Abstract
Autophagy is a key biological phenomenon conserved from yeast to mammals. Under basal conditions, activation of autophagy leads to the protein degradation as well as damaged organelles for maintaining cellular homeostasis. Deregulation of autophagy has been identified as a key mechanism contributing to the pathogenesis and progression of several liver diseases, including hepatocellular carcinoma (HCC), one of the most common and mortal types of cancer. Currently used treatment strategies in patients with HCC result in variable success rates. Therefore, novel early diagnosis and treatment techniques should be developed. Manipulation of autophagy may improve responses of cancer cell to treatments and provide novel targeted therapy options for HCC. In this review, we summarized how our understanding of autophagy-cell death connection may have an impact on HCC therapy.
Keywords: Autophagy, HCC, cell death, chemotherapy
INTRODUCTION
Autophagy is a catabolic response of cells to stress. During this process, cargo is delivered to the lysosomes for degradation, supporting new building block synthesis and allowing cells to maintain homeostasis. Autophagy is active at a basal level in cells, and it may further be upregulated in response to several types of stresses that disturb cellular homeostasis, including low cellular ATP levels, nutrient and growth factor deprivation, hypoxic conditions, endoplasmic reticulum (ER) stress, pathogen entry, or anticancer drug treatment (1). Autophagy products feed into cellular energy-generation pathways, facilitating cell survival under stressful conditions. In contrast, overactivation of autophagy may indeed lead to cell death through so far not well understood mechanisms as an alternative nonapoptotic programmed cell death mechanism, “autophagic cell death” has been reported to be responsible for killing cells in a number of scenarios (2–4).
Abnormalities related to autophagy are known to be related to various human pathologies ranging from neurodegenerative diseases to cancer, including hepatocellular carcinoma (HCC) (5). Moreover, autophagy has been described as one of the central pathways for liver health and disease. In starved animals, a grand majority of total protein and glycogen degradation in the liver depends on autophagic degradation (6). On the other hand, autophagy is related to several liver diseases, including fatty liver disease and HCC (7,8). For instance, blockage of autophagy and autophagolysosomal degradation in mice using genetic tools resulted in hepatosteatosis and hepatomegaly (9).
The role of autophagy in cancer-related processes is currently under investigation. Yet, a picture started to emerge. A number of studies showed that during transitions from normal cells to cancer cells, autophagy either plays a tumor-suppressor role or prevents cancer formation. In contrast, exploitation of autophagy to deal with hypoxia and energy crisis may allow fast-growing and poorly-vascularized tumors to survive and expand. Therefore, a comprehensive understanding of autophagy pathways that are operational in HCCs may be most rewarding, allowing development of new diagnosis and treatment techniques. In this review, we will briefly introduce the basic autophagic machinery and autophagy-cell death connections and summarize implication of autophagy-related cell death and survival for HCC management.
Autophagy mechanisms
The basic autophagy mechanism is conserved from yeast to man. It is tightly regulated by almost 40 different ATG (Autophagy) genes. Following the initial description of the pathway in the yeast, function of ATG genes and their products were studied under several physiological and pathological conditions.
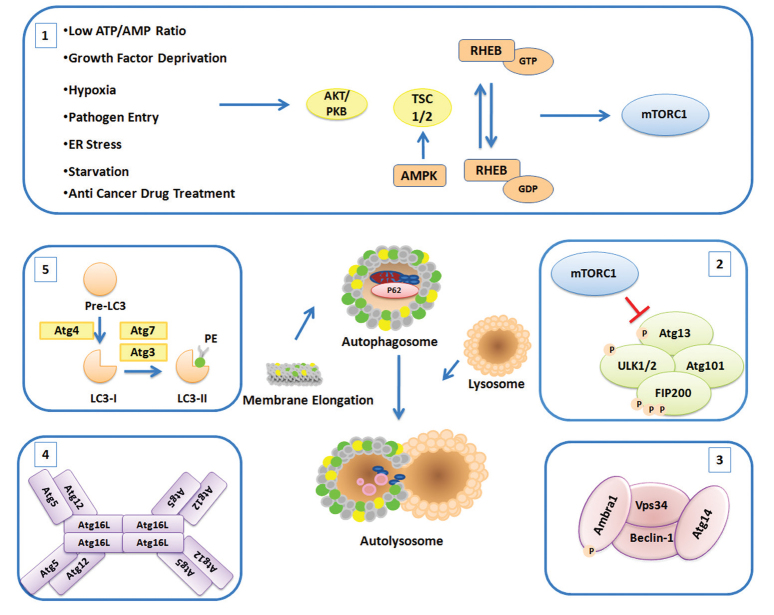
Autophagosome (or autophagic vesicle) and autolysosome formation is a result of well-studied sequential stages, including induction, vesicle nucleation, lysosome fusion, and degradation. Here we will briefly overview autophagosome formation stages and the role of major proteins involved in the machinery (Figure 1). Autophagosomal membrane lipids that are contributing to de novo autophagosome membrane synthesis appear to originate from various pre-existing membrane structures, such as plasma membrane, ER, or mitochondrial membranes (10).
Figure 1.
Schematic representation of the autophagosome formation stages and major proteins and complexes involved in the process
1: Upstream effectors; 2: ULK complex; 3: PI3K complex; 4: ATG5-12-16 complex; 5: LC3 lipidation
The most important upstream regulators of autophagy are the mammalian target of rapamycin complexes (mTORC1 and 2). A central serine/threonine kinase, the mTOR kinase, is the essential component of both mTOR protein complexes. These protein complexes play key roles in the regulation of cellular growth, cell-cycle progression, cell migration, and protein synthesis as well as the coordination of the catabolic autophagy activation with the activity of these essential cellular anabolic pathways.
When the growth conditions are favorable, mTOR complexes are active and the autophagic machinery is shut down. mTORC1 regulates the downstream Atg1/Ulk1 autophagy-related kinase complex (11). Under nutrient-rich conditions, mTOR phosphorylates ATG13 and ULK1/2, and their activity is inversely correlated with FIP200 phosphorylation. On the other hand, under nutrient deprivation, mTOR targets are dephosphorylated and ATG13 binds to ULK1/2 and FIP200. Then, ULK1/2 phosphorylates FIP200 and FIP200-ULK1-ATG13 complex (12). Hence, activated Atg1/ULK1 complex regulates the activity of a second complex named as class-III phosphatidylinositol 3-kinase (PI3K) complex, which contains the lipid kinase Vps34. The PI3K complex consists of Vps34, Vps15, Atg6, and Atg14 in the yeast. The mammalian counterparts of this complex include Beclin 1 (BECN1), ATG14L (Barkor), AMBRA1, hVps34, and p150 (13). Formation of phosphatidylinositol 3-phosphate (PI3P) molecules on cellular membranes creates a landing pad for the recruitment of other proteins and complexes that are required for autophagosome formation (1).
During the autophagosome membrane elongation step, two ubiquitination-like conjugation systems, namely ATG12-5-16L1 and ATG8 systems, are required. In the first conjugation system, ATG12 is conjugated to ATG5 by the help of ATG7 (E1-like enzyme) and ATG10 (E2-like enzyme) proteins. Covalent conjugation of ATG12 to the lysine 130 residue (K130) of ATG5 is followed by the addition of ATG16L protein to the complex. Oligomerization of ATG16L proteins results in the formation of an autophagy-related 800-kDa protein complex (11). ATG12-5-16L1 complexes possess an E3-like enzyme activity that is required for the second ubiquitination-like conjugation system. The second system involves the conjugation of ATG8/LC3 to a lipid molecule, generally to a phosphatidylethanolamine (PE). After cleavage of the carboxyl-terminus of LC3 protein by Atg4 cysteine proteases, a glycine residue is exposed. In this form, the LC3 protein is called LC3-I, a free cytosolic form of the protein.
Then, LC3-I is conjugated to a PE by the help of ATG7 and ATG3 E2-like enzymes, resulting in the appearance of a membrane-bound autophagic LC3-II form. Of note, the LC3-II form is associated with mature autophagosomes, and it is commonly used as a marker of autophagy, and it represents the number and distribution of autophagosomes during autophagic activity analyses. ATG18/WIPI proteins are other important players in autophagosome formation. ATG18/WIPI proteins are WD-repeat containing proteins that are able to recognize PI3P at the nascent autophagosome and they regulate autophagic activity through recruitment of two ubiquitin-like recruitment systems. In the yeast, ATG2 protein interacts with ATG18, this interaction was shown to be important for the membrane localization of ATG18 and elongation of autophagosome membranes. Studies in mammalian cells have also underlined the importance of WIPI proteins for autophagy. ATG9, a multi-pass transmembrane protein localized to late endosomes and the trans-Golgi network, is involved in the transport of membranes to forming autophagosomes. After completion and closure of autophagic vesicles, the last stage involves their fusion with late endosomes or lysosomes. Several membrane fusion events connect these two distinct compartments. and RAB proteins, SNAP receptor machinery, and dynein-mediated transport of autophagosomes along the microtubules are required for the fusion process to occur. Finally, the cargo inside the autophagosome is delivered to the lysosomal lumen and degraded by the action of hydrolytic enzymes in this compartment.
Initially, autophagy was described as a nonselective degradation pathway (14). However, recent studies showed that different autophagy receptors that are capable of recognizing specific cargo targets were identified, underlining the fact that autophagy may be selective (15,16). Autophagy receptors include SQSTM1/p62, NBR1, NDP52 (also known as a CALCOCO2), OPTN, and NIX (also known as BNIP3L) (17–21). Some of these receptors are able to bind and ubiquitinate targets. Moreover, several receptors share motifs called LIR (LC3-interacting region), allowing bridging between LC3 on the autophagosomes selective autophagy targets. Because autophagy receptors are also delivered to autolysosomes together with the cargo, their cellular levels are generally downregulated following autophagy activation. Hence, degradation of autophagy receptors is also another commonly used marker of autophagic activity.
Autophagy in hepatocellular carcinoma
The role of autophagy in cancer is complex [see (22) for a comprehensive review of the topic]. There is experimental evidence that in early phases of cancer formation, autophagy functions as an anticancer pathway, preventing malignant transformation of normal cells to cancer cells. On the other hand, autophagy is involved in various stages of cancer progression and metastasis. Especially, survival of fast-growing tumors has been correlated with their autophagic activity. A large collection of articles implicating autophagy in drug resistance exist as well. Here, we will summarize the role of autophagy in the context of liver cancer.
Liver cancer formation has been observed in a number of autophagy mice models. ATG6/BECN1 (Beclin 1) is a key gene in the autophagy pathway. BECN1 deletion is observed in 40%–75% human cancers (23,24). Interestingly, a heterozygous deletion of atg6/becn1 in mice resulted in increased tumorigenesis in multiple tissues, including the liver (23,24). Moreover, becn1 deletion accelerated hepatitis B virus (HBV)-related HCCs, underlining the importance of atg6/becn1 gene in liver cancer formation (23). Deletion of other autophagy genes, such as atg5 and atg7, leads to the formation of benign liver adenomas in mice models (25). In addition, liver-specific atg7 deletion results in hepatomegaly and hepatic failure, underlining the role of autophagy in liver homeostasis, disturbance of which may be the cause of HCC. Strikingly, additional p62 deletion in a liver-specific atg7 deficient background alleviated tumor burden, indicating that an important role of autophagy in this context is to eliminate cellular protein aggregates in a p62-dependent manner (26). Similarly, deletion of autophagy-related genes Uvrag enhanced susceptibility to HCC development in mice (27,28). Therefore, an important role of autophagy-related proteins and the autophagy pathway in liver cells is the preservation of liver homeostasis and prevention of HCC development (Figure 2).
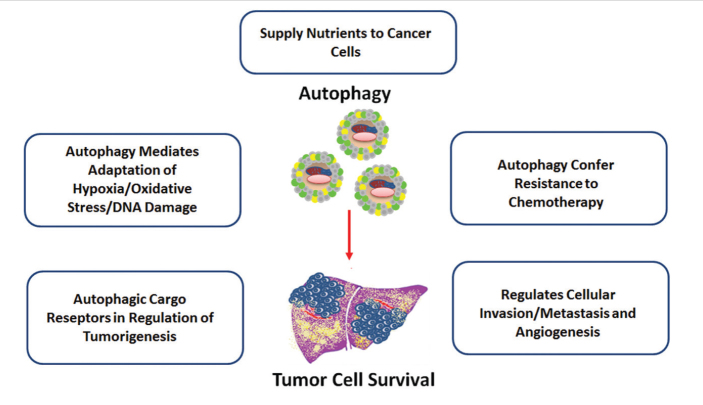
Figure 2.
Tumor-promoter roles of autophagy in HCC
HCC: hepatocellular carcinoma
Cancer-preventing effects of autophagy may be related to its role in clearing damaged mitochondria, elimination of abnormal and mutant proteins and protein aggregates, and specific elimination of proliferation-related proteins (5,29). Disturbances in autophagic activity result in higher levels of reactive oxygen species (ROS) and increase their susceptibility to DNA damage and genomic instability (30,31). First, damaged mitochondria and accumulation of protein aggregates boost ROS burden in cells. Moreover, other autophagy-related antioxidant mechanisms exist as well. For example, activation of NRF2, a key transcription factor in antioxidant defense, has been found to be regulated by autophagy (32). Under normal conditions, Keap1, an adaptor protein of Cullin-3 ubiquitin ligase, allows ubiquitination and degradation of NRF2. ROS accumulation results in the oxidation of Keap1 and its dissociation from NRF2, leading to its stabilization and nuclear migration. Another mechanism of Keap1 elimination is selective autophagy. Competitive binding of the autophagy receptor p62 to Keap1 followed by their selective autophagic degradation activates NRF2, triggering an antioxidant transcriptional pathway. p62 accumulation has been found to drive liver cancer formation in a number of mice models (25,33,34).
In contrast, autophagy is described as an important mechanism for cancer progression in established malignancies (Figure 3). For example, basal autophagy is elevated in hypoxic regions of some solid tumor types and found to be an essential role for tumor cell survival in experimental models (35). Tumor neovascularization may not always result in a homogenous vessel network, and especially in fast-growing tumors, regions that have limited access to nutrients and oxygen exist (36). Thus, cancer cells in these regions may be more dependent on autophagy than normal-growing cells. Indeed, autophagy has been shown to promote HCC growth in experimental studies (37–39). Autophagy is also believed to support the survival of cancer cells and contribute to metastasis and chemotherapy resistance.
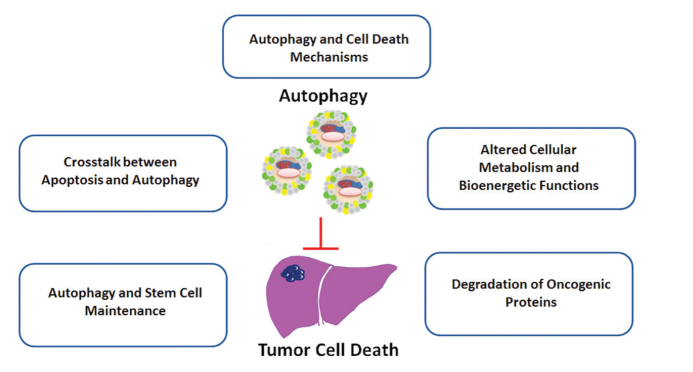
Figure 3.
Tumor-suppressor roles of autophagy in HCC
HCC: hepatocellular carcinoma
In summary, although autophagy may act as an antitumor pathway preventing early stages of cancer development in established tumors, it may protect cancer cells from various stress conditions, including starvation, oxidative stress, hypoxia, and chemotherapy, and it may contribute to the growth and spread of cancerous cells (13,40)
Autophagy and cell death
Autophagy is generally considered as a stress response and a cell-survival mechanism. It is frequently observed that dying cells exhibit autophagy activation. Whether this autophagic activity is a failing attempt to rescue stressed cells or conversely contributes to cell death is a matter of scientific debate. Yet under certain conditions, blockage of autophagy using chemicals or genetic tools may rescue cells from death. Moreover, autophagy activation is observed in a number of necrotic-like programmed cell death types, including necroptosis and autosis; however, the contribution of autophagy to these novel death pathways has not been thoroughly analyzed (41). Nevertheless, several independent articles showed the existence of a nonapoptotic cell death type that depended on autophagic activity (2,42–46).
In the context of cancer, autophagic cell death is shown to limit clonogenic survival. For example, H-ras, one of the most commonly mutated proteins in various cancers, is found to increase cellular levels of the autophagy protein Beclin 1 and induce caspase-independent cell death with autophagic characteristics (42). In multiple myelomas, cleavage of autophagic cell death inducer BCLAF1 by caspase-10 is required for cancer cell survival (43). In addition, several tumor-suppressor-related and cell-death-related proteins, including DAPK, DRP1, ZIP, p19ARF, and GBA, triggered autophagic cell death (2,45–48). Therefore, although autophagy allows cells to survive stressful conditions that cancer cells are facing during various stages of cancer, excessive autophagy and autophagic cell death may kill cancer cells and limit their progression and metastasis.
Autophagy and hepatocellular carcinoma therapy
Hepatocellular carcinoma is one of the most common cancer types. It is the third leading cause of cancer deaths worldwide (49). History of chronic liver disease and cirrhosis is among the factors that predispose patients to HCC development. Understanding the molecular mechanisms of HCC development and the contribution of autophagy deregulation to these mechanisms are among important challenges of modern medicine. Therefore, in this section, we will summarize preclinical and clinical studies that focused on autophagy in an HCC treatment context (Table 1).
Table 1.
Autophagy modulating therapeutics in HCC
| Therapeutics | Autophagy status | Autophagy effect on chemotherapy | Tested cell lines | Reference | |
|---|---|---|---|---|---|
| Conventional chemotherapeutics | Oxaliplatin | Increase | Chemoresistance | Huh-7 SMMC-7721 | (52) |
| Oxaliplatin | Increase | Chemoresistance | HepG2 | (51) | |
| Adriamycin | Increase | Chemoresistance | HepG2 | (56) | |
| Cisplatin | Increase | Chemoresistance | SMMC-7721 Hep3B HepG2 |
(53) | |
| 5-FU | Increase | Chemoresistance | SMMC-7721 Hep3B HepG2 |
(53) | |
| Epirubicin | Increase | Chemoresistance | HA22T/VGH | (54) | |
| Pemetrexed | Increase | Chemoresistance | HepG2 | (55) | |
| Small molecules | Sorafenib | Increase | Chemoresistance | PLC/PRF/5 Hep3B HepG2 |
(63) |
| Sorafenib | Increase | Chemosensitivity | Sk-Hep-1 PLC/PRF/5 Hep3B HepG2 |
(65) | |
| Panobinostat | Decrease | Chemosensitivity | Hep3B HepG2 Huh-7 |
(64) | |
| Bevacizumab | Increase | Chemoresistance | SMMC-7721 Hep3B |
(68) | |
| Linifanib | Increase | Chemoresistance | HepG2 Bel-7404 |
(69) | |
| SC-2001 | Increase | Chemosensitivity | Sk-Hep-1 PLC/PRF/5 Hep3B HepG2 |
(73) | |
| ABT-737 | Increase | Chemoresistance | Huh-7 PLC/PRF/5 Hep3B HepG2 |
(70) | |
| Salinomycin | Increase | Chemosensitivity | HepG2 | (71) | |
| Natural products | Baicalin | Increase | Chemosensitivity | SMMC-7721 | (74) |
| Galangin | Increase | Chemosensitivity | HepG2 | (75) | |
| Cannabinoids | Increase | Chemoresistance | HepG2 Huh-7 |
(76) | |
| Berberine | Increase | Chemosensitivity | SMMC-7721 HepG2 |
(77) | |
| Allicin | Increase | Chemosensitivity | HepG2 Hep3B |
(78) | |
| Matrine | Increase | Chemosensitivity | HepG2 Bel-7402 |
(79) | |
| Glycyrrhetinic acid | Increase | Chemosensitivity | HepG2 Hep3B |
(80) | |
| SCB | Increase | Chemosensitivity | Hep3B ML-1 |
(81) | |
| 20(S)-Ginsenoside Rg3 | Decrease | Chemosensitivity | Sk-Hep-1 HepG2 |
(82) | |
| Arenobufagin | Increase | Chemoresistance | HepG2 | (83) | |
| Bufalin | Increase | Chemoresistance | Huh-7 HepG2 LO2 |
(84) | |
| Noncoding RNAs | miR-199a-5p | Decrease | Chemoresistance | Huh-7 HepG2 |
(86) |
| miR-375 | Decrease | Chemoresistance | Huh-7 Hep3B |
(87) | |
| miR-101 | Decrease | Chemosensitivity | HepG2 HepG2 Hep3B SNU-182 Huh-7 PLC/PRF/5 HepaRG |
(90,91) | |
| miR-21 | Decrease | Chemoresistance | HepG2 Huh-7 |
(92) | |
| PTENP1 | Increase | Chemoresistance | Mahlavu | (94) | |
| Other therapies | NVP-BEZ235 | Increase | Chemoresistance | Hep3B PLC/PRF/5 |
(97) |
| MK-2206 | Decrease | Chemosensitivity | Mahlavu PLC SNU387 SNU449 SNU475 |
(98) | |
| GD0068 | Decrease | Chemosensitivity | HepG2 Huh-7 |
(99) | |
| OSU-03012 | Increase | Chemoresistance | Huh-7 | (101) | |
| Meloxicam | Increase | Chemoresistance | HepG2 Bel 7402 Huh-7 SMMC-7721 SMMC 7402 |
(103) | |
| SAHA | Increase | Chemoresistance | HepG2 Hep3B Huh-7 |
(105) | |
| Radiotherapy | Increase | N.D. | Sk-Hep-1 | (107) | |
| Radiotherapy | Increase | Chemoresistance | Sk-Hep-1 | (108) | |
| ADI-PEG20 | Increase | Chemoresistance | HepG2 SMMC-7721 |
(109) | |
| CD133/ Prominin-1 | Increase | Chemoresistance | HepG2 LO2 Hep3B Huh-7 |
(94) |
HCC: hepatocellular carcinoma; ND: not determined
Conventional chemotherapeutics
Chemotherapeutic agents were shown to activate autophagy in a number of cancer types. Oxaliplatin is a platinum-based chemotherapy agent that is widely used in the treatment of HCC (50,51). Indeed, oxaliplatin treatment led to autophagic activation in both HCC cells and xenografts (52). Inhibition of autophagy under these conditions increases the cytotoxicity of oxaliplatin, suggesting that autophagy may be an important player in the resistance in HCC to oxaliplatin toxicity (51,52). In another study, cisplatin and 5-FU were shown to induce the formation of autophagosomes in three different HCC cell lines, and attenuation of autophagy enhanced the cisplatin and 5-FU-induced cell death under both in vitro and in vivo conditions (53). The role of autophagy in chemoresistance of HCC to epirubicin has also been investigated. Combination of progesterone was found to overcome autophagy-related chemoresistance and allowed effective cancer cell elimination (54). Moreover, Yongxi et al. (55) showed that pemetrexed-induced autophagy in HepG2 HCC cell line blocked apoptosis activated by ERK inhibition. On the other hand, another chemotherapeutic agent, adriamycin, was found to induce mitochondrial depolarization and autophagy, and its combination with curcumin to block autophagy further decreased the level of proliferation in comparison with adriamycin alone (56).
Targeted small molecules
Sorafenib is an FDA-approved tyrosine kinase inhibitor (TKI) used in the treatment of HCC (57). The drug increased overall survival even in patients with advanced disease (57,58). Sorafenib induces both apoptosis and autophagy in HCC cells. Moreover, studies revealed that ER stress may be involved in sorafenib cytotoxicity (59). Modulation of proteasomal degradation also influenced sorafenib effects on cell fate. Combination with proteasome inhibitors significantly increased HCC cell death compared with sorafenib alone (60). Inhibition of mTOR and accumulation of autophagosomes were reported upon sorafenib treatment of HCC cells (61). Concomitantly, combination of sorafenib and chloroquine (CQ, a drug that prevents autophagosome maturation) had synergistic effects on tumor growth suppression (61,62). In line with this, sorafenib has been found to kill more cells when autophagy is attenuated using CQ or following genetic suppression by a specific siRNA (small interfering RNA) against Beclin 1 or ATG5 (63). In another study, panobinostat, a pan HDAC inhibitor, was found to enhance the effect of sorafenib by blocking autophagy (64). Moreover, a derivative of sorafenib, SC-59, that has a more potent effect on cancer cell viability than sorafenib, was shown to downregulate p-Stat3 levels and induce strong autophagy activation in HCC cell lines (65). Besides sorafenib, another TKI, nilotinib, stimulated autophagy in HCC cells through AMPK phosphorylation and regulation of PP2A (66). Combination with another agent, FTY720 (a potent sphingosine-1-phosphate receptor agonist), enhanced sorafenib-induced cytotoxicity in HCC cells (67).
Other targeted drugs also have autophagy-activating effects. For example, targeting vascular endothelial growth factor (VEGF) by bevacizumab triggered autophagy. Moreover, combinatory inhibition of autophagy together with bevacizumab elevated apoptosis levels in HCC cells (68). Another inhibitor of VEGF, linifanib, also induced autophagy in HCC cells, and similarly, its cytotoxic effects were further enhanced on autophagy suppression (69). Ni et al. provided evidence that resistance to the bcl-2 inhibitor ABT-737 is a result of the activation of a ROS-JNK-autophagy pathway in HCC cells (70). Moreover, salinomycin-mediated suppression of autophagy in HCC cells has been reported to result in their cell death through defective mitochondria accumulation and ROS accumulation (71). In another study, inhibition of Hsp90 by 17-AAG was shown to sensitize HCC cells against gossypol induced apoptosis through suppression of cytoprotective autophagy (72).
In contrast with these findings, a study by Tai et al. showed that sorafenib enhanced autophagy-dependent cell death in HCC both in vitro and in vivo (65). In line with this, SC-2001, an analog of the bcl-2 inhibitor obatoclax, induced autophagic cell death in HCC cells (73). It is possible that autophagy levels in these set-ups were more robust than those in other cited studies, converting chemoprotective autophagy to a cell death-inducing pathway.
Natural products
A number of natural products have been shown to have autophagy-related effects on the growth and survival of HCC cells. For example, baicalin is a natural flavonoid obtained from the Chinese herb Scutellaria baicalensis, and it exerts an apoptosis and autophagy-dependent inhibitory effect on HCC (74). Alpinia officinarum-derived Galangin is another natural flavonoid that induces autophagy in HCC cells through the activation of TGFb receptor/Smad axis (75). Various cannabinoid derivatives showed antitumor effects against HCC that depend on intact autophagic activity. Blockage of autophagy attenuates antitumor effects, thus supporting the idea that autophagic cell death is active under these circumstances (76). Similarly, berberine, allicin, matrine, and glycyrrhetinic acid are plant-derived molecules that show their antitumor effects through induction of either apoptosis and/or autophagy in HCC cells (77–80). In another study, administration of soybean fermentation products containing live bacteria (SCB) was shown to suppress HBV-related HCC tumor growth; under these conditions, SCB induced both apoptosis and autophagy (81). On the other hand, steroidal saponin 20(S)-Ginsenoside Rg3 has been shown to block autophagy and promotes doxorubicin sensitivity in HCC cells and tumors (82).
In addition to plant-derived natural products, venoms are another group of natural products that have been evaluated for cancer treatment. Arenobufagin, a venom isolated from toads, shows significant antineoplastic efficacy against both naive HepG2 cells and their multidrug resistant clones. Inhibition of autophagy is reported to enhance the level of apoptosis in this context (83). Another toad venom, bufalin, also has an antitumor activity on HCC cells, and its efficacy has been found to increase under autophagy-attenuated conditions (84).
Noncoding RNAs
MicroRNAs are associated with various cellular phenomena including cell death, differentiation, and diseases. Dysregulation of miRNAs is linked to cellular abnormalities and carcinogenesis, and changes in microRNA levels affect tumor growth and progression. As explained in detail above, autophagy abnormalities are also associated with cancer. Therefore, changes in the levels of a subset of miRNAs that control the autophagic activity may have important outcomes on cancer cell survival and drug responses (85). For example, levels of drug resistance-associated miR-199a-5p were found to be significantly decreased in patients with HCC following treatment with cisplatin. In fact, miR-199a-5p has been shown to be responsible from the attenuation of cisplatin-induced autophagy in HCC cell lines through ATG7 targeting. Inhibition of autophagy in HCC cells blocked miR-199a-5p downregulation-induced cell proliferation and cisplatin resistance (86). Another ATG7 targeting miRNA, miR-375, has been found to be downregulated in HCCs and decreases HCC cell viability under hypoxic conditions (87,88). Another miRNA, miR-224, is one of the most studied miRNAs in HCC, and it has been shown to target Smad4. Strikingly, high miR-224 levels were associated with lower Atg5 levels as well as lower Smad4 levels, and these findings significantly correlated with HBV infection and poor overall survival in patients with HCC (89). Interestingly under these conditions, autophagy was shown to limit miR-224 levels through the direct degradation of the miRNA, hence resulting in liver tumor suppression (89). MiR-101 has been characterized as an autophagy-inhibitory miRNA, and this effect has been shown to sensitize HCC cells against cisplatin, doxorubicin, and 5-FU (90,91). MiRNAs were also involved in sorafenib resistance in HCC. For instance, Mir-21 is found to suppress autophagy via PTEN/Akt axis and lead to sorafenib resistance (92). Sorafenib-induced miRNAs were also used for determining prognosis and follow-up. In a study, miR-423-5p was described as a positive regulator of autophagy in HCC cells. Levels of this miRNA in patient sera months after sorafenib treatment indicated a response to treatment, indicating the prognostic value of an autophagy-related miRNA in HCC (93).
Long noncoding RNAs (lncRNAs) have been associated with HCC as well. For example, PTENP1 is identified in a screen of lncRNAs targeting PTEN. In fact, PTENP1 acted as a competitor of several autophagy-regulating miRNAs, such as miR-17, miR-19b, and miR-20a, which target PTEN and PHLPP as well as autophagy genes ULK1, ATG7, and p62. Injection of a PTENP1-expressing virus to mice has been shown to stimulate autophagy and attenuated HCC tumor growth (94).
Other approaches
Recent studies indicate that autophagy regulator mTOR signaling is upregulated in a significant proportion of HCCs (95). Thus, mTOR pathway may be exploited as a drug target in HCC. For instance, RAD001 and BEZ235 are characterized as PI3K/mTOR-inhibitor drugs. Combination of these two drugs has been shown to suppress HCC growth both in vitro cell culture and in vivo in mice experiments (96). Moreover, orally available BEZ235 in a combination with autophagy blockage is also more effective in HCC treatment (97). Another molecule, the Akt inhibitor MK-2206, has been found to trigger cell death, and suppression of autophagy under these experimental conditions has been shown to further enhance the efficacy of the inhibitor in HCC cells (98). Another Akt inhibitor called GD0068 has shown synergistic effects with sorafenib and even suppresses the growth of sorafenib-resistant HCC cells converting cytoprotective autophagy to autophagic cell death (99).
Some studies on nonsteroidal anti-inflammatory drugs (NSAIDs) revealed that inhibition of COX-2, which may be highly expressed in some tumor types, is the underlying mechanism for the cancer-preventive effects attributed to these drugs (100). One of the derivatives of the NSAID celecoxib, OSU-03012, has been found to exert antitumor activities. Gao et al. revealed that autophagy levels were elevated in HCC cells upon OSU-03012 treatment. Blockage of autophagy decreased OSU-03012-induced cell death under both in vitro and in vivo conditions indicating that autophagic cell death is important in the effects of the drug in HCC cells (101). Yet in another study, suppression of autophagy by 3-MA was found to promote NSAID meloxicam-induced apoptosis in HCC (102,103).
Histone acetylation has been linked to cancer through aberrant regulation of cancer-related genes. Interestingly, HDAC1 has been reported to be overexpressed in HCC; yet, HDAC6 has been found to be decreased in HCCs compared with adjacent control tissues, and this observation is associated with poor prognosis (104). Nevertheless, HDAC inhibitors are tested as promising drugs against cancer, and several members of this group of drugs were also found to induce autophagy and even autophagic cell death in some contexts. SAHA, an important HDAC inhibitor, has been shown to induce autophagic cell death in HCC cells (105). In another study, HDAC1 inactivation inhibited proliferation of tumor cells and activate caspase-independent autophagic cell death (106). On the other hand, HDAC inhibitors OSU-HDAC42 and SAHA were both found to induce autophagy in HCC cells. Moreover, inhibition of autophagy decreases SAHA-induced cell death indicative of autophagic cell death activation in HCC (105).
In another study, irradiation was shown to kill HCC cells, which was further enhanced by the combination of oxaliplatin. In addition to this, when apoptosis was attenuated by a PARP inhibitor combination treatment, autophagic activation was observed and cell death responses were more robust (107). In a follow-up study, the same group showed that when HCC cells were treated with high LET irradiation, cells died in an autophagy-dependent manner under both in vitro and in vivo conditions (108). In an additional study, mTOR-inhibitor RAD001 was found to enhance high LET radiation-induced cytotoxicity in HCC cells (109). Altogether, high LET radiation-drug combinations have therapeutic effects against HCC, and autophagy appears to take part in the mechanism of action of these combinations.
Argininosuccinate synthetase (ASS) has been reported to be low in HCC cells. Thus, at least some HCC tumors may be auxotrophic for arginine and require arginine supply from extracellular sources (110). Consequently, autophagy and cell death were activated in HCC cells when they were exposed to a modified form of the arginine-degrading enzyme arginine deaminase (ADI-PEG20) (111). Moreover, an arginine-modifying enzyme, the enzyme peptidylarginine deiminase IV, has been reported to be related to chemoresistance in HCC through regulation of autophagy (112).
CONCLUSION
The above-cited studies underline the importance of autophagy for health and disease in the liver. In particular, with the advance of studies on autophagy cancer, the role of autophagy in HCC development and management becomes clearer. Especially, studies on the contribution of autophagy and related mechanisms to HCC chemoresistance are of special interest. There are several studies correlating autophagic activity with resistance to chemotherapeutic agents, including sorafenib. Several independent labs are currently working on finding novel small molecules that will be capable of manipulating autophagy for treatment purposes. Further studies, including clinical studies, are required to fully reveal the potential of the abovementioned strategies and these potential new drugs, alone or in combination with classical drugs, for the treatment of liver diseases and HCC.
Moreover, a strong connection between autophagy and liver pathologies, including nonalcoholic steatohepatitis, HBV, and hepatitis C virus infection and cirrhosis, has been reported (113). For instance, autophagy constitutes a major clearance of mechanism for intracellular pathogens, such as viruses. However, some viruses, including HBV and HCC, may hijack autophagic membranes during their replication (114). In addition, several autophagy deficient animal models have been shown to suffer from hepatic steatosis, and independent studies have consistently demonstrated that autophagy is involved in lipid and glycogen metabolism. Evidently, these and other abnormalities of liver function and pathologies are also closely related to HCC development. Thus, a better understanding of mechanisms underlying autophagy, its abnormalities, and its connection with liver diseases and disease-causing factors will certainly improve current diagnosis, treatment, follow-up, and prevention strategies for HCC. Autophagy constitutes one of the important medical fields that already started to provide examples of bench-to-bedside transitions. Hence, following this novel but fast-growing field will be most rewarding for both basic scientists and clinical researchers and practitioners.
Acknowledgements
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) 1001 project number 114Z982. D.G. is a recipient of an EMBO Strategical Development and Installation Grant (EMBO-SDIG), Turkish Academy of Sciences (TUBA) GEBIP Award, IKU Prof. Dr. Önder Öztunalı Science Award and TGC Sedat Simavi Health Sciences Award, Elginkan Foundation Technology Award and Parlar Foundation technology Incentive Award. Y.A. is supported by a TUBITAK-BIDEB 2211 scholarship for PhD studies.
Footnotes
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - Y.A., D.G.; Design - Y.A., D.G.; Supervision - D.G.; Data Collection and/or Processing - Y.A.; Analysis and/or Interpretation - Y.A., D.G.; Literature Search - Y.A., D.G.; Writing - Y.A., D.G.; Critical Reviews - D.G.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) 1001 project number 114Z982.
REFERENCES
- 1.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. https://doi.org/10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H, et al. DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ. 2008;15:1875–86. doi: 10.1038/cdd.2008.121. https://doi.org/10.1038/cdd.2008.121 [DOI] [PubMed] [Google Scholar]
- 3.Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–45. doi: 10.1016/S0070-2153(06)78006-1. https://doi.org/10.1016/S0070-2153(06)78006-1 [DOI] [PubMed] [Google Scholar]
- 4.Oral O, Akkoc Y, Bayraktar O, Gozuacik D. Physiological and pathological significance of the molecular cross-talk between autophagy and apoptosis. Histol Histopathol. 2016;31:479–98. doi: 10.14670/HH-11-714. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. https://doi.org/10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madrigal-Matute J, Cuervo AM. Regulation of Liver Metabolism by Autophagy. Gastroenterology. 2016;150:328–39. doi: 10.1053/j.gastro.2015.09.042. https://doi.org/10.1053/j.gastro.2015.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czaja MJ. Function of Autophagy in Nonalcoholic Fatty Liver Disease. Dig Dis Sci. 2016;61:1304–13. doi: 10.1007/s10620-015-4025-x. https://doi.org/10.1007/s10620-015-4025-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–906. doi: 10.1038/sj.onc.1207521. https://doi.org/10.1038/sj.onc.1207521 [DOI] [PubMed] [Google Scholar]
- 9.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12( Suppl 2):1542–52. doi: 10.1038/sj.cdd.4401765. https://doi.org/10.1038/sj.cdd.4401765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–5. doi: 10.1038/ncb0910-831. https://doi.org/10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. https://doi.org/10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. https://doi.org/10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. https://doi.org/10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. doi: 10.1038/cr.2013.169. https://doi.org/10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–41. doi: 10.1038/ncb0910-836. https://doi.org/10.1038/ncb0910-836 [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Yang L, Xu S, Zhang Z, Cao Y. The receptor proteins: pivotal roles in selective autophagy. Acta Biochim Biophys Sin (Shanghai) 2015;47:571–80. doi: 10.1093/abbs/gmv055. https://doi.org/10.1093/abbs/gmv055 [DOI] [PubMed] [Google Scholar]
- 17.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. https://doi.org/10.1083/jcb.200507002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–90. doi: 10.4161/cc.8.13.8892. https://doi.org/10.4161/cc.8.13.8892 [DOI] [PubMed] [Google Scholar]
- 19.von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012;48:329–42. doi: 10.1016/j.molcel.2012.08.024. https://doi.org/10.1016/j.molcel.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korac J, Schaeffer V, Kovacevic I, Clement AM, Jungblut B, Behl C, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013;126:580–92. doi: 10.1242/jcs.114926. https://doi.org/10.1242/jcs.114926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46. doi: 10.1038/cdd.2009.16. https://doi.org/10.1038/cdd.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karakas HE, Gozuacik D. Autophagy and cancer. Turkish Journal of Biology. 2014;38:720–39. https://doi.org/10.3906/biy-1408-16 [Google Scholar]
- 23.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. https://doi.org/10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. https://doi.org/10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. https://doi.org/10.1101/gad.2016211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. doi: 10.1016/j.cell.2007.10.035. https://doi.org/10.1016/j.cell.2007.10.035 [DOI] [PubMed] [Google Scholar]
- 27.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–99. doi: 10.1038/ncb1426. https://doi.org/10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–51. doi: 10.1038/ncb1634. https://doi.org/10.1038/ncb1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White E, Karp C, Strohecker AM, Guo Y, Mathew R. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol. 2010;22:212–7. doi: 10.1016/j.ceb.2009.12.008. https://doi.org/10.1016/j.ceb.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. https://doi.org/10.1101/gad.1565707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. https://doi.org/10.1101/gad.1545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katsuragi Y, Ichimura Y, Komatsu M. Regulation of the Keap1-Nrf2 pathway by p62/SQSTM1. Current Opinion in Toxicology. 2016;1:54–61. https://doi.org/10.1016/j.cotox.2016.09.005 [Google Scholar]
- 33.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, et al. p62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell. 2016;29:935–48. doi: 10.1016/j.ccell.2016.04.006. https://doi.org/10.1016/j.ccell.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler SM, Laggai S, Barghash A, Schultheiss CS, Lederer E, Artl M, et al. IMP2/p62 induces genomic instability and an aggressive hepatocellular carcinoma phenotype. Cell Death Dis. 2015;6:e1894. doi: 10.1038/cddis.2015.241. https://doi.org/10.1038/cddis.2015.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. https://doi.org/10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid MA, Kong M. Dealing with hunger: Metabolic stress responses in tumors. J Carcinog. 2013;12:17. doi: 10.4103/1477-3163.119111. https://doi.org/10.4103/1477-3163.119111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalik MA, Perra A, Ledda-Columbano GM, Ippolito G, Piacentini M, Columbano A, et al. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules. Oncotarget. 2016;7:5788–99. doi: 10.18632/oncotarget.6810. https://doi.org/10.18632/oncotarget.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, et al. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–2. doi: 10.1126/science.1096645. https://doi.org/10.1126/science.1096645 [DOI] [PubMed] [Google Scholar]
- 39.Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, et al. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–34. doi: 10.1038/cdd.2014.201. https://doi.org/10.1038/cdd.2014.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends Cell Biol. 2015;25:37–45. doi: 10.1016/j.tcb.2014.09.001. https://doi.org/10.1016/j.tcb.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Shoji-Kawata S, Sumpter RM, Jr, et al. Autosis is a Na+, K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–71. doi: 10.1073/pnas.1319661110. https://doi.org/10.1073/pnas.1319661110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–7. doi: 10.1073/pnas.0511288103. https://doi.org/10.1073/pnas.0511288103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. https://doi.org/10.1016/j.molcel.2011.02.009 [DOI] [PubMed] [Google Scholar]
- 44.Lamy L, Ngo VN, Emre NC, Shaffer AL, 3rd, Yang Y, Tian E, et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell. 2013;23:435–49. doi: 10.1016/j.ccr.2013.02.017. https://doi.org/10.1016/j.ccr.2013.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arakawa S, Tsujioka M, Yoshida T, Tajima-Sakurai H, Nishida Y, Matsuoka Y, et al. Role of Atg5-dependent cell death in the embryonic development of Bax/Bak double-knockout mice. Cell Death Differ. 2017;24:1598–608. doi: 10.1038/cdd.2017.84. https://doi.org/10.1038/cdd.2017.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasari SK, Bialik S, Levin-Zaidman S, Levin-Salomon V, Merrill AH, Jr, Futerman AH, et al. Signalome-wide RNAi screen identifies GBA1 as a positive mediator of autophagic cell death. Cell Death Differ. 2017;24:1288–302. doi: 10.1038/cdd.2017.80. https://doi.org/10.1038/cdd.2017.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–68. doi: 10.1083/jcb.200109094. https://doi.org/10.1083/jcb.200109094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. doi: 10.1016/j.molcel.2006.04.014. https://doi.org/10.1016/j.molcel.2006.04.014 [DOI] [PubMed] [Google Scholar]
- 49.Shani G, Marash L, Gozuacik D, Bialik S, Teitelbaum L, Shohat G, et al. Death-associated protein kinase phosphorylates ZIP kinase, forming a unique kinase hierarchy to activate its cell death functions. Mol Cell Biol. 2004;24:8611–26. doi: 10.1128/MCB.24.19.8611-8626.2004. https://doi.org/10.1128/MCB.24.19.8611-8626.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–73 e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrelli F, Coinu A, Borgonovo K, Cabiddu M, Ghilardi M, Lonati V, et al. Oxaliplatin-based chemotherapy: a new option in advanced hepatocellular carcinoma. a systematic review and pooled analysis. Clin Oncol (R Coll Radiol) 2014;26:488–96. doi: 10.1016/j.clon.2014.04.031. https://doi.org/10.1016/j.clon.2014.04.031 [DOI] [PubMed] [Google Scholar]
- 52.Du H, Yang W, Chen L, Shi M, Seewoo V, Wang J, et al. Role of autophagy in resistance to oxaliplatin in hepatocellular carcinoma cells. Oncol Rep. 2012;27:143–50. doi: 10.3892/or.2011.1464. [DOI] [PubMed] [Google Scholar]
- 53.Ding ZB, Hui B, Shi YH, Zhou J, Peng YF, Gu CY, et al. Autophagy activation in hepatocellular carcinoma contributes to the tolerance of oxaliplatin via reactive oxygen species modulation. Clin Cancer Res. 2011;17:6229–38. doi: 10.1158/1078-0432.CCR-11-0816. https://doi.org/10.1158/1078-0432.CCR-11-0816 [DOI] [PubMed] [Google Scholar]
- 54.Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–9. doi: 10.1016/j.canlet.2012.03.002. https://doi.org/10.1016/j.canlet.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 55.Chang WT, Cheng HL, Hsieh BS, Chiu CC, Lee KT, Chang KL. Progesterone increases apoptosis and inversely decreases autophagy in human hepatoma HA22T/VGH cells treated with epirubicin. ScientificWorldJournal. 2014;2014:567148. doi: 10.1155/2014/567148. https://doi.org/10.1155/2014/567148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong Y, Huang H, Pan H. Inhibition of MEK/ERK activation attenuates autophagy and potentiates pemetrexed-induced activity against HepG2 hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2015;456:86–91. doi: 10.1016/j.bbrc.2014.11.038. https://doi.org/10.1016/j.bbrc.2014.11.038 [DOI] [PubMed] [Google Scholar]
- 57.Qian H, Yang Y, Wang X. Curcumin enhanced adriamycin-induced human liver-derived Hepatoma G2 cell death through activation of mitochondria-mediated apoptosis and autophagy. Eur J Pharm Sci. 2011;43:125–31. doi: 10.1016/j.ejps.2011.04.002. https://doi.org/10.1016/j.ejps.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 58.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. https://doi.org/10.1056/NEJMoa0708857 [DOI] [PubMed] [Google Scholar]
- 59.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. https://doi.org/10.1016/S1470-2045(08)70285-7 [DOI] [PubMed] [Google Scholar]
- 60.Shi YH, Ding ZB, Zhou J, Hui B, Shi GM, Ke AW, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy. 2011;7:1159–72. doi: 10.4161/auto.7.10.16818. https://doi.org/10.4161/auto.7.10.16818 [DOI] [PubMed] [Google Scholar]
- 61.Honma Y, Harada M. Sorafenib enhances proteasome inhibitor-mediated cytotoxicity via inhibition of unfolded protein response and keratin phosphorylation. Exp Cell Res. 2013;319:2166–78. doi: 10.1016/j.yexcr.2013.05.023. https://doi.org/10.1016/j.yexcr.2013.05.023 [DOI] [PubMed] [Google Scholar]
- 62.Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–57. doi: 10.1002/ijc.26374. https://doi.org/10.1002/ijc.26374 [DOI] [PubMed] [Google Scholar]
- 63.Guidetti A, Carlo-Stella C, Locatelli SL, Malorni W, Pierdominici M, Barbati C, et al. Phase II study of sorafenib in patients with relapsed or refractory lymphoma. Br J Haematol. 2012;158:108–19. doi: 10.1111/j.1365-2141.2012.09139.x. https://doi.org/10.1111/j.1365-2141.2012.09139.x [DOI] [PubMed] [Google Scholar]
- 64.Yuan H, Li AJ, Ma SL, Cui LJ, Wu B, Yin L, et al. Inhibition of autophagy signi fi cantly enhances combination therapy with sorafenib and HDAC inhibitors for human hepatoma cells. World J Gastroenterol. 2014;20:4953–62. doi: 10.3748/wjg.v20.i17.4953. https://doi.org/10.3748/wjg.v20.i17.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lachenmayer A, Toffanin S, Cabellos L, Alsinet C, Hoshida Y, Villanueva A, et al. Combination therapy for hepatocellular carcinoma: additive preclinical efficacy of the HDAC inhibitor panobinostat with sorafenib. J Hepatol. 2012;56:1343–50. doi: 10.1016/j.jhep.2012.01.009. https://doi.org/10.1016/j.jhep.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, et al. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. doi: 10.1038/cddis.2013.18. https://doi.org/10.1038/cddis.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu HC, Lin CS, Tai WT, Liu CY, Shiau CW, Chen KF. Nilotinib induces autophagy in hepatocellular carcinoma through AMPK activation. J Biol Chem. 2013;288:18249–59. doi: 10.1074/jbc.M112.446385. https://doi.org/10.1074/jbc.M112.446385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmed D, de Verdier PJ, Ryk C, Lunqe O, Stal P, Flygare J. FTY720 (Fingolimod) sensitizes hepatocellular carcinoma cells to sorafenib-mediated cytotoxicity. Pharmacol Res Perspect. 2015;3:e00171. doi: 10.1002/prp2.171. https://doi.org/10.1002/prp2.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo XL, Li D, Sun K, Wang J, Liu Y, Song JR, et al. Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. J Mol Med (Berl) 2013;91:473–83. doi: 10.1007/s00109-012-0966-0. https://doi.org/10.1007/s00109-012-0966-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan H, Wang Z, Jiang L, Sui X, You L, Shou J, et al. Autophagy inhibition sensitizes hepatocellular carcinoma to the multikinase inhibitor linifanib. Sci Rep. 2014;4:6683. doi: 10.1038/srep06683. https://doi.org/10.1038/srep06683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ni Z, Wang B, Dai X, Ding W, Yang T, Li X, et al. HCC cells with high levels of Bcl-2 are resistant to ABT-737 via activation of the ROS-JNK-autophagy pathway. Free Radic Biol Med. 2014;70:194–203. doi: 10.1016/j.freeradbiomed.2014.02.012. https://doi.org/10.1016/j.freeradbiomed.2014.02.012 [DOI] [PubMed] [Google Scholar]
- 72.Klose J, Stankov MV, Kleine M, Ramackers W, Panayotova-Dimitrova D, Jager MD, et al. Inhibition of autophagic flux by salinomycin results in anti-cancer effect in hepatocellular carcinoma cells. PLoS One. 2014;9:e95970. doi: 10.1371/journal.pone.0095970. https://doi.org/10.1371/journal.pone.0095970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang B, Chen L, Ni Z, Dai X, Qin L, Wu Y, et al. Hsp90 inhibitor 17-AAG sensitizes Bcl-2 inhibitor (−)-gossypol by suppressing ERK-mediated protective autophagy and Mcl-1 accumulation in hepatocellular carcinoma cells. Exp Cell Res. 2014;328:379–87. doi: 10.1016/j.yexcr.2014.08.039. https://doi.org/10.1016/j.yexcr.2014.08.039 [DOI] [PubMed] [Google Scholar]
- 74.Su JC, Tseng PH, Hsu CY, Tai WT, Huang JW, Ko CH, et al. RFX1-dependent activation of SHP-1 induces autophagy by a novel obatoclax derivative in hepatocellular carcinoma cells. Oncotarget. 2014;5:4909–19. doi: 10.18632/oncotarget.2054. https://doi.org/10.18632/oncotarget.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X, Tang X, Liu H, Li L, Hou Q, Gao J. Autophagy induced by baicalin involves downregulation of CD147 in SMMC-7721 cells in vitro. Oncol Rep. 2012;27:1128–34. doi: 10.3892/or.2011.1599. https://doi.org/10.3892/or.2011.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Wu J, Lin B, Li X, Zhang H, Ding H, et al. Galangin suppresses HepG2 cell proliferation by activating the TGF-beta receptor/Smad pathway. Toxicology. 2014;326:9–17. doi: 10.1016/j.tox.2014.09.010. https://doi.org/10.1016/j.tox.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 77.Vara D, Salazar M, Olea-Herrero N, Guzman M, Velasco G, Diaz-Laviada I. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: role of AMPK-dependent activation of autophagy. Cell Death Differ. 2011;18:1099–111. doi: 10.1038/cdd.2011.32. https://doi.org/10.1038/cdd.2011.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou Q, Tang X, Liu H, Tang J, Yang Y, Jing X, et al. Berberine induces cell death in human hepatoma cells in vitro by downregulating CD147. Cancer Sci. 2011;102:1287–92. doi: 10.1111/j.1349-7006.2011.01933.x. https://doi.org/10.1111/j.1349-7006.2011.01933.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chu YL, Ho CT, Chung JG, Raghu R, Lo YC, Sheen LY. Allicin induces anti-human liver cancer cells through the p53 gene modulating apoptosis and autophagy. J Agric Food Chem. 2013;61:9839–48. doi: 10.1021/jf403241s. https://doi.org/10.1021/jf403241s [DOI] [PubMed] [Google Scholar]
- 80.Wang L, Gao C, Yao S, Xie B. Blocking autophagic flux enhances matrine-induced apoptosis in human hepatoma cells. Int J Mol Sci. 2013;14:23212–30. doi: 10.3390/ijms141223212. https://doi.org/10.3390/ijms141223212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang ZH, Li T, Chang LL, Zhu H, Tong YG, Chen XP, et al. Glycyrrhetinic Acid triggers a protective autophagy by activation of extracellular regulated protein kinases in hepatocellular carcinoma cells. J Agric Food Chem. 2014;62:11910–6. doi: 10.1021/jf503968k. https://doi.org/10.1021/jf503968k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Su CL, Chen FN, Won SJ. Involvement of apoptosis and autophagy in reducing mouse hepatoma ML-1 cell growth in inbred BALB/c mice by bacterial fermented soybean products. Food Chem Toxicol. 2011;49:17–24. doi: 10.1016/j.fct.2010.08.017. https://doi.org/10.1016/j.fct.2010.08.017 [DOI] [PubMed] [Google Scholar]
- 83.Kim DG, Jung KH, Lee DG, Yoon JH, Choi KS, Kwon SW, et al. 20(S)-Ginsenoside Rg3 is a novel inhibitor of autophagy and sensitizes hepatocellular carcinoma to doxorubicin. Oncotarget. 2014;5:4438–51. doi: 10.18632/oncotarget.2034. https://doi.org/10.18632/oncotarget.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang DM, Liu JS, Deng LJ, Chen MF, Yiu A, Cao HH, et al. Arenobufagin, a natural bufadienolide from toad venom, induces apoptosis and autophagy in human hepatocellular carcinoma cells through inhibition of PI3K/Akt/mTOR pathway. Carcinogenesis. 2013;34:1331–42. doi: 10.1093/carcin/bgt060. https://doi.org/10.1093/carcin/bgt060 [DOI] [PubMed] [Google Scholar]
- 85.Hu F, Han J, Zhai B, Ming X, Zhuang L, Liu Y, et al. Blocking autophagy enhances the apoptosis effect of bufalin on human hepatocellular carcinoma cells through endoplasmic reticulum stress and JNK activation. Apoptosis. 2014;19:210–23. doi: 10.1007/s10495-013-0914-7. https://doi.org/10.1007/s10495-013-0914-7 [DOI] [PubMed] [Google Scholar]
- 86.Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-Regulating microRNAs and Cancer. Front Oncol. 2017;7:65. doi: 10.3389/fonc.2017.00065. https://doi.org/10.3389/fonc.2017.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F, et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826–31. doi: 10.1016/j.bbrc.2012.06.048. https://doi.org/10.1016/j.bbrc.2012.06.048 [DOI] [PubMed] [Google Scholar]
- 88.Chang Y, Yan W, He X, Zhang L, Li C, Huang H, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–87 e8. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 89.He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357–69. doi: 10.1038/onc.2011.500. https://doi.org/10.1038/onc.2011.500 [DOI] [PubMed] [Google Scholar]
- 90.Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505–17. doi: 10.1002/hep.26659. https://doi.org/10.1002/hep.26659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019–24. doi: 10.3892/or.2013.2338. https://doi.org/10.3892/or.2013.2338 [DOI] [PubMed] [Google Scholar]
- 92.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590–8. doi: 10.1016/j.jhep.2013.10.028. https://doi.org/10.1016/j.jhep.2013.10.028 [DOI] [PubMed] [Google Scholar]
- 93.He C, Dong X, Zhai B, Jiang X, Dong D, Li B, et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867–81. doi: 10.18632/oncotarget.4814. https://doi.org/10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stiuso P, Potenza N, Lombardi A, Ferrandino I, Monaco A, Zappavigna S, et al. MicroRNA-423–5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated With Sorafenib. Mol Ther Nucleic Acids. 2015;4:e233. doi: 10.1038/mtna.2015.8. https://doi.org/10.1038/mtna.2015.8 [DOI] [PubMed] [Google Scholar]
- 95.Chen H, Luo Z, Dong L, Tan Y, Yang J, Feng G, et al. CD133/prominin-1-mediated autophagy and glucose uptake beneficial for hepatoma cell survival. PLoS One. 2013;8:e56878. doi: 10.1371/journal.pone.0056878. https://doi.org/10.1371/journal.pone.0056878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. 83 e1–11. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thomas HE, Mercer CA, Carnevalli LS, Park J, Andersen JB, Conner EA, et al. mTOR inhibitors synergize on regression, reversal of gene expression, and autophagy in hepatocellular carcinoma. Sci Transl Med. 2012;4:139ra84. doi: 10.1126/scitranslmed.3003923. https://doi.org/10.1126/scitranslmed.3003923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang Z, Shi G, Jin J, Guo H, Guo X, Luo F, et al. Dual PI3K/mTOR inhibitor NVP-BEZ235-induced apoptosis of hepatocellular carcinoma cell lines is enhanced by inhibitors of autophagy. Int J Mol Med. 2013;31:1449–56. doi: 10.3892/ijmm.2013.1351. https://doi.org/10.3892/ijmm.2013.1351 [DOI] [PubMed] [Google Scholar]
- 99.Simioni C, Martelli AM, Cani A, Cetin-Atalay R, McCubrey JA, Capitani S, et al. The AKT inhibitor MK-2206 is cytotoxic in hepatocarcinoma cells displaying hyperphosphorylated AKT-1 and synergizes with conventional chemotherapy. Oncotarget. 2013;4:1496–506. doi: 10.18632/oncotarget.1236. https://doi.org/10.18632/oncotarget.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhai B, Hu F, Jiang X, Xu J, Zhao D, Liu B, et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol Cancer Ther. 2014;13:1589–98. doi: 10.1158/1535-7163.MCT-13-1043. https://doi.org/10.1158/1535-7163.MCT-13-1043 [DOI] [PubMed] [Google Scholar]
- 101.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6:130–40. doi: 10.1038/nrc1801. https://doi.org/10.1038/nrc1801 [DOI] [PubMed] [Google Scholar]
- 102.Gao M, Yeh PY, Lu YS, Hsu CH, Chen KF, Lee WC, et al. OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma. Cancer Res. 2008;68:9348–57. doi: 10.1158/0008-5472.CAN-08-1642. https://doi.org/10.1158/0008-5472.CAN-08-1642 [DOI] [PubMed] [Google Scholar]
- 103.Dong X, Li R, Xiu P, Dong X, Xu Z, Zhai B, et al. Meloxicam executes its antitumor effects against hepatocellular carcinoma in COX-2- dependent and -independent pathways. PLoS One. 2014;9:e92864. doi: 10.1371/journal.pone.0092864. https://doi.org/10.1371/journal.pone.0092864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhong J, Dong X, Xiu P, Wang F, Liu J, Wei H, et al. Blocking autophagy enhances meloxicam lethality to hepatocellular carcinoma by promotion of endoplasmic reticulum stress. Cell Prolif. 2015;48:691–704. doi: 10.1111/cpr.12221. https://doi.org/10.1111/cpr.12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, et al. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56:644–57. doi: 10.1002/hep.25699. https://doi.org/10.1002/hep.25699 [DOI] [PubMed] [Google Scholar]
- 106.Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6:1057–65. doi: 10.4161/auto.6.8.13365. https://doi.org/10.4161/auto.6.8.13365 [DOI] [PubMed] [Google Scholar]
- 107.Xie HJ, Noh JH, Kim JK, Jung KH, Eun JW, Bae HJ, et al. HDAC1 inactivation induces mitotic defect and caspase-independent autophagic cell death in liver cancer. PLoS One. 2012;7:e34265. doi: 10.1371/journal.pone.0034265. https://doi.org/10.1371/journal.pone.0034265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altmeyer A, Jung AC, Ignat M, Benzina S, Denis JM, Gueulette J, et al. Pharmacological enhancement of autophagy induced in a hepatocellular carcinoma cell line by high-LET radiation. Anticancer Res. 2010;30:303–10. [PubMed] [Google Scholar]
- 109.Altmeyer A, Ignat M, Denis JM, Messaddeq N, Gueulette J, Mutter D, et al. Cell death after high-LET irradiation in orthotopic human hepatocellular carcinoma in vivo. In Vivo. 2011;25:1–9. [PubMed] [Google Scholar]
- 110.Altmeyer A, Josset E, Denis JM, Gueulette J, Slabbert J, Mutter D, et al. The mTOR inhibitor RAD001 augments radiation-induced growth inhibition in a hepatocellular carcinoma cell line by increasing autophagy. Int J Oncol. 2012;41:1381–6. doi: 10.3892/ijo.2012.1583. https://doi.org/10.3892/ijo.2012.1583 [DOI] [PubMed] [Google Scholar]
- 111.Wu L, Li L, Meng S, Qi R, Mao Z, Lin M. Expression of argininosuccinate synthetase in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:365–8. doi: 10.1111/jgh.12043. https://doi.org/10.1111/jgh.12043 [DOI] [PubMed] [Google Scholar]
- 112.Kuo MT, Savaraj N, Feun LG. Targeted cellular metabolism for cancer chemotherapy with recombinant arginine-degrading enzymes. Oncotarget. 2010;1:246–51. doi: 10.18632/oncotarget.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fan T, Zhang C, Zong M, Zhao Q, Yang X, Hao C, et al. Peptidylarginine deiminase IV promotes the development of chemoresistance through inducing autophagy in hepatocellular carcinoma. Cell Biosci. 2014;4:49. doi: 10.1186/2045-3701-4-49. https://doi.org/10.1186/2045-3701-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nordenstedt H, White DL, El-Serag HB. The changing pattern of epidemiology in hepatocellular carcinoma. Dig Liver Dis. 2010;42( Suppl 3):S206–14. doi: 10.1016/S1590-8658(10)60507-5. https://doi.org/10.1016/S1590-8658(10)60507-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453–6. doi: 10.1128/JVI.06064-11. https://doi.org/10.1128/JVI.06064-11 [DOI] [PMC free article] [PubMed] [Google Scholar]