ABSTRACT
Post-transcriptional control of messenger RNA (mRNA) is an important layer of gene regulation that modulates mRNA decay, translation, and localization. Eukaryotic mRNA decay begins with the catalytic removal of the 3′ poly-adenosine tail by deadenylase enzymes. Multiple deadenylases have been identified in vertebrates and are known to have distinct biological roles; among these proteins is Nocturnin, which has been linked to circadian biology, adipogenesis, osteogenesis, and obesity. Multiple studies have investigated Nocturnin’s involvement in these processes; however, a full understanding of its molecular function remains elusive. Recent studies have provided new insights by identifying putative Nocturnin-regulated mRNAs in mice and by determining the structure and regulatory activities of human Nocturnin. This review seeks to integrate these new discoveries into our understanding of Nocturnin’s regulatory functions and highlight the important remaining unanswered questions surrounding its regulation, biochemical activities, protein partners, and target mRNAs.
KEYWORDS: Nocturnin, NOCT, deadenylase, RNA decay, translational control
Introduction
Cells must sense and respond appropriately to a variety of external cues including hormones, nutrients, stress, and circadian inputs. These cues may trigger broad changes in the gene expression program to promote survival, such as those seen during nutrient starvation; alternatively, these changes may be targeted to specific pathways, e.g., the inflammatory response following injury or infection. Changes in gene expression may be exerted at multiple levels including messenger RNA (mRNA) synthesis, processing, and translation. Although regulation of mRNA transcription has received significant attention in the past, the importance of post-transcriptional control over gene expression has become increasingly apparent in recent years [1–4]. A multitude of cis- and trans-acting factors exert influence at this level, including those that modulate RNA stability and translation by shortening mRNA poly-adenosine (poly(A)) tails [5,6].
In vertebrates, Nocturnin (NOCT; also known as Noct, Noc, CCR4L, CCRN4L, and Ccr4c) was originally identified in Xenopus retina as a circadian-expressed RNA with homology to the yeast deadenylase CCR4 [7,8]. NOCT orthologs have been identified in invertebrates and vertebrates including Drosophila, fish, amphibians, and mammals, suggesting NOCT has an evolutionarily conserved function in metazoans [8–17]; however, it is absent from other organisms such as C. elegans and yeast. NOCT has been most extensively studied in mice, where loss of NOCT confers resistance to high-fat diet-induced obesity [9]. In addition to regulation of body mass, NOCT affects the regulation of intestinal lipid trafficking, and the differentiation of bone marrow mesenchymal stem cells into either adipocytes or osteoblasts (Fig. 1) [18–21]. As a putative RNA decay factor, NOCT has been proposed to contribute to these processes by destabilizing mRNAs encoding proteins with important metabolic and developmental functions. Several studies sought to understand NOCT function by characterizing its biochemical activity and identifying the mRNAs whose misregulation may explain NOCT knockout mouse phenotypes. Thus far these studies have had limited success and our understanding of NOCT function has remained unclear; however, recent reports provide new insights into NOCT biology, structure, and function. Consequently, it is now necessary to integrate these new results with previous data and to clearly define the remaining unanswered questions surrounding NOCT biology.
Figure 1.
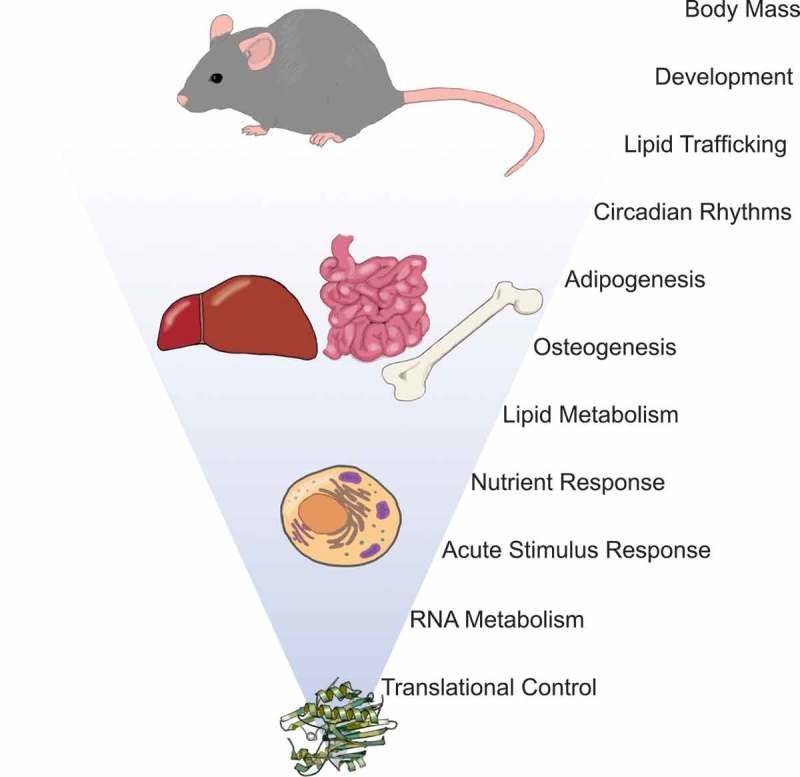
Molecular through physiological roles of NOCT. NOCT-mediated regulation of mRNA translation and decay at the molecular level affects multiple biological processes and modulates tissue development and function, circadian processes, and ultimately regulation of body mass.
Post-transcriptional control of mRNA abundance and stability by specialized deadenylases
The terminal 5′ 7-methylguanosine cap and 3′ poly(A) tail modifications found on mature mRNAs serve to both promote transcript association with the translation machinery and protect transcripts from exonuclease-mediated decay [22], and their enzymatic removal inhibits translation and destabilizes mRNAs. As the first and rate limiting step in mRNA decay, deadenylation is therefore an important regulatory nexus. Poly(A) tail removal is catalyzed by members of the deadenylase class of enzymes, twelve of which have been identified in mammals through sequence homology to either the yeast POP2 or CCR4 deadenylases. Six deadenylases are related to POP2 and share a DEDD-type catalytic domain (Pfam PF04857), while the remaining deadenylases including NOCT share homology with CCR4, a member of the exo-nuclease-endonuclease-phosphatase (EEP) superfamily (Pfam PF03372) [13].
Since all of the deadenylases could theoretically catalyze the same 3′-5′–directed removal of adenosines from an mRNA substrate, it is unsurprising that functional redundancy has been observed [13,23]. However, the collection of deadenylases is larger than expected if all deadenylases were fully interchangeable. Although a comprehensive review of the specific functions attributed to each deadenylase is beyond the scope of this article (readers are instead referred elsewhere [13,21,24–26]), several lines of evidence support functional specialization amongst these proteins, even among the most closely-related of these enzymes. The strongest support for this idea comes from analyses of vertebrate NOCT, CNOT7, and CAF1z loss-of-function models that exhibit distinct phenotypes, indicating unique functions for these enzymes [9,27–30].
Functional specificity among deadenylases could arise in part through tissue-, cell type- and developmental stage-specific expression as has been observed for the CCR4-NOT (CNOT) complex nuclease subunits CNOT6 and CNOT6L, which form mutually exclusive heterodimer complexes with CNOT7 and CNOT8, and have distinct roles even when co-expressed [23,26,31–34]. Additional functional specialization among deadenylases is likely mediated by divergent amino acid motifs within each protein, even among those sharing catalytic domain structure. Alignment of the amino acid sequences of human CCR4-type deadenylases reveals that although the EEP motif defining this class of enzymes is shared, the CNOT6/CNOT6L and ANGEL1/ANGEL2 paralog pairs exhibit the most extensive conservation while others members are more divergent (Fig. 2). The unique domains of each protein likely mediate subcellular localization, substrate, and regulatory partner specificity, as has been observed for CAF1z [30,35,36], PDE12 [37–39], PARN [36,40–42], and PNLDC1 [43–46]. NOCT biological function is likely further defined by its expression pattern and subcellular localization.
Figure 2.

NOCT is related to CCR4-type deadenylases and contains a conserved EEP catalytic domain. A) Multiple sequence alignment of human CCR4-type deadenylases. The shared characteristic EEP domain is boxed in pink; leucine-rich repeats found in CNOT6 and CNOT6L (boxed in green) are absent from NOCT. Putative NOCT catalytic residues (*) are conserved among EEP family members. Amino acid sequences were aligned using Clustal Omega and colored according to extent of chemical property similarity (charge, hydrophobicity, etc.) using Chroma [107,108]. Darker background shading indicates greater similarity, with strictly conserved positions indicated by black background and white text. Uniprot accession numbers: ANGEL1 Q9UNK9, ANGEL2 Q5VTE6, PDE12 Q6L8Q7, NOCT Q9UK39, CNOT6 Q9ULM6, CNOT6L Q965LI5. B) Phylogenetic tree demonstrating the relationships between human CCR4-type deadenylases and pair-wise amino acid identities. Phylogenetic relationships were determined using Clustal Omega and visualized using Interactive Tree of Life [107,109].
NOCT expression and biological functions
As in Xenopus, circadian fluctuations in mRNA abundance have been observed for NOCT orthologs in sea sponge [15], zebrafish [17], mice [10–12,47,48], and humans [49]. NOCT expression has been best characterized in mice, where NOCT mRNA abundance varies by tissue, and by time of day in several tissues [12,19,47]. Circadian variations in NOCT mRNA abundance are most dramatic in liver, followed by smaller fluctuations in heart and kidney; in each case, peak expression occurs at or soon after light offset [12,47,48]. These daily changes in transcript abundance suggested that NOCT may have functions relating to the circadian clock (Fig. 1). Consistent with this idea, the Drosophila NOCT ortholog curled has been implicated in the circadian light response [50]; however, NOCT knockout mice exhibit normal circadian behaviors, indicating that NOCT may function as a downstream effector of circadian signals in vertebrates [9].
The circadian clock may regulate NOCT transcription; however, the mechanism and consequences of this regulation are unclear. The core transcriptional regulators CLOCK and BMAL1 are recruited to the NOCT promoter in humans and mice through the action of a conserved E-box transcriptional element. Evidence from human cells suggests that CLOCK and BMAL1 associate with the NOCT promoter regardless of circadian time [51]; while BMAL1 ChIP-seq analysis in mice revealed time-dependent BMAL1 association with the NOCT promoter [52]. Interestingly, NOCT mRNA abundance in liver remains rhythmic in clock-deficient mice that lack a global circadian clock, as well as in the absence of a local circadian clock, suggesting that NOCT transcription is regulated by both local and global cues [53,54]. Furthermore, NOCT mRNA levels do not appear to be rhythmic in white adipose tissue from ad lib fed mice, suggesting that other cues may override clock function in certain contexts [55]. Consistent with this, there is evidence that mammalian NOCT transcription can be controlled by other transcription factors. Sequence analysis indicates the presence of binding sites for CRE [55], RevERBα[51], NFκB [10,56], and PPARγ[57]; and functional data support regulation by FoxO [58], PPARγ [57,59], STAT3 [60] and Nanog [60–62]. Furthermore, CLOCK-independent control of NOCT transcription is known in Xenopus photoreceptor cells and is mediated in part by CREB through a cis-regulatory element termed the Nocturnin Element [63,64]; however, this activity has not been extensively characterized.
The temporal alignment of peak liver NOCT mRNA abundance and mouse feeding behavior, and the increased NOCT protein and mRNA abundance in cultured cells following serum shock led to the hypothesis that NOCT may regulate gene expression in response to nutrient availability (Fig. 1) [65]. Surprisingly, the temporal profile of liver NOCT mRNA expression is not affected by fasting, nor is it altered in response to fasting followed by daytime refeeding when NOCT mRNA levels are low [55]. These data suggest that NOCT mRNA expression in liver is typically independent of nutrient availability; however, the timing of peak NOCT mRNA abundance in liver does shift when food availability is limited to daytime, suggesting NOCT mRNA levels can be regulated by nutrient availability under some circumstances [55]. Consistent with this idea, changes in NOCT mRNA abundance in response to feeding may be tissue specific. NOCT mRNA levels are constant in white adipose tissue from ad lib fed mice but become rhythmic in response to daytime feeding, and dietary fat induces NOCT mRNA expression in the proximal small intestine [19,55]. The connection between feeding and NOCT mRNA expression in certain contexts appears to be evolutionarily conserved, as several tissues in the goldfish Carassius auratus exhibit increased noc-a mRNA expression following a meal [16].
In some contexts, NOCT mRNA expression can also be induced under certain starvation conditions: in mice subject to daytime feeding, NOCT mRNA levels increase in white adipose tissue – but not liver – upon missing a feeding [55]. Starvation has also been connected with increased curled mRNA abundance in Drosophila [14], as well as time- and tissue-dependent induction of noc-b and repression of noc-a mRNA expression in C. auratus [16]. The links between NOCT expression and nutrient availability suggest a role for NOCT in modulating metabolic activity, perhaps regulating the expression of genes involved in energy uptake, storage, and utilization [9,15,16,19,55]. Whether nutrient availability and feeding regulate NOCT expression in humans remains to be explored.
NOCT has also been implicated in dietary lipid absorption and trafficking in the small intestine of mice (Fig. 1) [19]. In addition to induction in response to dietary fat, NOCT mRNA exhibits circadian fluctuations in abundance in the proximal portion of the small intestine where dietary lipids are absorbed, and nighttime NOCT protein abundance is highest in this region. Consistent with a role for NOCT in mediating lipid absorption and trafficking, intestines from NOCT-deficient mice accumulate excess lipids, and isolated enterocytes exhibit defects in lipid absorption and trafficking [19]. Together, these data indicate that NOCT activity may promote dietary lipid absorption and secretion, potentially forming the basis of NOCT knockout mouse resistance to high-fat diet-induced obesity and liver steatosis [9]; however, the mechanistic details of NOCT involvement in these processes is unclear. Moreover, the fate of unabsorbed dietary lipids is unclear as fecal lipid contents were reportedly unchanged between wild-type and NOCT knockout mice [19]. This finding remains paradoxical in light of the decreased intestinal lipid absorption and secretion [19] and reduced body temperature of NOCT knockout mice [9], as well as the absence of differences between NOCT knockout and wild-type mouse metabolic activity [9].
NOCT also functions during embryo development and later during mesenchymal stem cell differentiation (Fig. 1). Although NOCT is dispensable for mouse embryo viability, depletion of NOCT mRNA temporarily slows early development while overexpression leads to developmental arrest [66]. Later in development, STAT3 and Nanog-mediated NOCT transcription may promote stem cell pluripotency by repressing differentiation along mesoderm and endoderm lineages [60]. Developmental requirements for NOCT function have also been observed in Xenopus embryos, where manipulation of NOCT mRNA expression affects somite size, number, and organization [67,68]. Currently, NOCT’s function during Xenopus somitogenesis is unclear, as is whether NOCT is required for embryo viability. These data suggest that NOCT modulates the stability of developmentally important mRNAs in both mouse and Xenopus embryos; consequently, identification of NOCT target transcripts in embryos would provide significant insight into its function during early development.
NOCT also functions during adipose and bone tissue differentiation (Fig. 1). Adipocytes and osteoblasts are derived from a common mesenchymal stem cell precursor [69], and NOCT has been suggested to influence the balance of these cell types through its pro-adipogenic and anti-osteogenic activities. Mice lacking NOCT have reduced fat pad mass [9], and work in cultured cells demonstrated that NOCT expression promotes adipocyte differentiation from pre-adipocyte precursors and bone marrow mesenchymal stem cells [18,70]. Consistent with these roles, NOCT mRNA and protein expression increase during early and late adipogenesis in 3T3-L1 preadipocytes [18,70]. The mechanism by which NOCT promotes adipogenesis is currently unclear, but has been suggested to occur through increased nuclear entry of peroxisome proliferator-activated receptor gamma (PPARγ), the key transcriptional regulator of fat metabolism and adipogenesis, via an association between NOCT and PPARγ at the nuclear periphery [18]. Mutation of a conserved Mg2+ coordinating residue, E193, did not affect this interaction, leading the authors to conclude that this role is independent of NOCT catalytic activity [18]; however, mutation of the corresponding residue in human NOCT does not abrogate mRNA decay activity in cultured cells, indicating retention of mRNA regulatory activity by this mutant [71]. Another hypothesis for NOCT’s involvement in adipogenesis is that NOCT facilitates mitotic clonal expansion (MCE) of pre-adipocytes during adipocyte differentiation. In cultured mouse cells, NOCT depletion is associated with reduced MCE and Cyclin D1 abundance [70]; however, how NOCT depletion leads to these reductions is unclear. In addition to reduced adiposity, mice lacking NOCT exhibited increased bone mass, suggesting that NOCT negatively regulates osteogenesis [18]. Consistent with an anti-osteogenic role, NOCT mRNA and protein expression is downregulated during osteogenic differentiation, and studies in bone marrow mesenchymal stem cells demonstrated an inverse relationship between NOCT expression and osteogenic differentiation [18]. NOCT-mediated repression of osteogenesis is thought to occur in part through downregulation of osteogenic transcripts [18,72,73]. Moreover, NOCT knockout mice are protected against reductions in bone density induced by rosiglitazone (a PPARγ agonist), although it is not clear if this results from the lost induction of pro-adipogenic genes and/or lost repression of pro-osteogenic genes [73].
NOCT may also function during cellular responses to acute stimuli. Following stimulation of mouse cells with serum [65], mitogens [65], or lipopolysaccharide [56], NOCT mRNA abundance rapidly increases and displays expression dynamics similar to those observed for primary response genes [65,74,75]. Mitogen-inducible NOCT mRNA and protein expression may be unusual among deadenylases; however, only four other deadenylases have been tested for changes in mRNA abundance in response to serum stimulation, and protein levels have only been measured for one additional deadenylase [65]. The functional consequences of increased NOCT expression in these cases are almost entirely untested; however, these observations suggest that induction of NOCT expression by acute stimuli would result in degradation of cohort of mRNAs, thereby remodeling the transcriptome. Thus far, NOCT protein expression has only been linked with increased stability of inducible nitric-oxide synthase (iNOS) mRNA, and knockout mice injected with LPS exhibit increased survival compared to wild-type mice [56]; at present, the mechanisms underlying these outcomes are unstudied.
Limited data is currently available regarding NOCT function and regulation in humans. Rhythmic changes in NOCT mRNA expression are observed in synchronized human liver cells [51]; however, only two reports have suggested rhythmic expression of NOCT mRNA in human oral mucosa [76] and skeletal muscle [49]. Currently, insight into potential biological roles for human NOCT is limited to findings from genome-wide association studies (GWAS) and mRNA expression studies. One GWAS in Chinese men observed a correlation between the intronic tag SNP rs9684900 and body mass index [77]; however, it is not clear whether this SNP affects NOCT function, or whether this SNP or a closely-linked locus is the source of the association. NOCT mRNA abundance has been correlated with increased body mass index, suggesting a possible but unverified role in adipogenesis similar to that observed in mice [77]. Another GWAS study suggested a link between the intronic NOCT SNP rs3805213 and non-small cell lung cancer [78], while yet another study observed a correlation between NOCT expression and increased survival among small cell lung cancer patients [79]. These associations are interesting, but whether or not NOCT activity directly causes these phenotypes remains untested.
In contrast with transcriptional regulation, post-transcriptional control of NOCT expression has received little attention. This is surprising because circadian and non-circadian inputs result in substantial NOCT mRNA induction exceeding the subsequent increases observed for the protein [56,65], suggesting that NOCT mRNA may be subject to post-transcriptional regulation or posttranslational control of protein stability. Further, the observed oscillations in NOCT mRNA level necessitate control of mRNA decay in addition to the aforementioned transcriptional control. Indeed, a recent study indicated that NOCT mRNA levels may be regulated by both rhythmic transcription and degradation in mouse liver [80]. Thus far only the liver-specific microRNA miR-122 has been shown to fine-tune the NOCT mRNA abundance profile in mouse liver [81].
Nocturnin has unique structure and regulatory activity
Sequence alignment of CCR4 orthologs reveals that they share substantial amino acid sequence identity and similarity within the EEP catalytic domain; in contrast, the N-terminus is less conserved and is unique to NOCT orthologs (Fig. 2 and 3). At present the function of the N-terminus is unclear; however, it may be important for NOCT-specific functions including protein-protein interaction domains, and/or contain sites for posttranslational modifications that may modulate catalytic activity. Recently reported NOCT crystal structures (6BT1 and 6BT2; Fig. 4) [71] and an independently determined structure (6DIP) [82] revealed that the catalytic domain structure closely resembles the CNOT6L and PDE12 catalytic domains, which all possess the characteristic hydrolase fold found in EEP family members and are active in vitro [39,83,84]. Conserved residues that are predicted to have catalytic functions cluster in the active site (Fig. 4B), and their arrangement is nearly identical to that of corresponding residues in CNOT6L and PDE12 [71]. Together, these data suggest that NOCT possesses exoribonuclease – and potentially deadenylase – activity. Beyond the active site, the structures of CNOT6L, PDE12, and NOCT are more divergent. One striking feature is that NOCT possesses a basic patch near the active site that has been suggested to function in substrate binding (Fig. 4C, D) [71,82]; however, this hypothesis has not yet been experimentally tested.
Figure 3.
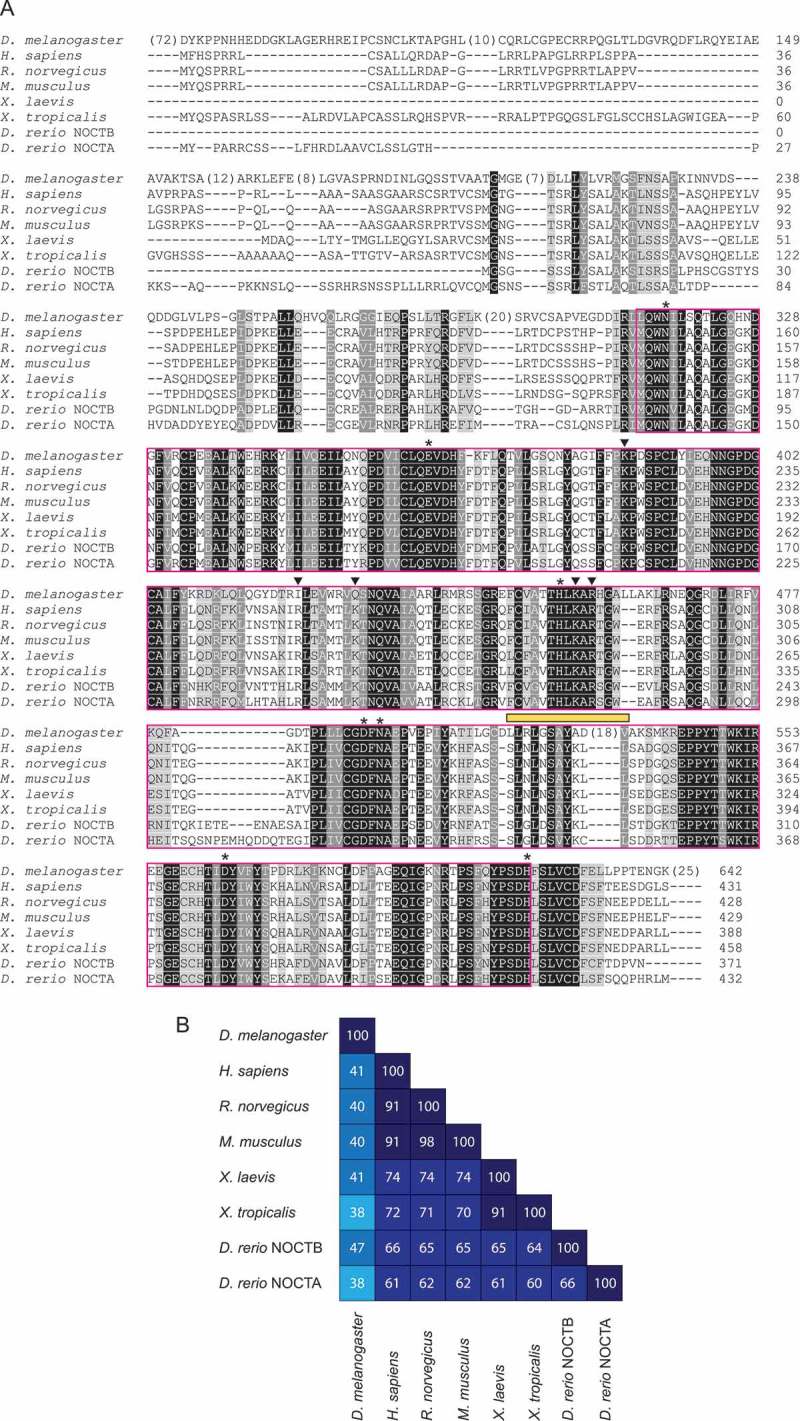
NOCT orthologs share a conserved EEP domain but have divergent N-termini. A) Amino acid sequence alignment of NOCT orthologs from various model organisms. Sequences were aligned and colored as in Fig. 2; darker shading indicates greater functional group similarity (charge, hydrophobicity, etc.). Strictly conserved positions are in white text with black background. Putative hNOCT catalytic (*) and basic patch residues (inverted triangles) are indicated. The putative PPARγ-interacting peptide is highlighted by a yellow bar. Uniprot accession numbers: D. melanogaster A8JQX3, H. sapiens Q9UK39, R. norvegicus Q9ET55, M. musculus O35710, X. laevis P79942, X. tropicalis Q28CV0, D. rerio NOCTB A0A0G2KRI0, D. rerio NOCTA E7F177. B) Pair-wise amino acid identities among NOCT orthologs.
Figure 4.
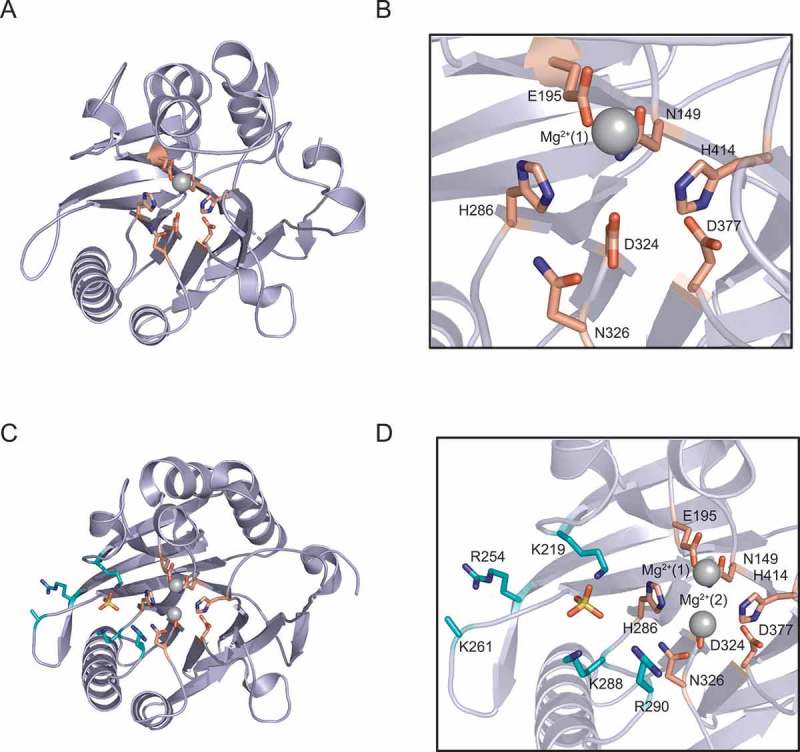
The structure of the hNOCT EEP catalytic domain (aa 120–431) reveals a conserved hydrolase fold. A) Overview of the 1.48 Å resolution structure (6BT1) [71]. Active site residues are shown; one Mg2+ ion (gray) is coordinated to E195 and ordered water molecules (not pictured). B) Magnified view of active site from (A). C) Overview of 2.48 Å resolution structure (6BT2) [71] with active site (salmon) and basic patch (cyan) residues shown. Two Mg2+ ions (gray), coordinated to E195 and D324, and one SO4 2–ion (yellow) bound to lysine side chains adjacent to the active site are shown. D) Magnified view of active site and adjacent basic patch shown in (C).
Despite the high degree of active site conservation among CNOT6L, PDE12 and NOCT (Fig. 2), new data indicate that highly purified, recombinant human and mouse NOCT are inactive against poly(A) RNA substrates in biochemical assays, whereas CNOT6L purified under the same conditions exhibited robust exonuclease activity [71]. These new observations contradict earlier studies performed using partially purified recombinant mouse and Xenopus NOCT that suggested that these enzymes catalyze degradation of poly(A) RNAs [56,65,85]. This discrepancy may be explained by differences in the purification strategies used to obtain the recombinant proteins, as RNA degradation assays are highly susceptible to RNase contamination [71]. Indeed, recent efforts to characterize mouse and human NOCT exonuclease activity under published reaction conditions were unsuccessful in detecting activity [71]. Moreover, NOCT substrate specificity remains unknown, and recombinant NOCT is not active against a range of phosphorylated substrates including nucleic acids, phospholipids, or phosphosugars [71]. Therefore, the enzymatic activity of pure recombinant NOCT in biochemical assays remains unknown.
While NOCT is not active in biochemical assays, the ability of NOCT to regulate mRNAs is supported by in vivo experiments. When experimentally directed to a target reporter mRNA in a tethered function assay in HEK293 cells, human NOCT reduced reporter protein and mRNA expression [71]. Repression of reporter protein expression is diminished by mutations in the conserved residues N149, D377, and H414, which are located in the NOCT active site (Fig. 4A, B), while mRNA degrading activity is diminished by mutation of H414 [71]. Together these data indicate that NOCT catalytic function contributes to mRNA translational repression and degradation in cells. Surprisingly, mutation of a conserved glutamate in the human NOCT active site (E195A) does not relieve reporter repression [71]. This result is unexpected because this residue is involved in Mg2+ coordination (Fig. 4) [18,65,71,85], and the corresponding residue in CNOT6L (E240) is required for catalytic activity [71,83]. The basis for this unexpected result is presently unclear, but may indicate that other residues can provide compensatory Mg2+ coordination. The observation that the NOCT E195 mutant retains RNA decay activity also is significant because mutation of the analogous residue in mouse NOCT was previously used as a tool for analyzing exoribonuclease-independent activity [18]. In light of human NOCT E195A activity, it is uncertain whether the mouse E193A mutation can accurately discriminate exoribonuclease-dependent and -independent activities, confounding the interpretation of data suggesting that NOCT performs a deadenylase-independent function [57].
NOCT activity in vivo against substrate mRNAs requires the presence of an accessible 3′ end, as NOCT does not repress expression of a reporter mRNA terminating in a highly-structured MALAT1 triple helix [71]. Lack of repression in this context, as well as the absence of RNA decay intermediates, is consistent with exoribonuclease activity. At this time it is unclear whether NOCT activity requires that the accessible 3′ end be composed of poly(A), or whether NOCT can act on any accessible 3′ sequence. In summary, cell-based analyses indicate that NOCT association with an mRNA can cause translational repression and mRNA decay, and that the active site residues contribute to these activities. Important insights will be gained by further exploring NOCT activity in vivo through the identification of its natural substrate RNAs and any protein partners that may be necessary for NOCT regulatory activity.
Do NOCT protein partners control its function?
Many RNA-degrading enzymes function in multisubunit complexes, and their enzymatic activity and substrate specificity can be dramatically altered by protein partners [13,86,87]. The predicted involvement of partner molecules – particularly RNA-binding proteins (RBPs) – in controlling NOCT activity is intriguing because such a partner could modulate the specificity and affinity of NOCT for target mRNAs as has been observed for other deadenylases (Fig. 5) [13,88]. Moreover, the discrepancy between NOCT in vivo and in vitro activity suggests that one or more protein partners is required for NOCT function; such a requirement has been observed previously for the PAN2 deadenylase [89,90]. Thus far, NOCT-interacting partners have not been comprehensively characterized, although one report using co-immunoprecipitation experiments in mammalian cells overexpressing tagged proteins demonstrated a physical interaction between mouse NOCT and PPARγ1 and PPARγ2 [18]. Subsequent analysis indicated that NOCT and PPARγ2 coexpression enhances PPARγ2 nuclear entry, and a ten amino acid region of NOCT (aa341–351) was proposed to mediate this interaction. Introduction of this peptide into cultured cells disrupts this interaction [18]; however, the NOCT-PPARγ interaction has not been directly tested via mutational analysis, nor is it clear whether this is a direct protein-protein interaction. Mapping this peptide onto the human NOCT catalytic domain structure reveals that few amino acid side chains in this peptide are surface-exposed and accessible for protein-protein interactions (Fig. 6), although structural rearrangements could theoretically increase their accessibility. Moreover, the functional effects of NOCT expression on PPARγ-regulated genes are also unclear. Interestingly, a genetic interaction was recently reported between Drosophila curled and the nuclear receptor HR4 [91], suggesting that NOCT orthologs may have a conserved role in modulating nuclear receptor-mediated gene expression; however, it is not known if this interaction occurs at the protein level.
Figure 5.
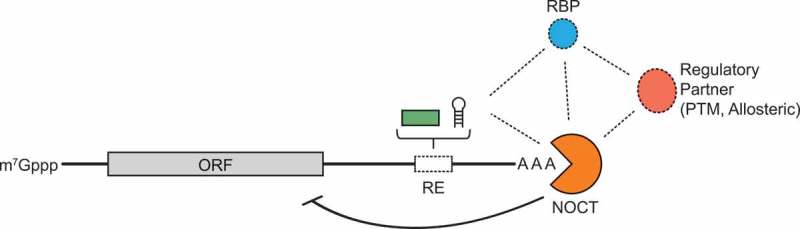
Hypothetical model for NOCT-mediated regulation of target mRNA transcripts. NOCT (orange) association with target transcripts may be direct or indirect. RNA-binding proteins (blue) and/or additional trans-acting factors (red) or post-transcriptional modifications (PTM) may be required for NOCT-mediated repression of targets. Cis-acting sequences regulatory element (RE) and/or structural elements may recruit NOCT or RNA-binding proteins (RBPs) to target transcripts. Association of NOCT complexes with substrate mRNAs represses expression of the encoded gene via RNA decay and possibly translational repression.
Figure 6.
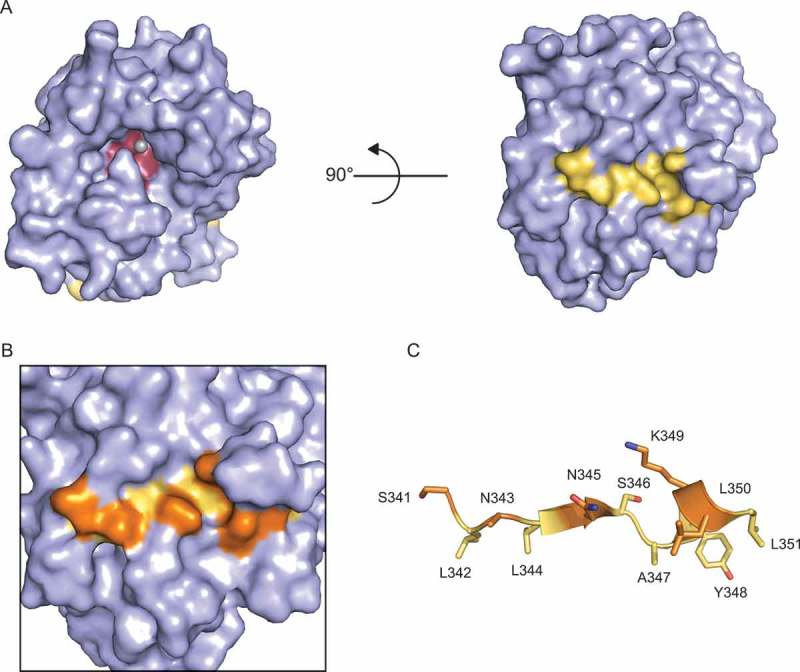
Amino acid side chains in the proposed PPARγ-interacting peptide are largely inaccessible. A) Surface rendering of NOCT 120–431 (6BT1; 1.48 Å resolution) [71]. The PPARγ-interacting peptide is highlighted in yellow and the active site is colored in red. One Mg2+ ion is coordinated in the active site. B) Magnified view of putative PPARγ-interacting peptide with solvent-accessible amino acid side chains highlighted in orange. C) Cartoon diagram view of the PPARγ-interacting peptide with solvent-exposed amino acid side chains colored in orange.
NOCT may also interact with the multimeric CNOT complex. Biochemical evidence from Drosophila and mammalian cell culture experiments using overexpressed, tagged proteins suggests that components of the CNOT deadenylase complex associate with NOCT [31,35,92]. Like NOCT knockout mice, CNOT3+/- and CNOT7-/- mice resist high-fat diet-induced obesity [33,93]; however, it is difficult to assess the extent of phenotypic similarity among these mice as they have not been directly compared under common conditions. The structural basis for any potential interaction between NOCT and members of the CNOT complex is also unclear, because NOCT lacks the leucine-rich repeats that mediate interaction of other CCR4 orthologs with the CNOT complex [11,94] (Fig. 2). Consequently, any interaction between NOCT and the CNOT complex would likely involve a novel mode of interaction and potentially represent a new subclass of CNOT complexes. An understanding of the full repertoire of NOCT-interacting proteins will be essential to our understanding of NOCT function, as such partners may control target mRNA selection and regulatory activities, as well as protein localization and stability. In the future, characterization of these interactions should include analysis of the effects of depleting these partners on NOCT-mediated regulation and whether the interaction is direct or indirect (Fig. 5).
Is NOCT activity spatially restricted?
Several deadenylases exhibit specific subcellular distribution [35,36–38,40–43], raising the question of whether NOCT activity may be spatially restricted. Studies of overexpressed tagged constructs found Xenopus NOCT to be cytoplasmic in photoreceptor cells [95], while mouse NOCT is perinuclear in human HEK293 cells [57] and is excluded from stress granules in mouse NIH3T3 cells [56]. Importantly, overexpression and the inclusion of tags could potentially lead to aberrant localization; consequently, it is important to also examine endogenous protein localization. Analysis of endogenous NOCT localization indicated both nuclear and cytoplasmic distribution in mouse embryos [66], and cytoplasmic localization in Xenopus retina [85]. Importantly, none of these studies were performed with sufficient resolution to confidently assign NOCT to a particular subcellular compartment, and its localization has not been confirmed by subcellular fractionation.
We investigated the potential for NOCT localization using sequence analysis tools to identify potential localization signals in NOCT. Analysis of the NOCT orthologs in Fig. 3 using NucPred [96], cNLS Mapper [97–99], and SeqNLS [100] tools did not predict nuclear localization, whereas PSORTII [101,102] predicted nuclear localization for Drosophila, X. laevis, and D. rerio NOCTB orthologs. Mitochondrial localization is predicted for several NOCT orthologs. PSORTII [101,102], TargetP [103], and MitoProt [104] all predict mitochondrial localization of X. tropicalis, R. norvegicus, H. sapiens, M. musculus, and D. rerio NOCTA. TargetP and MitoProt predictions indicate that D. rerio NOCTB is likely to be mitochondrial; however, this conflicts with the nuclear localization predicted by PSORTII. Neither TargetP nor MitoProt identify D. melanogaster or X. laevis NOCT as mitochondrial [103,104]. Although useful in predicting possible NOCT functions, these programs do not account for shuttling among subcellular compartments. Furthermore, it will be necessary to experimentally evaluate these predictions using multiple methods including high-resolution microscopy and cellular fractionation to examine endogenous NOCT distribution, as well as the identification and mutagenesis of localization signals.
What are the direct targets of NOCT?
A major remaining hurdle to understanding NOCT function is the identification of its target RNAs. As a putative deadenylase, NOCT is predicted to associate with target transcripts and catalyze removal of the poly(A) tail, triggering transcript decay. If NOCT functioned as a general deadenylase, NOCT depletion would be expected to result in widespread poly(A) tail length changes; however, no such effects have been observed [105], suggesting that NOCT activity is directed towards specific transcripts. Selection of target mRNAs may occur through direct target recognition by NOCT, or through an interaction with an RBP (Fig. 5). Regardless of the mechanism of target recognition, it is reasonable to expect that bona fide NOCT exoribonuclease targets should meet two general criteria regarding their abundance and association with NOCT. First, trends in target transcript abundance and/or poly(A) tail length should be inversely correlated with NOCT protein expression. Second, NOCT should associate with the target transcript in vivo either directly or indirectly via interaction with an RBP or other partner.
Initial attempts to identify NOCT targets utilized candidate-based approaches that were informed by the lean phenotype of NOCT knockout mice, and examined the rhythmicity of PPARγ and sterol regulatory element binding transcription factor 1c (srebp-1c) transcripts and the overall abundance of stearoyl-Coenzyme A desaturase 1 (scd1), sterol regulatory element binding transcription factor 1a (srebp-1a), and liver specific fatty acid binding protein 1 (l-fabp) in livers from wild-type and knockout mice fed standard and high-fat diets. Unexpectedly, none of these transcripts exhibited changes consistent with direct NOCT targeting; instead, PPARγ expression remained constant in NOCT knockout mice fed a high-fat diet, whereas scd1 and l-fabp levels were either unchanged or reduced in NOCT knockout mice fed either diet [9]. Similarly, analysis of candidate transcripts involved in regulating small intestine lipid dynamics revealed only transcripts that were either unaffected or decreased in abundance in NOCT knockout mice [19]. At present, physical associations between NOCT and these transcripts have not been tested. As the effects of NOCT-depletion on these transcripts are inconsistent with those expected of direct targets, it is likely that these transcripts are indirectly regulated by NOCT.
Other candidate-based approaches have sought to clarify NOCT pro-adipogenic and anti-osteogenic activities by identifying relevant target mRNAs. One study in mice examined Igf1 mRNA, an important factor in regulating bone density, and found that Igf1 may be regulated by NOCT in tissues outside the liver [72]. NOCT protein expression is inversely correlated with Igf1 mRNA abundance in cultured cells and in vivo, reporter constructs bearing Igf1 3ʹ untranslated region fragments were responsive to NOCT levels, and Igf1 mRNA physically associated with FLAG-tagged NOCT. Interestingly, NOCT differentially regulates Igf1 reporter constructs bearing long and short Igf1 3ʹ UTRs, indicating mRNA isoform-specific targeting. Although these data support classification of Igf1 as a NOCT target transcript, reporters bearing analogous Igf1 3ʹ UTR fragments from a different mouse strain are not regulated by NOCT, suggesting that Igf1 is not universally regulated by NOCT. Other studies of NOCT anti-osteogenic activity observed an inverse correlation between NOCT expression and abundance of alkaline phosphatase [18], osteocalcin [18,73], runt-related transcription factor 2 (Runx2) [18,73], activating transcription factor 4 (atf4) [73]; and osterix [73]; however, physical associations between NOCT and these transcripts have not been tested.
Several transcriptome-wide studies in mice have sought to identify NOCT target RNAs on the basis of differential mRNA abundance, poly(A) tail length, and circadian expression following NOCT depletion. Microarray analysis of differentiated mouse 3T3-L1 adipocytes with and without NOCT-directed shRNA identified 273 mRNAs with more than two-fold change in abundance in response to NOCT depletion; however, only 89 were upregulated in the absence of NOCT [70]. At this time, there have been no further studies validating these 89 transcripts as potential NOCT targets. Another study compared transcript abundance among mRNAs with long and short poly(A) tails in wild-type and NOCT knockout mice at times of high and low NOCT protein abundance [105]. This analysis identified 319 mRNAs with altered poly(A) tail length in NOCT knockout livers; among these transcripts, 213 had longer poly(A) tails in NOCT knockout livers at either timepoint but only ten transcripts had longer poly(A) tails coincident with peak NOCT abundance [105]. Moreover, most transcripts identified in this analysis did not exhibit circadian changes in abundance or polyadenylation in wild-type mice as might be expected of a transcript subject to circadian deadenylation and/or decay [105].
The most recent study investigating NOCT targets used RNA-seq to compare the change in abundance between minimum and maximum circadian expression in wild-type and NOCT knockout mice [106]. A group of mRNAs encoding proteins involved in cholesterol and lipid biosynthesis exhibited a greater increase in abundance in NOCT knockout livers compared with wild-type, and had a peak abundance coincident with peak NOCT protein levels. Close analysis of these transcripts revealed changes in abundance, modest changes to poly(A) tail length distribution, and one transcript with significantly altered median poly(A) tail length. Consistent with the involvement of these target genes in lipid biosynthesis, NOCT knockout mice have increased plasma triglyceride concentrations at the time of peak NOCT protein expression; in contrast, plasma cholesterol is not significantly affected. Interestingly, although expression of these mRNAs coincided with NOCT mRNA expression in response to fast and refeeding, they were not significantly more abundant in refed NOCT knockout mice as would be expected for NOCT targets. Even so, the increased abundance of these mRNAs in the absence of NOCT coincident with peak NOCT protein expression in wild-type mice establishes these transcripts as the best candidate targets to date. Future studies aimed at validating these transcripts as direct NOCT target mRNAs should include analysis of NOCT interaction with the mRNAs and measurement of NOCT mediated repression of the encoded proteins. Moreover, the cis-acting sequences and/or secondary structures within these transcripts that may serve as NOCT specificity determinants should be explored.
Despite multiple attempts to identify NOCT targets using candidate and genome-wide approaches, at this time only Igf1 meets both requirements for identification as a direct NOCT target; however, the strain-dependent nature of targeting indicates that it is not a universal target. One explanation for the apparent difficulty in identifying NOCT targets may the underlying assumption that because NOCT mRNA levels vary over the course of a day, NOCT protein levels and activity must also do so. Circadian changes in NOCT mRNA abundance have been well-documented for several species; however, NOCT protein levels are rarely examined and are less substantial than changes in mRNA abundance [56,72,81]. Additionally, whether NOCT catalytic activity is constitutive or dynamically regulated remains unknown. Alternatively, perhaps NOCT acts to degrade other RNA species. Recent reports have shown that certain deadenylases – although capable of deadenylating mRNAs – actually target noncoding RNAs [36,39]. If NOCT orthologs do indeed target mRNAs, it is likely that differential mRNA isoform and NOCT partner protein expression will intersect to produce tissue- and cell type-specific NOCT targeting; consequently, de novo identification of NOCT targets in relevant contexts will be necessary.
Concluding remarks
Over the last two decades, our understanding of NOCT function has expanded dramatically from its identification as a circadian-expressed transcript to its implication in metabolic regulation, development, and differentiation. While substantial progress has been made, many questions about NOCT’s in vivo roles remain, and human NOCT remains largely unstudied. Based on the phenotype of NOCT knockout mice, it is probable that NOCT regulates a subset of metabolically and developmentally important transcripts in relevant tissues including the digestive tract, liver, and adipose tissues. Specificity in NOCT targeting may be achieved either through direct or indirect recognition of cis-acting sequences and/or structures in target mRNAs (Fig. 5). Upon association with these targets, NOCT may inhibit mRNA translation and promote transcript decay, most likely in collaboration with additional partner proteins. At present, the identities of such specificity determinants and protein partners are unknown, and are likely to be unique to specific tissues and conditions. Furthermore, the contributions of NOCT catalytic site residues to target regulation remain unclear and will require testing within the context of endogenous NOCT repression complexes with natural substrate RNAs. Detailed analysis of factors controlling NOCT ortholog transcription, stability, and translation will also provide substantial insight into NOCT function and may aid in the design of experiments aimed at identifying NOCT targets and interacting partners. Similarly, it will be important to determine whether NOCT is subject to post-translational control, as such modifications could modulate NOCT activity and stability in response to various stimuli. Insights into these aspects of NOCT biology will not only deepen our understanding of deadenylase functions in gene regulation, but also the molecular mechanisms underlying diet-induced obesity, potentially revealing therapeutic targets for preventing and/or reducing obesity.
Funding Statement
This research was supported by [R01GM105707] from the National Institute of General Medical Sciences, National Institutes of Health to ACG; American Heart Association Predoctoral Fellowship [16PRE2670002] and NIH Chemistry-Biology Training Program Fellowship 5T32GM008597 to ETA.
Acknowledgments
We sincerely apologize to colleagues whose work we were unable to include due to space limitations. We thank Dr. Raymond Trievel and Dr. Peter Freddolino for helpful discussions and ideas pertaining to this work. We also wish to acknowledge R. Arvola, I. Enwerem, and K. McKenney for critical reading of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Schwanhausser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. [DOI] [PubMed] [Google Scholar]
- [2].Ghazalpour A, Bennett B, Petyuk VA, et al. Comparative analysis of proteome and transcriptome variation in mouse. Plos Genet. 2011;7:e1001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rabani M, Levin JZ, Fan L, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Corbett AH. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol. 2018;52:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. [DOI] [PubMed] [Google Scholar]
- [6].Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Green CB, Besharse JC. Use of a high stringency differential display screen for identification of retinal mRNAs that are regulated by a circadian clock. Brain Res Mol Brain Res. 1996;37:157–165. [DOI] [PubMed] [Google Scholar]
- [8].Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc Natl Acad Sci U S A. 1996;93:14884–14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Green CB, Douris N, Kojima S, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:9888–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dupressoir A, Barbot W, Loireau MP, et al. Characterization of a mammalian gene related to the yeast CCR4 general transcription factor and revealed by transposon insertion. J Biol Chem. 1999;274:31068–31075. [DOI] [PubMed] [Google Scholar]
- [11].Dupressoir A, Morel AP, Barbot W, et al. Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics. 2001;2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang Y, Osterbur DL, Megaw PL, et al. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev Biol. 2001;1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. [DOI] [PubMed] [Google Scholar]
- [14].Gronke S, Bickmeyer I, Wunderlich R, et al. Curled encodes the Drosophila homolog of the vertebrate circadian deadenylase Nocturnin. Genetics. 2009;183:219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Muller WE, Wang X, Grebenjuk VA, et al. Nocturnin in the demosponge Suberites domuncula: a potential circadian clock protein controlling glycogenin synthesis in sponges. Biochem J. 2012;448:233–242. [DOI] [PubMed] [Google Scholar]
- [16].Blanco AM, Gomez-Boronat M, Madera D, et al. First evidences of nocturnin in fish: two isoforms in goldfish differentially regulated by feeding. Am J Physiol Regul Integr Comp Physiol. 2017;314:R304–R312. ajpregu 00241 2017. [DOI] [PubMed] [Google Scholar]
- [17].Yang X, Fu J, Wei X. Expression patterns of zebrafish nocturnin genes and the transcriptional activity of the frog nocturnin promoter in zebrafish rod photoreceptors. Molecular Vision. 2017;23:1039–1047. [PMC free article] [PubMed] [Google Scholar]
- [18].Kawai M, Green CB, Lecka-Czernik B, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-gamma nuclear translocation. Proc Natl Acad Sci U S A. 2010;107:10508–10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Douris N, Kojima S, Pan X, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol. 2011;21:1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stubblefield JJ, Terrien J, Green CB. Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol Metab. 2012;23:326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Godwin AR, Kojima S, Green CB, et al. Kiss your tail goodbye: the role of PARN, Nocturnin, and Angel deadenylases in mRNA biology. Biochim Biophys Acta. 2013;1829:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Winkler GS, Balacco DL. Heterogeneity and complexity within the nuclease module of the Ccr4-Not complex. Front Genet. 2013;4:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Inada T, Makino S. Novel roles of the multi-functional CCR4-NOT complex in post-transcriptional regulation. Front Genet. 2014;5:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Temme C, Simonelig M, Wahle E. Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front Genet. 2014;5:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shirai YT, Suzuki T, Morita M, et al. Multifunctional roles of the mammalian CCR4-NOT complex in physiological phenomena. Front Genet. 2014;5:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Washio-Oikawa K, Nakamura T, Usui M, et al. Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J Bone Miner Res. 2007;22:1217–1223. [DOI] [PubMed] [Google Scholar]
- [28].Ogawa T, Ito C, Nakamura T, et al. Abnormal sperm morphology caused by defects in Sertoli cells of Cnot7 knockout mice. Arch Histol Cytol. 2004;67:307–314. [DOI] [PubMed] [Google Scholar]
- [29].Nakamura T, Yao R, Ogawa T, et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet. 2004;36:528–533. [DOI] [PubMed] [Google Scholar]
- [30].Lardelli RM, Schaffer AE, Eggens VR, et al. Biallelic mutations in the 3ʹ exonuclease TOE1 cause pontocerebellar hypoplasia and uncover a role in snRNA processing. Nat Genet. 2017;49:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lau NC, Kolkman A, van Schaik FM, et al. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–453. [DOI] [PubMed] [Google Scholar]
- [32].Chen C, Ito K, Takahashi A, et al. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem Biophys Res Commun. 2011;411:360–364. [DOI] [PubMed] [Google Scholar]
- [33].Takahashi A, Adachi S, Morita M, et al. Post-transcriptional stabilization of Ucp1 mRNA protects mice from diet-induced obesity. Cell Rep. 2015;13:2756–2767. [DOI] [PubMed] [Google Scholar]
- [34].Faraji F, Hu Y, Yang HH, et al. Post-transcriptional control of tumor cell autonomous metastatic potential by CCR4-NOT deadenylase CNOT7. Plos Genet. 2016;12:e1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol Cell Biol. 2007;27:1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Son A, Park JE, Kim VN. PARN and TOE1 constitute a 3ʹ end maturation module for nuclear non-coding RNAs. Cell Rep. 2018;23:888–898. [DOI] [PubMed] [Google Scholar]
- [37].Rorbach J, Nicholls TJ, Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Res. 2011;39:7750–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Poulsen JB, Andersen KR, Kjaer KH, et al. Human 2ʹ-phosphodiesterase localizes to the mitochondrial matrix with a putative function in mitochondrial RNA turnover. Nucleic Acids Res. 2011;39:3754–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pearce SF, Rorbach J, Van Haute L, et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yamashita A, Chang TC, Yamashita Y, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. [DOI] [PubMed] [Google Scholar]
- [41].Berndt H, Harnisch C, Rammelt C, et al. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. Rna. 2012;18:958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Fong KW, Li Y, Wang W, et al. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol. 2013;203:149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Anastasakis D, Skeparnias I, Shaukat AN, et al. Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development. Nucleic Acids Res. 2016;44:8908–8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ding D, Liu J, Dong K, et al. PNLDC1 is essential for piRNA 3ʹ end trimming and transposon silencing during spermatogenesis in mice. Nat Commun. 2017;8:819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Y, Guo R, Cui Y, et al. An essential role for PNLDC1 in piRNA 3ʹ end trimming and male fertility in mice. Cell Res. 2017;27:1392–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nishimura T, Nagamori I, Nakatani T, et al. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Barbot W, Wasowicz M, Dupressoir A, et al. A murine gene with circadian expression revealed by transposon insertion: self-sustained rhythmicity in the liver and the photoreceptors. Biochim Biophys Acta. 2002;1576:81–91. [DOI] [PubMed] [Google Scholar]
- [48].Menet JS, Rodriguez J, Abruzzi KC, et al. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Perrin L, Loizides-Mangold U, Chanon S, et al. Transcriptomic analyses reveal rhythmic and CLOCK-driven pathways in human skeletal muscle. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nagoshi E, Sugino K, Kula E, et al. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li R, Yue J, Zhang Y, et al. CLOCK/BMAL1 regulates human nocturnin transcription through binding to the E-box of nocturnin promoter. Mol Cell Biochem. 2008;317:169–177. [DOI] [PubMed] [Google Scholar]
- [52].Rey G, Cesbron F, Rougemont J, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biology. 2011;9:e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Oishi K, Miyazaki K, Kadota K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. [DOI] [PubMed] [Google Scholar]
- [54].Kornmann B, Schaad O, Bujard H, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biology. 2007;5:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gilbert MR, Douris N, Tongjai S, et al. Nocturnin expression is induced by fasting in the white adipose tissue of restricted fed mice. PLoS One. 2011;6:e17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Niu S, Shingle DL, Garbarino-Pico E, et al. The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PLoS One. 2011;6:e26954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kawai M, Green CB, Horowitz M, et al. Nocturnin: a circadian target of Pparg-induced adipogenesis. Ann N Y Acad Sci. 2010;1192:131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Paik JH, Kollipara R, Chu G, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hamza MS, Pott S, Vega VB, et al. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PLoS One. 2009;4:e4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bourillot PY, Aksoy I, Schreiber V, et al. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells. 2009;27:1760–1771. [DOI] [PubMed] [Google Scholar]
- [61].Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. [DOI] [PubMed] [Google Scholar]
- [62].Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. [DOI] [PubMed] [Google Scholar]
- [63].Liu X, Green CB. A novel promoter element, photoreceptor conserved element II, directs photoreceptor-specific expression of nocturnin in Xenopus laevis. J Biol Chem. 2001;276:15146–15154. [DOI] [PubMed] [Google Scholar]
- [64].Liu X, Green CB. Circadian regulation of nocturnin transcription by phosphorylated CREB in Xenopus retinal photoreceptor cells. Mol Cell Biol. 2002;22:7501–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Garbarino-Pico E, Niu S, Rollag MD, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nishikawa S, Hatanaka Y, Tokoro M, et al. Functional analysis of nocturnin, a circadian deadenylase, at maternal-to-zygotic transition in mice. J Reprod Dev. 2013;59:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Curran KL, Allen L, Porter BB, et al. Circadian genes, xBmal1 and xNocturnin, modulate the timing and differentiation of somites in Xenopus laevis. PLoS One. 2014;9:e108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Curran KL, LaRue S, Bronson B, et al. Circadian genes are expressed during early development in Xenopus laevis. PLoS One. 2008;3:e2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Pino AM, Rosen CJ, Rodriguez JP. In osteoporosis, differentiation of mesenchymal stem cells (MSCs) improves bone marrow adipogenesis. Biol Res. 2012;45:279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hee SW, Tsai SH, Chang YC, et al. The role of nocturnin in early adipogenesis and modulation of systemic insulin resistance in human. Obesity (Silver Spring). 2012;20:1558–1565. [DOI] [PubMed] [Google Scholar]
- [71].Abshire ET, Chasseur J, Bohn JA, et al. The structure of human Nocturnin reveals a conserved ribonuclease domain that represses target transcript translation and abundance in cells. Nucleic Acids Res. 2018;46(12):6257–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kawai M, Delany AM, Green CB, et al. Nocturnin suppresses Igf1 expression in bone by targeting the 3ʹ untranslated region of Igf1 mRNA. Endocrinology. 2010;151:4861–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Guntur AR, Kawai M, Le P, et al. An essential role for the circadian-regulated gene nocturnin in osteogenesis: the importance of local timekeeping in skeletal homeostasis. Ann N Y Acad Sci. 2011;1237:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Tullai JW, Schaffer ME, Mullenbrock S, et al. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem. 2007;282:23981–23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bahrami S, Drablos F. Gene regulation in the immediate-early response process. Adv Biol Regul. 2016;62:37–49. [DOI] [PubMed] [Google Scholar]
- [76].Zieker D, Jenne I, Koenigsrainer I, et al. Circadian expression of clock- and tumor suppressor genes in human oral mucosa. Cell Physiol Biochem. 2010;26:155–166. [DOI] [PubMed] [Google Scholar]
- [77].Chang YC, Chiu YF, Liu PH, et al. Genetic variation in the NOC gene is associated with body mass index in Chinese subjects. PLoS One. 2013;8:e69622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Couto P, Miranda D, Vieira R, et al. Association between CLOCK, PER3 and CCRN4L with non-small cell lung cancer in Brazilian patients. Mol Med Rep. 2014;10:435–440. [DOI] [PubMed] [Google Scholar]
- [79].Maragozidis P, Papanastasi E, Scutelnic D, et al. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Mol Cancer. 2015;14:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Wang J, Symul L, Yeung J, et al. Circadian clock-dependent and -independent posttranscriptional regulation underlies temporal mRNA accumulation in mouse liver. Proc Natl Acad Sci U S A. 2018;115:E1916–E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kojima S, Gatfield D, Esau CC, et al. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:e11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Estrella MA, Du J, Korennykh A. Crystal structure of human nocturnin catalytic domain. BioRxiv [Preprint] 2018. June 5; 330514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang H, Morita M, Yang X, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J. 2010;29:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wood ER, Bledsoe R, Chai J, et al. The role of phosphodiesterase 12 (PDE12) as a negative regulator of the innate immune response and the discovery of antiviral inhibitors. J Biol Chem. 2015;290:19681–19696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Baggs JE, Green CB. Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol. 2003;13:189–198. [DOI] [PubMed] [Google Scholar]
- [86].Jonas S, Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Gene Dev. 2013;27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. [DOI] [PubMed] [Google Scholar]
- [88].Van Etten J, Schagat TL, Hrit J, et al. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–36383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Brown CE, Tarun SZ Jr., Boeck R, et al. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wolf J, Valkov E, Allen MD, et al. Structural basis for Pan3 binding to Pan2 and its function in mRNA recruitment and deadenylation. EMBO J. 2014;33:1514–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Ou Q, Zeng J, Yamanaka N, et al. The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep. 2016;16:247–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Temme C, Zhang L, Kremmer E, et al. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA. 2010;16:1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Morita M, Oike Y, Nagashima T, et al. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/- mice. EMBO J. 2011;30:4678–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Mittal S, Aslam A, Doidge R, et al. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol Biol Cell. 2011;22:748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Baggs JE, Green CB. Functional analysis of nocturnin: a circadian clock-regulated gene identified by differential display. Methods Mol Biol. 2006;317:243–254. [DOI] [PubMed] [Google Scholar]
- [96].Brameier M, Krings A, MacCallum RM. NucPred–predicting nuclear localization of proteins. Bioinformatics. 2007;23:1159–1160. [DOI] [PubMed] [Google Scholar]
- [97].Kosugi S, Hasebe M, Entani T, et al. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem Biol. 2008;15:940–949. [DOI] [PubMed] [Google Scholar]
- [98].Kosugi S, Hasebe M, Matsumura N, et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. [DOI] [PubMed] [Google Scholar]
- [99].Kosugi S, Hasebe M, Tomita M, et al. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106:10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lin JR, Hu J. SeqNLS: nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS One. 2013;8:e76864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. [DOI] [PubMed] [Google Scholar]
- [102].Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Emanuelsson O, Nielsen H, Brunak S, et al. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. [DOI] [PubMed] [Google Scholar]
- [104].Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. [DOI] [PubMed] [Google Scholar]
- [105].Kojima S, Gendreau KL, Sher-Chen EL, et al. Changes in poly(A) tail length dynamics from the loss of the circadian deadenylase Nocturnin. Sci Rep. 2015;5:17059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Stubblefield JJ, Gao P, Kilaru G, et al. Temporal control of metabolic amplitude by nocturnin. Cell Rep. 2018;22:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Goodstadt L, Ponting CP. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–846. [DOI] [PubMed] [Google Scholar]
- [109].Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]


