ABSTRACT
A key step in pre-mRNA splicing is the recognition of 3ʹ splicing sites by the U2AF large and small subunits, a process regulated by numerous trans-acting splicing factors. How these trans-acting factors interact with U2AF in vivo is unclear. From a screen for suppressors of the temperature-sensitive (ts) lethality of the C. elegans U2AF large subunit gene uaf-1(n4588) mutants, we identified mutations in the RNA binding motif gene rbm-5, a homolog of the tumor suppressor gene RBM5. rbm-5 mutations can suppress uaf-1(n4588) ts-lethality by loss of function and neuronal expression of rbm-5 was sufficient to rescue the suppression. Transcriptome analyses indicate that uaf-1(n4588) affected the expression of numerous genes and rbm-5 mutations can partially reverse the abnormal gene expression to levels similar to that of wild type. Though rbm-5 mutations did not obviously affect alternative splicing per se, they can suppress or enhance, in a gene-specific manner, the altered splicing of genes in uaf-1(n4588) mutants. Specifically, the recognition of a weak 3ʹ splice site was more susceptible to the effect of rbm-5. Our findings provide novel in vivo evidence that RBM-5 can modulate UAF-1-dependent RNA splicing and suggest that RBM5 might interact with U2AF large subunit to affect tumor formation.
KEYWORDS: RNA splicing, U2AF, RBM5, 3ʹ splice site, transcriptome
Introduction
Pre-mRNA splicing (RNA splicing) is a highly regulated process that generates mature mRNAs in eukaryotic cells by removing intervening non-coding introns from pre-mRNA transcripts and simultaneously joins coding exons [1,2]. RNA splicing is carried out in a coordinated two-step trans-esterification reaction that is catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs) [1–3], and facilitated by hundreds of splicing factors [4]. Using different 5ʹ or 3ʹ splice sites and by including or skipping introns and exons from a single form of pre-mRNA transcript, alternative splicing can generate multiple distinct mRNA isoforms, which increases the proteome size of an organism and is considered a driving force for the biological complexity of metazoans [1,5,6]. Defects in RNA splicing are the causes of numerous human diseases [7,8].
RNA splicing are affected by numerous trans splice factors, cis splice elements, transcription, chromatin structures and histone modifications [1,4,9]. Hundreds of protein factors related to RNA splicing have been identified [10–13]. However, it is unclear how these factors interact to affect the splicing of various genes.
RNA recognition motif (RRM) is an abundant protein domain that can bind both RNAs and proteins [14]. The RNA-binding motif protein 5 gene (RBM5/LUCA-15/H37) encodes a protein containing two RRMs, two zinc fingers, one arginine-serine-rich domain (RS) and one glycine patch (G-patch) [15]. RBM5 and its paralog RBM6 are located in the human chromosome 3p21.3 region and were identified as candidate tumor suppressors of lung cancers and other solid tumors [16–22]. RBM10, another paralog of RBM5, is also frequently mutated in lung adenocarcinoma samples [23]. In mice and human, RBM5 can reduce lung cancer progression [24,25].
RBM5 can increase the expression of proapoptotic protein BAX and reduce that of anti-apoptotic proteins including BCL-2 and BCL-XL [26–28]. It could also affect the expression of other apoptosis and cell cycle genes [29]. RBM5 might regulate the splicing of apoptosis- and cancer-related genes by affecting the recognition of 3ʹ splice sites (3ʹ SS) [30–33]. The in vivo mechanism underlying the function of RBM5 is unclear.
C. elegans is a model organism for studying a variety of biological processes [34]. We previously identified mutations affecting the C. elegans orthologs of the U2AF large subunit (UAF-1 in C. elegans, U2AF65 or U2AF2 in mammals), splicing factor one (SFA-1) [35] and the splicing factor microfibrillar associated protein 1 (MFAP-1) [36] by screening for essential gene suppressors of a unc-93(gf) rubberband phenotype [37]. Studies of these mutants provided new insights into the in vivo regulation of alternative splicing [35,36,38] and suggest a previously unknown interaction of uaf-1 with the C. elegans spinal muscular atrophy gene smn-1 in affecting locomotion and lifespan [39]. The uaf-1(n4588) mutation causes temperature-sensitive (ts) lethality [35]. A screen for suppressors of the ts-lethal phenotype identified intragenic suppressors of uaf-1 and potentially extragenic suppressors [35].
In this study, we describe the genetic and molecular characterization of two extragenic suppressors of uaf-1(n4588). We found that they affect the C. elegans rbm-5. We also analyzed how rbm-5 and uaf-1 interact to affect gene expression and alternative splicing.
Results
The uaf-1(n4588) mutation causes embryonic lethality and sterility at high temperatures
We previously isolated the uaf-1(n4588) missense mutation from a screen for essential gene suppressors of the ‘rubberband’ Unc phenotype caused by the unc-93(e1500) gain-of-function mutation [35]. uaf-1(n4588) mutants were inviable at 25 °C and grew like wild type at 15 °C [35]. At 20 °C, uaf-1(n4588) mutants grew slightly slower and have partial protruding vulva (Pvl) and/or sterility (Ste) phenotypes (Figure S1). We also found that uaf-1(n4588) mutants were inviable at 22.5 °C (this study).
To examine how temperatures affect the embryonic development of uaf-1(n4588) mutants, eggs laid at 15 °C were shifted to 20 °C, 22.5 °C or 25 °C. With these treatments, most eggs would hatch in 24 hrs (Figure 1(a)). We next shifted L4 uaf-1(n4588) mutants grown at 15 °C to higher temperatures, allowing them to grow into adults and lay eggs for 24 hrs. Most eggs laid at 20 °C and 22.5 °C would hatch, while only ~ 10% eggs laid at 25 °C did so (Figure 1(b)).
Figure 1.
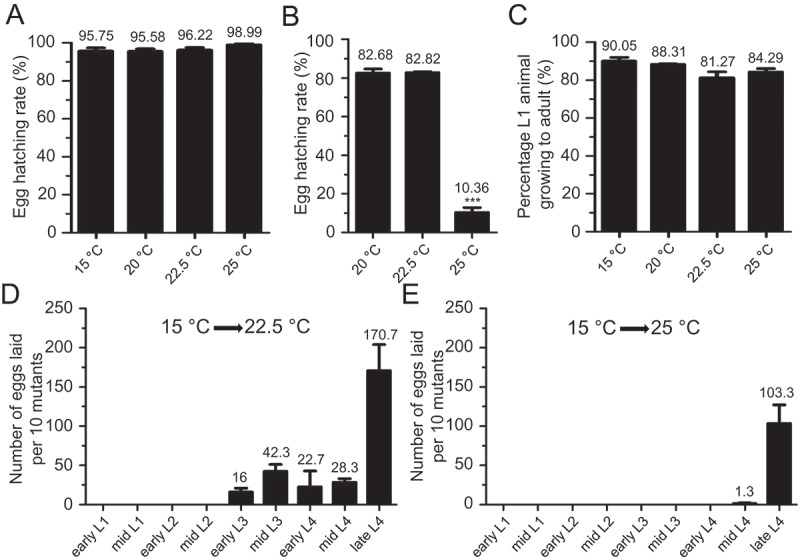
uaf-1(n4588) mutants exhibited embryonic lethality and sterility at high temperatures.
(a) uaf-1(n4588) mutant eggs laid at 15 °C were shifted to higher temperatures and the hatching rates were quantified. (b) L4 uaf-1(n4588) animals grown at 15 °C where shifted to higher temperatures, allowed to lay eggs for 24 hrs, and the egg-hatching rates were quantified. (c) L1 uaf-1(n4588) animals grown at 15 °C were shifted to higher temperatures and the percentages of animals that grew to adults were quantified. (d, e) uaf-1(n4588) mutants of various developmental stages grown at 15 °C were shifted to 22.5 °C or 25 °C, respectively, and allowed to develop to the mid L4 stage. 10 L4 animals were picked to new plates and allowed to grow and lay eggs for 24 hrs. The numbers of eggs were counted. Results were from 3 to 5 biological replicates. For A, B and C, 90 to 150 eggs or L1 animals were analyzed in each biological replicate. The value of each column is on top. Statistics: paired two-tailed Student’s t-test. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
To understand how high temperatures affect the larval development of uaf-1(n4588) mutants, we shifted L1 larva grown at 15 °C to higher temperatures, at which most larvae could grow into adults (Figure 1(c)). However, adults growing up at 22.5 °C and 25 °C were sterile.
To determine whether high temperatures affect specific developmental stages in causing sterility, we grew uaf-1(n4588) mutants to various larval stages at 15 °C and shifted them to 22.5 °C or 25 °C, allowing the larvae to grow into adults and lay eggs. When shifted to 22.5 °C, L2 or younger larvae would become sterile adults (Figure 1(d)), while at 25 °C mid-L4 or younger larvae would become so (Figure 1(e)). Together these results suggest that uaf-1(n4588) might cause lethality by affecting both embryonic development and fertility at high temperatures.
Extragenic mutations can partially suppress the sterility and embryonic lethality of uaf-1(n4588) mutants at high temperatures
A previous screen for suppressors of uaf-1(n4588) ts-lethality at 25 °C identified four intragenic suppressors of uaf-1 [35]. From the screen we also isolated seven potential suppressors that might be caused by extragenic mutations [35]. When reexamined, two isolates (n5130, n5131) exhibited extremely slow growth and severe sterility and therefore were not further analyzed.
We examined the five healthier extragenic suppressors (n5119, n5121, n5122, n5128 and n5132) in more details. At all temperatures, these mutations can strongly suppress the egg-laying defect of uaf-1(n4588) mutants (Figure 2(a-c)). At 20 °C, uaf-1(n4588) did not cause an apparent egg-hatching defect and the hatching rates were similar between uaf-1(n4588) single and uaf-1(n4588); sup double mutants (Figure 2(a’)). At 22.5 °C, uaf-1(n4588) caused a slightly defective egg-hatching rate, which was suppressed by n5119, n5121, n5122 and n5132 (Figure 2(b’)). At 25 °C, uaf-1(n4588) mutants had a severe egg-hatching defect, which was strongly suppressed by all five mutations (Figure 2(c’)). In addition, the Pvl and Ste phenotypes of uaf-1(n4588) mutants were also strongly suppressed by these mutations at 20 °C (Figure S1).
Figure 2.
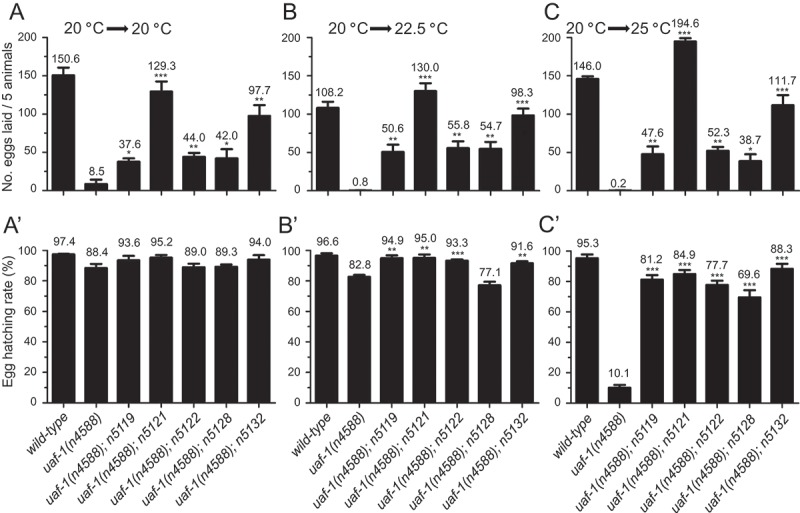
n5119, n5121, n5122, n5128 and n5132 can suppress the fertility and egg-hatching defects of uaf-(n4588) mutants at high temperatures.
Synchronized L4 animals grown at 20 °C were kept at 20 °C or moved to 22.5 °C and 25 °C. Eggs laid in 24 hours were counted and the hatching rates of the eggs 24 hrs after were quantified. (a, a’) At 20 °C, uaf-1(n4588) mutants laid significantly fewer eggs, which was improved in five double mutants. Most eggs laid by uaf-1(n4588) single mutants or the double mutants could hatch. (b, b’) Egg-laying and egg-hatching rates at 22.5 °C. The phenotypes are similar to those at 20 °C. (c, c’) Egg-laying and egg-hatching rates at 25 °C. The total numbers of eggs laid by five animals (a, b, c) and the hatching rates of these eggs (a’, b’, c’) are indicated on top of each column. Genotypes are indicated at the bottom. Results were based on 3 to 6 biological replicates. Because uaf-1(n4588) mutants lay few eggs, we used a massive number of mutants to generate enough eggs for the egg-hatching analysis in A’, B’ and C’. Statistics: paired two-tailed Student’s t-test. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
At 22.5 °C, uaf-1(n4588) single mutants would become sterile and eventually die, while uaf-1(n4588); sup double mutants were fertile and could propagate. At 25 °C, both uaf-1(n4588) single mutants and uaf-1(n4588); sup double mutants would die but the double mutants appeared healthier than the single mutants. Therefore, these mutations are partial suppressors of uaf-1(n4588) probably isolated as escapers in the original screen, which was performed at 25 °C [35].
Using visible genetic markers, we mapped n5119 and n5132 to Chr. I, n5121 and n5128 to Chr. III, and n5122 to Chr. V. n5119 might be a dominant suppressor, since both n5119/+ and n5119/n5119 could suppress the ts-lethality of uaf-1(n4588) at 22.5 °C. n5132 appeared to be a recessive suppressor because n5132/n5132 could suppress the ts-lethality of uaf-1(n4588) at 22.5 °C, while n5132/+ failed to do so. In this study, we analyzed the gene affected by n5119 and n5132.
Mutations in the RNA-binding motif protein 5 (RBM) gene rbm-5 can suppress the ts-lethality of uaf-1(n4588) mutants
To identify genes affected by n5119 and n5132, we used whole-genome sequencing to survey the coding sequences and splice sites of genes in the two mutants (see Materials and Methods). From the filtered sequence variants, we identified 12 candidate genes for n5119 and 5 for n5132 on Chr. I. Interestingly the RNA-binding motif protein gene rbm-5 (T08B2.5) is the only candidate mutated in both n5119 and n5132 mutants (Figure 3(a) and supplementary Table S1).
Figure 3.
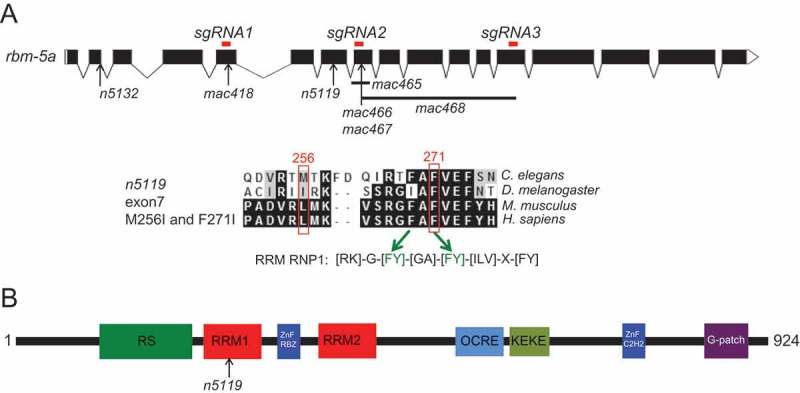
rbm-5a gene structure, RBM-5A protein domains and positions of rbm-5 mutations.
(a) Exon-intron structures of the rbm-5a isoform (designed using the Exon-Intron Graphic Maker software at www.wormweb.org according to gene sequences at www.wormbase.org). Exonic regions targeted by the CRISPR/Cas9 method are labeled on top as red bars. n5132 and n5119 mutations are indicated. The conserved RRM RNP1 sequence is shown, in which the F271I mutation corresponds to the second [FY] residue [43,44]. mac418 was obtained using sgRNA1, mac465, mac466 and mac467 were obtained using sgRNA2. mac468 was obtained using sgRNA2 and sgRNA3. (b) RBM-5 protein domains and positions of the n5119 missense mutation.
rbm-5 encodes the C. elegans homolog of the mammalian RBM5/Luca-15/H37 [15] (Figure S2). RBM5 was frequently deleted or mutated in lung cancers [19] and can affect alternative splicing, cell proliferation and apoptosis [19,30,32,40]. In C. elegans, the function of rbm-5 is unclear. In the WormBase (www.wormbase.org, WS258), nine rbm-5 transcript isoforms (rbm-5a, rbm-5b, rbm-5c, rbm-5d, rbm-5e, rbm-5f.1, rbm-5f.2, rbm-5g.1 and rbm-5g.2) are annotated (Figures 3(a) and S3), representing seven distinct encoding isoforms (rbm-5a, b, c, d, e, f and g) (Figures 3(a) and S3). In n5119 mutants, the less conserved Met256 (L131 in human) and the conserved Phe271 (F144 in human) encoded by rbm-5a are changed to isoleucines (Figures 3(a) and S2). Met256 and Phe271 are two residues in the first RNA recognition motif of RBM-5A (Figures 3(b) and S2), implying that the changes might affect RNA binding by the RBM-5 protein. In n5132 mutants, the 5ʹ splice site in rbm-5a intron 2 was mutated from gt to at (Figures 3(a) and S3). This mutation is predicted to affect the splicing of at least five isoforms (rbm-5a, b, d, e and f.1) (Figures 3(a) and S3).
To verify that rbm-5 mutations can suppress the ts-lethality of uaf-1(n4588) mutants, we used a modified CRISPR/Cas9 method [41] to generate new mutations in rbm-5 (Figure 3(a), sgRNA1, sgRNA2, sgRNA3). By this approach, we obtained five deletion mutations in rbm-5 (Figure 3(a) and Table S2). mac418 and mac467, both 4-bp deletions, affect exons 5 and 8 and were predicted to cause frameshifts (Figure 3(a) and Table S2). The mac465 deletion (98 bp) spans from intron 7 and exon8 and was predicted to disrupt the splicing and coding of rbm-5a (Figure 3(a) and Table S2). mac466 is a tri-nucleotide deletion predicted to cause a single amino acid deletion in RRM1 (Figure 3(a) and Table S2). mac468 is a 940-bp deletion spanning from exon 8 to exon 13 and was predicted to cause a 238-amino acid deletion if rbm-5 pre-mRNA was spliced correctly (Figure 3(a) and Table S2). Except mac466, the other four mac alleles (mac418, mac465, mac467, mac468) are likely null mutations by disrupting the splicing of rbm-5 and/or generating truncated RBM-5 proteins (Figure 3(a)). We found that homozygous mutants of mac465 and mac468 were viable and exhibited no obvious defects (Table 1), suggesting that rbm-5 is not essential for C. elegans survival.
Table 1.
rbm-5 mutations can suppress the ts-lethality of uaf-1(n4588) mutants at 22.5°C.
| Strains | Survival at 22.5 °C | Survival at 25 °C |
|---|---|---|
| uaf-1(n4588) | No | No |
| rbm-5(n5119) | Yes | Yes |
| rbm-5(n5132) | Yes | Yes |
| rbm-5(mac465) | Yes | Yes |
| rbm-5(mac468) | Yes | Yes |
| rbm-5(n5119); uaf-1(n4588) | Yes | No |
| rbm-5(n5119)/+; uaf-1(n4588) | Yes | No |
| rbm-5(n5132); uaf-1(n4588) | Yes | No |
| rbm-5(n5132)/+; uaf-1(n4588) | No | No |
| rbm-5(mac418); uaf-1(n4588) | Yes | No |
| rbm-5(mac465); uaf-1(n4588) | Yes | No |
| rbm-5(mac466); uaf-1(n4588) | No | No |
| rbm-5(mac467); uaf-1(n4588) | Yes | No |
| rbm-5(mac468); uaf-1(n4588) | Yes | No |
| uaf-1(n4588 n5127) | Yes | Yes (sick) |
| rbm-5(n5119); uaf-1(n4588 n5127) | Yes | Yes |
| rbm-5(n5132); uaf-1(n4588 n5127) | Yes | Yes |
mac418, mac465, mac467 and mac468 all could recessively suppress the ts-lethality of uaf-1(n4588) mutants at 22.5 °C (Table 1). mac466 failed to suppress uaf-1(n4588) (Table 1), suggesting that this mutation does not disrupt RBM-5 function severely enough. None of the mac alleles could suppress the lethality of uaf-1(n4588) mutants at 25 °C (Table 1). We found that mac418 failed to complement n5132 in suppressing the ts-lethality of uaf-1(n4588) mutants, suggesting that n5132 also causes a loss of function in rbm-5.
Both n5119 heterozygous and homozygous mutations can suppress uaf-1(n4588) (Table 1). We speculate that n5119 might function by a dominant-negative or haploinsufficient mechanism. n5119 carries two missense mutations in the RRM1 domain of RBM-5A that change the less conserved M256 and the conserved F271 to isoleucine (Figure 3(a)). F271 is comparable to F199 of the human U2AF large subunit RRM1 [42,43], a key aromatic amino acid essential for RNA binding in most RRMs (Figure 3(a), RRM RNP1) [43,44]. An F199A mutation can weaken the interaction of U2AF large subunit RRM1 with polypyrimidine consensus sequence by 9 folds [43]. Hence, it is possible that the n5119 F271I mutation caused reduced or altered affinity of RBM-5 for its RNA targets.
We previously isolated uaf-1(n5127) as an intragenic suppressor of uaf-1(n4588) that enables the animals to survive at 25 °C in an unhealthy state [35]. n5119 and n5132 can completely suppress the unhealthiness of uaf-1(n4588 n5127) mutants at 25 °C (Table 1), suggesting that these mutations can interact with uaf-1 in an allele-independent manner.
rbm-5 functions in neurons to affect the ts-lethality of uaf-1(n4588) mutants
Using an 1183-bp rbm-5 endogenous promoter (see Materials and Methods) to drive GFP expression, we observed fluorescent signals primarily in multiple head neurons, ventral nerve cord, the CAN neuron, an intestinal cell and a muscle cell in the posterior part of the transgenic animals (Figure 4(a-d)). However, we failed to obtain stable transgenic lines in the wild-type, n5119 or n5132 background using an rbm-5a cDNA transgene, probably due to the toxicity of the transgene. To overcome this problem, we used the ubiquitous eft-3 promoter [45] to drive an rbm-5a cDNA::GFP fusion transgene and obtained stable transgenic lines. The RBM-5A::GFP fusion protein was exclusively localized in the nuclei of the numerous visible cells (Figure 4(e,f)), suggesting that RBM-5 is a nuclear protein.
Figure 4.
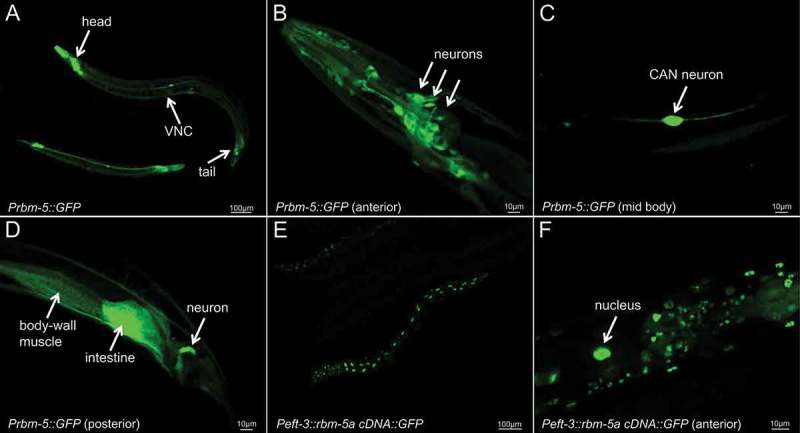
Fluorescent pictures of animals expressing a Prbm-5::GFP transcriptional fusion transgene and a Peft-3::rbm-5a cDNA::GFP translational fusion transgene.
(a) A low resolution picture of Prbm-5::GFP transgenic animals showing GFP expression in the anterior, posterior and ventral nerve cord (VNC). High resolution picture of the anterior (b), mid body (c) and posterior (d) of a Prbm-5::GFP transgenic animal. Low (e) and high (f) resolution pictures of a Peft-3::rbm-5a cDNA::GFP transgenic animal showing that the RBM-5A::GFP fusion protein was exclusively localized in the nuclei. We determined the identities of Prbm-5::GFP-positive cells by examining their anatomical positions, morphologies, patterns and neighboring cells based on the descriptions at www.wormatlas.org.
The Peft-3::rbm-5a transgene itself does not affect the survival of animals at different temperatures (Table 2). When placed in the rbm-5(mac468); uaf-1(n4588) background, this transgene caused slow growth and reduced fertility at 15 °C, while it led to lethality at 20 °C and 22.5 °C (Table 2). The transgene further caused lethality in the uaf-1(n4588) single mutant background even at 15 °C (Table 2). These results suggest that the Peft-3::rbm-5a transgene can rescue the suppression of the uaf-1(n4588) ts-lethality by the rbm-5(mac468) lf mutation and rbm-5 overexpression can enhance the lethality caused by uaf-1(n4588).
Table 2.
The rbm-5a cDNA::GFP transgene under control of the ubiquitous eft-3 promoter or the neuron-specific unc-119 promoter could rescue the suppression of the uaf-1(n4588) ts-lethality by the rbm-5(mac468) mutation.
| Strains | Survival at 15 °C | Survival at 20 °C | Survival at 22.5 °C |
|---|---|---|---|
| uaf-1(n4588) | Yes | Yes (sick) | No |
| rbm-5(mac468); uaf-1(n4588); | Yes | Yes | Yes |
| Peft-3::rbm-5a cDNA::GFP Tg #1 | Yes | Yes | Yes |
| Peft-3::rbm-5a cDNA::GFP Tg #2 | Yes | Yes | Yes |
|
rbm-5(mac468); uaf-1(n4588); Peft-3::rbm-5a cDNA::GFP Tg #1 |
Yes (sick) | No | No |
|
rbm-5(mac468); uaf-1(n4588); Peft-3::rbm-5a cDNA::GFP Tg #2 |
Yes (sick) | No | No |
|
uaf-1(n4588); Peft-3::rbm-5a cDNA::GFP Tg #1 |
No | No | No |
|
uaf-1(n4588); Peft-3::rbm-5a cDNA::GFP Tg #2 |
No | No | No |
| Punc119::rbm-5a cDNA::GFP Tg #1 | Yes (sick) | Yes (sick) | Yes (sick) |
| Punc119::rbm-5a cDNA::GFP Tg #2 | Yes (sick) | Yes (sick) | Yes (sick) |
| rbm-5(mac468); uaf-1(n4588); Punc119::rbm-5a cDNA::GFP Tg #1 | Yes (sick) | No | No |
| rbm-5(mac468); uaf-1(n4588); Punc119::rbm-5a cDNA::GFP Tg #2 | Yes (sick) | No | No |
Since the rbm-5 endogenous promoter drives reporter expression in multiple neurons (Figure 4(a-d)), we also tested whether neuron-specific expression of rbm-5 could affect the ts-lethality of uaf-1(n4588) mutants by introducing an rbm-5a transgene under control of the neuron-specific unc-119 promoter [46,47]. In wild-type background, the Punc-119::rbm-5a::GFP transgene caused slow growth and apparently reduced fertility at all temperatures tested (Table 2). When placed in the rbm-5(mac468); uaf-1(n4588) background, the transgene led to lethality at 20 °C and 22.5 °C, similar to the effect of the Peft-3::rbm-5a transgene (Table 2). These results suggest that rbm-5 functions in neurons to promote the deleterious phenotypes caused by uaf-1(n4588).
rbm-5 mutations can reduce the number of differentially expressed genes and reverse some altered gene expression in uaf-1(n4588) mutants to levels similar to wild type
Mammalian RBM5 can affect the splicing of different genes [30–32,40]. We examined whether rbm-5 mutations can affect RNA splicing in C. elegans and how rbm-5 interacts with uaf-1 in doing so.
We performed whole transcriptome shotgun sequencing (RNA-Seq) experiments and compared the embryonic transcriptomes of wild-type, uaf-1(n4588) single mutants and rbm-5(mut); uaf-1(n4588) double mutants grown at 20 °C. At 20 °C, uaf-1(n4588) could produce enough eggs for extracting total RNAs.
We found that rbm-5 mutations only affected the expression of a small number of genes (3 up, 0 down, 3 total for n5119; 9 up, 13 down, 22 total for n5132) in embryos (Figure 5(a)). The number of genes affected was dramatically increased to 194 in uaf-1(n4588) embryos (120 up and 74 down) (Figure 5(a)). Interestingly, rbm-5 mutations significantly reduced this number. For example, rbm-5(n5119); uaf-1(n4588) double mutants had 74 genes affected (56 up and 18 down) and rbm-5(n5132); uaf-1(n4588) had 52 affected (36 up and 16 down) (Figure 5(a)).
Figure 5.
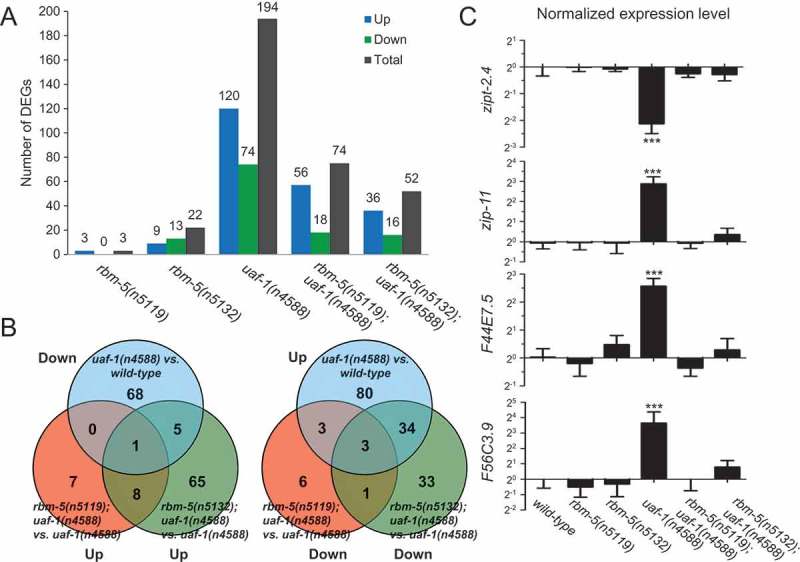
RNA-Seq identified genes differentially expressed in uaf-1(n4588) and rbm-5; uaf-1(n4588) mutants.
(a) Numbers of up- and down-regulated genes in mutants compared to wild type. Genotypes are at bottom. (b) Venn diagram comparison of genes down-regulated in uaf-1(n4588) mutants and genes up-regulated in rbm-5; uaf-1(n4588) double mutants compared to uaf-1(n4588) single mutants, and vice versa. (c) qRT-PCR verification of the expression of four genes predicted by RNA-Seq to be altered by uaf-1(n4588) that were reversed by rbm-5 mutations. Statistics: Bonferroni test with one-way ANOVA. ***: p < 0.001.
KEGG (www.genome.jp/kegg/) enrichment analyses of the differentially expressed genes (DEGs) suggest that genes in 40 pathways are affected by uaf-1(n4588) (Table S3). These pathways include the metabolism of multiple biomolecules, diseases, synapse functions, signaling pathways and spliceosome, etc, suggesting that uaf-1 might affect embryonic development by regulating the expression and/or splicing of genes primarily involved in these processes. Compared to wild type, rbm-5(n5119) single mutation did not alter gene expression in any of these pathways (Table S3). rbm-5(n5132) single mutation only affected gene expression in four pathways, including chemical carcinogenesis, cytochrome P450-related metabolism of xenobiotics and drugs, and Parkinson’s disease (Table S3).
Analyses of double mutants suggest that rbm-5(n5119) could reverse the gene expression in 35 of the 40 pathways affected by uaf-1(n4588) to levels similar to that of wild type. rbm-5(n5132) could reverse the gene expression in 16 of the 35 pathways (Table S3). Though it is unclear how these pathways are involved in C. elegans embryonic development, it is plausible that the corrected expression/splicing of genes in these pathways can sufficiently mitigate the deleterious effects of uaf-1(n4588).
We further looked into the DEGs (> 2-fold) in uaf-1(n4588) and compared that to rbm-5; uaf-1(n4588) double mutants (Result S1). Among 194 genes affected by uaf-1(n4588), rbm-5(n5119) reversed the expression of 144 (including 78 up-regulated and 66 down-regulated genes) to levels closer to that of wild type, while rbm-5(n5132) reversed that of 177 genes (including 111 up-regulated and 66 down-regulated ones) to levels closer to that of wild type (Result S1).
We identified four DEGs in uaf-1(n4588) mutants that were reversed by both rbm-5 mutations (Figure 5(b)), including zipt-2.4 (down in n4588), zip-11 (up), F44E7.5 (up) and F56C3.9 (up). qRT-PCR verified altered expression of these genes in uaf-1(n4588) mutants and the reversion by rbm-5 mutations as predicted by RNA-seq (Figure 5(c)).
rbm-5 can modulate the altered splicing caused by uaf-1(n4588)
To understand how rbm-5 interacts with uaf-1(n4588) in affecting alternative splicing, we used RT-PCR to examine 80 genes alternatively spliced in uaf-1(n4588) mutants with the highest statistical significance predicted by RNA-Seq (Table S4). Among these genes, only three (flp-1, mbk-2, zipt-2.4) were verified (Figure 6(a)). The splicing of these genes represents three distinct patterns: alternative 3ʹ SS (flp-1), exon skipping (mbk-2), and alternative 5ʹ SS (zipt-2.4) (Figure 6(a)). rbm-5 mutations could apparently suppress the altered splicing of mbk-2 and zipt-2.4 in uaf-1(n4588) mutants but not that of flp-1 (Figure 6(a)).
Figure 6.
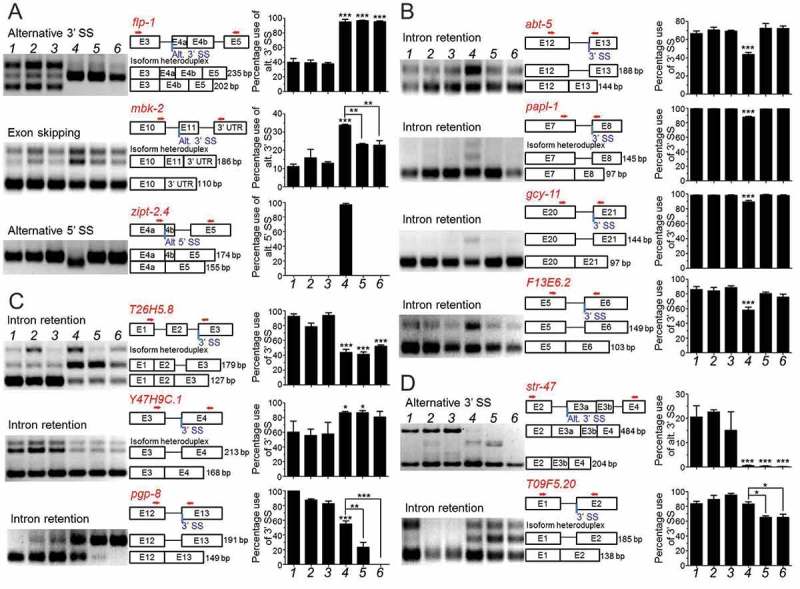
RT-PCR analyses of genes with altered splicing in uaf-1(n4588) single and/or rbm-5; uaf-1(n4588) double mutants.
(a) Three genes with altered splicing in uaf-1(n4588) as predicted by RNA-Seq were verified. (b) The altered splicing of four genes with the ‘gtag’ 3’SS in uaf-1(n4588) mutants can be suppressed by rbm-5 mutations. (c) The altered splicing of T26H5.8 and Y47H9C.1 with the ‘gtag’ 3’SS in uaf-1(n4588) mutants was not affected by rbm-5 mutations and that of pgp-8 was enhanced by rbm-5 mutations. (d) For genes with the ‘ttag’ 3ʹ SS, rbm-5 mutations did not affect the altered splicing of str-47 in the uaf-1(n4588) background while functioned synthetically with uaf-1(n4588) to cause altered splicing of T09F5.20. From left to right, genotypes are shown in numerical order: wild type (1), rbm-5(n5119) (2), rbm-5 (n5132 (3), uaf-1(n4588) (4), rbm-5(n5119); uaf-1(n4588) (5) and rbm-5(n5132); uaf-1(n4588) (6). Exon-intron structures and lengths of PCR fragments are indicated. Red arrows: PCR primers. Quantifications are based on three biological replicates. Statistics: Bonferroni test with one-way ANOVA. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
We previously found that uaf-1(n4588) could increase the recognition of the ‘gcag’ 3ʹ SS and the ‘gtag’ 3ʹ SS (conserved splice acceptor underlined) in the C. elegans unc-93(e1500) pre-mRNA and tos-1 pre-mRNA [35,38], respectively, in which the ‘g’ nucleotide at position −4 might play a critical role for the increased recognition [35]. We also found that the splicing of strong 3ʹ SS in these genes was not affected by uaf-1(n4588) [35,38]. In mbk-2, the 3ʹ SS of intron 10 is gtag. uaf-1(n4588) could increase the recognition of this 3ʹ SS (Figure 6(a)).
To understand how ‘gtag’ affects alternative splicing in other genes, we examined 20 genes containing this sequence as 3ʹ SS that were predicted by RNA-Seq to be alternatively spliced in uaf-1(n4588) mutants (Table S4). The splicing of seven genes, including abt-5, papl-1, gcy-11, F13E6.2, T26H5.8, Y47H9C.1 and pgp-8, was verified to be altered by uaf-1(n4588) (Figure 6(b,c)). In six genes (abt-5, papl-1, gcy-11, F13E6.2, T26H5.8 and pgp-8), the altered splicing was caused by a decreased recognition of the ‘gtag’ 3ʹ SS (Figure 6(b,c), Table 3). In one gene (Y47H9C.1), the altered splicing was caused by an increased recognition of this 3ʹ SS (Figure 6(c) and Table 3). rbm-5 mutations exerted different effects on these splicing events: they suppressed the decreased recognition of this 3ʹ SS in four genes (abt-5, papl-1, gcy-11, F13E6.2) (Figure 6(b), Table 3), did not obviously affect the altered recognition in two genes (T26H5.8, Y47H9C.1) and enhanced the decreased recognition in one gene (pgp-8) (Figure 6(c), Table 3). Therefore, uaf-1(n4588) can either decrease or increase the recognition of the ‘gtag’ 3ʹ SS and rbm-5 mutations could suppress or enhance the effects of uaf-1(n4588) in a gene-specific manner.
Table 3.
Effects of rbm-5 mutations on the recognition of SS altered by uaf-1(n4588).
| SS recognition in different mutants |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | Splice sites | Sequence | rbm-5(n5119) | rbm-5(n5132) | uaf-1(n4588) | rbm-5(n5119); uaf-1(n4588) | rbm-5(n5132); uaf-1(n4588) |
| Alt 3ʹ SS gtag | |||||||
| mbk-2 | 3ʹ SS | cttgtag | WT | WT | Up | Suppressed | Suppressed |
| abt-5 | 3ʹ SS | attgtag | WT | WT | Down | Suppressed | Suppressed |
| papl-1 | 3ʹ SS | tttgtag | WT | WT | Down | Suppressed | Suppressed |
| F13E6.2 | 3ʹ SS | tttgtag | WT | WT | Down | Suppressed | Suppressed |
| gcy-11 | 3ʹ SS | tttgtag | WT | WT | Down | Suppressed | Suppressed |
| T26H5.8 | 3ʹ SS | attgtag | WT | WT | Down | No effect | No effect |
| Y47H9C.1 | 3ʹ SS | attgtag | WT | WT | Up | No effect | No effect |
| pgp-8 | 3ʹ SS | tttgtag | WT | WT | Down | Enhanced | Enhanced |
| Alt 3ʹ SS ttag | |||||||
| flp-1 | 3ʹ SS | cttttag | WT | WT | Up | No effect | No effect |
| str-47 | 3ʹ SS | aatttag | WT | WT | Down | No effect | No effect |
| T09F5.20 | 3ʹ SS | aatttag | WT | WT | WT | Synthetic (Down) | Synthetic (Down) |
| Alt 5ʹ SS | |||||||
| zipt-2.4 | 5ʹ SS | ATgtactgc | WT | WT | Up | Suppressed | Suppressed |
‘ttag’ is the 3ʹ SS in intron 3 of the flp-1 gene that was alternatively recognized in uaf-1(n4588) mutants (Figure 6(a) and Table 3). ‘ttag’ is more frequently found in the C. elegans genome than ‘gtag’ [48,49]. We used RT-PCR to examine 20 genes with ‘ttag’ 3ʹ SS that RNA-Seq predicted to be alternatively spliced in uaf-1(n4588) mutants (Table S4). Only the splicing of one gene (str-47) was verified to be altered in uaf-1(n4588) mutants due to a decreased recognition of this 3ʹ SS (Figure 6(d) and Table 3). rbm-5 mutations did not further affect this altered recognition (Figure 6(d) and Table 3). The splicing of another gene, T09F5.20, was not altered in uaf-1(n4588) single mutants (Figure 6(d)). However, it was altered in rbm-5; uaf-1(n4588) double mutants as a result of a decreased recognition of the 3ʹ SS, suggesting a synthetic interaction of rbm-5 and uaf-1 (Figure 6(d) and Table 3).
An examination of the sequences of the genes that we verified (Figures 5(c), 6 and S4) suggests that altered splicing of zipt-2.4 and abt-5 resulted in frameshift and premature stop codons, which might trigger nonsense-mediated decay (NMD) [50] and caused their reduced expression. The reduced expression of papl-1 probably did not result from NMD as its altered splicing did not cause frameshift or a premature stop codon. The expression levels of seven genes, including flp-1, mbk-2, gcy-11, F13E6.2, T26H5.8, pgp-8 and T09F5.20, were not affected by any single or double mutations (Figure S4), while that of Y47H9C.1 was significantly increased in uaf-1(n4588) single and rbm-5; uaf-1(n4588) double mutants (Figure S4). Therefore, a majority of the altered splicing did not lead to reduced gene expression. For zipt-2.4 and abt-5, rbm-5 mutations probably rescued their expression (Figures 5(c) and S4) by correcting the defective splicing caused by uaf-1(n4588) (Figure 6).
It is intriguing that uaf-1(n4588) led to the recognition of a cryptic 5ʹ SS in the zipt-2.4 gene, which was suppressed by rbm-5 mutations (Figure 6(a)). Mammalian RBM5, RBM6 and RBM10 can bind exonic sequences close to 5ʹ SS of numerous genes [40], raising a question whether the altered recognition at this cryptic 5ʹ SS might reflect an in vivo function of rbm-5 and uaf-1 in regulating splicing at 5ʹ SS. Alternatively, such a change might result from uaf-1(n4588)-induced secondary effect, e.g., the altered splicing of a UAF-1 target gene that was required for the recognition of 5ʹ SS.
Discussion
In this study, we identified mutations in the RNA-binding motif gene rbm-5 as suppressors of the ts-lethality caused by uaf-1(n4588). rbm-5 interacts with uaf-1 to affect gene expression and alternative splicing in C. elegans. We suggest that the tumor suppressor function of human RBM5 might involve splicing of genes regulated by the U2AF large subunit.
C. elegans uaf-1 mutations reveal novel features of the U2AF large subunit
We previously isolated splicing factor gene mutations, uaf-1(n4588), sfa-1(n4562) and mfap-1(n4564 n5214) [35,36], as suppressors of the ‘rubberband’ Unc phenotype of unc-93(e1500gf) animals. These genes affect alternative splicing in a similar manner [35,36,38], implying a functional interaction in vivo.
uaf-1(n4588) is predicted to change the conserved Thr180 to isoleucine in the UAF-1 protein [35]. Four intragenic suppressive mutations of uaf-1(n4588) (M157I, P177L, V179M and T180F) [35] exhibited differential effects on the recognition of 3ʹ SS in vivo [35,38]. In mammalian U2AF large subunit, the corresponding residues of the four amino acids are M110, S142, M144 and T145, respectively [35,51]. M110 is a conserved residue in the UHM-ligand motif (ULM) [52], which is also known as the ‘U2AF small subunit-interacting domain’ [35,53]. S142, M144 and T145 are located in a small region between ULM and RRM1 of the U2AF large subunit, the function of which is unclear.
Recently, a structure-function study of U2AF large subunit suggests that residues of this region (a.a. 141 to 147 in mammalian U2AF large subunit) [54] are important for high-affinity binding to the polypyrimidine tract. A Q147A change could reduce the affinity of U2AF large subunit for the polypyrimidine tract by 5 folds and this region might coordinate with the C-terminal region of RRM2 and the inter-RRM linker region to affect the affinity and specificity of RNA binding [54]. This study corroborates our findings that residues in the region between ULM and RRM1 are important for 3ʹ SS recognition and alternative splicing in C. elegans [35,38]. Specifically, the n4588 T180I mutation can lead to, in a gene-specific manner, increased or decreased recognition of weak 3ʹ SS with a ‘g’ nucleotide at −4 position, suggesting that this mutation might cause UAF-1 to acquire altered affinity for the short pyrimidine tract of C. elegans 3ʹ SS.
rbm-5 mutations suppress the ts-lethality of uaf-1(n4588) mutants probably by correcting the altered expression and/or splicing of multiple genes
We found that uaf-1(n4588) led to altered expression and/or splicing of numerous genes in C. elegans embryos (Figures 5 and 6). rbm-5 mutations can reverse the abnormal expression of multiple genes altered by the uaf-1(n4588) mutation.
Of the 11 genes that exhibited altered splicing in uaf-1(n4588) mutants (Figure 6, excluding T09F5.20), rbm-5 mutations did not affect the splicing of any one. However, rbm-5 mutations can suppress the altered splicing of six genes, enhance that of one and did not apparently modulate that of the other four genes in the uaf-1(n4588) background. The 12th gene, T09F5.20, exhibited altered splicing only in rbm-5; uaf-1(n4588) double mutants (Figure 6(d)) but was not affected by any single mutations. Five genes can affect fertility, embryonic development, larval development or the germline (Table S5: zipt-2.4, F44E7.5, mbk-2, papl-1 and F13E6.2) (www.wormbase.org). We postulate that rbm-5 mutations might suppress uaf-1(n4588) by correcting the abnormal expression and/or splicing of these and other genes.
RBM5 interacts with U2AF large subunit to affect RNA splicing
In mammals, RBM5 can affect splicing especially at weak 3ʹ SS [30,32,40], which might depend on RBM5 binding to multiple different cis RNA sequences [31,32,40,55]. RBM5 was isolated as an interacting protein of U2AF large subunit and their interaction requires the RS domain of U2AF large subunit and the C-terminal region of RBM5 [30]. RBM5 co-localizes with U2AF large subunit in the cell nuclei [30]. RBM5 also affects the transition from pre-spliceosomal to spliceosomal complexes [30,33], a possible mechanistic explanation of how RBM5 affects splicing [56].
In our study, though rbm-5 did not apparently affect RNA splicing of the tested genes by itself, rbm-5 mutations can suppress or enhance, in a gene-specific manner, the altered recognition of weak 3ʹ SS caused by uaf-1(n4588) (Table 3). Therefore, the cellular and molecular studies in mammals and the genetic studies in C. elegans suggest that RBM5 act together with U2AF large subunit to regulate the splicing at weak 3ʹ SS.
RBM-5 is a modulator of UAF-1 function
In mouse, RBM5 is essential for embryonic development and the survival of newborns [25]. It is also required for spermatid differentiation and male fertility probably by affecting spermatid RNA splicing [57]. C. elegans rbm-5 is primarily expressed in neurons and not essential for survival or fertility. rbm-5 mutations can suppress the ts-lethality and sterility of uaf-1(n4588) mutants at 22.5 °C but not at 25 °C, suggesting that rbm-5 is a partial suppressor.
Though U2AF large subunit is ubiquitously expressed, its effects on RNA splicing are probably cell type-dependent. The ts-lethality of uaf-1(n4588) mutants is likely caused by collective defects in multiple tissues, e.g., neurons, muscles, the germline etc. It is plausible that amelioration of a subset of these defects can lead to better survival of the mutants at non-permissive temperatures but not result in a full recovery. The partial suppression of uaf-1(n4588) lethality by rbm-5 mutations at 22.5 °C may be caused by corrected RNA splicing only in neurons. However, the defective splicing of non-neuronal genes or rbm-5-independent genes likely remain un-rescued in rbm-5; uaf-1(n4588) double mutants, which may underlie the lethality at 25 °C.
The C. elegans genome contains ~ 100 genes encoding RNA recognition motif proteins (www.wormbase.org) [58] with unknown functions. It is possible that one or more of these genes can act redundantly with rbm-5, which might explain why rbm-5 mutants are viable. Such a redundancy could also underlie the apparent lack of effect of rbm-5 on the splicing of the genes that we tested (Figure 6). Since we only examined the effects of rbm-5 on splicing at the embryonic stage, we cannot exclude the possibility that rbm-5 can apparently affect splicing in larval or adult stages. It is also possible that RBM-5 only affects RNA splicing subtly, which might be detected by more sensitive and robust approaches than the RT-PCR method we used. This subtle effect on splicing is probably important for C. elegans to live in the wild but not essential for survival in a laboratory environment.
In mammals, RBM5 can physically bind U2AF large subunit [30] and may also compete with U2AF large subunit for binding to the polypyrimidine tract [32]. RBM5 also binds multiple different cis RNA sequences [31,32,40,55]. Recent genome-wide studies suggest that several RNA-binding proteins affect splicing by modulating the binding of U2AF large subunit to alternative 3ʹ SS [59,60]. It is plausible that in C. elegans RBM-5 modulates UAF-1 binding to 3ʹ SS by interacting with the RS domain of UAF-1 or competing for binding the pyrimidine tract preceding 3ʹ SS. Considering that the defects in recognizing weak 3ʹ SS by UAF-1(n4588) were rescued by rbm-5 mutations for several genes, it is interesting to postulate a potential proofreading regulation of alternative splicing by reducing rbm-5 expression. In addition, rbm-5 mutations promoted defective alternative splicing of two other genes in uaf-1(n4588) background (Figure 6), implying that this hypothetical proofreading might also be achieved by increasing rbm-5 expression. Since weak 3ʹ SS were primarily affected in our studies (Figure 6) [35,38], this modulation of UAF-1 activity by RBM-5 might lead to a flexible regulation of alternative splicing without impinging upon the constitutive activity of UAF-1 on strong sites. Together, these findings suggest that RBM5 and some other RNA binding proteins can affect alternative splicing by modulating the binding of U2AF large subunit to weak 3ʹ SS in a tissue-specific manner.
uaf-1(n4588) provides a sensitized background for analyzing the in vivo function of RBM-5
uaf-1(n4588) causes both a loss of function and an altered function in UAF-1 [35,38]. Considering that U2AF subunits are essential for animal survival [35,61–63], the viability of uaf-1(n4588) mutants at permissive temperatures provides a critical reagent for understanding the genetic interactions between uaf-1 and other splicing factors. The effects of rbm-5 mutations on alternative splicing were only obvious in the presence of uaf-1(n4588) (Figure 6), suggesting that the sensitized genetic background provided by uaf-1(n4588) can be used for studying the in vivo functions of other similar splicing factors. These splicing factors might modulate alternative splicing that is difficult to be reliably detected with current techniques in wild-type backgrounds and/or laboratory settings.
RBM5 might interact with U2AF large subunit in cancer pathogenesis
Recurrent mutations in several splicing factors, including U2AF1 (U2AF35, U2AF small subunit), SF3B1, SRSF2, ZRSR2 were found to be associated with different cancers [64]. The genetic features of these mutations and mouse model studies suggest that U2AF1, SF3B1 and SRSF2 promote cancer formation by gain of function or altered function mechanisms and therefore are potentially proto-oncogenes [64]. Recurrent mutations in U2AF1 and the RBM5 paralog RBM10 were also associated with lung adenocarcinomas [23]. In addition, 220 somatic mutations in RBM5 have been identified in various cancers as annotated in the Cancer Genome Atlas (TCGA) (www.cancergenome.nih.gov). These mutations include premature stop codons, deletions, splice site changes or intronic/exonic changes that potentially affect splicing, consistent with the idea that RBM5 might function as a tumor suppressor.
We found that 116 somatic mutations in the U2AF large subunit gene have been identified in multiple cancers as annotated at TCGA, with more than two thirds of them found in seven types of cancers, which include uterine corpus endometrial carcinoma, stomach adenocarcinoma, colon adenocarcinoma, bladder urothelial carcinoma, skin cutaneous melanoma, liver hepatocellular carcinoma and lung squamous cell carcinoma. Interestingly, an M110I mutation corresponding to the n5127 M157I mutation in C. elegans UAF-1 [35] was identified in a case of uterine corpus endometrial carcinoma. Mutations in the U2AF large subunit gene and the splicing factor one gene (sfa-1) have also been proposed to be potential cancer drivers based on a comprehensive analysis of the mutation profiles of over 400 splicing factors in various cancers [65].
In mammals, RBM5 can interact with the U2AF large subunit [30,32], the spliceosomal SmN/B/B’ proteins [30,33] and the DExD/H-box protein DHX15 [66]. How these interactions contribute to the tumor suppressor function of RBM5 is unclear. That rbm-5 can modulate UAF-1-dependent RNA splicing provides in vivo evidence that the interaction of RBM5 and the U2AF large subunit is conserved. It further suggests that the tumor suppressor function of RBM5 might be related to the U2AF large subunit and potentially the U2AF small subunit (U2AF1), a proto-oncogene. Further analyses of the underlying mechanism should provide new insights into the misregulation of RNA splicing in cancers.
Materials and methods
Strains
C. elegans strains were grown at 20 °C unless otherwise indicated. N2 (Bristol) is the reference wild type strain [67]. Strains used in this study include:
CSM600 rbm-5(n5119) I (backcrossed 4 times, derived from MT17914)
CSM599 rbm-5(n5132) I (backcrossed 4 times, derived from MT17927)
CSM601 rbm-5(n5119) I; uaf-1(n4588 n5127) III
CSM602 rbm-5(n5132) I; uaf-1(n4588 n5127) III
CSM603 rbm-5(n5119) I; uaf-1(n4588) III
CSM604 rbm-5(n5132) I; uaf-1(n4588) III
CSM911 rbm-5(mac465) I
CSM912 rbm-5(mac468) I
CSM725 rbm-5(mac418) I; uaf-1(n4588) III
CSM838 rbm-5(mac465) I; uaf-1(n4588) III
CSM839 rbm-5(mac466) I; uaf-1(n4588) III
CSM840 rbm-5(mac467) I; uaf-1(n4588) III
CSM847 rbm-5(mac468) I; uaf-1(n4588) III
MT14846 uaf-1(n4588) III
MT17922 uaf-1(n4588 n5127) III
MT17914 uaf-1(n4588) III; n5119
MT17916 uaf-1(n4588) III; n5121
MT17917 uaf-1(n4588) III; n5122
MT17923 uaf-1(n4588) III; n5128
MT17927 uaf-1(n4588) III; n5132
CSM949 macEx523[Prbm-5::GFP]
CSM841 macEx448[Peft-3::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #1
CSM842 macEx449[Peft-3::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #2
CSM976 rbm-5(mac468) I; uaf-1(n4588) III; macEx448[Peft-3::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #1
CSM977 rbm-5(mac468) I; uaf-1(n4588) III; macEx449[Peft-3::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #2
CSM990 macEx533[Punc-119::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #1
CSM991 macEx534[Punc-119::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #2
CSM992 rbm-5(mac468) I; uaf-1(n4588) III; macEx533[Punc-119::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #1
CSM993 rbm-5(mac468) I; uaf-1(n4588) III; macEx534[Punc-119::rbm-5a cDNA::GFP; Pmyo-2::mCherry] #2
Temperature shift experiments
Eggs laid or synchronized animals of various developmental stages grown at 15°C or 20°C were moved to incubators with higher temperatures, including 20°C, 22.5°C and 25°C. The egg hatching rate, larval development and/or egg-laying were quantified.
For egg-hatching, 90 ~ 150 eggs laid at 15 or 20 °C were shifted to higher temperatures. Unhatched eggs were counted after 24 hours. For larval development, 90 ~ 150 L1 animals grown at 15 °C were shifted to higher temperatures and the numbers of adults were counted after three days. For egg-laying, animals were allowed to develop to various larval stages at 15 or 20 °C and then moved to higher temperatures to grow into mid L4 larvae. 5 or10 L4 animals were picked to new plates and allowed to develop and lay eggs for 24 hrs at higher temperatures. The numbers of eggs were counted.
Identification of n5119 and n5132 using genetic mapping and whole-genome sequencing
n5119 and n5132 were mapped to Chr. I using visible genetic markers. Similarly n5121 and n5128 were mapped to Chr. III and n5122 to Chr. V.
Animals were washed from NGM plates and starved for several hours in M9. Genomic DNAs were extracted by proteinase K digestion followed by RNase A treatment and two rounds of phenol–chloroform extraction. Three genomic DNA libraries (380 bp inserts) were constructed by Berry Genomics Co., Ltd (Beijing) using Illumina’s paired-end protocol and paired-end sequencing (100-bp reads) was performed on the Illumina HiSeq 2000. Over 4G clean bases were mapped to the N2 genome (Wormbase release 220) after removal of duplicated reads. SNP calling was performed using Genome Analysis Toolkit (GATK) with the N2 genome as reference. 2,517 (n5119), 2,888 (n5121), 2602 (n5122), 2,650 (n5128), and 3,108 (n5132) SNPs were detected for these strains. SNPs shared among strains were excluded as they were likely derived from common ancestors. To enhance the stringency for mutation identification, we set the depth of reference base (WT) to be < 6 in these mutants. Exon or splice site SNPs with a variant quality greater than 30 were selected. Based on these criteria, we obtained 83 (n5119), 107 (n5121), 81 (n5122), 126 (n5128), and 83 (n5132) SNPs overall.
Since n5119 and n5132 were mapped to Chr. I, we postulated that a single gene might be affected by both mutations. Comparing SNP variants on Chr. I (12 SNPs n5119 and 5 for n5132) (Table S1) identified T08B2.5 (rbm-5) as the only candidate gene that was mutated in both isolates. The identification of n5121, n5122 and n5128 is in progress.
Generation of rbm-5 mutations using the CRISPR/Cas9 method
We followed the method by Farboud et al. [41] with minor modifications. Plasmids for microinjection were purified using OMEGA’s Midi Plasmid Purification kit (Omega Bio-tek). The following DNA mixture was injected: 50 ng/μl pPD162 (Peft-3::Cas9-SV40_NLS), 25 ng/μl PU6::sgRNA, and 20 ng/μl pPD95_86 (Pmyo-3::GFP) plasmid as co-injection marker.
F1 animals with GFP signals in body-wall muscles were picked to individual plates and the progeny were analyzed for mutations at or near the target sequences by sequencing.
We tested eight sgRNAs that target different rbm-5 exons (Table S6) and found that three sgRNAs (Figure 3(a)) were efficient in inducing deletions at or near the target regions (Table S6, red target sequences). Two sgRNAs generated four deletions in exons 5 and 8 (mac418, mac465, mac466 and mac467) (sgRNA1 and sgRNA2, Figure 3(a) and Table S2). We co-injected sgRNA2 and sgRNA3 to generate large deletions and isolated a 940-bp deletion (mac468) that covered a region from RRM1 to the OCRE domain of the RBM-5A protein (Figure 3(a) and Table S2)
Plasmids
To construct the Prbm-5::GFP plasmid, a PCR-amplified rbm-5 promoter (1183 bp upstream of the rbm-5 start codon) were subcloned to pPD95_79 using SbfI/XmaI restriction sites.
To construct the Peft-3::rbm-5a cDNA::GFP plasmid, a PCR-amplified eft-3 promoter (a 597-bp (−16 to −612 base) fragment upstream of the eft-3 start codon) [45] was subcloned to pPD95_79 using SbfI/XmaI sites. The rbm-5a cDNA was amplified and subcloned to the pPD95_79-Peft-3 backbone using XmaI/KpnI sites. To construct the Punc-119::rbm-5a cDNA::GFP plasmid, a promoter of the neuron-specific unc-119 gene [46,47] (2188 bp upstream of the unc-119a start codon) was amplified and subcloned to Peft-3::rbm-5a cDNA::GFP by replacing Peft-3 using SbfI/XmaI sites. PCR primers are listed in Table S7.
Transgenes were crossed to different mutants to test the effects on survival.
Transgene experiments
Germline transgene experiments were performed as described [68]. For Prbm-5::GFP transcriptional reporter, a transgene solution containing 20 ng/μl Prbm-5::GFP was injected to wild type. For Peft-3::rbm-5a cDNA::GFP reporter, a mixture containing 1 ng/μl of the transgene and 2.5 ng/μl of the pCFJ90 (Pmyo-2::mCherry) plasmid as co-injection marker was injected to wild type. For Punc-119::rbm-5a cDNA::GFP reporter, a mixture containing 0.2 ng/μl of the transgene and 2.5 ng/μl of pCFJ90 was injected to wild type. We tried but failed to obtain stable lines with a Prbm-5::rbm-5a cDNA transgene under control of the 1183-bp rbm-5 endogenous promoter at concentrations of 0.1, 1, 10 or 50 ng/μl in wild-type or double mutant backgrounds, suggesting that overexpressing rbm-5 using its endogenous promoter is highly toxic. Transgenic animals were observed using a Leica TCS SP5 II laser confocal microscope.
Identification of rbm-5 -expressing cells
We determined the identities of Prbm-5::GFP-positive cells by examining their anatomical positions, morphologies, patterns and neighboring cells and compare those to the descriptions at www.wormatlas.org. Neurons were determined by their positions and neurite morphologies. Body-wall muscles have typical rhomboid shapes and rows of dense bodies on plasma membrane that are visible under microscope. C. elegans intestine is comprized of 20 large epithelial cells that are identified by the tube shape they form and the alignment from the pharynx to the rectum.
Whole transcriptome shotgun sequencing (RNA-Seq)
Synchronized adult animals were bleached to obtain enough eggs, which were washed three times with H2O. Total RNA was prepared using Trizol (Invitrogen), treated with RNase-Free DNase I (New England Biolabs) and incubated at 75 °C for 10 min to inactivate DNase I. RNA-Seq was performed by in-house scientists at Annoroad Gene Technology Corporation (Beijing)
Raw data was processed with Perl scripts to ensure the quality of data used in further analysis. Bowtie2 was used for building the genome index, and Clean Data was mapped to the WBcel235 alignments using TopHat v2.0.12. The Integrative Genomics Viewer was used to analyze the mapping results by the heatmap, histogram, scatter plot or other styles.
Fragments counts were obtained with HTSeq v0.6.0 and WBcel235 Ensembl annotation. Gene expression analysis was performed using DEGSeq 1.18.0. The q-value was assigned to each gene and adjusted by the Benjamini and Hochberg normalization for controlling the false discovery rate (FDR). Genes with q ≤ 0.05 and |log2_ratio|≥ 1 were identified as differentially expressed genes (DEGs). Alternative splicing analysis was performed with Asprofile 1.0.4 and Cuffcompare 2.2.1 by constructing de novo annotation from the Ensembl input and merged alignment files. KEGG pathway analysis of differentially expressed genes was carried out using Hypergeometric test.
RT-PCR experiments
Total RNAs were prepared from eggs collected from bleached adults using Trizol (Invitrogen), treated with RNase-Free DNase I (New England Biolabs) and incubated at 75 °C for 10 min to inactivate DNase I. First-strand cDNA was synthesized with random hexamer oligonucleotides using Maxima First Strand cDNA Synthesis Kit (Thermo Fisher). For alternative splicing, 3 biological replicates of each strain were analyzed and the proportion of each splice isoform was quantified using the NIH ImageJ software.
qRT–PCR was performed on 3 biological replicates of each strain using the Maxima SYBR Green qPCR Master Mix (Thermo Scientific). Fluorescence signals were detected using LightCycler® 96 Instrument (Roche). Each 30 μl PCR reaction contained 1 to 10 ng RT template, 0.5 mM PCR primers and 15 μl 2 x SYBR Green PCR Master Mix. After a preincubation step (95 °C for 10 min), two-step amplification was performed using 40 cycles of denaturation (95 °C for 15 s) and annealing (60 °C for 30 s).
The genotype examined include wild type, rbm-5(n5119), rbm-5(n5132), uaf-1(n4588), rbm-5(n5119); uaf-1(n4588) and rbm-5(n5132); uaf-1(n4588). The genes examined include zipt-2.4, zip-11, F44E7.5, F56C3.9, flp-1, mbk-2, abt-5, papl-1, gcy-11, F13E6.2, T26H5.8, Y47H9C.1, pgp-8 and T09F5.20. We used the crt-1 gene as loading control as its expression was abundant in embryos and its FPKM (fragments per kilobase of gene per million mapped reads) was similar across the samples based on RNA-Seq results. PCR primers for detecting expression levels or alternative splicing are listed in Table S7.
Data availability
All RNA-Seq datasets generated and analyzed during the current study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE115695. All other data are available from the corresponding author upon reasonable request.
Statistic analysis
P values were determined by Paired two-tailed Student’s t-test or Bonferroni’s multiple comparison test using GraphPad Prism 5.0 software.
Funding Statement
This work was supported by the National Key R&D Program of China [2016YFC1201805], National Natural Science Foundation of China [31371253] and National Natural Science Foundation of China [31571045] to LM; National Institutes of Health grant numbers R01NS094564 and R21NS106307 to YCM.
Acknowledgments
This study was initiated in the laboratory of H. Robert Horvitz (HRH). H.R.H. is the David H. Koch Professor of Biology at the Massachusetts Institute of Technology and an Investigator of the Howard Hughes Medical Institute. We thank members of the Ma laboratory for suggestions. The study is supported by National Natural Science Foundation of China grants (No. 31371253, No. 31571045) and a MOST grant (2016YFC1201805) to LM. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions
Conceived and designed the experiments: CZ, XG, LM. Performed the experiments: CZ, XG, SH, WG, JX, LM. Analyzed the data: CZ, YM, LM. Wrote the paper: CZ, YM, LM.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Maniatis T, Tasic B.. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418(6894):236–243. PubMed PMID: 12110900. [DOI] [PubMed] [Google Scholar]
- [2].Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12(3):340–345. PubMed PMID: 10801464. [DOI] [PubMed] [Google Scholar]
- [3].Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253(5016):157–163. PubMed PMID: 1853200. [DOI] [PubMed] [Google Scholar]
- [4].Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12(1):5–14. [DOI] [PubMed] [Google Scholar]
- [5].Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17(2):100–107. PubMed PMID: 11173120. [DOI] [PubMed] [Google Scholar]
- [6].Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–388. PubMed PMID: 10916158. [DOI] [PubMed] [Google Scholar]
- [7].Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. PubMed PMID: 11967553. [DOI] [PubMed] [Google Scholar]
- [8].Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749–761. PubMed PMID: 17726481. [DOI] [PubMed] [Google Scholar]
- [9].Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Neubauer G, King A, Rappsilber J, et al. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20(1):46–50. PubMed PMID: 9731529. [DOI] [PubMed] [Google Scholar]
- [11].Rappsilber J, Ryder U, Lamond AI, et al. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12(8):1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stevens SW, Ryan DE, Ge HY, et al. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9(1):31–44. PubMed PMID: 11804584. [DOI] [PubMed] [Google Scholar]
- [13].Zhou Z, Licklider LJ, Gygi SP, et al. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419(6903):182–185. [DOI] [PubMed] [Google Scholar]
- [14].Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18(3):290–298. PubMed PMID: 18515081. [DOI] [PubMed] [Google Scholar]
- [15].Sutherland LC, Rintala-Maki ND, White RD, et al. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94(1):5–24. PubMed PMID: 15514923. [DOI] [PubMed] [Google Scholar]
- [16].Edamatsu H, Kaziro Y, Itoh H. LUCA15, a putative tumour suppressor gene encoding an RNA-binding nuclear protein, is down-regulated in ras-transformed Rat-1 cells. Genes Cells. 2000;5(10):849–858. PubMed PMID: 11029660. [DOI] [PubMed] [Google Scholar]
- [17].Maarabouni MM, Williams GT. The antiapoptotic RBM5/LUCA-15/H37 gene and its role in apoptosis and human cancer: research update. ScientificWorldJournal. 2006;6:1705–1712. PubMed PMID: 17195868; PubMed Central PMCID: PMCPMC1825760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. PubMed PMID: 12469122. [DOI] [PubMed] [Google Scholar]
- [19].Sutherland LC, Wang K, Robinson AG. RBM5 as a putative tumor suppressor gene for lung cancer. J Thorac Oncol. 2010;5(3):294–298. Epub 2010/02/27. PubMed PMID: 20186023. [DOI] [PubMed] [Google Scholar]
- [20].Timmer T, Terpstra P, van den Berg A, et al. An evolutionary rearrangement of the Xp11.3-11.23 region in 3p21.3, a region frequently deleted in a variety of cancers. Genomics. 1999;60(2):238–240. PubMed PMID: 10486216. [DOI] [PubMed] [Google Scholar]
- [21].Wei MH, Latif F, Bader S, et al. Construction of a 600-kilobase cosmid clone contig and generation of a transcriptional map surrounding the lung cancer tumor suppressor gene (TSG) locus on human chromosome 3p21.3: progress toward the isolation of a lung cancer TSG. Cancer Res. 1996;56(7):1487–1492. PubMed PMID: 8603390. [PubMed] [Google Scholar]
- [22].Welling DB, Lasak JM, Akhmametyeva E, et al. cDNA microarray analysis of vestibular schwannomas. Otol Neurotol. 2002;23(5):736–748. PubMed PMID: 12218628. [DOI] [PubMed] [Google Scholar]
- [23].Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. PubMed PMID: 22980975; PubMed Central PMCID: PMCPMC3557932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jamsai D, Watkins DN, O’Connor AE, et al. In vivo evidence that RBM5 is a tumour suppressor in the lung. Sci Rep. 2017;7(1):16323 PubMed PMID: 29176597; PubMed Central PMCID: PMCPMC5701194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Loiselle JJ, Roy JG, Sutherland LC. RBM5 reduces small cell lung cancer growth, increases cisplatin sensitivity and regulates key transformation-associated pathways. Heliyon. 2016;2(11):e00204 PubMed PMID: 27957556; PubMed Central PMCID: PMCPMC5133678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mourtada-Maarabouni M, Sutherland LC, Williams GT. Candidate tumour suppressor LUCA-15 can regulate multiple apoptotic pathways. Apoptosis. 2002;7(5):421–432. PubMed PMID: 12207175. [DOI] [PubMed] [Google Scholar]
- [27].Oh JJ, Razfar A, Delgado I, et al. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 2006;66(7):3419–3427. Epub 2006/04/06. PubMed PMID: 16585163. [DOI] [PubMed] [Google Scholar]
- [28].Sutherland LC, Lerman M, Williams GT, et al. LUCA-15 suppresses CD95-mediated apoptosis in Jurkat T cells. Oncogene. 2001;20(21):2713–2719. PubMed PMID: 11420683. [DOI] [PubMed] [Google Scholar]
- [29].Mourtada-Maarabouni M, Keen J, Clark J, et al. Candidate tumor suppressor LUCA-15/RBM5/H37 modulates expression of apoptosis and cell cycle genes. Exp Cell Res. 2006;312(10):1745–1752. PubMed PMID: 16546166. [DOI] [PubMed] [Google Scholar]
- [30].Bonnal S, Martinez C, Forch P, et al. RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition. Mol Cell. 2008;32(1):81–95. Epub 2008/10/15. PubMed PMID: 18851835. [DOI] [PubMed] [Google Scholar]
- [31].Fushimi K, Ray P, Kar A, et al. Up-regulation of the proapoptotic caspase 2 splicing isoform by a candidate tumor suppressor, RBM5. Proc Natl Acad Sci USA. 2008;105(41):15708–15713. PubMed PMID: 18840686; PubMed Central PMCID: PMCPMC2572934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jin W, Niu Z, Xu D, et al. RBM5 promotes exon 4 skipping of AID pre-mRNA by competing with the binding of U2AF65 to the polypyrimidine tract. FEBS Lett. 2012;586(21):3852–3857. Epub 2012/09/29. PubMed PMID: 23017209. [DOI] [PubMed] [Google Scholar]
- [33].Mourao A, Bonnal S, Soni K, et al. Structural basis for the recognition of spliceosomal SmN/B/B’ proteins by the RBM5 OCRE domain in splicing regulation. Elife. 2016;5 PubMed PMID: 27894420; PubMed Central PMCID: PMCPMC5127646 DOI: 10.7554/eLife.14707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. 2015;200(2):387–407. PubMed PMID: 26088431; PubMed Central PMCID: PMCPMC4492366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ma L, Horvitz HR. Mutations in the Caenorhabditis elegans U2AF large subunit UAF-1 alter the choice of a 3ʹ splice site in vivo. PLoS Genet. 2009;5(11):e1000708 PubMed PMID: 19893607; PubMed Central PMCID: PMCPMC2762039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma L, Gao X, Luo J, et al. The Caenorhabditis elegans gene mfap-1 encodes a nuclear protein that affects alternative splicing. PLoS Genet. 2012;8(7):e1002827 Epub 2012/07/26. PubMed PMID: 22829783; PubMed Central PMCID: PMC3400559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96(1):147–164. PubMed PMID: 6894129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ma L, Tan Z, Teng Y, et al. In vivo effects on intron retention and exon skipping by the U2AF large subunit and SF1/BBP in the nematode Caenorhabditis elegans. Rna. 2011;17(12):2201–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gao X, Teng Y, Luo J, et al. The survival motor neuron gene smn-1 interacts with the U2AF large subunit gene uaf-1 to regulate Caenorhabditis elegans lifespan and motor functions. RNA Biol. 2014;11(9):1148–1160. Epub 2014/12/09. PubMed PMID: 25483032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bechara EG, Sebestyen E, Bernardis I, et al. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52(5):720–733. PubMed PMID: 24332178. [DOI] [PubMed] [Google Scholar]
- [41].Farboud B, Meyer BJ. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015;199(4):959–971. PubMed PMID: 25695951; PubMed Central PMCID: PMCPMC4391549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ito T, Muto Y, Green MR, et al. Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)). Embo J. 1999;18(16):4523–4534. PubMed PMID: 10449418; PubMed Central PMCID: PMCPMC1171527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sickmier EA, Frato KE, Shen H, et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell. 2006;23(1):49–59. PubMed PMID: 16818232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Daubner GM, Clery A, Allain FH. RRM-RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol. 2013;23(1):100–108. PubMed PMID: 23253355. [DOI] [PubMed] [Google Scholar]
- [45].Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14(17):2173–2184. PubMed PMID: 10970881; PubMed Central PMCID: PMCPMC316897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141(3):977–988. PubMed PMID: 8582641; PubMed Central PMCID: PMCPMC1206859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Maduro M, Pilgrim D. Conservation of function and expression of unc-119 from two Caenorhabditis species despite divergence of non-coding DNA. Gene. 1996;183(1–2):77–85. PubMed PMID: 8996090. [DOI] [PubMed] [Google Scholar]
- [48].Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 2000;10(8):1115–1125. PubMed PMID: 10958630. [DOI] [PubMed] [Google Scholar]
- [49].Wang F, Huang S, Ma L. Caenorhabditis elegans operons contain a higher proportion of genes with multiple transcripts and use 3ʹ splice sites differentially. PLoS One. 2010;5(8):e12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].He F, Jacobson A. Nonsense-Mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet. 2015;49:339–366. PubMed PMID: 26436458; PubMed Central PMCID: PMCPMC4837945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zamore PD, Patton JG, Green MR. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355(6361):609–614. PubMed PMID: 1538748. [DOI] [PubMed] [Google Scholar]
- [52].Corsini L, Bonnal S, Basquin J, et al. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat Struct Mol Biol. 2007;14(7):620–629. PubMed PMID: 17589525. [DOI] [PubMed] [Google Scholar]
- [53].Rudner DZ, Kanaar R, Breger KS, et al. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol. 1998;18(4):1765–1773. PubMed PMID: 9528748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Agrawal AA, Salsi E, Chatrikhi R, et al. An extended U2AF(65)-RNA-binding domain recognizes the 3ʹ splice site signal. Nat Commun. 2016;7:10950 PubMed PMID: 26952537; PubMed Central PMCID: PMCPMC4786784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nguyen CD, Mansfield RE, Leung W, et al. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol. 2011;407(2):273–283. PubMed PMID: 21256132. [DOI] [PubMed] [Google Scholar]
- [56].Kotlajich MV, Hertel KJ. Death by splicing: tumor suppressor RBM5 freezes splice-site pairing. Mol Cell. 2008;32(2):162–164. Epub 2008/ 10/28 PubMed PMID: 18951082. [DOI] [PubMed] [Google Scholar]
- [57].O’Bryan MK, Clark BJ, McLaughlin EA, et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 2013;9(7):e1003628 PubMed PMID: 23935508; PubMed Central PMCID: PMCPMC3723494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lorkovic ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30(3):623–635. PubMed PMID: 11809873; PubMed Central PMCID: PMCPMC100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Howard JM, Lin H, Wallace AJ, et al. HNRNPA1 promotes recognition of splice site decoys by U2AF2 in vivo. Genome Res. 2018;28(5):689–698. PubMed PMID: 29650551; PubMed Central PMCID: PMCPMC5932609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sutandy FXR, Ebersberger S, Huang L, et al. In vitro iCLIP-based modeling uncovers how the splicing factor U2AF2 relies on regulation by cofactors. Genome Res. 2018;28(5):699–713. PubMed PMID: 29643205; PubMed Central PMCID: PMCPMC5932610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kanaar R, Roche SE, Beall EL, et al. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science. 1993;262(5133):569–573. PubMed PMID: 7692602. [DOI] [PubMed] [Google Scholar]
- [62].Rudner DZ, Kanaar R, Breger KS, et al. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci USA. 1996;93(19):10333–10337. PubMed PMID: 8816800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. Rna. 1999;5(4):487–494. PubMed PMID: 10199565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dvinge H, Kim E, Abdel-Wahab O, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–430. PubMed PMID: 27282250; PubMed Central PMCID: PMCPMC5094465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Seiler M, Peng S, Agrawal AA, et al. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1):282–96 e4. PubMed PMID: 29617667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Niu Z, Jin W, Zhang L, et al. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012;586(7):977–983. PubMed PMID: 22569250. [DOI] [PubMed] [Google Scholar]
- [67].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. PubMed PMID: 4366476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mello CC, Kramer JM, Stinchcomb D, et al. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10(12):3959–3970. PubMed PMID: 1935914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq datasets generated and analyzed during the current study are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE115695. All other data are available from the corresponding author upon reasonable request.


