Abstract
Background
Multicolor flow cytometry (MFC) analysis is widely used to identify minimal residual disease (MRD) after treatment for acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). However, current manual interpretation suffers from drawbacks of time consuming and interpreter idiosyncrasy. Artificial intelligence (AI), with the expertise in assisting repetitive or complex analysis, represents a potential solution for these drawbacks.
Methods
From 2009 to 2016, 5333 MFC data from 1742 AML or MDS patients were collected. The 287 MFC data at post-induction were selected as the outcome set for clinical outcome validation. The rest were 4:1 randomized into the training set (n = 4039) and the validation set (n = 1007). AI algorithm learned a multi-dimensional MFC phenotype from the training set and input it to support vector machine (SVM) classifier after Gaussian mixture model (GMM) modeling, and the performance was evaluated in The validation set.
Findings
Promising accuracies (84·6% to 92·4%) and AUCs (0·921–0·950) were achieved by the developed algorithms. Interestingly, the algorithm from even one testing tube achieved similar performance. The clinical significance was validated in the outcome set, and normal MFC interpreted by the AI predicted better progression-free survival (10·9 vs 4·9 months, p < 0·0001) and overall survival (13·6 vs 6·5 months, p < 0·0001) for AML.
Interpretation
Through large-scaled clinical validation, we showed that AI algorithms can produce efficient and clinically-relevant MFC analysis. This approach also possesses a great advantage of the ability to integrate other clinical tests.
Fund
This work was supported by the Ministry of Science and Technology (107-2634-F-007-006 and 103–2314-B-002-185-MY2) of Taiwan.
Keywords: Acute myeloid leukemia, Myelodysplastic syndrome, Multicolor flow cytometry, Minimal residual disease, Artificial intelligence
Research in context.
Evidence before this study
Multiparameter flow cytometry (MFC) has been utilized extensively to detect minimal residual disease (MRD) and risk stratification for hematological malignancies, such as AML (acute myeloid leukemia) and MDS (myelodysplastic syndrome). However, current MFC interpretation is through subjective manual gating which has unavoidable drawbacks including individual idiosyncrasy and time-consuming. Although research endeavors have been put into computational method development for universal automated MFC analysis, they were not developed from bone marrow samples, which is clinically essential but complex for analysis. Furthermore, none of them are from large-scaled real-world datasets, nor effectively validated in clinical settings.
Added value of this study
In this study, we utilized two artificial intelligence (AI) techniques to develop a MFC interpretation algorithm for MRD detection using a real-world cohort of over 1000 AML and MDS patients with over 5000 MFC data on bone marrow samples. High clinical validity of the algorithm was demonstrated, through successful outcome prediction in the post-induction setting.
Implications of all available evidence
We demonstrated the algorithms developed via AI could accomplish classification task in a very short time (merely 7 s) with about 90% accuracies on MRD detection on AML and MDS. In addition, the results of predicting outcome in the post-induction setting demonstrated a high prognostic significance of the AI algorithms.
Alt-text: Unlabelled Box
1. Introduction
Acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) are characterized by abnormal proliferation of myeloid progenitors and subsequent bone marrow failure [1]. Existence of minimal (or measurable) residual disease (MRD), which refers to leukemic cells detected below the threshold for morphological recognition (about 5%), is a valuable marker for evaluating the response after treatment, and now serves as an important prognostic indicator for AML [2]. The European LeukemiaNet (ELN) MRD Working group consensus report recommends MRD testing as part of the standard of care for AML patients [3]. Studies have demonstrated that multiparameter flow cytometry (MFC) can effectively detect minimal residual disease (MRD) and stratify prognosis in AML and MDS after therapy [[4], [5], [6], [7], [8], [9], [10]]. However, current MFC presents drawbacks such as lack of inter-lab standardization [11], and painstaking manual gating process involving serial projections of two dimensional attributes [12]. Two main MFC analysis approaches for leukemia MRD detection are used now [13]. Leukemia-associated aberrant immune-phenotype (LAIP) approach assays MRD under the assumption that the residual disease possesses the phenotype identical to the initial one, and therefore is highly dependent on individually selected antibody combination panels according to leukemia phenotype identified at diagnosis [13,14]. Instead, “difference from normal” approach uses a standardized panel of antibodies for all specimens and distinguishes abnormal residual leukemic cells from normal ones with established immunophenotypic profiles, and therefore does not require knowledge of the phenotype at diagnosis for the MRD detection [13,14]. Although more biologically reasonable, the LAIP approach risks in higher false negative MRD rates due to altered antigen expression from clonal evolution during disease progression [14,15]. Furthermore, the quality of both approaches depends highly on experienced physicians, and individual idiosyncrasy inevitably affects diagnostic reproducibility and objectivity. In addition, manual gating is time-consuming and infeasible to obtain information from the multivariate measurement data due to it observational nature [12,16]. A reliable automated MFC analysis can benefit and improve the healthcare quality by providing rapid clinical decision support.
Supervised machine learning (SML), a branch of artificial intelligence (AI), operates by learning from data and expert labels to generate reliable automated inference [[17], [18], [19]]. Rather than using predefined model, SML performs inference by learning the underlying patterns (functional mapping) between measurement data and desirable outcome variable with large-scale data [20]. In recent years, a growing number of breakthroughs utilizing AI in clinical research have been reported regarding automatic disease pattern recognition and outcome stratification [[21], [22], [23]]. For instance, expert-level accuracy can be achieved by applying SML approach on images for skin cancer diagnosis [21,22], or diabetic retinopathy identification [24,25]. SML approach in estimating mortality within 100 days after hematopoietic stem cell transplant (HSCT) using alternating decision tree model has been studied on retrospective registry data [23]. Although several SML-based approaches have been developed for automated MFC analysis and its visualization tools in AML or MDS [12,[26], [27], [28], [29], [30], [31], [32], [33], [34]], they either suffer from small case number without large-scaled clinical validation, or use MFC data derived from peripheral blood, an approach with high false negative MRD rates and therefore not commonly used in clinical settings. Furthermore, none of them attempts to correlate with patient outcome. In this work, we applied SML techniques in analyzing MFC dataset to develop an automated MFC interpretation algorithm for detecting MRD objectively in AML and MDS patients, and we validate it with large-scaled clinical data and patient survival, the most relevant clinical outcome.
2. Materials and methods
2.1. Study population and variables
From 2009 to 2016, 1742 AML or MDS patients who were treated at National Taiwan University Hospital were enrolled retrospectively. A total of 5333 MFC data for bone marrow aspiration from them were included for analysis (Supp. Table S1). To illustrate prognostic impacts, 287 AML patients with available post-induction bone marrow MFC data (MFC performed from day+0 to day+45 after the initiation date of induction chemotherapy) and clinical outcome were included in the survival analysis. Their cytogenetic and gene mutation analysis were used for risk stratification by the 2017 European LeukemiaNet (ELN) recommendation [35]. This study, along with the policy to waive informed consents, was approved by the Research Ethic Committee of the National Taiwan University Hospital (No. 201705016RINA).
2.2. MFC measurement
MFC was performed on each enrolled bone marrow aspirate samples with a myeloid panel consisting of markers listed in Supp. Table S2, and the antibodies used were listed in Supp. Table S3. A total of 100,000 events were collected for each tube within the panel. Two different flow cytometers were used in different time periods: 2574 MFC were performed on FASCalibur (Calibur) (Becton Dickinson Bioscience) from Sep 2009 to Oct 2013 and 2759 MFC on FASCanto-II (Canto-II) (Becton Dickinson Bioscience) from Oct 2013 to Dec 2016.
2.3. Cytogenetic and molecular testing
Trypsin-Giemsa technique was used for banding metaphase chromosomes, and cytogenetic was karyotyped according to the International System for Human Cytogenetic Nomenclature, as described earlier [36]. Genetic mutations including NPM1, FLT3-LTD, CEBPA, RUNX1, and CBFB-MYH11 mutations were examined also as described previously [37,38]. The cytogenetic and genetic mutation analyses conducted at diagnosis were included for risk stratification.
2.4. MFC labeling for SML algorithm training
Each MFC data had been manually analyzed using the “different-from-normal” approach, and the results were categorized into 3 groups: “AML” for freshly diagnosed AML and residual AML cells after treatments, “MDS” for freshly diagnosed MDS and residual MDS cells after treatment, and “normal” represents specimens without diseased cells. The labels are mutually exclusive for each MFC data.
2.5. Outcome set, training set and validation set sample selection
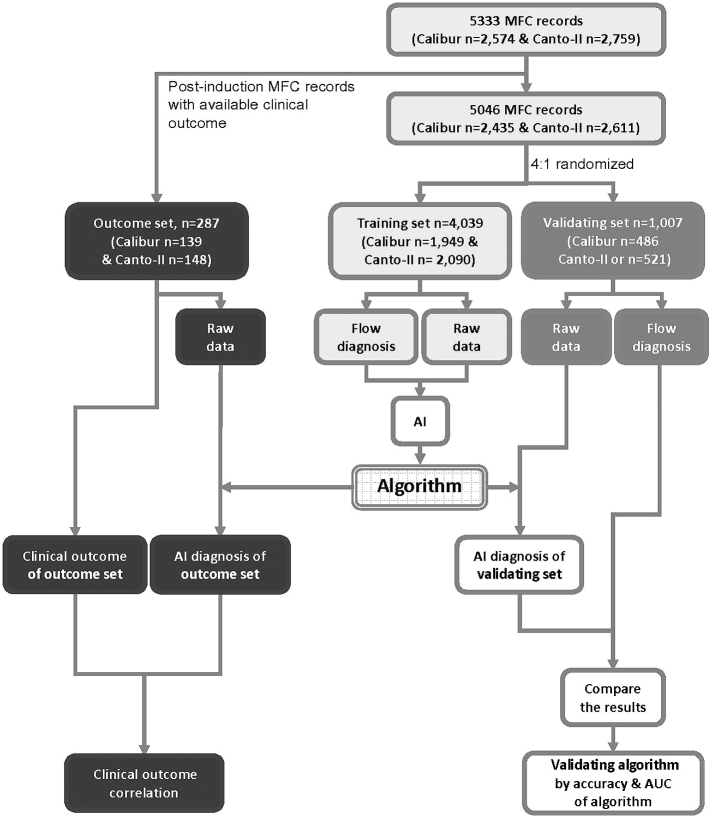
After leaving 287 MFC data out from 287 AML patients with available post-induction bone marrow MFC data and clinical outcome as the outcome set for survival analysis, the rest of the MFC data were 4:1 randomized into the training set and the validation set, consisting 4039 and 1007 MFC data, respectively (Fig. 1). The training set was used for training and tuning the SML algorithm, and the validation set for evaluating the performance. Manual analytical results were blinded when MFC data in the outcome set was analyzed by SML algorithm. Algorithms for pair-wise recognition (AML-vs-normal, MDS-vs-normal and abnormal (AML + MDS)-vs-normal) were developed independently. Algorithms were also separately developed for MFC data from Calibur and Canto-II, and an independent algorithm was generated for the combined MFC sub-datasets after we convert MFC values from Calibur with the conversion formula: Canto-II = Calibur MFI × (218/10,000) provided by the manufacturer. We used a five-fold cross-validation evaluation scheme.
Fig. 1.
Training, validation, and outcome sets for algorithm development.
The 287 post-induction MFC data of 287 AML patients were assigned in the outcome set first, and the rest of MFC data were randomly assigned to the training set and validation set with 4:1 ratio respectively. The raw data consisting of one 100,000 (events) *6 (channels) matrix for each tube (as in Supp. Table S2) together with the flow diagnosis label in the training set are used to train the classification algorithm. The accuracy is determined by comparing the concordance rate between the flow diagnosis label and AI diagnosis for each given sample in the validation set. Flow diagnosis label is the manual interpretation results.
Abbreviation: AI, artificial intelligence; AUC, area under the receiver operating characteristic (ROC) curve
2.6. SML algorithm development
The recorded raw values from the 6 fluorescent channels of each tube were max-min normalized. We then derived a MFC feature vector to characterize these raw cells attributes, and diagnostic classification was performed by support vector machine (SVM) [39]. The phenotype representation was derived via two steps: first, we modeled each tube's raw attributes values with a generative probability distribution; then we derived a high-dimensional vectorized representation by computing the Fisher gradient score with respect to the learned model parameters for each tube sample. Finally, the concatenation of multiple tube-level vectors provided a joint representation to characterize each MFC data, termed the MFC feature vector. An SVM was further trained to classify the diagnoses on these MFC feature vectors (Supp. Fig. S1). Specifically, each of the tubes was statistically-modeled as a multivariate Gaussian mixture model (GMM). The GMM was trained in an unsupervised manner using maximum likelihood estimation to derive the model parameters, which include the following:
| (1) |
where ωk; μk; σk were weight, mean and covariance respectively and K indicated how many clusters there were in the GMM. Using the learned GMM with parameter set λ (including weight, mean, and covariance), we can derive the tube-level feature vector:
| (2) |
be a set of T FC cell samples in each tube, and the gradient of log likelihood was termed as the Fisher score function: ∇λlogP(X| λ), where likelihood for a given GMM was defined as
| (3) |
Then, the tube-level feature vector was derived as the first and second order statistics of the gradient function (the gradient function indicated the direction of λ for the original GMM to better fit the data sample X).
| (4) |
| (5) |
Where
| (6) |
was the posterior data likelihood.
Each tube-level feature vector was a vector of [gμkXgσkX].
These tube-level feature vectors were concatenated and L2-normalized:
| (7) |
where R was the tube-level feature vector with d dimensions. The normalization of the vector was important to ensure that each feature vector was of unit-norm in order to provides better numerical representation that can be used in the SVM classification. Each normalized tube-level feature vector for a patient's measurement was concatenated together, which forms the final feature dimensions.
The use of GMM model as the generative probabilistic representation with Fisher scoring to derive vectorized representation combined the advantage of both generative and discriminative properties in compactly representing the high-dimensional information in the raw FC samples. In summary, the original raw cell attributes of each tube were encoded into a tube-level feature vector. Vectors of each tube formed the final high-dimensional (Dim = 2*K*D, where K was the number of Gaussian components and D is the dimension of raw data) input to the supervised machine learning classifier. We used VLfeat open source python toolbox for the Fisher-vector GMM encoding [40], and scikit-learn, another open source package, for the support vector machine (SVM) with linear kernel function to perform linear SVM classification, which operated by finding a hyper-plane to maximize the classification margin [41]. Both the number of Gaussian components of the GMM model and the penalty factor C of the SVM were obtained by grid search. All the experiments were conducted in a device equipped with Intel i7-6700 @ 3.40 GHz and 64GB random access memory (RAM).
The pseudo code of the algorithm is illustrated below:
Input data {X1, X2, …, XN} ∈ XT×D
Input initial GMM, λt ∈ ΛtK×D,
| (8) |
For t in T:
Train tube-level GMM:
Use {X1, t, X2, t, …, XN, t} and EM algorithm
Update λt ← λt′
With GMM λt, compute tube-level feature vector:
| (9) |
End
| (10) |
for i = 1, …, N
| (11) |
Output {ϕ1, ϕ2, …, ϕN}
Input feature vectors {ϕ1, ϕ2, …, ϕN}
Input labels {Y1, Y2, …, YN}
SVM classifier for {(ϕi, Yi)i=1, …, n}
2.7. Sensitivity-specificity and tube importance evaluation
To evaluate the classification performance, accuracy (ACC) was used and defined as the concordance rate between the diagnoses made from manual and AI interpretations. Furthermore, the test sensitivity and specificity were assessed using AUC (area under receiver operating characteristic (ROC) curve).
2.8. Survival analysis
To predict survival is one ultimate clinical application for MRD detection. In order to validate the clinical effectiveness of our SML algorithm in detecting MRD, we proposed to examine the correlation of SML interpretation results and the survival in AML patients. Survival analysis was performed on the 287 AML patients in the outcome set, with blinded manual interpretation results at analysis. Overall survival (OS) was measured from the date of MFC data to the date of allogeneic HSCT (allo-HSCT), or the date of last follow-up, or death of any cause, whichever comes first. Progression–free survival (PFS) was measured from the date of MFC data to the date of first relapse, to the date of allo-HSCT, or to the date of last follow-up, whichever comes first. The Kaplan-Meier method was used to estimate OS and PFS. Cox proportional hazard models were used to estimate hazard ratios (HRs) for univariate and multivariable analyses of OS and PFS. AI-diagnosis of each MFC data, genetic risk group, age, gender, and induction chemotherapy were used as covariates. All statistical analyses were conducted using survival package in R and Kaplan-Meier curves were plotted using survminer package in R (R Core Team) [42].
3. Results
3.1. Patient characteristics for MFC data
The characteristics of 5333 enrolled MFC data were listed in Supp. Table S1. For Caliber, 2574 MFC data from 908 patients were collected, and 2759 MFC data from 1046 patients for Canto-II. As much as 31·5% (1683/5333) of MFC data were interpreted as abnormal (AML or MDS). AML was interpreted in 26·8% Calibur and 22·9% Canto-II MFC data, and MDS in 5·3% Calibur and 8·2% Canto-II data.
3.2. Algorithm performance
We generated classification algorithms in 9 different comparative scenarios: AML-vs-normal, MDS-vs-normal and Abnormal (AML or MDS)-vs-Normal on Calibur, Canto-II, and Calibur+Canto-II respectively.
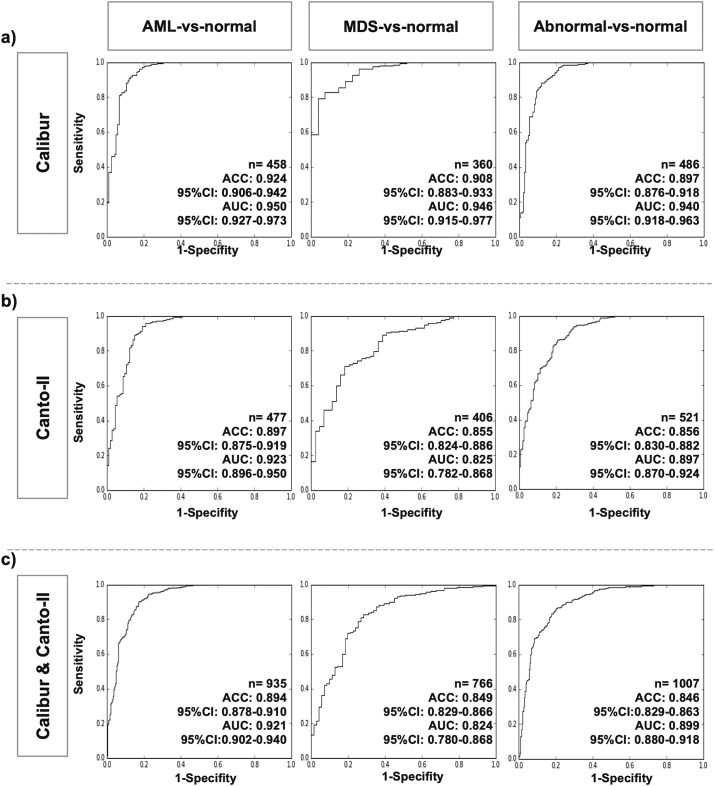
The algorithm performance was illustrated in Fig. 2, and the change in accuracy and AUC as function of different number of Gaussian components was shown in Supp. Table S4. The AML-vs-normal classification accuracy achieved scores ranging from 89·4% to 92·4% in different scenarios, whereas the accuracy of MDS-vs-normal classification achieved 84·9% to 90·8%. For abnormal-vs-normal classification, the accuracy ranged from 84·6 to 89·7%. Based on AUC and the shape of ROC curves, the AML-vs-normal classifier had the highest performance, followed by the abnormal-vs-normal and the MDS-vs-normal. Moreover, the overall classifier performance for Calibur sub-dataset was higher than that for Canto-II sub-dataset, which was relatively equivalent to that for Calibur+Canto-II sub-dataset (Fig. 2A–C). The ACC and AUC for the five-fold cross-validation in the validation set were illustrated in Supp. Table S5. The whole training process was completed within 13 h, and the average running time was 7 s for conducting analysis in single MFC data with developed SML algorithm.
Fig. 2.
Algorithm performance assessment on the validation set.
Binary classification performance for the AML-vs-normal, MDS-vs-normal and abnormal-vs-normal groups: (A) Calibur sub-dataset, (B) Canto-II sub-dataset, (C) Calibur & Canto-II sub-dataset. The “n” value indicates the number of MFC data in the analysis for each column. The five-fold cross-validation were performed on five independent validation sets with non-overlapping MFC data and shown in Supp. Table S5.
Abbreviation: ACC: accuracy, equal to the concordance rate with the flow diagnosis of multi-color flow cytometry data; AUC: Areas under the receiver operating characteristic (ROC) curves. MFC, multi-color flow cytometry
3.3. Feature selection analysis
Feature selection analysis was performed to find the relative importance of markers in the automated algorithm. In the first round, we trained the algorithms with data from just one tube, and then we found the best tube with highest AUC. The abnormal-vs-normal algorithm was used for analysis for Calibur and Canto-II, with two-fold cross-validation. As shown in Table 1, we found that learning from one single tube could yield a reliable AUC (ranging from 0·898 to 0·943 for Calibur, and from 0·829 and 0·886 for Canto-II), although we noted that the tubes with the best performance was not the same (5th tube (CD16/CD13/CD45) for Calibur and 2nd tube (HLA-DR/CD11b/CD45) for Canto-II).
Table 1.
Single tube feature selection analysis with two-fold validation.
| Datasets | AUC of each individual tube |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st | 2nd | 3rd | 4th | 5th | 6th | 7th | 8th | 9th | 10th | 11th | 12th | 13th | ||
| Calibur | Fold 1 | 0·898 | 0·924 | 0·913 | 0·917 | 0·902 | 0·917 | 0·914 | 0·911 | 0·931 | 0·931 | 0·924 | – | – |
| N | 2014 | 2016 | 2016 | 2016 | 2011 | 2011 | 2010 | 2010 | 1876 | 1908 | 2012 | – | – | |
| Fold 2 | 0·920 | 0·932 | 0·928 | 0·934 | 0·933 | 0·931 | 0·943 | 0·939 | 0·937 | 0·940 | 0·934 | |||
| N | 2070 | 2071 | 2072 | 2072 | 2068 | 2069 | 2069 | 2069 | 1927 | 1964 | 2070 | |||
| Canto-II | Fold 1 | 0·829 | 0·840 | 0·850 | 0·844 | 0·847 | 0·857 | 0·844 | 0·832 | 0·870 | 0·863 | 0·860 | 0·841 | 0·841 |
| N | 2149 | 2150 | 2149 | 2150 | 2149 | 2149 | 2149 | 2149 | 2147 | 1386 | 1290 | 798 | 815 | |
| Fold 2 | 0·843 | 0·848 | 0·849 | 0·858 | 0·869 | 0·859 | 0·859 | 0·821 | 0·874 | 0·858 | 0·841 | 0·844 | 0·886 | |
| N | 2197 | 2197 | 2196 | 2197 | 2196 | 2196 | 2196 | 2196 | 2193 | 1424 | 1322 | 816 | 828 | |
Markers measure in each tube are the same as that in Supplement Table S2.
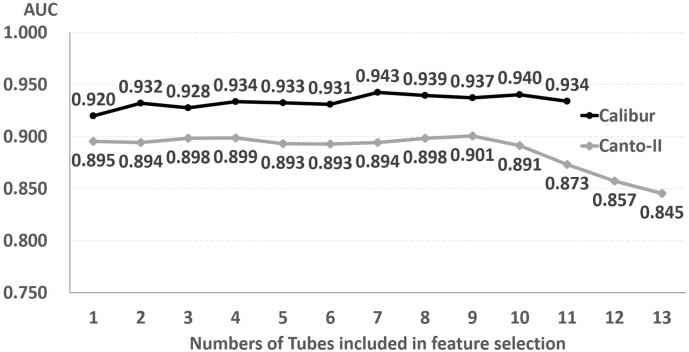
Next, we trained the algorithm by adding data from each of the remained tubes to that from previous selected tube(s), and we found the best 2-tube combination with highest AUC; the process was repeated until data from all tubes were included. The tubes selected in each round and the resultant AUCs were listed (Supp. Table S6), and the fold1 resultant AUCs were illustrated in Fig. 3. Interestingly, including data from more than the best 2-tubes did not significantly improve AUC scores in Calibur (from 0·932 (2 tubes) to 0·934 (11 tubes)). In Canto-II, data from 4 tubes seemed adequate to obtain high AUC scores (from 0·899 (4 tubes) to 0·845 (13 tubes)). These findings suggested that the SML approach could execute a binary classification well with just a fraction of tubes in the whole myeloid panel.
Fig. 3.
Feature selection analysis of abnormal vs normal classifier.
Tube combinations: 1: Calibur-5th tube (CD16, CD13 & CD45), Canto-II-2nd tube (HLA-DR, CD11b & CD45); 2: Calibur-5th & 11th tube (HLA-DR, CD34 & CD45), Canto-II-2nd & 4th tube (CD46, CD38 & CD45); 3: Calibur-5th & 9th (CD34, CD38 & CD45) & 11th tube, Canto-II-2nd & 4th & 7th tube (CD14, CD33 & CD45); 4: Calibur-2nd, 5th, 9th & 11th, Canto-II-2nd, 4th, 7th & 9th tube; 5: Calibur-2nd,4th,5th,9th & 11th tube, Canto-II-2nd, 4th, 6th (CD15, CD34 & CD45), 7th & 9th tube. Tubes included in each feature selection analysis experiments are listed in Supp. Table S5.
3.4. Prognostic value of AI-diagnosis of MFC data on clinical outcome
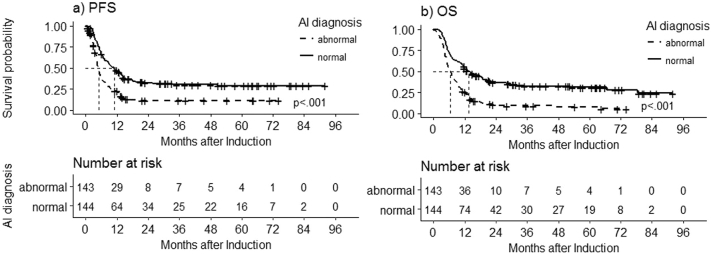
To evaluate the prognostic significance of the binary classification by AI, survival analysis was conducted on 287 AML patients in the outcome set. The median follow-up was 21·3 (ranging 1·0–96·1) months, and their demographics were shown in Table 2. Majority of them received standard induction chemotherapy (n = 262, 91·3%), and 144 (49·8%) had received allo-HSCT. Based on the genetic risk stratification [32], the adverse, intermediate and favorable risk categories took 19·5% (n = 56), 60·6% (n = 174) and 19·5% (n = 56) of the patients, respectively. The patients with abnormal post-induction MFC by AI had significant worse prognosis compared to those with normal one (median PFS: 4·9 (95% confidence interval (CI) 4·4–5·6) vs 10·9 (8·2–14·0) months, p < 0·0001 (Fig. 4A); median OS: 6·5 (95% CI 5·4–8·0) vs 13·6 (11·2–18·8) months, p < 0·0001 (Fig. 4B). In the univariate analysis, genetic risk groups and MFC diagnosis by AI had impacts on both PFS and OS (Table 3). Furthermore, multivariate analysis confirmed genetic risk groups and MFC diagnosis by AI were also independent prognostic factors (Table 3). These results were also illustrated by survival curve stratified by genetic risk groups (Supp. Fig. S2). For AML patients with favorable genetic risk, those with abnormal post-induction MFC by AI had significant worse PFS and OS than those with normal one (median PFS 5·3 (95% CI 4·8- not reached) vs 15·4 (12·9-not reached) months, p = 0·049; median OS 9·1 (95% CI 6·2-not reached) vs 28·1 (18·0-not reached) months, p = 0·031); this was also true for AML patients with intermediate genetic risk (median PFS 5·5 (95% CI 4·5–7·5) vs 10·6 (8·2–14·1) months, p < 0·001; median OS 6·7 (95% CI 5·3–9·1) vs 14·4 (11·2–22·0) months, p < 0·001). However, no significant differences were noted for AML patients with adverse genetic risk.
Table 2.
Patient demographics of the outcome set.
| Patient characteristics | N (%) |
|---|---|
| All patients | 287 (100·0%) |
| Gender (n = 287) | |
| Male | 132 (46·0%) |
| Female | 155 (54·0%) |
| Age (y) (n = 287) | |
| <30 | 21 (7·3%) |
| 30–39 | 61 (21·3%) |
| 40–49 | 57 (19·9%) |
| 50–59 | 63 (22·0%) |
| ≥60 | 85 (29·6%) |
| Induction chemotherapy (n = 287) | |
| Standard | 262 (91·3%) |
| Non-standard | 25 (8·7%) |
| Genetic groupa (n = 287) | |
| Adverse | 56 (19·5%) |
| Intermediate | 174 (60·6%) |
| Favorable | 56 (19·5%) |
| NA | 1 (0·3%) |
| HSCT (n = 287) | |
| Yes | 144 (49·8%) |
| No | 143 (50·2%) |
Abbreviation: HSCT, Hematopoietic stem cell transplant.
Genetic group assigned following 2017 European LeukemiaNet (ELN) recommendations.
Fig. 4.
Kaplan-Meier curves of progression-free survival (PFS) and overall survival (OS) by post-induction AI diagnosis in patients with AML.
(A) Significant longer post-induction PFS observed in the “AI diagnosis: normal” group (median PFS 10·9 months (95% CI 8·2–14·0 months), n = 144) compared to the “AI diagnosis: abnormal” group (median PFS 4·9 months (95% CI 4·4–5·6 months), n = 143), log-rank P < ·001. b) Significant longer post-induction OS was observed in the “AI diagnosis: normal” group (median OS 13·6 months (95% CI 11·2–18·8 months), n = 144) compared to the “AI diagnosis: abnormal” group (median OS 6·5 months (95% CI 5·4–8·0 months), n = 143), log-rank P < ·001
Abbreviation: AI, artificial intelligence; CI, confidence interval
Table 3.
Prognostic significance of variables in PFS and OS by univariate and Multivariate Cox proportional hazards regression analysis.
| PFS analysis subgroups | Univariate Cox analysis |
Multivariate Cox analysis |
||
|---|---|---|---|---|
| HR (95%CI) | P value | HR (95% CI) | P value | |
| Gender (Male: Female) | 1·00 (0·77–1·32) | 0·977 | – | – |
| Age (>50 y: ≤50 y) | 1·19 (0·87–1·63) | 0·288 | – | – |
| Genetic group (favorable: adverse) | 0·27 (0·17–0·42) | <001a | 0·31 (0·20–0·49) | 3·6 × 10−7 |
| Genetic group (intermediate: adverse) | 0·42 (0·30–0·59) | <001a | 0·42 (0·30–0·59) | 4·9 × 10−7 |
| Induction (Standard: non-standard) | 0·63 (0·39–1·01) | 0·057 | – | – |
| AI Diagnosis (no abnormality: abnormal) | 0·48 (0·37–0·63) | <001a | 0·52 (0·39–0·69) | 4·4 × 10−6 |
| OS analysis subgroups | Univariate Cox analysis |
Multivariate Cox analysis |
||
|---|---|---|---|---|
| HR (95%CI) | P value | HR (95% CI) | P value | |
| Gender (Male: Female) | 1·02 (0·78–1·33) | 0·885 | – | – |
| Age (>50 y: ≤50 y) | 1·14 (0·84–1·56) | 0·395 | – | – |
| Genetic group (favorable: adverse) | 0·25 (0·16–0·39) | <001a | 0·31 (0·19–0·48) | 3·8 × 10−7 |
| Genetic group (intermediate: adverse) | 0·46 (0·33–0·64) | <001a | 0·49 (0·35–0·68) | 2·0 × 10−5 |
| Induction (Standard: non-standard) | 0·75 (0·44–1·17) | 0·205 | – | – |
| AI Diagnosis (no abnormality: abnormal) | 0·44 (0·34–0·58) | <001a | 0·51 (0·38–0·67) | 1·4 × 10−6 |
Abbreviation: PFS, progression free survival; OS, overall survival, HR: hazard ratio, CI: confidence interval.
Included in multivariate Cox analysis.
4. Discussion
In this study, we showed that a SML approach combining GMM-based phenotype representation with SVM supervised models trained on a large amount of MFC data can rapidly classify specimens with high accuracy, and the results are of high prognostic significance for AML patients after induction chemotherapy. Furthermore, the average time for the algorithm to accomplish the task on one sample was roughly 7 s, in contrast with 20 min estimated to be required for manual gating by an experienced hematologist. Therefore, this study demonstrated that SML algorithm can be clinically-useful in supporting physicians to conduct MFC interpretation with high efficiency and fidelity. The time, manpower and training requirement for MFC interpretation can be significantly reduced.
Detecting MRD plays an important role in guiding decisions in treating hematological malignancies, because persistent detectable MRD usually indicates inadequate treatment and therefore implies poor prognosis [2]. In myeloid leukemia, e.g. AML and MDS, although detecting MRD with MFC is proved to be of prognostic significance for survival [[4], [5], [6], [7], [8], [9], [10]], the methodology is still evolving and no best strategy is identified yet, probably because the MFC expression profiles of normal bone marrow elements and their disease counterpart are significantly overlapped. Considering the nature of potential antigen expression alteration during AML disease progression, we used the expert labeling from the “different-from-normal” approach for our SML algorithm development; we also used “pooled” non-leukemic bone marrow as the normal template, instead of pre-set immunophenotypic phenotypes from experiences. Stressed bone marrow, therefore, can probably be more efficiently separated from true MRD-positive bone marrow samples.
Although the manual gating is still the mainstream of MFC interpretation in clinical service, interpersonal variability during gating has been shown as a major factor affecting outcome prediction in flow-cytometry based experiments [43]. Moreover, with modern MFC platforms measuring >100 parameters on a single-cell level [26], conventional 2D-plot manual gating is becoming an infeasible means to comprehensively present the information acquired in the measured MFC data to the physicians. Numerous groups have developed computational methods to accelerate the MFC data analysis to address this issue [12,[27], [28], [29],44]. SVM-based approaches have been shown to achieve great performance in leukemia vs non-leukemia cell classification. For instance, A SVM based model developed by Toedling et al. was able to distinguish acute lymphoblastic leukemia (ALL) from non-ALL cells with 99·78% specificity and 98·87% sensitivity [30]. However, the model was developed on a small cohort of 37 patients and the specimen sources include both peripheral blood and bone marrow. Distributional-based clustering approach has also been proposed in improving the visualization process, e.g., a non-parametric Bayesian model [45] or Gaussian Mixture Model (GMM) [46]. The GMM-based approach uses non-negative matrix factorization to derive lower dimension feature space in an unsupervised manner and could be effective for cell clustering purposes [46]. Since our goal is to distinguish the abnormal samples from normal samples not abnormal cells from normal cells within one sample, the supervised feature selection can directly extract effective feature dimensions.
An AutoFLOW project has been established and a software package developed supervised GMM approach to assist the ALL MRD assessment [31]. This study demonstrated that both SVM-based and GMM-based models are very promising to become a next-generation automated MFC analysis tool. However, they were developed on ALL disease with relative stable immunophenotypic compare to AML, which may have antigenic shift from diagnosis to relapse. In addition, in the bone marrow environment, the presence of normal myeloid lineages cells is mixed with malignant myeloid leukemia cells of AML patients, therefore, the approach that is successful for ALL specimens won't necessary applicable to AML. Hence the performance on classification of AML vs non-AML diseases should be investigated in separate studies.
Several SML approaches have been reported to have promising performance in analyzing AML MFC data. For instance, Thomas et al. used viSNE clustering to help improve visualization in the manual gating process to achieve better sensitivity in recognizing AML samples [47]. A LIBSVM model has shown to achieve 0·986 efficiency between automated and conventional analysis in AML MRD cell fraction assessment [32]. However, this model was trained on a small cohort (159 data from 36 patients), which can raise concerns about their representation of the heterogeneity of the disease in real-world setting. In addition to cell type classification models, the Flow-CAP project has identified multiple computational MFC analysis methods with great sample classification performance (accuracy 0·92–1·00). However, concerns about the representative sampling for the small cohort (only 43 AML out of 359 peripheral blood samples) still exist [12]. Another non-parametric Bayesian-GMM model has been shown to be able to recognize differences between normal and AML samples, and also the direction of change in disease progression [33]. The Bayesian-GMM model was developed using hyper-parameters prior to determining the number of clusters, which would be inefficient if the data and the clusters do not fit to assumption of prior. Compared to the posterior probability values as the phenotype vector approach used in this study [33], we used fisher-scoring phenotype vector to further include the gradient of probability function which provides strong discriminative power. Furthermore, the model was trained using 100 AML vs 100 non-AML peripheral blood and 49 stage I lymphoma vs 100 AML bone marrow specimens [33], both of which are small cohort and the comparison weren't being able to simulate that in clinical practice. Another potential drawback for above approaches is that their classification models were developed in peripheral blood samples, while evaluation on samples from bone marrow is still the mainstay in clinical practice. As the bone marrow environment contains many cell types at various developmental stages while majority cells in peripheral blood are fully developed and differentiated, classification models for these two specimens should be developed separately if we were to apply them in clinical setting, and to establish classification models in bone marrow would be more difficult [30]. Nowadays, many open sourced automated MFC analysis tools have been released, but it remains a challenge to perform comparisons across different subjects, time points, and experimental conditions [34,48]. For instance, continuous changes from premalignant MDS to AML make it hard to develop a distinct biology-based classification system because of their significant morphological and genetic diversity. Due to the heterogeneity of the MDS and AML as well as the complex composition of the bone marrow specimens, there hasn't been an algorithm developed for AML and MDS MRD detection and clinically validated. In our study, we addressed this issue by utilizing a large number of real-world samples consisting of both normal (non-diseased) and abnormal (diseased) clinical phenotypes to develop classification algorithms, which allows more flexibility when making a diagnosis.
GMM was used as our background generative model, and then a probabilistic gradient-based approach, i.e., the Fisher scoring vector [49], for deriving the high-dimensional MFC feature vector representation. This particular approach is both generative and discriminative. The feature vector captures the variabilities and interacting information on the multi-measurements per sample. The use of vectorized approach is important in achieving strong supervised classifier training on a large-scale data samples, and is important in speed up the computation. The remarkable performance can be attributed to fundamentally different approaches in terms of automating the diagnosis procedure. Deriving a phenotype representation that captures inherent variabilities in a high dimensional space in combination with maximum-boundary based optimization used in the SVM naturally provides a better predictive power.
In our study, three binary classification models for predicting AML-vs-normal, MDS-vs-normal and abnormal-vs-normal were constructed, instead of a multi-class classification model. This is because AML and MDS can represent as a continuous disease spectrum rather than two distinct diseases, as mentioned before. To address definite manual interpretation in MFC data from these cases would be of question, so that we still constructed three binary classification systems in our experiments. In all of our binary classification tasks, the algorithm performance reached over 0·85 ACC, suggesting a good consistency with manual analyses. Mismatching may be related to low-frequency aberrant phenotypes with inadequate training samples for algorithm, peripheral blood contamination in bone marrow samples, or even from misclassified manual gating due to interpersonal idiosyncrasy. Increasing training sample size, incorporating results from other MRD detecting methods for data labeling, or direct training with clinical endpoints may help to resolve this issue. Developing a multi-class classification model is also a potential future research direction.
We found that AUCs were generally slightly higher for Calibur sub-dataset compared to Canto-II sub-dataset and in Calibur+Canto-II sub-datasets (Supp. Table S5). The differences between Calibur and Canto-II sub-datasets originate not only from the machines but also in the collection of samples. The difference in the machine reading was mitigated by adopting the numerical transformation between the two machines given by the manufacturer, and we have further ensured the tube dimension was the same in our experiments. The pooled data together was to ensure the completeness of the experiments. However, it is evident that the differences between the two machines are not simply result of numerical reading but also additional factors (for example, the year for sample collection), which require future investigation study. This also further underscores the importance of our on-going effort in developing appropriate general/transferrable (or machine-appropriate) algorithms across sites and across machines.
Another interesting finding in our study is that the results of feature selection analysis support discarding the data dimensions on the tube level in order not only to reduce the computation loading on the classifier [50], and also provide understanding about the power of each tube in identifying the relevant disease. The study reported by Hassan et al. used the several statistics function to encode the raw feature as a vector while we used probability based clusters to encode raw features as our phenotype encoding vector method [50]. This approach allows more flexible and representative to describe the latent distribution compared to only using statistics values. Moreover, the SVM approach can be more discriminative in classification tasks compared to LR utilized in the Hassan et al. study [50].
We found that for Caliber sub-dataset, as few as one tube (3 markers) can achieve AUC of 0·920, while the AUC from all tubes was improved slightly to 0·934. The findings for Canto-II were similar. These results implicate that the tubes required for MRD detection could be greatly reduced with SML approach, and hence the time and cost of MFC running time. The biological implications of these findings are also worthy of further exploration.
In summary, machine learning is a powerful tool for automated MFC analysis on MRD detection in AML and MDS. It not only is a faster and reliable way for MFC data interpretation, but also possess a great advantage in its ability to integrate with other clinical tests including morphology, genomics, and cytogenetics for MRD detection and prognostic stratification. Although future research is still needed to validate the full spectrum of utilization in clinical practice, a clinical decision-making support system can be started with this scalable and reproducible approach.
Data sharing statement
The dataset and sub-datasets generated and/or analyzed in the current study are not publicly available because they contain historical patient data from National Taiwan University Hospital; but the de-identified parts (information excluding patient's identification, such as their ID or chart numbers) are available from the corresponding author upon reasonable request for research purpose after approval by the Research Ethic Committee of National Taiwan University Hospital.
The codes are available from the corresponding author upon reasonable request for research purpose after publication.
Acknowledgments
Acknowledgements
This work was supported by the Ministry of Science and Technology (107-2634-F-007-006 and 103-2314-B-002-185-MY2) of Taiwan.
The sponsors of this study are government departments that support science in general. They had no role in gathering, analyzing, or interpreting the data.
Declaration of interests
There is no conflict of interest to be declared for all authors.
Authors contributions
BSK, YFW, CC Lee and JLT designed the research; BSK., CC Li, PFW, SSH., HAH, HHH, MY, CTL, JHL, CHT, TCH, SJW, SYH, WCC, HFT and JLT interpreted the clinical and MFC data; BSK, YFW, JLL, CC Lee and JLT performed the experiments and analyzed the results; BSK, YFW, CC Lee and JLT wrote the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.042.
Contributor Information
Chi-Chun Lee, Email: cclee@ee.nthu.edu.tw.
Jih-Luh Tang, Email: tangjh@ntu.edu.tw.
Appendix A. Supplementary data
Supplementary material
References
- 1.Saultz J.N., Garzon R. Acute myeloid leukemia: A concise review. J Clin Med. 2016;5(3):33. doi: 10.3390/jcm5030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coltoff A., Houldsworth J., Keyzner A., Renteria A.S., Mascarenhas J. Role of minimal residual disease in the management of acute myeloid leukemia—a case-based discussion. Ann Hematol. 2018;97:1155–1167. doi: 10.1007/s00277-018-3330-9. [DOI] [PubMed] [Google Scholar]
- 3.Schuurhuis G.J., Heuser M., Freeman S. Minimal/measurable residual disease in AML: A consensus document from the European LeukemiaNet MRD Working Party. Blood. 2018;131:1275–1291. doi: 10.1182/blood-2017-09-801498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loken M.R., Alonzo T.A., Pardo L. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children's Oncology Group. Blood. 2012;120:1581–1588. doi: 10.1182/blood-2012-02-408336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grimwade D., Freeman S.D. Defining minimal residual disease in acute myeloid leukemia: Which platforms are ready for "prime time"? Blood. 2014;124:3345–3355. doi: 10.1182/blood-2014-05-577593. [DOI] [PubMed] [Google Scholar]
- 6.Kern W., Haferlach T. Quantification of minimal residual disease by multiparameter flow cytometry in acute myeloid leukemia. From diagnosis to prognosis. Med Klin (Munich) 2005;100:54–59. doi: 10.1007/s00063-005-1122-1. [DOI] [PubMed] [Google Scholar]
- 7.Terwijn M., van Putten W.L., Kelder A. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31:3889–3897. doi: 10.1200/JCO.2012.45.9628. [DOI] [PubMed] [Google Scholar]
- 8.Freeman S.D., Virgo P., Couzens S. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol. 2013;31:4123–4131. doi: 10.1200/JCO.2013.49.1753. [DOI] [PubMed] [Google Scholar]
- 9.Venditti A., Maurillo L., Buccisano F. Pretransplant minimal residual disease level predicts clinical outcome in patients with acute myeloid leukemia receiving high-dose chemotherapy and autologous stem cell transplantation. Leukemia. 2003;17:2178–2182. doi: 10.1038/sj.leu.2403138. [DOI] [PubMed] [Google Scholar]
- 10.Buccisano F., Maurillo L., Spagnoli A. Cytogenetic and molecular diagnostic characterization combined to postconsolidation minimal residual disease assessment by flow cytometry improves risk stratification in adult acute myeloid leukemia. Blood. 2010;116:2295–2303. doi: 10.1182/blood-2009-12-258178. [DOI] [PubMed] [Google Scholar]
- 11.Mosna F., Capelli D., Gottardi M. Minimal residual disease in acute myeloid leukemia: Still a work in progress? J Clin Med. 2017;6(6):57. doi: 10.3390/jcm6060057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aghaeepour N., Finak G., Flow C.A.P.C. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimwade D., Freeman S.D. Defining minimal residual disease in acute myeloid leukemia: Which platforms are ready for "prime time"? Hematology Am Soc Hematol Educ Program. 2014;2014:222–233. doi: 10.1182/asheducation-2014.1.222. [DOI] [PubMed] [Google Scholar]
- 14.Loken M.R. Residual disease in AML, a target that can move in more than one direction. Cytometry B Clin Cytom. 2014;86:15–17. doi: 10.1002/cyto.b.21140. [DOI] [PubMed] [Google Scholar]
- 15.Zeijlemaker W., Gratama J.W., Schuurhuis G.J. Tumor heterogeneity makes AML a "moving target" for detection of residual disease. Cytometry B Clin Cytom. 2014;86:3–14. doi: 10.1002/cyto.b.21134. [DOI] [PubMed] [Google Scholar]
- 16.Lugli E., Roederer M., Cossarizza A. Data analysis in flow cytometry: The future just started. Cytometry A. 2010;77:705–713. doi: 10.1002/cyto.a.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witten I.H., Frank E., Hall M.A., Pal C.J. 2016. Data Mining: Practical machine learning tools and techniques: Morgan Kaufmann. [Google Scholar]
- 18.Bishop C. 2006. Pattern recognition and machine learning: Springer-Verlag New York. [Google Scholar]
- 19.Mitchell T.M. Carnegie Mellon University; Pittsburgh, PA, USA: 2006. The discipline of machine learning. School of Computer Science, Carnegie Mellon University. [Google Scholar]
- 20.Hamet P., Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36–S40. doi: 10.1016/j.metabol.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A., Kuprel B., Novoa R.A. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteva A., Kuprel B., Novoa R.A. Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;546:686. doi: 10.1038/nature22985. [DOI] [PubMed] [Google Scholar]
- 23.Shouval R., Labopin M., Bondi O. Prediction of allogeneic hematopoietic stem-cell transplantation mortality 100 days after transplantation using a machine learning algorithm: A European group for blood and marrow transplantation acute leukemia working party retrospective data mining study. J Clin Oncol. 2015;33:3144–3151. doi: 10.1200/JCO.2014.59.1339. [DOI] [PubMed] [Google Scholar]
- 24.Gulshan V., Peng L., Coram M. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 25.Wong T.Y., Bressler N.M. Artificial intelligence with deep learning technology looks into diabetic retinopathy screening. JAMA. 2016;316:2366–2367. doi: 10.1001/jama.2016.17563. [DOI] [PubMed] [Google Scholar]
- 26.Mair F., Hartmann F.J., Mrdjen D., Tosevski V., Krieg C., Becher B. The end of gating? An introduction to automated analysis of high dimensional cytometry data. Eur J Immunol. 2016;46:34–43. doi: 10.1002/eji.201545774. [DOI] [PubMed] [Google Scholar]
- 27.Naim I., Datta S., Rebhahn J., Cavenaugh J.S., Mosmann T.R., Sharma G. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, part 1: Algorithm design. Cytometry A. 2014;85:408–421. doi: 10.1002/cyto.a.22446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosmann T.R., Naim I., Rebhahn J. SWIFT-scalable clustering for automated identification of rare cell populations in large, high-dimensional flow cytometry datasets, part 2: biological evaluation. Cytometry A. 2014;85:422–433. doi: 10.1002/cyto.a.22445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sorensen T., Baumgart S., Durek P., Grutzkau A., Haupl T. ImmunoClust--An automated analysis pipeline for the identification of immunophenotypic signatures in high-dimensional cytometric datasets. Cytometry A. 2015;87:603–615. doi: 10.1002/cyto.a.22626. [DOI] [PubMed] [Google Scholar]
- 30.Toedling J., Rhein P., Ratei R., Karawajew L., Spang R. Automated in-silico detection of cell populations in flow cytometry readouts and its application to leukemia disease monitoring. BMC Bioinform. 2006;7:282. doi: 10.1186/1471-2105-7-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takenga C., Dworzak M., Diem M. Lecture notes in computer science. Vol. 10209. Springer International Publishing; Cham: 2017. A clinical tool for automated flow cytometry based on machine learning methods; pp. 537–548. IWBBIO. [Google Scholar]
- 32.Ni W., Hu B., Zheng C. Automated analysis of acute myeloid leukemia minimal residual disease using a support vector machine. Oncotarget. 2016;7:71915–71921. doi: 10.18632/oncotarget.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajwa B., Wallace P.K., Griffiths E.A., Dundar M. Automated assessment of disease progression in acute myeloid leukemia by probabilistic analysis of flow cytometry data. IEEE Trans Biomed Eng. 2017;64:1089–1098. doi: 10.1109/TBME.2016.2590950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brinkman R.R., Aghaeepour N., Finak G., Gottardo R., Mosmann T., Scheuermann R.H. Automated analysis of flow cytometry data comes of age. Cytometry A. 2016;89:13–15. doi: 10.1002/cyto.a.22810. [DOI] [PubMed] [Google Scholar]
- 35.Dohner H., Estey E., Grimwade D. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tien H.F., Wang C.H., Lin M.T. Correlation of cytogenetic results with immunophenotype, genotype, clinical features, and ras mutation in acute myeloid leukemia. A study of 235 Chinese patients in Taiwan. Cancer Genet Cytogenet. 1995;84:60–68. doi: 10.1016/0165-4608(95)00084-4. [DOI] [PubMed] [Google Scholar]
- 37.Chou S.C., Tang J.L., Hou H.A. Prognostic implication of gene mutations on overall survival in the adult acute myeloid leukemia patients receiving or not receiving allogeneic hematopoietic stem cell transplantations. Leuk Res. 2014;38:1278–1284. doi: 10.1016/j.leukres.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Hou H.A., Lin C.C., Chou W.C. Integration of cytogenetic and molecular alterations in risk stratification of 318 patients with de novo non-M3 acute myeloid leukemia. Leukemia. 2014;28:50–58. doi: 10.1038/leu.2013.236. [DOI] [PubMed] [Google Scholar]
- 39.Corinna Cortes aVV . Vol. 20.3. 1995. Support-vector networks. Machine learning; pp. 273–297. [Google Scholar]
- 40.Vedaldi A., Fulkerson B. 2010. Vlfeat: An open and portable library of computer vision algorithms. Proceedings of the 18th ACM international conference on Multimedia; Firenze, Italy. 1874249: ACM; pp. 1469–1472. [Google Scholar]
- 41.Buitinck L., Louppe G., Blondel M. European conference on machine learning and principles and practices of knowledge discovery in databases. Czech Republic; Prague: 2013. API design for machine learning software: experiences from the scikit-learn project.https://hal.inria.fr/hal-00856511/document 2013-09-23. [Google Scholar]
- 42.R Core Team (2017) R: A language and environment for statistical computing: R foundation for statistical computing. https://www.R-project.org/ (Vienna, Austia.)
- 43.Maecker H.T., McCoy J.P., Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol. 2012;12:191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedreira C.E., Costa E.S., Lecrevisse Q., van Dongen J.J., Orfao A., Euroflow C. Overview of clinical flow cytometry data analysis: Recent advances and future challenges. Trends Biotechnol. 2013;31:415–425. doi: 10.1016/j.tibtech.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 45.Dundar M., Akova F., Yerebakan H.Z., Rajwa B. A non-parametric Bayesian model for joint cell clustering and cluster matching: identification of anomalous sample phenotypes with random effects. BMC Bioinform. 2014;15:314. doi: 10.1186/1471-2105-15-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiter M., Rota P., Kleber F., Diem M., Groeneveld-Krentz S., Dworzak M. Clustering of cell populations in flow cytometry data using a combination of Gaussian mixtures. Pattern Recog. 2016;60:1029–1040. [Google Scholar]
- 47.Köhnke T., Rechkemmer S., Bücklein V.L. Improved detection of minimal residual disease by flow cytometry in AML by combining manual gating and visne clustering. Blood. 2015;126 (2593–93) [Google Scholar]
- 48.Kvistborg P., Gouttefangeas C., Aghaeepour N. Thinking outside the gate: single-cell assessments in multiple dimensions. Immunity. 2015;42:591–592. doi: 10.1016/j.immuni.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perronnin F., Dance C. 2007. Fisher kernels on visual vocabularies for image categorization. 2007 IEEE Conference on Computer Vision and Pattern Recognition; Minneapolis, MN; pp. 1–8. [Google Scholar]
- 50.Hassan S.S., Ruusuvuori P., Latonen L., Huttunen H. Flow cytometry-based classification in cancer research: A view on feature selection. Cancer Inform. 2015;14:75–85. doi: 10.4137/CIN.S30795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material