ABSTRACT
Platysternon megacephalum is the sole living representative of the poorly studied turtle lineage Platysternidae. Their mitochondrial genome has been subject to gene rearrangement and control region duplication, resulting in a unique mitochondrial gene order in vertebrates. In this study, we sequenced the first full-length turtle (P. megacephalum) liver transcriptome using single-molecule real-time sequencing to study the transcriptional mechanisms of its mitochondrial genome. ND5 and ND6 anti-sense (ND6AS) forms a single transcript with the same expression in the human mitochondrial genome, but here we demonstrated differential expression of the rearranged ND5 and ND6AS genes in P. megacephalum. And some polycistronic transcripts were also reported in this study. Notably, we detected some novel long non-coding RNAs with alternative polyadenylation from the duplicated control region, and a novel ND6AS transcript composed of a long non-coding sequence, ND6AS, and tRNA-GluAS. These results provide the first description of a mtDNA transcriptome with gene rearrangement and control region duplication. These findings further our understanding of the fundamental concepts of mitochondrial gene transcription and RNA processing, and provide a new insight into the mechanism of transcription regulation of the mitochondrial genome.
KEYWORDS: Platysternon megacephalum, mtDNA, gene rearrangement, CR duplication, full-length transcriptome
Introduction
All vertebrate mitochondrial DNAs (mtDNAs) characterized to date are double-strand circular molecules about 16 kb in size, usually comprising coding regions for 13 proteins, two rRNAs, 22 tRNAs, and a major non-coding control region (CR) [1]. The first complete mtDNA sequence of the turtle Platysternon megacephalum was determined and shown to encode 37 genes, along with rearrangement of ND5, two CRs (reported to show high similarity at the 5′ end of 800 bp in all individuals), and three non-coding spacers (Figure 1) [2]. However, few studies have examined CR duplication and gene rearrangement in P. megacephalum mtDNA, except for some molecular phylogenetic research on mtCRs, which suggested an independent evolution pattern of the two mtCRs [3]. CR duplication has increasingly been reported in mtDNA from different species with the duplicated CRs showing identical or high similarity [4–6]. However, mtDNA is characterized by a high mutation rate [7], and they have been subject to strong selection pressure over a prolonged period, which suggest that the widespread duplicated CRs must have positive effects on gene expression and regulation. An interesting experiment using cultured human transmitochondrial cell lines showed that the cells initially contained a mixture of single-CR and multiple-CR mtDNA, but the mtDNA population eventually shifted towards multiple CRs, suggesting positive selection for cells with multiple CRs [8]. Furthermore, the mtCR contains the main regulatory sequences for transcriptional termination, transcription, initiation, and replication initiation in most animals [9]. The significance of CR duplication in certain species thus warrants attention; but indeed, information on duplicated CRs in the transcription area is currently lacking.
Figure 1.
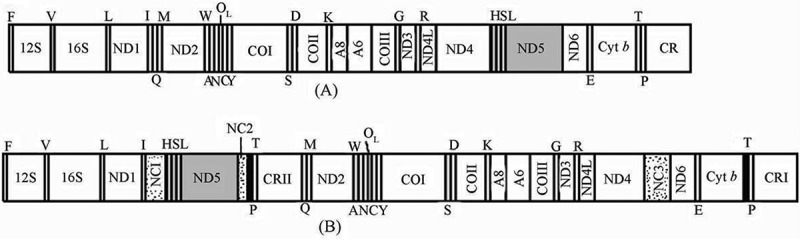
Comparison of mitochondrial gene arrangements between typical vertebrates (A) and P. megacephalum (B). ND1–6, and ND4L: subunits 1–6 and 4L of nicotine amid adenine dinucleotide dehydrogenase; A 6 and 8: ATP synthase F0 subunits 6 and 8; CO1–3: cytochrome c oxidase subunits 1–3; 12S and 16S: 12 and 16S rRNA. CR I and II: control region I and II; NC1–3: non-coding spacers 1–3; ØT: pseudo T; OL represents the replication origin of L strand [2].
The mitochondrial genome in animals can not only encode some coding RNAs, but also process the polycistronic precursors [10–12], and revealed a set of non-coding RNAs (lncRNAs) including lncND5, lncND6, lncCyt b RNA, sense mitochondrial non-coding RNA, and the long intergenic non-coding RNA predicting cardiac remodeling, some of which have been shown to be related to human diseases [10,11,13–15]. A recent study also revealed the two novel lncRNAs MDL1 (mitochondrial D-loop 1) and MDL1AS (mitochondrial D-loop 1 antisense) from mtCRs, predicted to be long transcription initiation RNAs in the human mitochondrial genome, while MDL1 was proposed to occur extensively in all animals [12]. The mtDNAs from many species have been sequenced and many of them have been shown to include special structures in terms of gene order and non-coding regions. It is therefore important to determine if these distinctive non-coding regions contain any novel transcripts, and to further to explore their function.
Progress in high-throughput sequencing technology have advanced research into gene transcription [16–18], and single-molecule real-time (SMRT) sequencing technology has been used to generate transcriptome data for Sorghum bicolor [19], Zea mays [20], and Anopheles stephensi [21], resulting in further insights in the complexity of gene transcription. In the current study, we sequenced the first full-length transcriptome from the liver of turtle P. megacephalum using SMRT technology, and constructed a quantitative transcripts map of the P. megacephalum mitochondrial genome. Identification of full-length transcripts allowed us to gather more comprehensive and accurate information on transcription, RNA processing, mRNA maturation, and other topics in relation to the mitochondrial genome. This study aimed to find out if the non-coding regions, especially in the two CRs, possess any novel transcripts, and to further analyze the distinct structural features of these transcripts in P. megacephalum mtDNA. We also aimed to determine the regulatory transcriptional functions of the two CRs and the molecular mechanisms of RNA processing and mRNA maturation, and to explore the influence of the gene rearrangement on mtDNA genetic transcription.
Results
Single-molecule transcripts and re-sequenced mitochondrial genome
Three Iso-Seq libraries with insert sizes of 1–2, 2–3, and 3–6 kb were sequenced with a total of seven PacBio SMRT cells. After removing low-quality regions and both end adapters, the raw reads were split into high-quality (accuracy ≥ 75%) sub-reads. All the sub-reads were processed into circular consensus sequencing reads that were then used to produce draft reads of insert transcripts (not containing 5′ primer, 3′ primer, or polyA tail). A library with insert sizes of 1–2 kb was generated by three smart cells and produced 274,152 draft reads of insert transcripts with a 2233-bp mean insert read length. Libraries of 2–3 kb and 3–6 kb were generated by two smart cells, respectively. The former produced 166,087 draft transcripts with a 3119-bp mean read length, and the latter produced 133,308 draft reads with a 4553-bp mean read length. The re-sequenced mitochondrial genome is annotated in Table 1, showing the same gene order and genetic makeup as described by Peng et al (Supplementary file 1) [2].
Table 1.
Annotation of the P. megacephalum mitochondrial genome.
| Name | Start | Stop | Strand | Length |
|---|---|---|---|---|
| tRNA-Phe | 1 | 70 | + | 70 |
| 12S | 71 | 1028 | + | 958 |
| tRNA-Val | 1028 | 1096 | + | 69 |
| 16S | 1097 | 2702 | + | 1607 |
| tRNA-Leu | 2702 | 2777 | + | 76 |
| ND1 | 2778 | 3743 | + | 966 |
| tRNA-Ile tRNA-His |
3749 3948 |
3819 4017 |
+ + |
71 70 |
| tRNA-Ser | 4018 | 4083 | + | 66 |
| tRNA-Leu | 4084 | 4154 | + | 71 |
| ND5 | 4156 | 5930 | + | 1771 |
| tRNA-Thr | 6087 | 6155 | + | 69 |
| tRNA-Pro | 6156 | 6224 | - | 69 |
| CRII | 6225 | 7628 | 1404 | |
| tRNA-Gln | 7629 | 7699 | - | 71 |
| tRNA-Met | 7699 | 7767 | + | 69 |
| ND2 | 7768 | 8802 | + | 1035 |
| tRNA-Trp | 8807 | 8885 | + | 79 |
| tRNA-Ala | 8887 | 8955 | - | 69 |
| tRNA-Asn | 8957 | 9030 | - | 74 |
| tRNA-Cys | 9059 | 9124 | - | 66 |
| tRNA-Tyr | 9125 | 9195 | - | 71 |
| COI | 9200 | 10735 | + | 1536 |
| tRNA-Ser | 10736 | 10806 | - | 71 |
| tRNA-Asp | 10810 | 10879 | + | 70 |
| COII | 10880 | 11560 | + | 681 |
| tRNA-Lys | 11568 | 11639 | + | 72 |
| ATP8 | 11641 | 11802 | + | 162 |
| ATP6 | 11811 | 12493 | + | 683 |
| COIII | 12494 | 13276 | + | 783 |
| tRNA-Gly | 13278 | 13347 | + | 70 |
| ND3 | 13348 | 13696 | + | 349 |
| tRNA-Arg | 13698 | 13766 | + | 69 |
| ND4L | 13767 | 14060 | + | 294 |
| ND4 | 14057 | 15433 | + | 1377 |
| ND6 | 16077 | 16598 | - | 522 |
| tRNA-Glu | 16599 | 16666 | - | 68 |
| Cytb tRNA-Thr |
16671 18815 |
17814 17884 |
+ + |
1144 70 |
| tRNA-Pro | 17886 | 17956 | - | 71 |
| CRI | 17957 | 19036 | 1080 |
Quantitative transcripts map of P. megacephalum mitochondrial genome
The full-length transcripts of the mRNAs, rRNAs, and tRNAs were identified by circular consensus sequencing reads to confirm their fidelity (Supplementary file 2). The draft transcripts without the 5′ end cDNA primers, 3′ end cDNA primers, and 3′ polyA sequences were used to construct a quantitative transcripts map of the mitochondrial genome (Figure 2). Alignment of the draft transcripts to the mitochondrial genome showed the visual quantitative transcripts with a mapping rate of 0.9% (5461/573,547). The transcript expression levels from the highest to the lowest were COI, COII, ATP8/6, COIII, ND4L/4, ND5, 16S, ND1, Cytb, ND3, 12S, ND2, and ND6. It is worth noting that the same phenomenon of low expression of ND2, and ND6 was also reported in the human mitochondrial genome and insect mitochondrial genome, thus we suggested that it’s a universal phenomenon of the low transcriptional level of these genes. The low expression of the ND6 could be the reason of ND6AS transcripts [12,22], but the down-regulated expression of ND2 could be a reason that ND2 mRNA may have a short half-life to maintain the low transcriptional level.
Figure 2.
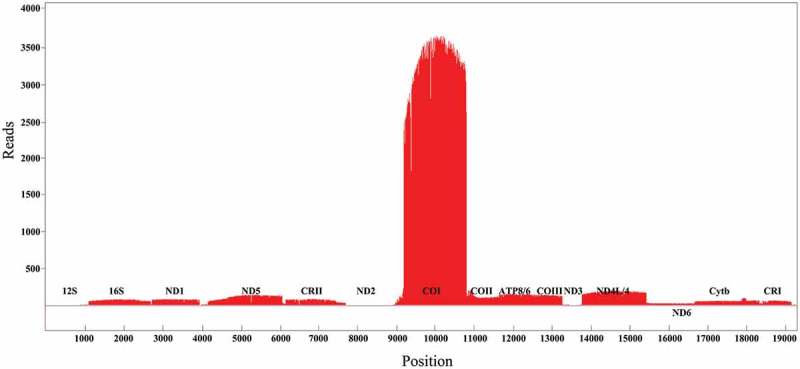
The quantitative transcription map of P. megacephalum mitochondrial genomes. Alignments of transcripts on Heavy strand in red color were piled along the positive y-axis. Alignments of transcripts on Light strand in Green color were piled along the negative y-axis. tRNAs were not represented.
Detection of lncRNAs from CRI and CRII
Previous studies have revealed novel transcripts in the mtCR MDL1 region [12,22]. The current results also detected polycistronic transcripts (Supplementary file 1) aligned forwardly to the P. megacephalum CRI. The long polycistronic transcripts covered Cytb/tRNA-Thr/tRNA-ProAS/CRI (ChrM:16,671–19,003), and was named tRNA-ProAS/CRI (ChrM:17,886–19,003) pm-MDL1 in accordance with the previous research. Interestingly, there were two kinds of lncRNAs related to CRII detected in P. megacephalum mtDNA. The one covered the antisense tRNA-Pro (tRNA-ProAS) gene and antisense tRNA-Gln (tRNA-GlnAS), named as tRNA-ProAS/CRII/tRNA-GlnAS (ChrM: 6156–7699), the other only covered the tRNA-Pro antisense gene and CRII, named pm-MDL2 (mitochondrial D-loop 2). And the pm-MDL2 also showed alternative polyadenylation, which generated three isoforms, named pm-MDL2-1 (ChrM:6156–7471), pm-MDL2-2 (ChrM:6156–7450), and pm-MDL2-3 (ChrM:6156–7425) (Figure 3).
Figure 3.
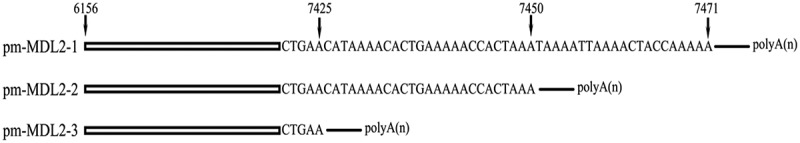
Transcript structures of LncRNAs pm-MDL2-1, pm-MDL2-2, pm-MDL2-3. Arrows represented the alternative polyadenylation sites.
CRI and CRII contained three conserved functional sections including a terminal-associated sequence, central conserved domain, and conserved sequence blocks (CSB) in all individuals [3]. CRI has a variable number tandem repeats at the 3′ end, while CRII does not have this kind of sequence feature. The position of the polyadenylated sites also differed between pm-MDL1 and pm-MDL2. In pm-MDL1, the polyadenylated sites were located between CSB1 and CSB2, but in pm-MDL2, and the polyadenylated sites were behind the 3′ end of CSB3 (Figure 4).
Figure 4.
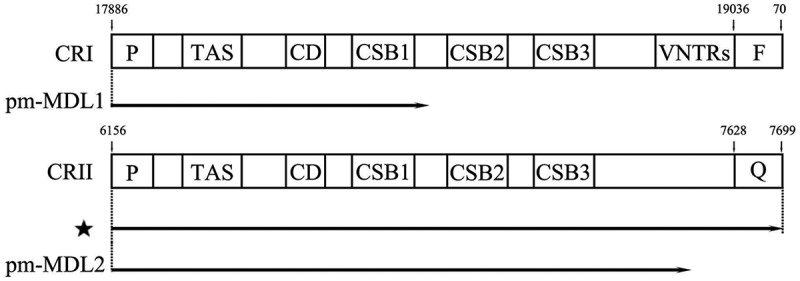
Comparison of transcription of pm-MDL1 and pm-MDL2. Prepresents tRNA-pro, F represents tRNA-Phe, Q represents tRNA-Gln, ⋆ represents the LncRNA of tRNA-ProAS/CRII/tRNA-GlnAS.
Monocistronic transcripts, polycistronic transcripts, RNA precursors, and RNA processing
Single-molecule long-read sequencing detected both monocistronic and polycistronic transcripts in P. megacephalum mtDNA. In this study, the mRNA transcripts of ND5 with a 3′ end untranslated region, COII, and 16S rRNA encoded by the major coding strand were generated as monocistronic transcripts. Two mature transcripts (ATP8/6 and ND4/4L) were identified as polycistronic mRNAs, ATP8/6/COIII (ChrM:11,641–13,277) and ND4L/4 (ChrM:13,767–15,433). Other types of polycistronic transcripts included CO1/tRNA-SerAS (ChrM:9194–10806) containing tRNA and a coding gene, tRNA-Leu/ND1/tRNA-Ile/Non-coding (ChrM:2711–3947) containing tRNA, a coding gene, and a non-coding sequence at the 3′ end, COII/tRNA-Lys/ATP8/6/COIII (ChrM:10880–13277), COI/tRNA-SerAS/tRNA-Asp/COII/tRNA-Lys/AT8/6/COIII (ChrM:9341–13277), and tRNA-Asp/COII/tRNA-Lys/ATP8/6/COIII (ChrM:11810–11277) containing more than two coding genes and tRNAs (Supplementary file 2). A diagram explaining the processing of the COI to COIII transcripts was shown in Figure 5.
Figure 5.
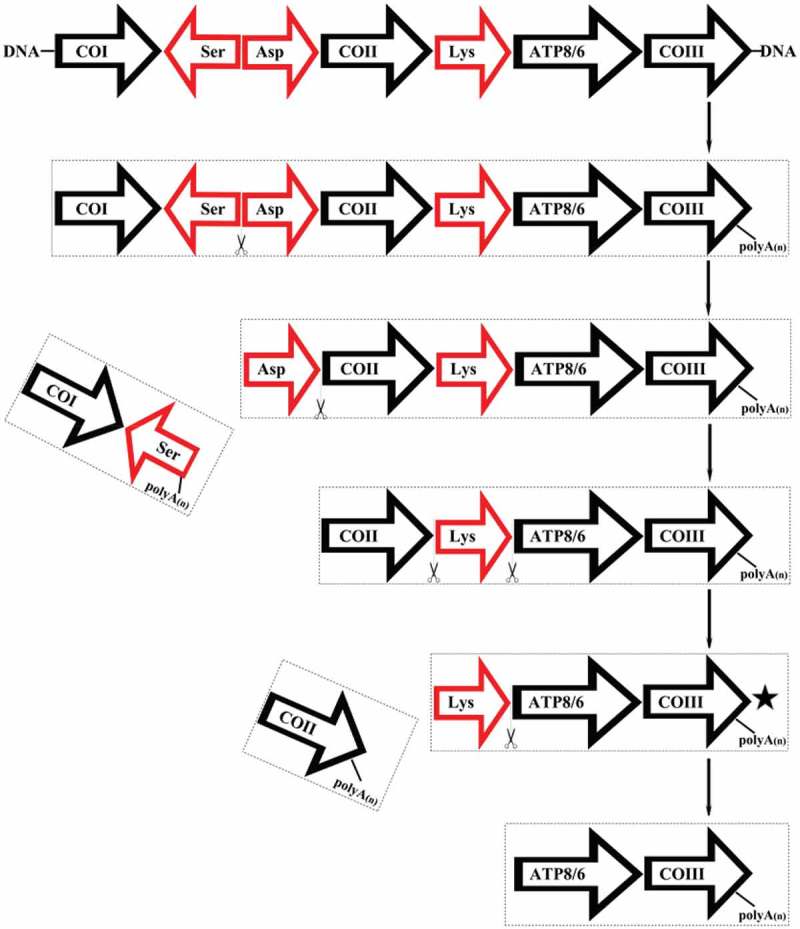
Models of processing COI to COIII. ⋆ represents putative transcripts, which were not obtained in this study.
Novel transcript of ND6AS
Gao et al. identified ND5/ND6AS/tRNA-GluAS as a single polycistronic transcript in the human mitochondrial genome [12]. A similar situation was observed in the turtle, Mauremys reevesii, which showed the same mitochondrial genome order as in humans (data not shown). However, rearrangement of the mitochondrial genome in P. megacephalum resulted in a change in the position of ND5, and the ND5/ND6AS/tRNA-GluAS transcripts could no longer be detected. Instead, we obtained two mature transcripts, ND5/non-coding (ChrM:4156–6084) and ND6AS/tRNA-GluAS (ChrM:15,434–16,666). Furthermore, we found a long non-coding sequence of 645 bp at position 15,434–16,076 in front of the 5′ end of ND6AS/tRNA-GluAS, and the non-coding sequences seemed not to be further cleaved from ND6AS/tRNA-GluAS. A 1234-bp novel transcript was therefore found in the P. megacephalum mitochondrial transcriptome. ORF Finder predicted four peptides in the novel transcript as follows: CFKNYLMQFQVMMAPMALAWHLALLTPMLPYYNTPHSSMKQHYSTILVQLF MSMSTELTMLNLNLPTTSRTKYMLL (ChrM:15,551–15,781), HLLMAPMTTNNLISLKINKDKNHITIYTWTMTKLTKSTYYSPTHAH (ChrM:15,887–16,027), LHKAPHETSHEPIPAQQMMLTMALNQQSKKLHYLNNKIMTLH (ChrM:16,079–16,207), and DMAPQPVQPNKQKLPMYPPNKLKTTLKKQKTPQHSPTPRTQKPPPGLNLKLRNKATDYKMRQPFKKQNKTQMKM (ChrM:16,364–16,588).
Discussion
In this study, we sequenced the first full-length liver of turtle transcriptome by single-molecule long-read sequencing on the PacBio platform. The main discoveries included differential gene expression, monocistronic and polycistronic transcripts, and RNA precursors of P. megacephalum mitochondrial genes. Notably, we identified some novel transcripts, including the lncRNAs of pm-MDL1, pm-MDL2-1, pm-MDL2-2, pm-MDL2-3, and tRNA-ProAS/CRII/tRNA-GlnAS, and a novel transcript of ND6AS/tRNA-GluAS. These findings may provide new insights into mitochondrial gene transcription in animal mitochondrial genomes showing gene rearragments and CR duplication.
Gao et al. identified ND5/ND6AS/tRNAGluAS as a single polycistronic transcript, and ND5 showed low expression together with ND6AS/tRNAGluAS [12]. However, in the current study, the order was rearranged in P. megacephalum, and ND5 showed higher expression than ND6AS/tRNAGluAS in liver. We proposed that ND6AS/tRNAGluAS existed at the 3′ end of ND5, and played a significant negative regulatory role in maintaining the expression of ND5 in most vertebrates without mitochondrial gene rearrangement. We detected a novel mtDNA-encoded ND6AS transcript with low abundance in P. megacephalum, but were unable to determine whether it was a lncRNA or not, owing to the sequence was predicted to encode polypeptides. Further studies are therefore needed to ensure if this novel transcript codes for some proteins or just a novel lncRNA. If the answer was the the former, we need to determine its function in regulating mitochondria, nuclei, or in both.
The mitochondrial CR has been reported to be related to mtDNA transcription and replication [23]. The current results identified several lncRNAs in CRI and the duplicated CRII in liver, raising the issue of the functions of CRI and CRII in regulating transcription initiation or termination of mtDNA. We suggested that both CRI and CRII contained a transcription termination region, given that the 3′ ends of pm-MDL1 and pm-MDL2 terminated respectively in the middle of CRI and CRII. However, we also detected numerous tRNA-ProAS/CRII/tRNA-GlnAS transcripts, which might indicate that the function of this transcription termination region in CRII was inefficient. And as well, the function of these unique lncRNAs in the liver of P. megacephalum warrants further investigation.
In conclusion, CR duplication and gene rearrangments in the mtDNA have been reported in increasing numbers of species [24–26], and we accordingly provided the first evidence of the mtDNA structure in the unique turtle specis P. megacephalum. But in this study, the analysis is based on a single individual with only one tissue of liver, so it cannot be ignored that tissue specificity has the effects on our results. The mitochondrial genome includes specific sequence structures in many species, and the current findings thus further our understanding of mitochondrial genome transcription and provide the basis for further exploration of mitochondrial CR duplication and gene rearrangments.
Materials and methods
Library preparation and full-length transcriptome sequencing
Procedures involving animals and their care were consistent with NIH guidelines (NIH Pub.No. 85–23, revised 1996) and in accordance with the approval of the Committee of Anhui Normal University under approval number #20170611.
One adult turtle (P. megacephalum) was collected from Yuehe pet market in Guangzhou, Guangdong province in the middle of April 2017 and kept 1 month at the provincial key laboratory of the conservation and exploitation research of biological resources in Anhui, China. In early June, the turtle was selected to extract liver for total RNA extraction with Trizol Universal Reagent (Tiangen). The agarose gel electrophoretic analysis showed degradation of the RNA. And the total RNA was eluted with nuclease-free H2O.
Then, the qualified total RNA was subjected to PacBio sequencing at Tianjin Biochip Corporation (Tianjin, China). The RNA was reverse transcribed using a The Clontech SMARTer PCR cDNA Synthesis Kit and amplified with 3ʹ SMART CDS Primer II A (5ʹ-AAGCAGTGGTATCAACGCAGAGTAC-T(30)-3ʹ) and SMARTer II A Oligonucleotide (5ʹ-AAGCAGTGGTATCAACGCAGAGTACATGGG-3ʹ). Three sequencing libraries (1–2, 2–3, 3–6 kb) were prepared according to the PacBio Iso-Seq protocol. The sequencing was performed on the PacBio RS II sequencer, seven SMRT cells with three SMRT cells for library of 1–2 kb, each two for libraries of 2–3 kb and 3–6 kb were run.
DNA sequencing of P. megacephalum mitochondrial genome
Two individuals (P. megacephalum), which were also collected from Yuehe pet market, were selected to re-sequencing the mtDNA using Sanger sequencing. The one was used as a specimen for determination of the complete mitochondrial sequence, and the other was used for determination of partial sequences to confirm the gene order or sequences in the mitochondrial genome. The primers and the methods followed Peng et al in 2006 [2]. The annotation of (P. megacephalum) mitochondrial genes was conducted using MITOS server (http://mitos.bioinf.uni-leipzig.de).
Data analysis
Analysis was performed using the PacBio SMART Analysis v2.3.0 (http://www.pacb.com/devnet/). The three libraries were analyzed separately. The circular consensus sequencing (CCS) reads were produced with parameters (Minimum Full Passes = 0, Minimum Predicted Accuracy = 75), then the draft reads of insert transcripts were produced with parameters (Minimum Sequence Length = 300) by removing the 5ʹ end cDNA primers, 3ʹ end cDNA primers and 3ʹ ployA sequences. We used Blastn to query all CCS to the complete mitochondrial genome reference and filtered out the high matching error rate CCS. Mecat2ref, one module of MECAT was used to align draft transcripts to the complete mitochondrial genome (https://github.com/xiaochuanle/mecat) [27]. And IGV v2.4.5 was used for data visualization alignment result (http://www.broadinstitute.org/igv) [28].
Funding Statement
This research was supported by the National Natural Science Foundation of China (NSFC, No. 31372198) and the Research Fund of the Key Laboratory of Biotic Environment and Ecological Safaty of Anhui province.
Acknowledgments
We would like to thank Genergy for providing sequencing service and primary data analysis. This research was supported by the National Natural Science Foundation of China (NSFC, No. 31372198) and the Research Fund of the Key Laboratory of Biotic Environment and Ecological Safaty of Anhui province. We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data for this article can be accessed here.
References
- [1].Clayton DA. Transcription and replication of animal mitochondrial DNAs Int Rev Cytol. 1992;141:217–232. [DOI] [PubMed] [Google Scholar]
- [2].Peng QL, Nie LW, Pu YG. Complete mitochondrial genome of Chinese big-headed turtle, Platysternon megacephalum, with a novel gene organization in vertebrate mtDNA. Gene. 2006;380:14–20. [DOI] [PubMed] [Google Scholar]
- [3].Zheng C, Nie L, Wang J, et al. Recombination and evolution of duplicate control regions in the mitochondrial genome of the asian big-headed turtle, Platysternon megacephalum. PLoS One. 2013;8:e82854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shao R, Barker SC, Mitani H, et al. Evolution of duplicate control regions in the mitochondrial genomes of metazoa: a case study with australasian ixodes ticks. Mol Biol Evol. 2005;22:620–629. [DOI] [PubMed] [Google Scholar]
- [5].Jiang ZJ, Castoe TA, Austin CC, et al. Comparative mitochondrial genomics of snakes: extraordinary substitution rate dynamics and functionality of the duplicate control region. BMC Evol Biol. 2007;7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cathryn LA, Michael CD, John HT, et al. An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses. Mol Ecol. 2005;14:3605–3613. [DOI] [PubMed] [Google Scholar]
- [7].Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci. 1979;76:1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tang Y, Manfredi G, Hirano M, et al. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol Biol Cell. 2000;11:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Silva PF, José AE, Montoya J. Replication and transcription of mammalian mitochondrial Dna. Exp Physiol. 2004;88:41–56. [DOI] [PubMed] [Google Scholar]
- [10].Rackham O, Shearwood AM, Mercer TR, et al. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. Rna-A Publ Rna Soc. 2011;17:2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Barshad G, Marom S, Cohen T, et al. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet. 2018;34:682–692. [DOI] [PubMed] [Google Scholar]
- [12].Gao S, Tian X, Chang H, et al. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion. 2018;38:41–47. [DOI] [PubMed] [Google Scholar]
- [13].Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dorn GW. LIPCAR, A mitochondrial lnc in the noncoding RNA Chain? Circ Res. 2014;114:1548–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lung B, Zemann A, Madej MJ, et al. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006;34:3842–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Clark WC, Evans ME, Dominissini D, et al. tRNA base methylation identification and quantification via high-throughput sequencing. RNA. 2016;22:1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Simpson RM, Bruno AE, Bard JE, et al. High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA. 2016;22:677–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lipovich L, Hou ZC, Jia H, et al. High‐throughput RNA sequencing reveals structural differences of orthologous brain‐expressed genes between western lowland gorillas and humans. J Comp Neurol. 2015;524:288–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abdel-Ghany SE, Hamilton M, Jacobi JL, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 2016;7:11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang B, Tseng E, Regulski M, et al. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun. 2016;7:11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jiang X, Hall AB, Biedler JK, et al. Single molecule RNA sequencing uncovers trans-splicing and improves annotations in Anopheles stephensi. Insect Mol Biol. 2017;26:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gao S, Ren Y, Sun Y, et al. PacBio full-length transcriptome profiling of insect mitochondrial gene expression. RNA Biol. 2016;13:820–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod. 2000;15:11–17. [DOI] [PubMed] [Google Scholar]
- [24].Eberhard JR, Wright TF, Bermingham E. Duplication and concerted evolution of the mitochondrial control region in the parrot genus amazona. Mol Biol Evol. 2001;18:1330–1342. [DOI] [PubMed] [Google Scholar]
- [25].Lee J-S, Miya M, Lee Y-S, et al. The complete DNA sequence of the mitochondrial genome of the self-fertilizing fish Rivulus marmoratus (Cyprinodontiformes, Rivulidae) and the first description of duplication of a control region in fish. Gene. 2001;280:1–7. [DOI] [PubMed] [Google Scholar]
- [26].Liu H, Li H, Song F, et al. Novel insights into mitochondrial gene rearrangement in thrips (Insecta: thysanoptera) from the grass thrips, Anaphothrips obscurus. Sci Rep. 2017;7:4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xiao CL, Chen Y, Xie SQ, et al. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods. 2017;14:1072. [DOI] [PubMed] [Google Scholar]
- [28].Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


