. Furthermore, nutrient deprivation and hypoxia have been linked to an increased capacity to migrate and invade the surrounding tissue [8]. Therefore, new approaches to decipher the role of the TME during DCIS evolution could provide clinicians with more comprehensive prognostic information.
As mounting evidence shows the relevance of the TME, new therapeutic opportunities arise to target these tumor-specific metabolic pathways [9,10]. However, there is a lack of relevant in vitro models to understand the complex interactions that influence patient response [11]. Mimicking the DCIS microenvironment requires the presence of multiple normal and tumor cell populations (i.e., cancer, stromal cells) with different metabolic phenotypes, embedded in a 3D environment that recapitulates the mammary duct structure. Although several in vitro models have been developed to study DCIS, most of them lack the of a mammary duct model with the tumor cells trapped inside [11]. On the other hand, animal models like the MIND model can provide a more complex environment compared with the classic in vitro models [12,13]. In the MIND model, patient-derived cells or human breast cancer cell lines are injected into the mouse mammary duct, mimicking in a much more precise way the human DCIS microenvironment. However, animal models rise ethical considerations and monitoring and controlling the microenvironment is more challenging. Microfluidic models offer the potential to more accurately mimic the complex in vivo components [14,15]. Several groups have developed several microfluidic models to generate luminal structures (e.g., blood vessels) embedded in a 3D extracellular matrix with/without stromal cells [[16], [17], [18], [19], [20]]. In one of our models, we mimicked the DCIS architecture using normal mammary cells to generate the mammary duct, whereas a DCIS model cell line (MCF10A-DCIS.com) was injected into the mammary duct [21]. In the present work, the model was modified to include two flanking lumens to perfuse media, metabolites or drugs; as well as to allow the retrieve of culture medium for downstream analysis. Using the model, DCIS cell behavior was scrutinized by nuclear magnetic resonance (NMR) spectroscopy, RT-qPCR, confocal microscopy, and multi-photon optical metabolic imaging (OMI). The results revealed that DCIS cells generated a microenvironment within the microdevice characterized by hypoxia and nutrient starvation. During DCIS invasion, the invading cells exhibited altered metabolism compared with the cells within the mammary duct. To demonstrate the utility of the model in evaluating cancer therapeutics, the system was exposed to a hypoxia-activated prodrug which selectively targeted the hypoxic DCIS cells, whereas normoxic cells remain viable.
2. Materials and methods
2.1. Reagents
Calcein AM (CAM) (C34851), propidium iodide (PI) (P1304MP), hypoxia reagent (H10498), 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (NBDG), cell tracker red CMTPX (C34552), green CMFDA (C7025), and blue (C2110), were purchased from Thermo Fisher and stock solutions were prepared following supplier instructions. Rhodamine B (Rho) (Sigma R6626), Doxorubicin (DOX) (Selleckchem, S1208) and Tirapazamine (TPZ) (Sigma, SML0552) were dissolved at 1 mg/ml, 100 mM, and 50 mM respectively in DMSO.
2.2. Cell culture
The human mammary epithelial cell line (MCF10A) was obtained from ATCC (ATCC® CRL-10317™) and maintained with DMEM/F12 medium (Thermo Fisher, 11,320,033) supplemented with 5% horse serum (Invitrogen, Carlsbad, CA, USA), 20 ng/ml epidermal growth factor (Peprotech, AF-100-15), 0.5 mg/ml hydrocortisone (Sigma-Aldrich, H0888), 100 ng/ml Cholera toxin (Sigma-Aldrich, C9903), 10 μg/ ml insulin (Sigma-Aldrich, I6634) and 1% Pen/Strep (Thermo Fisher, 15,070,063) on regular tissue culture flasks. MCF10A-DCIS.com cells (will be referred as DCIS cells for simplicity) were developed by Miller et al. and distributed by Asterand (Detroit, MI, USA) [22,23]. They were generated from a MCF10AT clone initiated from a xenograft that evolved to DCIS lesions. DCIS cells were maintained in MCF10A medium. Human mammary fibroblasts (HMFs) were obtained from Dr. Charlotte Kuperwasser (Tufts University) and maintained in DMEM supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 1% Pen/Strep.
2.3. Microdevice fabrication
The microdevices were fabricated by soft lithography following the protocol described in [16]. Briefly, a SU-8 template containing the geometry was fabricated using SU-8100 (Y131273, MICROCHEM) and polydimethylsiloxane (PMDS) (Dow Corning) was poured on top and polymerized for 4 h at 80 °C. PDMS microdevices were detached from the SU-8 wafer and assembled. The PDMS rods were fabricated by injecting liquid PDMS through 23-gauge needles (337 mm diameter) (BD Precision Glide); after PDMS polymerization, rods were placed into the microdevices. Finally, the assembled microdevices and 60 mm glass-bottom Petri dishes (P50G-1.5-30-F, MatTek) were treated with oxygen plasma and bonded together.
2.4. Cell culture within the microdevice
The microdevices were sterilized by UV light exposure for 20 min. To improve the collagen hydrogel attachment to the microdevice, microdevices were pretreated with 2% poly(ethyleneimine) (Sigma-Aldrich, 03880) diluted in water for 10 min and 0.4% glutaraldehyde (Sigma-Aldrich, G6257) diluted in water for 30 min. Microdevices were washed three times with sterile distilled water.
The collagen hydrogel was prepared as followed: 10.6 μl of 10× PBS (79,382, Sigma), 2.34 μl of 1 N NaOH (221,465, Sigma), 93.65 μl of 9.61 mg/ml collagen type I (354,249, Corning) and finally 93.4 μl of MCF10A growth medium (with or without 2 × 106 HMF cells/ml). 10 μl of this hydrogel mixture was injected though the gel loading port in each microdevice. Microdevices were placed into the incubator at 37 °C and 5% CO2 for 20 min to allow collagen polymerization. Once collagen was polymerized, the PDMS rods were removed using sterile tweezers, generating a tunnel through the collagen hydrogel. To recreate the mammary duct structure, 1.5 μl of 15 × 106 MCF10A cells/ml was injected through the central lumen. Microdevices were placed upside-down in the incubator for 15 min to allow the cells to attach to the top side of the lumen. Then the microdevices were flipped upside-up and left in the incubator for another 2 h. Finally, 5 μl of MCF10A medium was flushed through the lumen to wash out the non-attached cells and microdevices were left in the incubator overnight. The next day, DCIS cells were trypsinized and suspended at 100 × 106 cells/ml. 2 μl of DCIS cell suspension was injected through the MCF10A lumen to generate the DCIS model. MCF10A medium was refreshed through the lateral lumens daily.
2.5. Cell staining
In some experiments cell were fluorescently labeled with Red/Green/Blue cell tracker (Thermo Fisher, C34552, C7025, C2110 respectively). The stock solution was diluted 1 to 1000 in growth medium and cells were trypsinized and incubated in this medium for 30 min. Next, cells were washed twice with phosphate-buffered saline (PBS) (Lonza BE17-516F) to remove the excess cell tracker.
2.6. Cell viability staining
Stock solutions of 5 mg/ml CAM and 2 mg/ml PI were dissolved in DMSO and distilled water respectively. To test cell viability within microfluidic devices and in Petri dishes, stock solutions of CAM and PI were diluted to 5 and 4 μg/ml, respectively, in PBS. The CAM/PI solution was perfused through the lateral microchannels. Cell viability was evaluated using a Leica SP8 3× STED Super-resolution microscope.
2.7. Hypoxia profile
Hypoxia profile was analyzed within the microdevices using the hypoxia reagent. Stock hypoxia reagent solution was diluted in growth medium as well as the collagen hydrogel at 10 μM to ensure a homogeneous concentration. Hypoxia signal was visualized at different time points using a Leica SP8 3× STED Super-resolution microscope. A 488 nm white laser was used to excite the hypoxia-sensing dye and emission was detected using a 650 ± 50 nm photomultiplier tube.
2.8. Glucose penetration
To study whether glucose penetration in the lumen model was hindered by the epithelial cells, NBDG was dissolved at 200 mM in the appropriate epithelial growth medium for MCF10A and DCIS cells. The NBDG-supplemented medium was perfused through one of the lateral channels and NBDG diffusion was monitored under the Leica SP8 microscope (488 nm laser and 590 ± 30 detector).
2.9. NMR sample preparation and spectra acquisition
Metabolites from cell media were harvested using a protein precipitation with methanol extraction procedure as described in [24]. Briefly, 100 μL of media was mixed at a 1:2 (v/v) ratio with methanol, vortexed, and incubated for 20 min at −20 °C. media samples were then centrifuged at 11,093g for 30 min. The supernatant was collected and dried using a Vacufuge Plus (Eppendorf). The concentrated metabolite samples were reconstituted in 600 μL of phosphate buffered deuterium oxide (D2O) solution. Phosphate buffered D2O solution was comprised of 0.1 M D2O (Acros Organics), 0.5 mM 3-trimethylsilyl-propionate-2, 2, 3, 3,-d4 (TMSP, δ = 0.0 ppm, internal standard) and 0.2% w/v sodium azide. Samples were centrifuged at 17968g for 10 min and 550 μL of supernatant was collected into 5 mm NMR tubes (Norell Inc.).
1H NMR metabolomic analysis of media samples was performed as described in [25]. Media samples were analyzed using a 500 MHz Bruker Avance III spectrometer with a 5 mm cryogenic probe at a temperature of 298 K at the National Magnetic Resonance Facility at Madison (NMRFAM). One dimensional (1D) 1H NMR spectra were acquired using 1D Nuclear Overhauser Effect Spectroscopy with presaturation and spoil gradients (NOESYGPPR1D) pulse sequence with a relaxation delay of 2 s, a mixing time of 10 ms, and a pre-scan delay of 30 μs. Each spectrum consisted of 128 free induction decays (FIDs) and a spectral width of 12 ppm. Line broadening (LB) of the FIDs was set to 0.5 Hz. Using Bruker Top-Spin™ software (version 3.2.5), the chemical shifts were referenced to the TMSP peak (δ = 0.0 ppm).
2.10. Metabolomics data analysis
1H NMR spectra were imported into ACD/1D NMR Processor software (Advanced Chemistry Development) where phase and baseline corrections were adjusted and solvent region removal (water: 4.7–5 ppm) was performed. Metabolite concentrations were determined using ChenomX NMR Suite Profiler (version 7.7, ChenomX Inc.). TMSP was added to all samples as a reference compound to aid in determining metabolite concentrations. After metabolite identification and quantification, metabolite concentrations were exported to an Excel file. The coefficient of variation (CV) was calculated for all features and metabolites with high coefficient of variation (CV) values (CV > 0.30) were excluded from analysis. This was done to ensure that features with high variation between replicates would not influence metabolomics analysis. Some metabolites with consistent concentrations between replicates but high CV values were not excluded from the analysis given that due to their low concentration values, any small change in concentration would generate a high CV value. Metabolites unique to a particular group were included in the data interpretation and discussion but were excluded from principal component analysis (PCA) as this analysis is very sensitive to outliers. Glucose was excluded from multivariate statistical analysis due to high variability affecting normalization.
Prior to analysis, the Statistics, Enrichment, and Biomarker Analysis modules in Metaboanalyst was used to normalize metabolite concentration data by the total concentration for each sample and scale using auto scaling [[26], [27], [28]]. Briefly, PCA was performed to identify inherent patterns in the data and visually capture sample variance. Hierarchical clustering using Pearson correlation and Ward clustering algorithm was performed on the metabolite concentration data in order to generate a heatmap of metabolic profiles for all conditions. DCIS and mammary duct metabolic concentration data were used to generate a Pearson correlation plot. Metabolite concentrations identified as significant by one-way ANOVA with Tukey's HSD post-hoc analysis (p-value <0.05) were plotted in Excel for all four conditions analyzed. Differentially enriched metabolic pathways obtained from metabolite set quantitative enrichment analysis [29] were visually represented in network format using the Metscape 3 plugin in Cytoscape [30]. Classical univariate receiver operator characteristic (ROC) curve analyses were carried out to identify potential biomarkers [31].
2.11. mRNA extraction, RT-qPCR and clinical comparison
To study how cells responded to the changing microenvironment within the microfluidic model, the expression of a panel of genes was analyzed by RT-qPCR. Briefly, MCF10A cells were pipetted in the central lumen to mimic the mammary duct and DCIS cells were injected inside. mRNA was extracted after 3 and 24 h in cell culture using a Dynabeads™ mRNA DIRECT™ Purification Kit (61,011, Thermo Fisher). mRNA was reverse transcribed to cDNA using the RT2 First strand kit (330,401, Qiagen). cDNA was analyzed by RT-qPCR using a Qiagen RT2 profiler custom panel (CLAH25337, Qiagen) and data was analyzed using the Qiagen online software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). The RT-qPCR data was compared with the clinical data obtained in the METABRIC study, the largest breast cancer genomic profile study in the cBioPortal database (http://www.cbioportal.org/) [32,33]. The database was screened for the genes that were found to be significantly different in the model. The percentage of the different alterations identified in the study was calculated (i.e. amplification, mRNA upregulation, deep deletion, mRNA downregulation and multiple alterations). A genetic network including these genes was generated using the online software. The network included the affected genes and their intermediates.
2.12. NAD(P)H/FAD imaging
Fluorescence lifetime images and intensity were taken on a custom-built inverted multiphoton microscope (Bruker Fluorescence Microscopy, Middleton, WI), as described previously [[34], [35], [36]]. Briefly, the system consists of a titanium:sapphire laser (Spectra Physics, Insight DS-Dual), an inverted microscope (Nikon, Eclipse Ti), and a 40× water immersion (1.15NA, Nikon) objective. NAD(P)H and FAD images were acquired sequentially for the same field of view. NAD(P)H fluorescence was isolated using an excitation wavelength of 750 nm and an emission bandpass filter of 440/80 nm. FAD fluorescence was isolated using an excitation wavelength of 890 nm and an emission bandpass filter of 550/100 nm. Fluorescence lifetime images were collected using time correlated single photon counting electronics (SPC-150, Becker and Hickl) and a GaAsP photomultiplier tube (H7422P-40, Hamamatsu). A pixel dwell time of 4.8 μs was used to acquire a 512 × 512 pixel images over 60s total integration time. For intensity imaging a pixel dwell time of 4.8 μs was used to collect 1024 × 1024 images. The photon count rates were maintained at 1–2 × 105 photons/s to ensure adequate photon observations for lifetime decay fits, and no photobleaching. The instrument response function was measured from second harmonic generation of urea crystals excited at 900 nm, and the full width at half maximum (FWHM) was calculated to be 244 ps. A Fluoresbrite YG microsphere (Polysciences Inc.) was imaged as a daily standard for fluorescence lifetime. The lifetime decay curves were fit to a single exponential decay and the fluorescence lifetime was measured to be 2.1 ns (n = 7), which is consistent with published values.
2.13. Quantification of fluorescence lifetime components
NAD and NADPH (indicated as NAD(P)H) and FAD fluorescence lifetime images of cells were analyzed using SPCImage software (Becker & Hickl) as described previously [35]. Briefly, at each pixel, the fluorescence lifetime decay curve was deconvolved with the instrument response function and fit to a two-component exponential decay model, I(t) = α1*exp.(−t/τ1) + α2*exp.(−t/τ2) + C, where I(t) is the fluorescence intensity at time t after the laser excitation pulse, α represents the fractional contribution from each component, C accounts for background light, and τ represents the fluorescence lifetime of each component. A two-component model was used because both NAD(P)H and FAD can exist in two conformational states, bound or unbound to enzymes [37]. For NAD(P)H the short and long lifetime components correspond with the unbound and bound conformations respectively [37]. While the opposite is true for FAD, the short and long lifetime components reflect the bound and unbound conformations respectively [37]. The mean lifetimes were calculated using, τm = α1τ1+ α2τ2 for both NAD(P)H and FAD. The optical redox ratio was calculated from the NAD(P)H and FAD lifetime data by summing the photons detected at each pixel in the image to compute the total intensity. The intensity of NAD(P)H was then divided by the intensity of FAD for each pixel.
An automated cell segmentation pipeline was created in Cell Profiler. Briefly, a customized threshold code identified pixels belonging to nuclear regions. Cells were identified by propagating out from the nuclei within the image. An Otsu Global threshold was used to improve the propagation and prevent it from continuing into background pixels. The cell cytoplasm was defined as the cell borders minus the nucleus. Values for the τm, τ1, τ2, α1, and intensities of NAD(P)H and FAD as well as the redox ratio were averaged for all pixels within each cell cytoplasm.
2.14. Image and statistical analysis
Microscopy images were analyzed using FIJI® (www.FIJI.com). To analyze molecule diffusion and cell viability, a rectangle-shape region was drawn, and the intensity profile was calculated using the FIJI software. At least 100 cells per sample were analyzed to calculate the OMI variables of that sample. Every experiment was repeated at least three times. The normal distribution was tested by the Kolmogorov-Smirnov test. Statistical significance was set at p < 0.05. For nonparametric comparisons, a Kruskal-Wallis test was performed followed by the Mann-Whitney U test.
3. Results
3.1. Establishment of the DCIS model
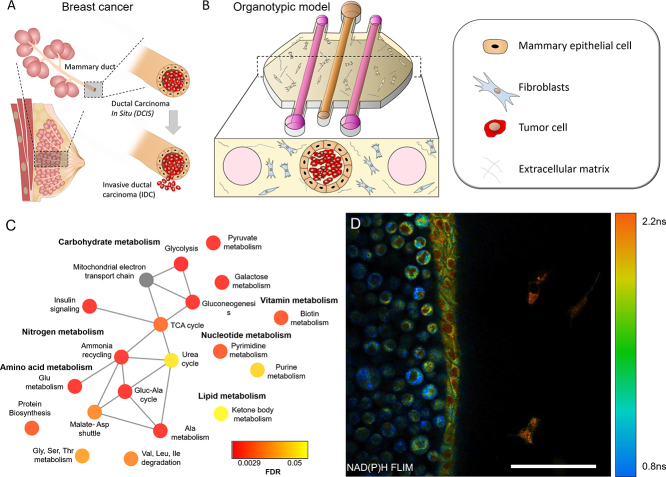
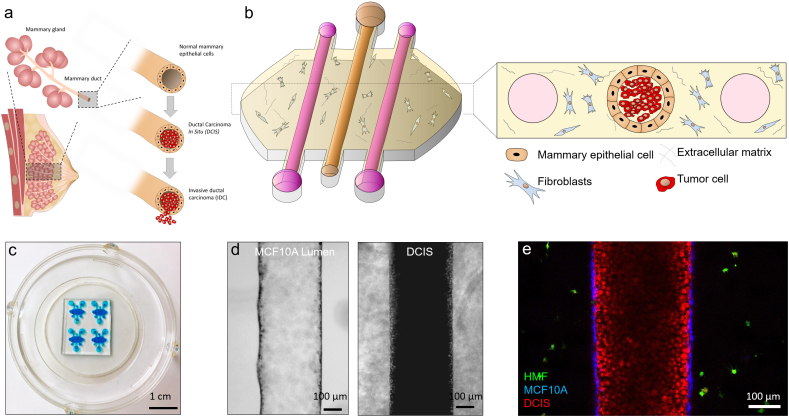
To generate a mammary duct model, PDMS-based microdevices with three lumens were fabricated (Fig. 1a–c). HMFs were embedded in the collagen hydrogel. Next, mammary epithelial cells (MCF10A) were seeded through the central lumen to generate the mammary duct model. After 24 h in culture, MCF10A cells generated a continuous epithelium and MCF10A or DCIS cells were injected through the central lumen (Fig. 1d and e).
Fig. 1.
a) Scheme of the DCIS structure. b) Scheme of the microfluidic model. c) Microdevice picture. Blue-colored water was introduced within the microdevices for visualization purposes. d) MCF10A empty lumen after 24 h in cell culture. DCIS cells were injected within the MCF10 lumen. e) Confocal image showing the HMF (1 × 106 cells/ml), MCF10A (15 × 106 cells/ml) and DCIS (100 × 106 cells/ml) labeled with cell tracker green, blue and red respectively.
3.2. Hypoxia and glucose diffusion
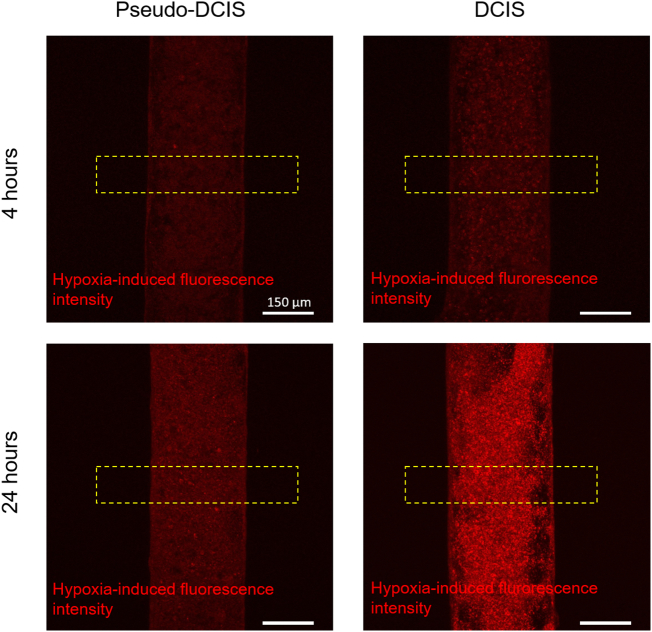
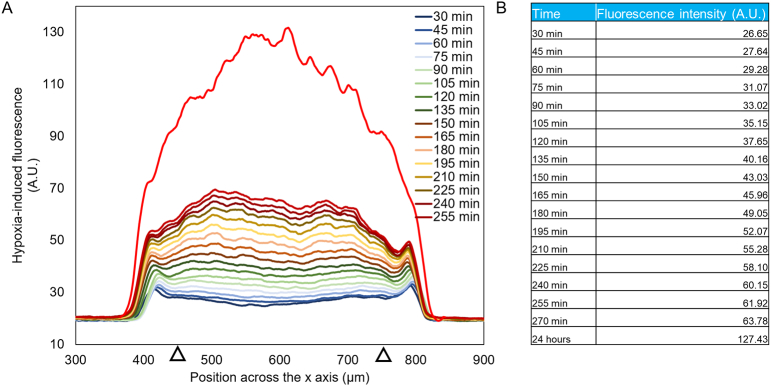
In order to study hypoxia, microdevices were divided into three groups: 1) mammary duct model, with MCF10A cells forming a hollow lumen; 2) DCIS model, with the MCF10A lumen full of DCIS cells; and 3) pseudo-DCIS, composed of a MCF10A lumen with MCF10A cells inside (Fig. 2a). Although this last condition seems biologically unlikely, since normal cells do not grow within the mammary duct; it allowed us to evaluate if the observed DCIS oxygen metabolism was a product of a higher cell density or due to specific metabolic alterations presence in the DCIS cells. To detect the levels of oxygen within the model, a hypoxia-sensing dye was added to the collagen hydrogel before hydrogel polymerization. This dye increases its fluorescence as oxygen tension decreases, particularly below 5%. The hypoxia sensor fluorescence progressively increased during the first 4 h in the DCIS model (Supplementary Movie 1), reaching maximum intensity after 24 h (Supplementary Fig. 1, Supplementary Fig. 2 and Fig. 2a). Conversely, in the mammary duct model (i.e., the MCF10A lumen with no other cells inside), no hypoxia signal was observed. When the mammary duct model was filled with MCF10A cells, the hypoxia signal observed after 24 h was lower compared with the DCIS model. Additionally, in the DCIS model, this hypoxia signal rapidly increased with penetration into the DCIS duct, reaching a maximum at the lumen center. However, in the lumen filled with MCF10A cells, the profile reached a plateau across the lumen. These results suggest that the DCIS model has a faster oxygen metabolism that leads to greater hypoxia in the center of the lumen.
Fig. 2.
a) Microdevices were divided in three groups: 1- Mammary duct, containing only fibroblasts and MCF10A cells; 2- Pseudo-DCIS, similar to group 1 but with the addition of MCF10A within the lumen; 3- DICS, similar to group 1 but with DCIS cells within the lumen. The hypoxia signal (in red) is observed when DCIS cells are present inside the mammary duct model. In a lumen without DCIS cells, there is no hypoxia signal. The graph shows the hypoxia profile along the yellow rectangle in the pictures. b) NBDG diffusion along the time in the mammary duct model. NBDG diffusion is clearly hindered by the luminal cells. The graph shows the diffusion profile along the yellow rectangles shown in the pictures. c) Rhodamine diffusion profile in the mammary duct lumen. Rhodamine is a hydrophobic compound that can diffuse through cell membrane, showing a much faster penetration through the lumen.
Supplementary Fig. 1.
Hypoxia profile evolution. MCF10A or DCIS were cultured inside the mammary duct model in the presence of the hypoxia-sensing dye. Hypoxia signal was visualized after 4 and 24 h respectively. In both conditions hypoxia was observed after 4 h and I increased after 24 h. The DCIS cells exhibited a higher hypoxia signal.
Supplementary Fig. 2.
Time-lapse hypoxia analysis. A) The graph shows the hypoxia profile at different time-points across the lumen. A hypoxia-induced fluorescence was observed 30 min after injecting the hypoxia-sensing dye and increased during the experiment. B) The table shows the hypoxia-induced fluorescence intensity values at different time points. To obtain these values, a section spanning from 450 μm to 750 μm (denoted by black triangles) was defined and the average intensity value was calculated. These values were used to fit the parameters used in the computational model.
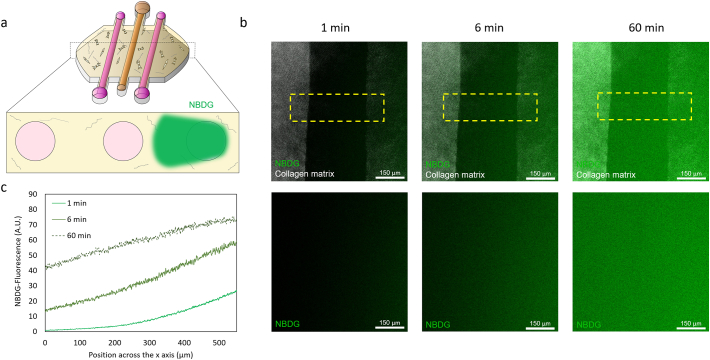
Next, the penetration of nutrients through the lumen was evaluated. The fluorescent glucose analogue NBDG was perfused through one of the lateral lumens (Fig. 2b and supplementary Fig. 3). NBDG is a small molecule (molecular weight < 1 kDa) that rapidly diffused through the hydrogel, and reached the central lumen after 1 min. Interestingly, when NBDG reached the lumen wall, its diffusion was significantly decreased. After 60 min, the NBDG fluorescence intensity at the lumen core was lower than 30%, compared with the intensity outside the lumen. Therefore, this experiment demonstrated that glucose penetration inside the lumen was not completely governed by passive diffusion; but was also regulated by active transport through the cells forming the lumen wall.
Supplementary Fig. 3.
NBDG diffusion. a) The fluorescent glucose analog was perfused through the right lateral lumen in a microdevice with no cells. b) Confocal images showing the NBDG diffusion (in green) through the collagen hydrogel (in grey) and the lumen. C) Diffusion profile across the yellow rectangle at different time-points.
Previous reports have shown that other nutrients (i.e., small hydrophobic molecules) can passively diffuse through the cell membrane [38], nourishing the cells into the lumen. To explore this idea, we injected rhodamine B, a small hydrophobic (<1 kDa) compound that naturally fluoresces in red, in the right lateral lumen (Fig. 2c). After 3 min, rhodamine B concentrated inside the cells, but also diffused through the MCF10A cells to penetrate inside the central lumen. Altogether, these observations demonstrate that nutrient transport inside the mammary duct model depends on the considered molecule, and transport can be modulated by the epithelial cells.
3.3. Metabolite characterization
Next, we set out to characterize the metabolite profile within the model. In this experiment, microdevices were divided into four groups: 1) control group, with no cells in the microdevices; 2) mammary duct model, with MCF10A cells forming an empty lumen; 3) DCIS model, with the MCF10A lumen full of DCIS cells; and 4) pseudo-DCIS, composed of a MCF10A lumen with MCF10A cells inside (Fig. 3a). After 24 h in culture, the media from the lateral lumens was retrieved and analyzed by 1H NMR spectroscopy and metabolomic analysis.
Fig. 3.
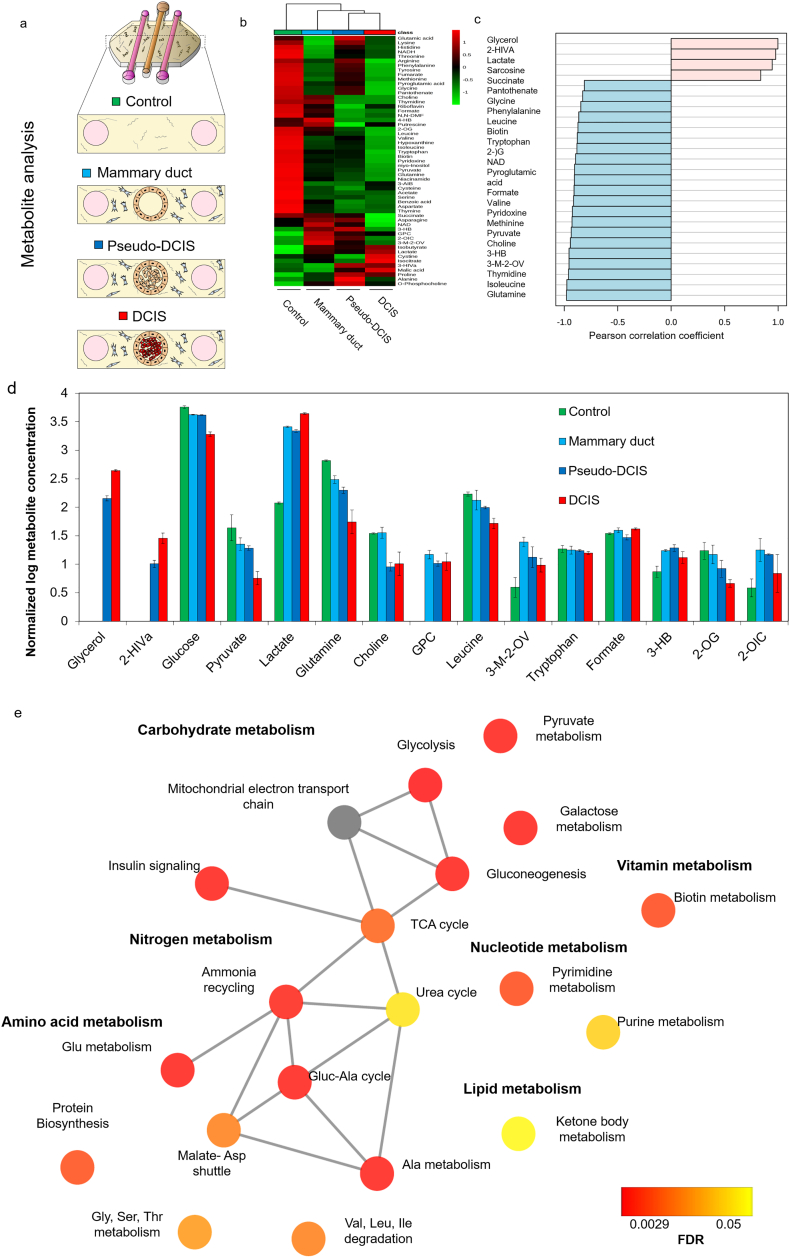
a) Schematic illustrating the four conditions from which media compositions were analyzed using 1H NMR metabolomics. b) Heatmap showing the metabolic profiles of all four conditions analyzed. Heatmap was generated by hierarchical clustering using Pearson correlation as a distance measurement and Ward clustering algorithm. Legend shows auto scaled values from 1 to −1 assigned to the relative metabolite concentrations. Individual metabolites are shown in the rows and group averages are shown in the columns of the heatmap. c) Pearson correlation coefficient plot portraying the top 25 metabolites correlated to DCIS compared to mammary duct control. d) Bar graph depicting the normalized metabolite concentrations of the top 14 significantly altered metabolites between the DCIS and mammary duct models. Pyruvate was not significantly different between these two conditions but was added since it is involved in glycolysis. Error bars represent the standard deviation. FDR < 0.05 [one-way ANOVA with Tukey's HSD post-hoc analysis]. e) Network representation of metabolite set enrichment analysis results comparing mammary duct vs. DCIS groups. Graph was generated using Metscape. Colour gradient from red to yellow represent FDR values indicating statistically significant change in pathways. Grey colour indicates there was no significant change for that particular pathway. Sample groups: Control, with no cells (green); mammary duct model, with MCF10A cells forming an empty lumen (cyan);Pseudo-DCIS, composed of MCF10A lumen with MCF10A cells inside (blue) and DCIS model, with the MCF10A lumen full of DCIS cells (green). Graphs in panels b and c were generated using Metaboanalyst. N = 3 biological replicates per sample group.
Principal Component Analysis (PCA) and hierarchical clustering revealed four clusters corresponding to the four sample groups (Supplementary Fig. 4 and Fig. 3b). To further identify which metabolites and metabolic pathways were affected, the top features that correlated with the DCIS model according to Pearson correlation coefficient values were identified (Fig. 3c). fourteen metabolites were significantly different between the mammary duct and the DCIS models (Fig. 3d). In the DCIS model, glucose and pyruvate concentrations in the media decreased while lactate levels increased relative to the other three conditions. These metabolite changes in the media are consistent with increased glycolysis in the DCIS model. Moreover, glutamine concentrations were lower in the DCIS model, which could result from increased glutaminolysis. Choline was reduced only in DCIS and pseudo-DCIS groups compared to control and mammary duct models, suggesting the higher cell density within the duct was modifying cell metabolism. In this context, Glycerol and 2-hydroxyisovalerate (2-HIVa) were only present in DCIS and pseudo-DCIS, but not in the mammary duct model or control conditions. This observation suggested again that there was a metabolic change that depended only on the cell density, rather than on genetic differences.
Supplementary Fig. 4.
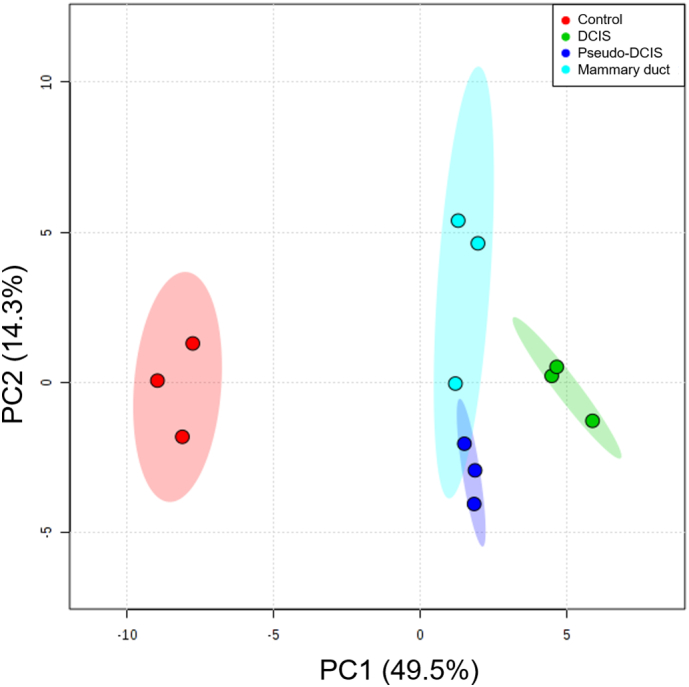
Principal component analysis (PCA) score plot showing variance of metabolic samples. Sample groups: Control, with no cells inside (red); DCIS model, with the MCF10A lumen full of DCIS cells (green); lumen control, composed of MCF10A lumen with MCF10A cells inside (blue); and mammary duct model, with MCF10A cells forming an empty lumen (cyan). N = 3 biological replicates (circles) per sample group were analyzed. Shaded ellipses represent the 95% confidence region for each sample group.
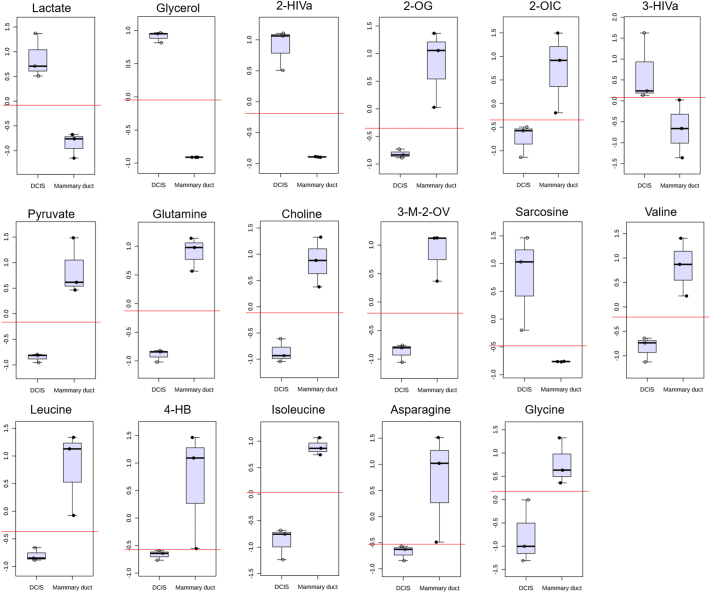
We further performed classical univariate ROC curve analysis on the metabolite concentration data to identify differences between the mammary duct and DCIS microdevice models (Supplementary Fig. 5). This analysis aimed to test the potential of the model to identify DCIS metabolomic biomarkers or targets. We identified 17 metabolites with an area under the curve (AUC) value of 1 and fold change values higher than ±0.5. These metabolites were significantly different between DCIS and mammary duct models and likewise strongly correlated with the DCIS model. Upon further validation, these metabolites, either individually or in combination, could be potential biomarkers used to diagnose aggressive DCIS leading to an IDC from the benign indolent DCIS.
Supplementary Fig. 5.
Box and Whisker plots of the 17 metabolites identified as potential biomarkers using classical univariate ROC curve analysis. The top and bottom whiskers show the highest and lowest data points, respectively. The top and bottom grey boxes describe the upper and lower quartile, respectively, while the dark black line inside the box shows the median of the data. Red lines depict the optimal classification cut-off. All metabolites included had AUC = 1 and fold change > ±0.5. N = 3 biological replicates per sample group.
Metabolite set enrichment analysis (Fig. 3e) revealed 19 metabolic pathways that were altered between the DCIS and mammary duct models, including pathways associated with: rapid proliferation; carbohydrate metabolism (e.g., glycolysis, gluconeogenesis and pyruvate metabolism), nucleotide metabolism (pyrimidine and purine metabolism), amino acid metabolism (glycine, serine, and threonine metabolism; glutamine metabolism, and malate-aspartate shuttle), lipid metabolism (ketone body metabolism) and nitrogen metabolism (urea cycle, ammonia recycling). Together, 1H NMR metabolomics analysis revealed significant differences and identified specific changes in metabolite concentrations and metabolic pathways between the DCIS and the other models.
3.4. Gene expression analysis
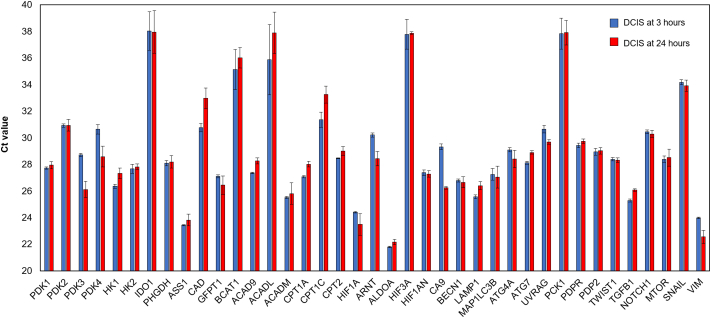
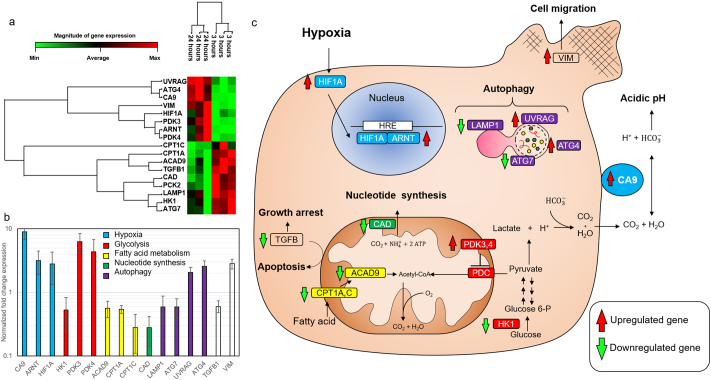
Next, we performed genetic analyses by RT-qPCR to further explore how DCIS cells respond to these microenvironmental conditions. The expression of 40 genes related to different metabolic pathways and cellular functions was analyzed after a 3 and 24 h within the microdevice (Supplementary Fig. 6). Among the genes analyzed, seventeen of them showed significant differences (only genes with ct values lower than 35 were considered). The non-supervised clustering algorithm showed these genes were divided into two clusters, where one cluster was upregulated after 24 h, whereas the other was downregulated (Fig. 4a). We grouped these genes in different pathways (i.e., hypoxia-related genes, glycolysis, fatty acid metabolism, nucleotide synthesis, and autophagy) (Fig. 4b). TGF-β and vimentin were not included in any of these groups. The hypoxia-related genes (HIF1A, ARNT, and CA9) were upregulated after 24 h, which suggests that cells adapted to the hypoxia generated within the DCIS model. Genes involved in fatty acid transportation into the mitochondria and fatty acid metabolism (ACAD9, CPT1A, and CPT1C) showed a downregulation; which could suggest decreased fatty acid consumption rate under hypoxic conditions [39].
Supplementary Fig. 6.
Gene expression analysis by RT-PCR. A set of different genes were analyzed in the DCIS model after 3 and 24 h in cell culture. The Ct values are shown as the average of at least three independent replicates and error bars are shown as the standard deviation.
Fig. 4.
Gene expression. a) Heat-map showing statistically significant gene expression changes within the microfluidic model after 3 and 24 h in cell culture. mRNA was extracted from the lumen and analyzed by RT-PCR using a Qiagen custom gene panel. Data was analyzed using Qiagen RT2 profiler software. The magnitude of gene expression change is showed in the horizontal bar. UV Radiation Associated Gene (UVRAG); Autophagy-Related Gene 4 (ATG4); Carbonic Anhydrase 9 (CA9); vimentin (VIM); Hypoxia-inducible Factor 1-alpha (HIF1A); Pyruvate Dehydrogenase Kinase 3 (PDK3); Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT, also known as HIF1B); Pyruvate Dehydrogenase Kinase 4 (PDK4); Carnitine Palmitoyl Transferase 1C (CPT1C); Acyl-Coenzyme A Dehydrogenase Family Member 9 (ACAD9); Transforming Growth Factor-beta 1 (TGFB1); Carbamoyl-Phosphate Synthetase 2, Aspartate Transcarbamylase, and Dihydroorotase (CAD); Lysosomal-associated membrane protein 1 (LAMP1); Hexokinase 1 (HK1); Autophagy-Related Gene 7 (ATG7). Each column represents one individual experiment. b) Bar chart showing the magnitude of gene change expression. Genes were grouped by metabolic pathways; excepting TGF-β1 and vimentin, which were not include into any group. Data are displayed in logarithmic scale as mean ± 95%confindent interval [Student's t-test]. c) Scheme illustrating the main role of the proteins whose mRNA expression was statistically significant.
Similarly, the downregulation of HK1 suggested a decrease in the glycolysis rate; whereas the overexpression of PDK3 and PDK4 indicated a shift towards an anaerobic glucose metabolism. Autophagy-related genes showed a complex pattern, two of them were upregulated whereas the other two were downregulated. Finally, we observed a downregulation of TGF-β and an upregulation of vimentin, suggesting activation of tumor survival response and migration, respectively. Altogether, these results demonstrate how DCIS cells adapt to the microenvironment generated by their metabolic activity. The results obtained in the microfluidic model were compared with genetic profiles obtained from DCIS patients in the clinic (METABRIC study, 2509 cases) [40]. The results showed that 56% of the patients showed alterations in at least one of the genes affected in the model. The most common alterations for CA9, ARNT, HIF1α (i.e., hypoxia-related genes), PDK4 and VIM genes were mRNA upregulation and amplification; which is good agreement with the overexpression obtained in the model for these genes (Fig. 5a). On the other hand, mRNA downregulation and deep deletion were the most common alterations for HK1 and LAMP1 genes; resembling the results observed in the model. Using the online software cBioPortal (http://www.cbioportal.org/) and the METABRIC database, a gene network including the 16 genes found to be significant was generated (Fig. 5b). The network describes the relationships between the different genes affected. Additionally, the software identified multiple drugs evaluated in the clinic against the affected pathways.
Fig. 5.
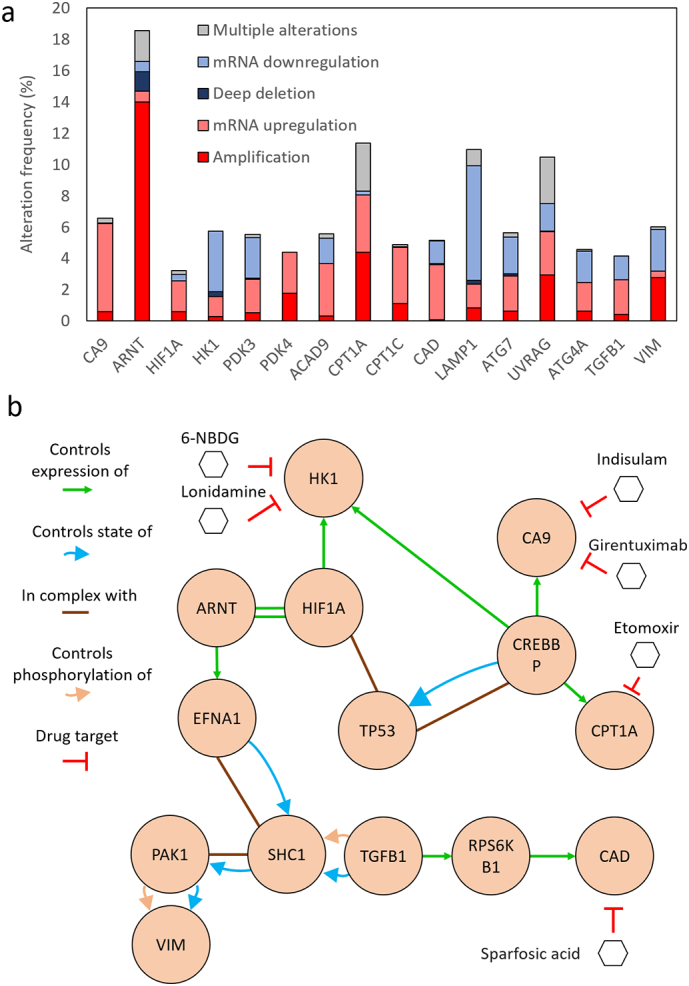
Patient genetic profile obtained in the METABRIC study. a) The graph shows the different genetic alterations identified in patients in the 16 genes that were significantly different in the model. b) Genetic network showing the main interactions between the affected genes and their intermediates. Drugs used in clinical trials targeting these pathways are also included.
3.5. Optical metabolic imaging during the invasion process
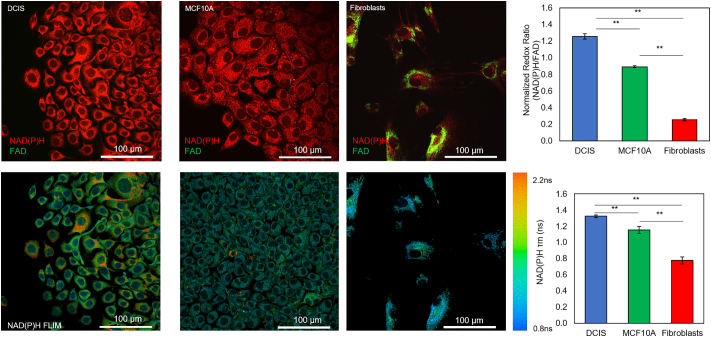
Next, we used multi-photon optical metabolic imaging to evaluate whether the hypoxia and nutrient gradients across the lumen could lead to the generation of different metabolic phenotypes within the lumen. OMI is based on the autofluorescence of NADH, NADPH and FAD cofactors and provides real-time monitoring of cell metabolism at a single cell level [36]. Both NADH and NADPH have similar fluorescence spectra, thus their combined fluorescence is expressed as NAD(P)H. Therefore, we analyzed the evolution of the redox ratio (i.e. NAD(P)H/FAD ratio) and NAD(P)H fluorescence lifetime (FLIM) (NAD(P)H τm) in the model. The redox ratio and NAD(P)H τm are affected by the intracellular metabolism and the intracellular microenvironment, allowing differences at a single cell level to be detected. MCF10A, MCF10-DCIS cells, and HMFs were seeded on flat glass bottom Petri dishes to evaluate their redox ratio and NAD(P)H τm after two days in 2D (Fig. 6). The results showed DCIS cells had a higher redox ratio compared with MCF10A and at least 7-fold higher than HMFs. This observation agrees with previous observations suggesting DCIS cells had a faster glycolytic metabolism; which leads to the accumulation of NAD(P)H in the cytoplasm. Additionally, the NAD(P)H τm was also statistically different between DCIS, MCF10A cells and HMFs, again suggesting the presence of a different intracellular metabolism (Fig. 6).
Fig. 6.
Optical metabolic imaging of multiple cell types seeded on 2D. NAD(P)H signal is shown in red, whereas FAD signal appears in green. In the graph, the ratio between the NAD(P)H and FAD signal is shown for comparison between the different cell types. Graphs show the mean ± Standard Error. ** denotes p-value <0.001 [one-way ANOVA].
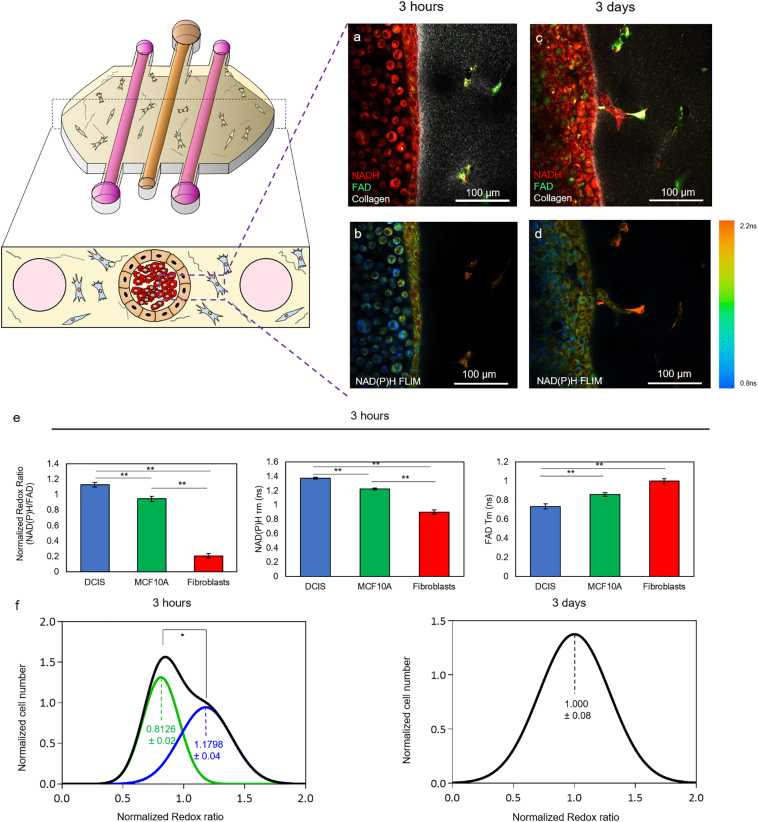
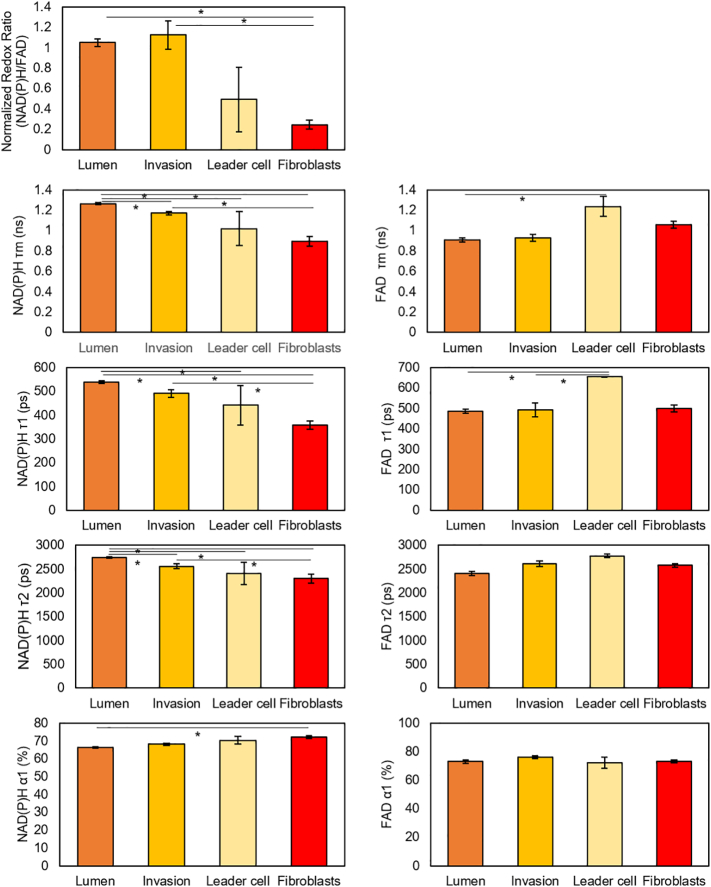
Consistent with these 2D results, after 3 h in within the device, DCIS, MCF10A, and HMF cells maintained distinct redox ratio and NADP(P)H τm (Fig. 7a–b, e and Supplementary Fig. 7). However, after 3 days, the redox ratio and NAD(P)H τm across the lumen developed into a gradient ranging from the lumen perimeter to the center (Fig. 7c–d). Furthermore, after 3 h in culture, the redox ratio of all the cells in the lumen (i.e. cells in suspension and cells forming the lumen wall) was analyzed and two different populations were identified (Fig. 7f, left graph, green and blue curves), likely corresponding to MCF10A and DCIS respectively. Interestingly, after 3 days only a single population could be identified within the lumen (Fig. 7f, right graph). This change suggests that the tumor and normal cells adapt their metabolism according to the different microenvironments present in the lumen core compared with the surrounding region. Additionally, 3 days after seeding the DCIS cells, cells started to invade the surrounding matrix by a collective movement (i.e. forming groups of cells where one or few cells lead the path and degrade the collagen matrix, whereas the other cells follow the path attached to the leader cells) (Fig. 7c–d). Interestingly, cells located within the lumen, the invading branch or at the tip of the invading branch showed several differences in NAD(P)H and NAD(P)H τm (Supplementary Fig. 8), suggesting invading cells relied on a different metabolism compared with those trapped within the duct.
Fig. 7.
Optical metabolic imaging of cells cultured within the microfluidic devices. a-d) 3 h after seeding the cells, HMF and epithelial cells (MCF10A and DCIS) exhibit different redox ratio as well as NAD(P) τm and FAD τm. After 3 days in cell culture, the cells within the lumen show a more heterogenous pattern and cells from the lumen started to invade the surrounding matrix. e) Graphs showing the Redox Ratio, NAD(P)H τm and FAD τm 3 h after injecting the DCIS in the central lumen. f) A population analysis was performed to quantify the redox ratio on the cells located in the lumen (including those at the lumen wall, black line). After 3 h in culture, the analysis revealed that the population can be deconvoluted in two populations (green and blue lines respectively) with a different redox ratio (p-value <0.05). After 3 days in culture, only one population was identified within the lumen and the redox was closer to the less reduced population identified after 3 h. Data shows the mean ± standard deviation. * denotes p-value <0.05 [one-way ANOVA].
Supplementary Fig. 7.
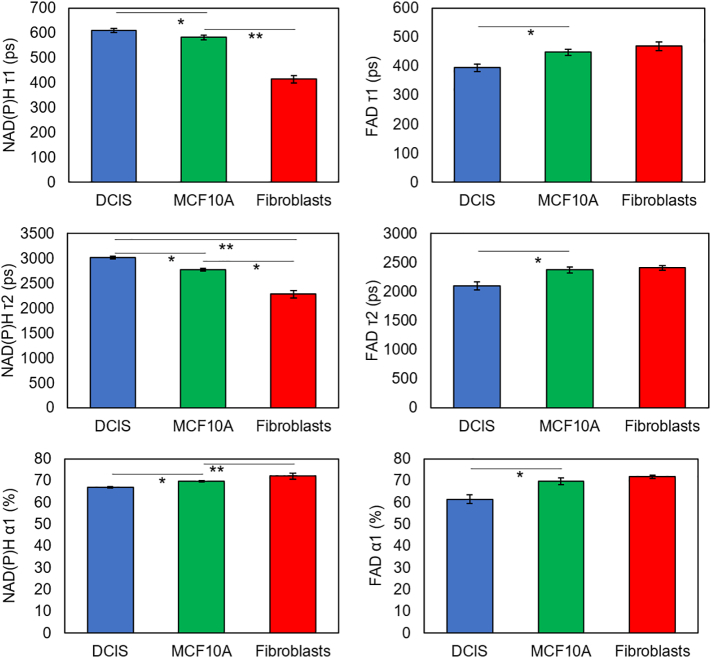
OMI analysis of the DCIS model after 3 h in culture. Normalized redox ratio, NAD(P)H τm, τ1, τ2, α1 and FAD τm, τ1, τ2, α1 were calculated for HMF, MCF10A and DCIS cells. The analysis reveals these different cell populations exhibit differences in the metabolic parameters analyzed. Data shows mean, and error bars indicate standard deviation. * denotes p-value <.05 [one-way ANOVA].
Supplementary Fig. 8.
OMI analysis of the DCIS model after 3 days in culture. Normalized redox ratio, NAD(P)H τm, τ1, τ2, α1 and FAD τm, τ1, τ2, α1 were calculated for HMF, cell within the lumen, invading cells at the invading branch and the leader cell at the tip of the invading branch. The analysis reveals these different cell populations exhibit multiple differences in the metabolic parameters analyzed. Data shows mean, and error bars indicate standard deviation. * denotes p-value <.05 [one-way ANOVA].
3.6. Targeting the DCIS microenvironment
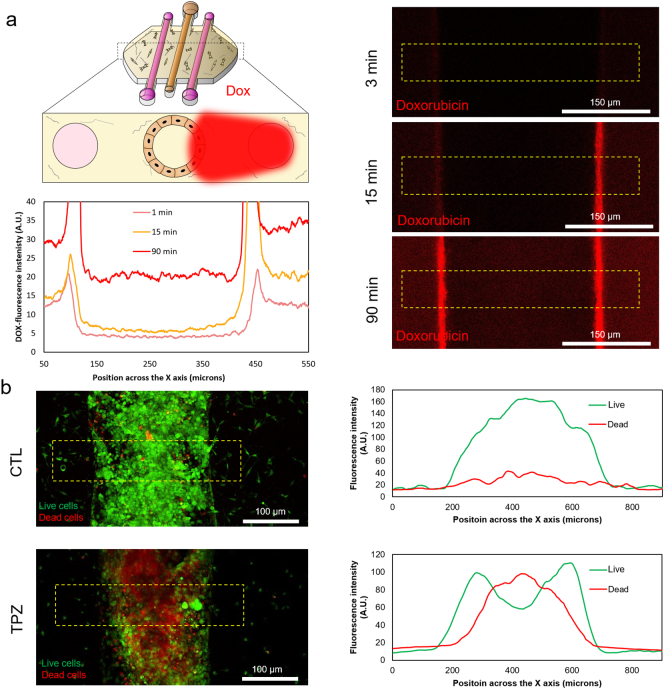
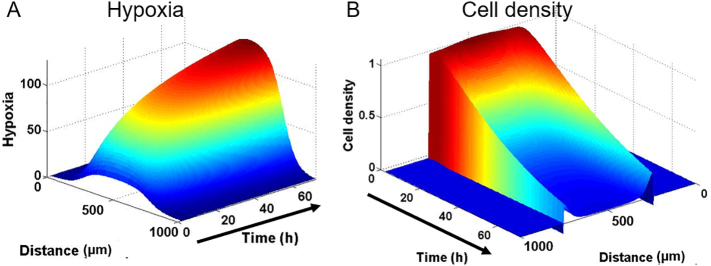
Recently, researchers have speculated that the altered microenvironment within a tumor can lead to metabolic vulnerabilities in tumor cells that can be used as targets for cancer therapeutics. To evaluate this hypothesis, we tested a drug that specifically targets hypoxic cells (i.e. Tirapazamine, TPZ); inducing DNA damage only under hypoxic conditions. First, we determined whether small hydrophobic drugs could penetrate through the lumen wall (Fig. 8a). Doxorubicin, a classic chemotherapeutic agent that is fluorescent in red, was perfused through the right lateral lumen. Doxorubicin, like Rhodamine B, rapidly penetrated through the MCF10A cells and reached the core of the empty lumen. 100 μM TPZ was added through the lateral lumens and cell viability was evaluated after 3 days. In the absence of TPZ, cells showed high viability (Fig. 8b). However, when cultured in the presence of TPZ, a region of dead cells appeared in the center of the lumen (Fig. 7b). This necrotic region was confined to the innermost part of the lumen, where hypoxia was more intense; whereas the edges of the lumen and the HMFs remained viable. These results demonstrate the relevance of the DCIS microenvironment for drug sensitivity and show how the presented DCIS model could be used to test new drugs. Furthermore, TPZ diffusion and cytotoxicity was simulated, showing a good agreement with the experimental data (Supplementary Fig. 9). The model was able to mimic the differential toxicity in the normoxic and hypoxic areas based on the TPZ diffusion coefficient and toxicity values obtained from the literature. This shows how computational simulations can be applied to study the complex tumor dynamics and drug interactions.
Fig. 8.
Drug testing. a) DOX, which is fluorescent in red, was perfused through the right lateral lumen. The DOX diffusion profile through an MCF10A central lumen was observed at different times. DOX rapidly diffused through the collagen and was uptaken by the MCF10A cells. After 90 m min DOX is clearly present inside the empty lumen. The graph shows the DOX diffusion profile across the yellow rectangle. b) TPZ was perfused through both lateral lumens and after 3 days in culture cell viability was evaluated using CAM/PI. The addition of TPZ caused an intense cell mortality. The analysis of the viable/dead cell profile showed that TPZ exerted a higher toxicity in the center of the lumen.
Supplementary Fig. 9.
Computational simulation of the hypoxia and cell density within the lumen in the presence of TPZ. A) Hypoxia profile simulation mimics the hypoxia gradient observed within the microdevice. B) Cell density in the presence of TPZ was modeled, rendering a gradient cell viability where the lowest viability was located at the center of the lumen.
4. Discussion
The mechanisms that allow for tumor cell survival within the harsh conditions of the mammary duct remain elusive. Recapitulating the TME in vitro is challenging because most current in vitro models rely on classic 2D Petri dishes where the TME is mostly absent. Here, we have developed a breast cancer model that mimics the DCIS structure and allows the co-culture of multiple cell types; recreating critical elements of the DCIS TME.
The model showed how DCIS cells use distinct metabolic pathways compared with normal mammary epithelial cells. The rapid consumption of glucose and glutamine supports the hypothesis that DCIS cells rely on glycolysis to enable their accelerated growth [41]. In vivo, the mammary gland selectively controls the diffusion of metabolites inside the duct. Glucose is actively transported within the cells via membrane transporters (GLUT) and then metabolized into lactose, which is finally excreted into the mammary duct during lactation. This glucose barrier effect was observed in the microfluidic model since NBDG diffusion was severely hindered by the MCF10A cells. Despite this barrier effect, the NMR data showed a significant glucose reduction in the media in the DCIS model, suggesting that this slow glucose penetration could be enough to support the DCIS cells. Additionally, hypoxia increases the secretion of lactose and other nutrients into the mammary duct in vivo [42]. Therefore, the hypoxia observed in the model could enhance glucose transport into the duct. Additionally, when glucose levels are low, cells can survive by undergoing gluconeogenesis and replenishing their glucose levels at the expense of using glucogenic amino acids [43]. In this context, metabolite set enrichment analysis (Fig. 3e) showed enrichment in gluconeogenesis and a decrease in glucogenic amino acids (e.g., asparagine, glutamine, glycine, valine, and isoleucine) in the DCIS model, thus supporting this hypothesis. This model could be used in the future to study glucose transport to the mammary duct and evaluate new DCIS therapies that target glucose transport. Additionally, some metabolic alterations seemed to be caused by environmental pressure (e.g., hypoxia and nutrient starvation due to high cell density) rather than genetic mutations. In this context, previous reports have also shown that mammary cells undergo profound metabolic changes when they grow under matrix detachment conditions [44]. Arguably, DCIS cells growing within the lumen may be affected similarly. Therefore, this model could also be used to study how malignant and non-malignant cells differ in their response to environmental factors like hypoxia, nutrient starvation, accumulation of waste products or matrix detachment. In response to the harsh microenvironment within the lumen, gene expression analysis showed that DCIS cells switched towards an anaerobic metabolism after 24 h. Under hypoxic conditions, HIF1A dimerizes in the cell nucleus with ARNT (also known as HIF1B) to induce the expression of numerous hypoxia-related genes (e.g., CA9) [45,46]. In this context, CA9 is known to be overexpressed in hypoxic tumor cells to regulate their acidic intracellular pH, allowing tumor cell survival under toxic pH conditions [47]. Increase in PDK3 and PDK4 levels, which phosphorylate and block pyruvate dehydrogenase complex, suggest a shift towards lactic acid fermentation; which causes intracellular and extracellular acidification through CA9 activity [48]. Additionally, reduction in CAD expression (the first rate-limiting step in pyrimidine and nucleotide synthesis) suggests a decreased proliferation rate, may be caused by the hypoxic and starving environment [49]. Moreover, vimentin is known to be involved in epithelial-mesenchymal transition and cell migration and was upregulated in the DCIS model. The increased vimentin expression could partially explain the migration that was observed in the DCIS model after 3 days in culture [50,51].
TGF-β plays a complex role in breast cancer since it can promote tumor growth or induce tumor suppression, while maintaining other functions such as immune escape. In the early stages of breast cancer (i.e. DCIS), TGF-β seems to play a tumor suppressive role, whereas in the later stages it may promote metastasis [52,53]. In our model, the reduction of TGF-β could be associated with the generation of a tumor-promoting environment. Finally, two of the genes related to autophagy were downregulated, whereas the other two were upregulated. Autophagy is a complex process that engages numerous factors. These results indicate that a subset of autophagy-related genes could be critical to support tumor cell survival under this hypoxic and starving microenvironment [54,55].
OMI showed that DCIS cells responded differently depending on their position inside the lumen or the invading branch (Fig. 7). In this context, cells located at the invading branch and the leader cell exhibited a lower NAD(P)H FLIM compared with cells located in the lumen. Previous reports have shown that NAD(P)H FLIM reduction indicates an increase in the amount of free NAD(P)H in the cell cytosol in these cells as a consequence of a more intense glycolysis [56,57]. This observation could indicate that as invading cells escape from the starving lumen, they have access to more glucose, accelerating their glycolytic metabolism. Additionally, other studies have shown that epithelial cells, including breast cancer, rely on collective movement to invade the surrounding tissue [58,59]. Interestingly, collective invasion requires a different molecular machinery compared with single cell invasion, which may offer some new alternatives to target cancer migration and invasion [60].
Finally, the DCIS model was used to evaluate the possibility of target tumor cells based on the surrounding microenvironment. TPZ showed a gradient of toxicity with a similar pattern to the oxygen gradient (Fig. 2, Fig. 8). Interestingly, the cells at the lumen periphery, as well as the fibroblasts, remained unaffected by the TPZ. Therefore, hypoxia-activated prodrugs could be combined with other drugs to target both normoxic and hypoxic DCIS cells.
In conclusion, the presented model better generates a more complex microenvironment compared with traditional 2D cell culture or regular 3D spheroids. The model allowed the observation of spatial-temporal metabolic and phenotypic gradients across the lumen. In this context, DCIS cells showed a heterogeneous metabolic response across the lumen, as well as heterogeneous response to TMZ. This observation suggested that the phenotype heterogeneity generated by nutrients gradients will probably require a complex treatment to destroy all the different DCIS cell populations within the lumen (e.g., normoxic vs. hypoxic cells). Therefore, this model could help to identify more accurate prognostic biomarkers for DCIS and uncover novel therapeutic strategies for DCIS in a much faster way than more simplistic in vitro models.
The following are the supplementary data related to this article.
Hypoxia profile was monitored in a time-lapse experiment in the DCIS model.
Supplementary video 2 Z-stack showing the collagen second harmonic generation.
Author contribution
JMA and KL fabricated the microdevices. JMA performed the cell culture experiments and confocal imaging. JMA and MM performed the RT-qPCR experiments and data analysis. SA and SPP analyzed the metabolite composition by NMR. JMA performed the OMI and TH and AG did the quantitative analysis. IG generated the computational simulation. KBW, MCS, DJB and JMA wrote the manuscript and all the authors reviewed it.
Acknowledgments
Acknowledgments
University of Wisconsin Carbone Cancer Center (AAB7173). Morgridge Research Institute and Boehringer-Ingelheim Foundation. NIH grants R01CA186134, R33CA225281, R01CA186134, R01 CA164492, R01 CA185747, R01 CA205101, Mary Kay Foundation grant 067-16, DoD BCRP grant W81XWH-13-1-0194, and NSF grant CBET-1642287. National Institutes of General Medical Sciences (P41GM103399 and P41GM66326). National Science Foundation Graduate Research Fellowship (DGE-1256259). EPA STAR STAR 83573701. T32 ES007015-39NIH NIEHS. University of Wisconsin Advanced Opportunity Fellowship through the Graduate Engineering Research Scholars (GERS) Program.
Competing financial interests
David J. Beebe is a board member and stockowner of Tasso, Inc. and a stockowner of Bellbrook Labs, LLC. David J. Beebe is a founder, stockowner, and consultant of Salus Discovery LLC. David J. Beebe is an advisor and stockowner of Lynx Biosciences, LLC, Onexio Biosystems, LLC, and Stacks to the Future, LLC. David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso Inc., Salus Discovery LLC, Stacks to the Future, LLC and Onexio Biosystems, LLC.
Funding
tUniversity of Wisconsin Carbone Cancer Center (AAB7173). Morgridge Research Institute and Boehringer-Ingelheim Foundation. NIH grants R01 CA164492, R01 CA185747, R01 CA205101, Mary Kay Foundation grant 067-16, DoD1 BCRP grant W81XWH-13-1-0194, and NSF grant CBET-1642287. National Institutes of General Medical Sciences (P41GM103399 and P41GM66326). National Science Foundation Graduate Research Fellowship (DGE-1256259).
Contributor Information
Jose M. Ayuso, Email: ayusodomingu@wisc.edu.
David J. Beebe, Email: djbeebe@wisc.edu.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. Ca-Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.ACS Cancer Facts and Figures 2015: Special Section Breast Carcinoma in situ. 2015. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2015/special-section-breast-carcinoma-in-situ-cancer-facts-and-figures-2015.pdf Available from.
- 3.Polyak K. Breast cancer: Origins and evolution. J Clin Invest. 2007;117(11):3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward E.M., Desantis C.E., Lin C.C., Kramer J.L., Jemal A., Kohler B. Cancer statistics: Breast cancer in situ. Ca-Cancer J Clin. 2015;65(6):481–495. doi: 10.3322/caac.21321. [DOI] [PubMed] [Google Scholar]
- 5.Nofech-Mozes S., Spayne J., Rakovitch E., Hanna W. Prognostic and predictive molecular markers in DCIS - A review. Adv Anat Pathol. 2005;12(5):256–264. doi: 10.1097/01.pap.0000184177.65919.5e. [DOI] [PubMed] [Google Scholar]
- 6.Espina V., Liotta L.A. What is the malignant nature of human ductal carcinoma in situ? Nat Rev Cancer. 2011;11(1):68–75. doi: 10.1038/nrc2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espina V., Wysolmerski J., Edmiston K., Liotta L.A. Attacking breast cancer at the preinvasion stage by targeting autophagy. Womens Health (Lond) 2013;9(2) doi: 10.2217/whe.13.5. [157-70. Epub 2013/03/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z.J., Semenza G.L., Zhang H.F. Hypoxia-inducible factor 1 and breast cancer metastasis. J Zhejiang Univ Sci B. 2015;16(1):32–43. doi: 10.1631/jzus.B1400221. [Epub 2015/01/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teicher B.A., Linehan W.M., Helman L.J. Targeting cancer metabolism. Clin Cancer Res. 2012;18(20):5537–5545. doi: 10.1158/1078-0432.CCR-12-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vander Heiden M.G. Targeting cancer metabolism: A therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 11.Brock E.J., Ji K., Shah S., Mattingly R.R., Sloane B.F. In vitro models for studying invasive transitions of ductal carcinoma In Situ. J Mammary Gland Biol Neoplasia. 2018 doi: 10.1007/s10911-018-9405-3. [Epub 2018/07/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kittrell F., Valdez K., Elsarraj H., Hong Y., Medina D., Behbod F. Mouse Mammary Intraductal (MIND) method for transplantation of patient derived primary DCIS cells and cell lines. Bio-Protocol. 2016;6(5) doi: 10.21769/bioprotoc.1744. [Epub 2016/07/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behbod F., Gomes A.M., Machado H.L. Modeling human ductal carcinoma In Situ in the mouse. J Mammary Gland Biol Neoplasia. 2018 doi: 10.1007/s10911-018-9408-0. [Epub 2018/08/27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sackmann E.K., Fulton A.L., Beebe D.J. The present and future role of microfluidics in biomedical research. Nature. 2014;507(7491):181–189. doi: 10.1038/nature13118. [Epub 2014/03/14] [DOI] [PubMed] [Google Scholar]
- 15.Esch E.W., Bahinski A., Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov. 2015;14(4):248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez-Torres J.A., Peery S.L., Sung K.E., Beebe D.J. LumeNEXT: A practical method to pattern luminal structures in ECM gels. Adv Healthc Mater. 2016;5(2):198–204. doi: 10.1002/adhm.201500608. [Epub 2015/11/27] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bischel L.L., Young E.W., Mader B.R., Beebe D.J. Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels. Biomaterials. 2013;34(5):1471–1477. doi: 10.1016/j.biomaterials.2012.11.005. [Epub 2012/11/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen D.H., Stapleton S.C., Yang M.T., Cha S.S., Choi C.K., Galie P.A. Biomimetic model to reconstitute angiogenic sprouting morphogenesis in vitro. Proc Natl Acad Sci U S A. 2013;110(17):6712–6717. doi: 10.1073/pnas.1221526110. [Epub 2013/04/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen M.B., Whisler J.A., Frose J., Yu C., Shin Y., Kamm R.D. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat Protoc. 2017;12(5):865–880. doi: 10.1038/nprot.2017.018. [Epub 2017/03/31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DTT Phan, Wang X., Craver B.M., Sobrino A., Zhao D., Chen J.C. A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip. 2017;17(3):511–520. doi: 10.1039/c6lc01422d. [Epub 2017/01/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bischel L.L., Beebe D.J., Sung K.E. Microfluidic model of ductal carcinoma in situ with 3D, organotypic structure. BMC Cancer. 2015;15 doi: 10.1186/s12885-015-1007-5. [12. Epub 2015/01/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tait L.R., Pauley R.J., Santner S.J., Heppner G.H., Heng H.H., Rak J.W. Dynamic stromal-epithelial interactions during progression of MCF10DCIS.com xenografts. Int J Cancer. 2007;120(10):2127–2134. doi: 10.1002/ijc.22572. [Epub 2007/02/03] [DOI] [PubMed] [Google Scholar]
- 23.Miller F.R., Santner S.J., Tait L., Dawson P.J. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92(14):1185–1186. doi: 10.1093/jnci/92.14.1185a. [Epub 2000/07/25] [DOI] [PubMed] [Google Scholar]
- 24.Gowda G.A.N., Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014;86(11):5433–5440. doi: 10.1021/ac5005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhute V.J., Palecek S.P. Metabolic responses induced by DNA damage and poly (ADP-ribose) polymerase (PARP) inhibition in MCF-7 cells. Metabolomics. 2015;11(6):1779–1791. doi: 10.1007/s11306-015-0831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J.G., Mandal R., Sinelnikov I.V., Broadhurst D., Wishart D.S. MetaboAnalyst 2.0-a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012;40(W1) doi: 10.1093/nar/gks374. (W127-W33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia J.G., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp356. (W652-W60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia J.G., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43(W1) doi: 10.1093/nar/gkv380. (W251-W7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia J.G., Wishart D.S. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq329. (W71-W7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karnovsky A., Weymouth T., Hull T., Tarcea V.G., Scardoni G., Laudanna C. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–380. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia J.G., Broadhurst D.I., Wilson M., Wishart D.S. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9(2):280–299. doi: 10.1007/s11306-012-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J.J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5) doi: 10.1158/2159-8290.CD-12-0095. [401-4. Epub 2012/05/17] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannon T.M., Shah A.T., Skala M.C. Autofluorescence imaging captures heterogeneous drug response differences between 2D and 3D breast cancer cultures. Biomed Opt Express. 2017;8(3):1911–1925. doi: 10.1364/BOE.8.001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah A.T., Diggins K.E., Walsh A.J., Irish J.M., Skala M.C. In Vivo autofluorescence imaging of tumor heterogeneity in response to treatment. Neoplasia. 2015;17(12):862–870. doi: 10.1016/j.neo.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh A.J., Cook R.S., Skala M.C. Functional optical imaging of primary human tumor organoids: Development of a personalized drug screen. J Nucl Med. 2017;58(9):1367–1372. doi: 10.2967/jnumed.117.192534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teale F.W.J. Principles of fluorescence spectroscopy - Lakowicz. J Nature. 1984;307(5950):486. [Google Scholar]
- 38.Gm Cooper. 2nd edition. Sinauer Associates; 2000. The cell a molecular approach. [Google Scholar]
- 39.Huang D., Li T.T., Li X.H., Zhang L., Sun L.C., He X.P. HIF-1-mediated suppression of Acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014;8(6):1930–1942. doi: 10.1016/j.celrep.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 40.Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7 doi: 10.1038/ncomms11479. [11479. Epub 2016/05/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantor J.R., Sabatini D.M. Cancer cell metabolism: One Hallmark, many faces. Cancer Discov. 2012;2(10):881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao F.Q. Biology of glucose transport in the mammary gland. J Mammary Gland Biol Neoplasia. 2014;19(1):3–17. doi: 10.1007/s10911-013-9310-8. [Epub 2013/11/14] [DOI] [PubMed] [Google Scholar]
- 43.Leithner K. PEPCK in cancer cell starvation. Oncoscience. 2015;2(10):805–806. doi: 10.18632/oncoscience.252. [Epub 2015/12/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schafer Z.T., Grassian A.R., Song L.L., Jiang Z.Y., Gerhart-Hines Z., Irie H.Y. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461(7260):109. doi: 10.1038/nature08268. -U18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dengler V.L., Galbraith M., Espinosa J.M. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49(1):1–15. doi: 10.3109/10409238.2013.838205. [Epub 2013/10/09] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semenza G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123(9):3664–3671. doi: 10.1172/JCI67230. [Epub 2013/09/04] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benej M., Pastorekova S., Pastorek J. Carbonic anhydrase IX: Regulation and role in cancer. Subcell Biochem. 2014;75:199–219. doi: 10.1007/978-94-007-7359-2_11. [Epub 2013/10/23] [DOI] [PubMed] [Google Scholar]
- 48.Zhang W., Zhang S.L., Hu X.H., Tam K.Y. Targeting tumor metabolism for cancer treatment: Is pyruvate dehydrogenase kinases (PDKs) a viable anticancer target? Int J Biol Sci. 2015;11(12):1390–1400. doi: 10.7150/ijbs.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howell J.J., Ricoult S.J.H., Ben-Sahra I., Manning B.D. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc T. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- 50.Kokkinos M.I., Wafai R., Wong M.K., Newgreen D.F., Thompson E.W., Waltham M. Vimentin and epithelial-mesenchymal transition in human breast cancer - Observations in vitro and in vivo. Cells Tissues Organs. 2007;185(1–3):191–203. doi: 10.1159/000101320. [DOI] [PubMed] [Google Scholar]
- 51.Liu C.Y., Lin H.H., Tang M.J., Wang Y.K. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6(18):15966–15983. doi: 10.18632/oncotarget.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel P.M., Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3(11):807–821. doi: 10.1038/nrc1208. [Epub 2003/10/15] [DOI] [PubMed] [Google Scholar]
- 53.Stampfer M.R., Yaswen P. Culture models of human mammary epithelial cell transformation. J Mammary Gland Biol Neoplasia. 2000;5(4):365–378. doi: 10.1023/a:1009525827514. [Epub 2004/02/20] [DOI] [PubMed] [Google Scholar]
- 54.Cheong H., Lu C., Lindsten T., Thompson C.B. Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol. 2012;30(7):671–678. doi: 10.1038/nbt.2285. [Epub 2012/07/12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji Y., Di W., Yang Q., Lu Z., Cai W., Wu J. Inhibition of autophagy increases proliferation inhibition and apoptosis induced by the PI3K/mTOR inhibitor NVP-BEZ235 in breast cancer cells. Clin Lab. 2015;61(8) doi: 10.7754/clin.lab.2015.150144. [1043-51. Epub 2015/10/03] [DOI] [PubMed] [Google Scholar]
- 56.Skala M.C., Riching K.M., Bird D.K., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K.W. In vivo multiphoton fluorescence lifetime imaging of protein-bound and free nicotinamide adenine dinucleotide in normal and precancerous epithelia. J Biomed Opt. 2007;12(2) doi: 10.1117/1.2717503. [024014. Epub 2007/05/05] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sameni S., Syed A., Marsh J.L., Digman M.A. The phasor-FLIM fingerprints reveal shifts from OXPHOS to enhanced glycolysis in Huntington disease. Sci Rep. 2016;6 doi: 10.1038/srep34755. [34755. Epub 2016/10/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedl P., Alexander S. Cancer invasion and the microenvironment: Plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [Epub 2011/11/29] [DOI] [PubMed] [Google Scholar]
- 59.Friedl P., Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [Epub 2009/06/24] [DOI] [PubMed] [Google Scholar]
- 60.Ilina O., Friedl P. Mechanisms of collective cell migration at a glance. J Cell Sci. 2009;122(Pt 18):3203–3208. doi: 10.1242/jcs.036525. [Epub 2009/09/04] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hypoxia profile was monitored in a time-lapse experiment in the DCIS model.
Supplementary video 2 Z-stack showing the collagen second harmonic generation.