Abstract
Background/Aims
Toll-like receptors (TLRs), particularly TLR2 and TLR4, take part in elicitation of immune responses against Helicobacter pylori (H. pylori). This study aimed to investigate the relationship between single nucleotide polymorphisms (SNP) rs3804099 in the TLR2 gene and rs4986790 in the TLR4 gene with H. pylori infection and peptic ulcer (PU).
Materials and Methods
Blood specimens were obtained from 350 individuals, including 100 H. pylori-infected patients with PU, 125 H. pylori-infected asymptomatic subjects (AS), and 125 non-infected healthy subjects (NHS). The DNA was extracted, and the SNPs were determined using ARMS-PCR method.
Results
The frequency of CT genotype at TLR2 SNP rs3804099 in both the PU and AS groups was significantly higher than in the NHS group (p<0.05). In total H. pylori-infected individuals (PU+AS), the frequency of the CT genotype at rs3804099 was also significantly higher than in the NHS group (p<0.005). The frequency of the CC genotype at rs3804099 in PU+AS was markedly lower than in the NHS group (p=0.066). PU patients carried CT genotype more frequently than total healthy individuals (AS+NHS) (p<0.03). The distribution of the TT genotype was lower, whereas the frequency of the CT genotype was higher in AS individuals infected with CagA+ strains than those infected with CagA- strains (p<0.03). No significant differences were found among the PU, AS, and NHS groups regarding the genetic differences at rs4986790 in the TLR4 gene.
Conclusion
These results provide evidence regarding the association of the rs3804099 in the TLR2 gene with H. pylori infection and PU. The rs3804099 may affect vulnerability to H. pylori infection, particularly to CagA+ strains of bacteria.
Keywords: Helicobacter pylori, peptic ulcer, TLR2, TLR4, gene polymorphism
INTRODUCTION
Colonization of the human stomach by Helicobacter pylori causes the development of peptic ulcers (PU), gastric cancer, and mucosa-associated lymphoid tissue lymphoma in approximately 15%–20%, 0.1%–3%, and less than 0.01% of H. pylori-infected individuals, respectively (1,2). Moreover, association of H. pylori infection with various extra-gastrointestinal diseases such as hematologic, cardiovascular, metabolic, neurologic, and dermatologic disorders has been also reported (3–5). The interplay between H. pylori-related virulence factors as well as genetic background of the host, such as immune system-related genes, has a prominent role in the clinical outcomes of H. pylori infection (2,6). The cytotoxin-associated gene A (CagA) is considered to be the strongest virulence parameter of H. pylori; therefore, the infection with CagA+ strains of bacteria is correlated with a higher risk of PU and gastric cancer development than the infection with CagA− strains (7–9).
Toll-like receptors (TLRs) are an important subset of the sensors of immune cells that recognize pathogen-associated molecular patterns (DAMP) (10,11). TLR-related immune response may influence the infection outcome in the direction of pathogen deletion or its persistence (12). H. pylori-derived components are recognized by some types of TLRs, particularly TLR2, TLR4, TLR5, and TLR9 (10,12).
Under physiological situations, the expression of TLR2 and TLR4 on the gastrointestinal epithelium is low; however, their expression is raised after infection with H. pylori (11). The binding of TLR2 and TLR4 to the H. pylori-derived components results in the quick production of various proinflammatory mediators, such as cytokines, chemokines, and nitric oxide (NO) (11,12). Enhanced production of different proinflammatory cytokines was indicated in gastric biopsies collected from H. pylori-positive patients (13).
The polymorphisms in a TLR gene may influence its expression, thereby altering its capability for binding to ligand and changing its related signaling pathways (14). Several single nucleotide polymorphisms (SNPs) in the TLR2 and TLR4 genes were linked with H. pylori-associated gastrointestinal diseases (14,15). The SNP +597T>C (rs3804099) is a synonymous polymorphism placed in the third exon of the TLR2 gene on the chromosome 4q32 (16). The association of the SNP rs3804099 with a number of infectious diseases such as bacterial meningitis and pulmonary tuberculosis was reported in previous studies (17,18).
The SNP +896T/C (rs4986790) is a non-synonymous polymorphism located in the third exon of the TLR4 gene on the chromosome 9, and it leads to the substitution of amino acids Asp299Gly. The replacement of Asp299Gly amino acids changes the extracellular part of TLR4 and may cause diminished ligand binding, low responsiveness to lipopolysaccharide, and reduced transporting of TLR4 to the cellular surface (19). The results from a meta-analysis study indicated that the SNP rs4986790 in TLR4 exhibits an increased risk, decreased risk, or no association with different infectious diseases (20). There are also controversies regarding the association of the SNP rs4986790 in TLR4 with H. pylori-related diseases because no association or positive association was reported in studies from different countries (19,21–24).
The polymorphisms in the TLR genes may affect the human immune responses against pathogens and influence their clinical outcomes (14). There is no report about the association of the SNPs rs3804099 in TLR2 gene and rs4986790 in TLR4 gene with H. pylori and its-related PU in Iranian populations. This investigation aimed was to evaluate the association of aforementioned SNPs with H. pylori and its-related PU in a population from southeastern Iranian, Kerman.
MATERIALS AND METHODS
Subjects
In total, 100 H. pylori-infected patients with PU (age: 40.6 ± 8.7 years) and 250 healthy individuals [including 125 H. pylori-infected asymptomatic (AS) carriers (age: 39.1±9.3 years) and 125 non-infected healthy individuals (age: 40±10.13 years)] were recruited in the study. The PU group was selected among patients referred to the Afzalipoor hospital affiliated with Kerman University of Medical Sciences (Kerman, Iran) from October 2016 to January 2017. The diagnostic procedures of PU were performed by digestive specialist, according to the upper gastrointestinal endoscopic observations, and none of the patients were taking medications (including nonsteroidal anti-inflammatory drugs) at the time of investigation. In PU patients, the status of H. pylori infection was assessed by rapid urease test (RUT) and seropositivity for bacterium-specific immunoglobulin G (IgG) was assessed using an enzyme-linked immunosorbent assay (ELISA) method. The RUT test was performed on a biopsy specimen taken from the gastric antrum during endoscopic observation. For PU patients, the status of H. pylori infection was considered positive if both tests (RUT and H. pylori-specific IgG) were reactive.
The AS carriers were seropositive for H. pylori-specific IgG, as described below. The NHS were seronegative for both specific IgG and IgA against H. pylori. The AS carriers as well as the NHS were recruited among blood donors and interviewed concerning the gastrointestinal clinical signs, and none of them had previous history of gastrointestinal or other relevant abnormalities. The Ethical Committee of the Kerman University of Medical Sciences evaluated and approved the protocol of investigation. Moreover, sample was collected after obtaining written informed consent from patients. A blood specimen of the peripheral blood (5 mL) was obtained from each individual, and the serum sample was separated and stored at −70°C until analyzed.
Measurement of H. pylori-specific antibodies
The serum levels of H. pylori-specific IgG and IgA were measured using the commercial ELISA kits (Euroimmun, Lübeck, Germany), according to the manufacturer’s instructions. The serum IgG levels against CagA were also assessed using the commercial ELISA kit (Diagnostic Bioprobes, Milano, Italy).
DNA extraction
The salting out protocol was used for genomic DNA separation from the peripheral blood leukocytes, as previously described by Miller et al. (25). The DNA concentrations and their purity were checked using a spectrophotometry system (Ependorf, Germany) according to the calculation of the optical density at 260 and 280 nm wavelengths. Notably, 200 μl sample of whole peripheral blood yields 5.70±2.40 μg of DNA with A260/A280 of 1.88±80.0. DNA specimens were stored at −20°C until used.
Determination of the TLR2 gene polymorphism at position of rs3804099
The TLR2 SNP rs3804099 was genotyped by the amplification refractory mutation system-polymerase chain reaction (ARMS-PCR) method (18).
The PCR reaction was prepared by mixing the following reagents in a microcentrifuge tube on ice: 10×Taq polymerase buffer (2.5 μL), 2 units of Taq DNA polymerase, 0.5 μL of MgCl2 (stock concentration 1.5 mM), 0.5 μL of each dNTP [dATP, dCTP, dGTP, and dTTP (stock concentration of 10 mM)], 1 μL of each primer, 1 μL of genomic DNA, and sterile double-distilled water to make up a total volume of 25 μL. The sequences of the used primers and the product sizes are demonstrated in Table 1.
Table 1.
Primers used for the determination of SNP rs4986790 and SNP rs3804099 in the TLR2 and TLR4 genes, respectively
| PCR products | Primer | Gene |
|---|---|---|
| TLR2 rs3804099 | Forward outer: 5-ATTGCAAATCCTGAGAGTGGGAA-3 | Common product size: 349 bp |
| Reverse outer: 5-CAAACTTTCATCGGTGATTTTCACA-3 | TT: 228 bp | |
| Forward inner(T allele): 5-CCAAAAAGTTTGAAGTCAATTCAGAAT-3 | CC:173 bp | |
| Reverse inner(C allele): 5-TCATATGAAGGATCAGATGACTTCCG-3 | CT: 228 bp and 173 bp | |
| TLR4 rs4986790 | Forward outer: 5′-TGAACCCTATGAACTTTATCC-3′ | Common product size: 383 bp |
| Reverse outer: 5′-GTTAACTAATTCTAAATGTTGCCATC-3′ | AA: 147 bp | |
| Forward inner(A allele): 5′-GCATACTTAGACTACTACCTCGATGA-3′ | GG:287 bp | |
| Reverse inner(G allele): 5′-CAAACAATTAAATAAGTCAATAATAC-3′ | AG:147 bp and 287 bp |
PCR: Polymerase chain reaction; SNP: Single nucleotide polymorphism; TLR: Toll-like receptors
The amplification program was designed as follows: an initial denaturation stage (at 94°C for 5 minutes), followed by 30 cycles of denaturation (at 95°C for 30 seconds), annealing (at 53°C for 30 seconds), and extension (at 72°C for 40 seconds). A final extension stage was also performed (at 72°C for 10 minutes) to terminate the PCR process.
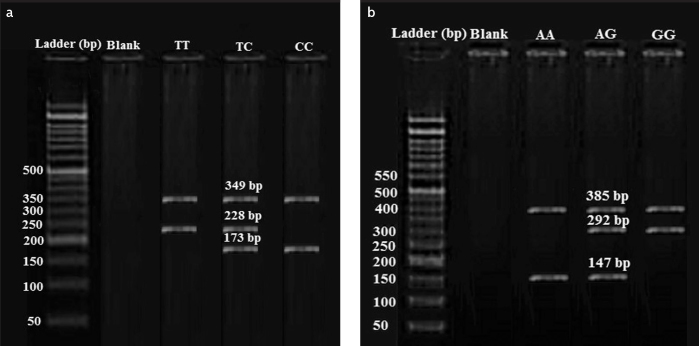
The product sizes of the PCR reaction were determined by electrophoresis on a 2% agarose gel. The common PCR control product was 349 bp, and the allele-specific products were 228 bp and 173 bp for the presence of T and C alleles, respectively (Figure 1a).
Figure 1. a, b.
Determination of TLR2 gene polymorphism at the position of rs3804099 using the ARMS-PCR method The first column shows a ladder pattern; the second column is blank; the third, fourth, and fifth columns represent the TT, TC, and CC genotypes, respectively (a); Detecting of TLR4 gene polymorphism at the position of rs4986790 using the ARMS-PCR method The first column shows a ladder pattern; the second column is blank; the third, fourth, and fifth columns represent the AA, AG, and GG genotypes, respectively (b)
Determination of the TLR4 gene polymorphism at position of rs4986790
The TLR4 SNP rs4986790 was also determined using ARMS-PCR, as described previously (25). The common PCR product was 385 bp, and the allele-specific products were 292 bp and 147 bp for the presence of G and A alleles, respectively (Table 1) (Figure 1b) (25).
Statistical analysis
The genotype and allele frequencies were computed in PU patients and control individuals by direct gene enumerating. The variables were compared using χ2 test, and a p-value of less than 0.05 was considered to be significant. The data were analyzed using the Statistical Package for Social Sciences (SPSS) software, version 22 (IBM Corp.; Armonk, NY, USA).
RESULTS
No significant difference was observed among the PU, PS, and NHS groups concerning their ages (p=0.32). The men/women ratios were similarly distributed in three groups [70 men (70.0%) and 30 women (30.0%) in PU patients, 87 men (69.6%) and 38 women (30.4%) in both AS and NHS groups].
H. pylori-specific immunoglobulins
The seroprevalence of anti-H. pylori IgG antibodies in PU patients and healthy subjects was 96.0% and 58.2%, respectively. The PU patients who were positive for both RUT and H. pylori-specific IgG tests were enrolled in the study. The seropositivity for anti-CagA IgG in H. pylori-infected PU patients and asymptomatic subjects was 85.0% and 62.3%, respectively. The determination of the anti-H. pylori IgA performed in seronegative subjects for IgG against H. pylori. The NHS were seronegative for both specific IgG and IgA against H. pylori.
Association of TLR2 SNP rs3804099 with PU and H. pylori infection
The frequencies of the TT, CC, and CT genotypes at rs3804099 in the TLR2 gene were 34.0%, 16.0%, and 50.0% in the PU group; 40.0%, 16.0%, and 44.0% in the AS group; and 44.8%, 24.0%, and 31.2% in the NHS group, respectively. A significant difference in genotype frequencies at SNP rs3804099 in the TLR2 gene was observed among the PU, AS, and NHS groups (p<0.05). The prevalence of the CT genotype at TLR2 SNP rs3804099 in both the PU and AS group was found to be significantly higher than that in the NHS group (p<0.05). The distribution of the CT genotype at SNP rs3804099 did not significantly differ between the PU and AS group (Table 2).
Table 2.
Frequencies of the genotypes and alleles at TLR2 SNP rs4986790 in PU, AS, and NHS, total infected subjects (PU+AS), and total healthy subjects (AS+NHS)
| Genotype | PU | AS | NHS | Total infected subjects (PU+AS) | Total healthy subjects (AS+NHS) | p |
|---|---|---|---|---|---|---|
| TT | 34 (34.0%) | 50 (40.0%) | 56 (44.8%) | 84 (37.3%) | 106 (42.4%) | *0.25; **0.17; ***0.14 |
| CT | 50 (50.0%) | 55 (44.0%) | 39 (31.2%) | 105 (46.7%) | 94 (37.6%) | *0.05; **0.005; ***0.03 |
| CC | 16 (16.0%) | 20 (16.0%) | 30 (24.0%) | 36 (16.0%) | 50 (20.0%) | *0.54; **0.06; ***0.38 |
| T | 118 (59.0%) | 155 (62.0%) | 151 (60.4%) | 273 (60.7%) | 306 (61.2%) | *0.80; **0.94; ***0.59 |
| C | 82 (41.0%) | 95 (38.0%) | 99 (39.6%) | 177 (39.3%) | 194 (38.8%) | *0.80; **0.94: ***0.59 |
comparison of genetic variations between the PU and NHS groups
comparison of genetic variations between total infected subjects (PU+AS)
comparison of genetic variations between total healthy subjects (AS+NHS) and the PU group
AS: asymptomatic subjects; NHS: non-infected healthy subjects; PU: peptic ulcer; SNP: single nucleotide polymorphism; TLR: Toll-like receptors
To evaluate the association of a given SNP with H. pylori infection, we compared the genetic variations in that SNP, between total H. pylori-infected subjects (PU+AS groups) and the NHS group. In total H. pylori-infected individuals (PU+AS groups), the frequencies of the TT, CC, and CT genotypes at rs3804099 in the TLR2 gene were 37.3%, 16.0%, and 46.7%, respectively (Table 3). In all H. pylori-infected individuals (PU+AS groups), the frequency of the CT genotype at SNP rs3804099 in the TLR2 gene was also significantly higher than that in the NHS group (p<0.005). However, the frequency of the CC genotype at SNP rs3804099 in the TLR2 gene was markedly lower in total H. pylori-infected subjects (PU+AS groups) than in the NHS group (p=0.066) (Table 3).
Table 3.
The frequencies of the genotypes and alleles at TLR2 SNP rs4986790 in H. pylori-infected subjects according to the CagA status of bacteria
| TLR2 gene polymorphism | Alleles | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Groups | CagA status | TT | CT | CC | Total | C | T | p |
| PU | CagA+ | 27 (38.6%) | 34 (48.6%) | 9 (12.9%) | 70 (100.0%) | 52 (37.1%) | 88 (62.9%) | *0.22 |
| CagA- | 7 (23.3%) | 16 (53.3%) | 7 (23.3%) | 30 (100.0%) | 30 (50.0%) | 30 (50.0%) | **0.09 | |
| Total | 34 (34.0%) | 50 (50.0%) | 16 (16.0%) | 100 (100.0%) | 82 (41.0%) | 118 (59.0%) | ||
| AS | CagA+ | 24 (31.6%) | 40 (52.6%) | 12 (15.8%) | 76 (100.0%) | 64 (42.1%) | 88 (57.9%) | *0.03 |
| CagA- | 26 (52.6%) | 15 (30.6%) | 8 (16.3%) | 49 (100.0%) | 31 (31.6%) | 67 (68.4%) | **0.09 | |
| Total | 50 (40.0%) | 55 (44.0%) | 20 (16.0%) | 125 (100.0%) | 95 (38.0%) | 155 (62.0%) | ||
| NHS | ---- | 56 (44.8%) | 39 (31.2%) | 30 (24.0%) | 125 (100.0%) | 99 (39.6%) | 151 | (0.4%) |
comparison of the genotypes and alleles distribution at SNP rs4986790 between subjects infected with CagA+ strains of H. pylori and those infected with CagA- strains of bacteria in specified group
AS: asymptomatic subjects; CagA: cytotoxin-associated gene A; NHS: non-infected healthy subjects; PU: peptic ulcer; SNP: single nucleotide polymorphism; TLR: Toll-like receptors
To evaluate the association of a given SNP with PU disease, we compared the genetic variations in that SNP between PU patients and total healthy subjects (AS+NHS groups). The comparison of the genetic variations at SNP rs3804099 in the TLR2 gene between PU patients and total healthy subjects (AS+NHS groups) was demonstrated in Table 4. The frequencies of the TT, CC, and CT genotypes at rs3804099 in the TLR2 gene were 42.4%, 20.0%, and 37.6% in total healthy subjects (AS+NHS groups), respectively. The CT genotype frequency at SNP rs3804099 in the TLR2 gene in PU group was significantly higher than total healthy subjects (AS+NHS groups) (p<0.03).
Table 4.
The frequencies of the genotypes and alleles at TLR4 SNP rs3804099 in PU, AS, and NHS, total infected subjects (PU+AS), and total healthy subjects (AS+NHS)
| Genotype | PU | AS | NHS | Total infected subjects (PU+AS) | Total healthy subjects (AS+NHS) | p |
|---|---|---|---|---|---|---|
| AA | 77 (77.0%) | 103 (82.4%) | 95 (76.5%) | 180 (80.0%) | 198 (79.2%) | *0.42; **0.38; ***0.65 |
| AG | 18 (18.0%) | 15 (12.0%) | 25 (20.0%) | 33(14.7%) | 40 (16.0%) | *0.21; **0.20; ***0.64 |
| GG | 5 (5.0%) | 7 (5.6%) | 5 (4.0%) | 12 (5.3%) | 12 (4.8%) | *0.83; **0.57; ***0.93 |
| A | 172 (86.0%) | 221 (88.4%) | 215 (86.0%) | 393 (87.3%) | 436 (87.2%) | *0.66; **0.61; ***0.67 |
| G | 28 (14.0%) | 29 (11.6%) | 35 (14.0%) | 57 (12.7%) | 64 (12.8%) | *0.66; **0.61; ***0.67 |
comparison of genetic variations between the PU and NHS groups
comparison of genetic variations between total infected subjects (PU+AS) and the NHS group
comparison of genetic variations between total healthy subjects (AS+NHS) and the PU group
AS: asymptomatic subjects; NHS: non-infected healthy subjects; PU: peptic ulcer; SNP: single nucleotide polymorphism; TLR: Toll-like receptors
The C and T allele frequencies at SNP rs3804099 in the TLR2 gene in the PU, AS, NHS, total H. pylori-infected subjects (PU+AS groups) and total healthy subjects (AS+NHS groups) are also summarized in Table 2. No association was found between the presence of the alleles C and T at SNP rs3804099 in the TLR2 gene with PU and H. pylori infection.
The genetic differences at SNP rs3804099 in the TLR2 gene in H. pylori-infected subjects according to the CagA status of bacteria are summarized in Table 3. Regarding the genotype and allele frequencies at SNP rs3804099, there was no significant difference identified between PU patients infected with H. pylori CagA+ strains and patients infected with CagA− strains. In the AS group, the distribution of the TT genotype was lower, whereas the prevalence of the CT genotype was higher in individuals infected with CagA+ strains than in those infected with CagA− strains of H. pylori (p<0.03).
Association of TLR4 SNP rs4986790 with PU and H. pylori infection
The AA, AG, and GG genotype frequencies and the A and G allele frequencies at SNP rs4986790 in the TLR4 gene in the PU, AS, and NHS groups are demonstrated in Table 4. The differences of genotype and allele frequencies at SNP rs4986790 in the TLR4 gene between the PU, AS, and NHS groups were not significant.
We also assessed the association between TLR4 SNP rs4986790 and H. pylori infection. The genetic differences at SNP rs4986790 in the TLR4 gene in total H. pylori-infected subjects (PU+AS group) and the NHS group are presented in Table 4. Regarding the genotype and allele frequencies TLR4 SNP, no significant differences were found between total H. pylori-infected subjects and the NHS group (Table 4).
The comparison of the genetic differences at SNP rs4986790 in the TLR4 gene between patients with PU and total healthy subjects (AS+NHS group) is demonstrated in Table 4. The distribution of genotypes and alleles in TLR4 SNP was similarly expressed in total healthy subjects and the patient with PU.
The genetic differences at SNP rs4986790 in the TLR4 gene in H. pylori-infected subjects according to the CagA status of bacteria are summarized in Table 5. In both the PU and AS groups, no significant differences were identified between individuals infected with CagA+ H. pylori strains and those infected with CagA− strains with respect to the genotype and allele distribution at TLR4 SNP rs4986790.
Table 5.
The frequencies of the genotypes and alleles at TLR4 SNP rs3804099 in H. pylori-infected subjects according to the CagA status of bacteria
| TLR4 gene polymorphism | Alleles | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Groups | CagA status | TT | CT | CC | Total | C | T | p |
| PU | CagA+ | 53 (75.7%) | 13 (18.6%) | 4 (5.7%) | 70 (100.0%) | 119 (85.0%) | 21 (15.0%) | *0.847 |
| CagA- | 24 (80.0%) | 5 (16.7%) | 1 (3.3%) | 30 (100.0%) | 53 (88.3%) | 7 (11.7%) | **0.535 | |
| Total | 77 (77.0%) | 18 (18.0%) | 5 (5.0%) | 100 (100.0%) | 172 (86.0%) | 118 (59.0%) | ||
| AS | CagA+ | 63 (82.9%) | 9 (11.8%) | 4 (5.3%) | 76 (100.0%) | 135 (88.8%) | 17 (11.2%) | *0.975 |
| CagA- | 40 (81.6%) | 6 (12.2%) | 3 (6.1%) | 49 (100.0%) | 86 (87.75%) | 12 (12.25%) | **0.798 | |
| Total | 103 (82.4%) | 15 (12.0%) | 7 (5.6%) | 125 (100.0%) | 221 (88.4%) | 29 (11.6%) | ||
| NHS | 95 (76.5%) | 25 (20.0%) | 5 (4.0%) | 125 (100.0%) | 215 (86.0%) | 35 (14.0%) | ||
comparison of the genotypes and alleles distribution at SNP rs4986790 between subjects infected with the CagA+ strains of H. pylori and those infected with CagA- strains of bacteria in specified group
AS: asymptomatic subjects; CagA: cytotoxin-associated gene A; NHS: non-infected healthy subjects; PU: peptic ulcer; SNP: single nucleotide polymorphism; TLR: Toll-like receptors
DISCUSSION
The results show an association between a SNP in the TLR2 gene (rs3804099) with H. pylori infection and PU. However, no significant relationship was found between TLR2 rs3804099 and H. pylori-associated gastritis in a study from Thailand (22). TLR2 is expressed by various lymphoid and non-lymphoid cells and binds to various microbial-derived components as a result of its capability to make heterodimers with TLR1, TLR6, and perhaps TLR10 (12). Various H. pylori-originated molecules, such as lipopolysaccharide (HP-LPS), heat shock protein 60 (HP-HSP60), and neutrophil-activating protein (HP-NAP), contribute to the induction of TLR2-linked responses. The stimulation of TLR2 results in the activation of NF-κB and induction of cytokine expression in epithelial cells, monocytes/macrophages, dendritic cells, neutrophils, and B cells that play a critical role in the regulation of immune response to H. pylori (10,12). The TLR2-related immune response elicited by H. pylori-originated components may play a determinative role in the outcome of the infection toward the eradication or persistence of bacteria or the occurrence of immunopathological consequences (12).
The actual mechanisms by which TLR2 rs3804099 influences the susceptibility to H. pylori and PU remain to be clarified in further studies. The TLR2 rs3804099 may affect the susceptibility to H. pylori and PU by influencing the immune responses. The stimulation of TLR2 leads to the induction of pro- or anti-inflammatory reactions that may influence the fate of H. pylori infection (12). It has been reported that PBMCs from healthy subjects with the CT/TT genotypes at TLR2 rs3804099 produced higher amounts of TNF-α, IL-1β, and IL-6 after the Legionella pneumophila stimulation, representing a stronger immune response to bacterial antigens (26). Chen et al. (27) reported that the peripheral leukocytes from traumatic patients who have the C allele in the position of rs3804099 produce higher amounts of IL-10, IL-8, and TNF-α than those having T allele. Therefore, it is possible that TLR2 rs3804099 influence vulnerability to H. pylori infection through the cytokine production.
The SNP rs3804099 may also influence the expression of TLR2. The peptidoglycan-stimulated PBMCs from patients with Behcet’s disease who carried rs3804099 TT genotype expressed higher amounts of TLR2 mRNA than those who carried the CC or CT genotypes after stimulation with peptidoglycan (28). The influences of TLR2 rs3804099 on the TLR2-related immunomodulatory and inflammatory properties during H. pylori infection need to be considered in future studies. The stimulation of PBMCs from individuals with various genotypes at rs3804099 by H. pylori-derived antigens and then after the detection of the cytokine profiles in the supernatant of cells would clear the possible influences of this SNP on the immune response to bacteria.
The results of this investigation also demonstrated that the distribution of the TT genotype was lower, whereas the prevalence of the CT genotype was higher in AS individuals infected with H. pylori CagA+ strains than those infected with CagA− strains. These findings represent an association between TLR2 SNP rs3804099 and strain type of H. pylori (CagA+ or CagA−) that infects a person. Therefore, the TLR2 rs3804099 CT genotype can be considered as a risk factor for PU by increasing the susceptibility to infection with H. pylori CagA+ strains. We have observed that the existence of a homozygosity situation (CT) at TLR2 rs3804099 was related to H. pylori infection and PU disease. These observations represent the presence of C and T alleles in a Trans situation with each other may have reciprocal effects that lead to higher risk of H. pylori infection and PU development.
In this study, no association was found between the genetic variations in TLR4 SNP rs4986790 with H. pylori infection and PU. As mentioned, there are controversies about the relationship between the TLR4 SNP rs4986790 and H. pylori-related disorders. The results of the studies from India and Hungary showed no association between TLR4 SNP rs4986790 and gastritis or PU diseases (21,22). However, a positive association was indicated between TLR4 SNP rs4986790 and H. pylori infection, PU, and gastric cancer in other studies from Finland, India, and Brazil (19,23,24). Both TLR2 and TLR4 are involved in the induction of the initial immune response to H. pylori (10, 12). TLR2 may play more important roles than TLR4 in regulating the immune responses against H. pylori (12). There are some differences between TLR2 and TLR4 during H. pylori infection. In opposition to TLR4, which forms homodimer, the transmission of signals from TLR2 initiates after its homodimerization as well as its heterodimerization with either TLR1 or TLR6, and possibly TLR10, providing more capability to recognize more ligands molecules and induce different responses (10,12). Hence, more H. pylori-originated components, such as HP-LPS, HP-NAP, HP-HSP60, Urease, and CagA bind to TLR2, while only HP-LPS is identified through TLR4 (12). Moreover, there are some differences between TLR2 and TLR4 in signaling pathways so that the TLR2-related signaling pathways may exhibit more plasticity regarding the outcome of signals toward inflammatory or anti-inflammatory responses (12). It was indicated that H. pylori-induced IL-8 synthesis needs TLR2 rather than TLR4. In addition, the H. pylori-induced COX-2 production is suppressed in TLR2-negative cells but not in TLR4-deficient cells, showing that bacteria induce the COX-2 expression via a TLR2-linked manner (12,29). The neutralizing antibodies targeting the TLR2 but not TLR4 also diminish the H. pylori-induced cytokine release from the gastric epithelial cells (30). Therefore, it appears that TLR2 is more prominent than TLR4 in regulating of the immune responses to H. pylori. The significant plasticity in the TLR2-related signals during H. pylori infection is attributable to the variation in the components that act as TLR2 ligands (12).
As mentioned, there are several SNPs in the TLR2 and TLR4 genes. Evaluating the other SNPs give better insights regarding the possible association of the TLR2- and TLR4-linked genetic variations with H. pylori and relevant diseases.
In conclusion, our findings in a sample of Iranian population provide evidence that polymorphism +597T>C (rs3804099) in the TLR2 gene [but not SNP +896T/C (rs4986790) in the TLR4 gene] with H. pylori infection and PU disease. TLR2 SNP rs3804099 may influence the vulnerability to H. pylori infection, especially to CagA+ strains of bacteria.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the Ethical Committee of the Kerman University of Medical Sciences.
Informed Consent: Written informed consent was obtained from patients who participated in this study.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - A.J.; Design - A.J.; Supervision - A.J.; Resource - A.J.; Materials - A.K., M.M.H.; Data Collection and/or Processing - E.M., M.N.; Analysis and/or Interpretation - A.J.; Literature Search - A.J.; Writing - A.J.; Critical Reviews - A.J.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: This study was supported by Kerman University of Medical Sciences, Kerman, Iran.
REFERENCES
- 1.Jafarzadeh A, Salari M. Seroprevalence of anti-Helicobacter pylori and anti-CagA antibodies in peptic ulcer and healthy subjects in the city of Rafsanjan. J Res Med Sci. 2006;11:285–91. [Google Scholar]
- 2.Datta De D, Roychoudhury S. To be or not to be: The host geneticfactor and beyond in Helicobacter pylori mediated gastro-duodenal diseases. World J Gastroenterol. 2015;21:2883–95. doi: 10.3748/wjg.v21.i10.2883. https://doi.org/10.3748/wjg.v21.i10.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jafarzadeh A, Nemati M, Tahmasbi M, Ahmadi P, Rezayati MT, Sayadi AR. The association between infection burden in Iranian patients with acute myocardial infarction and unstable angina. Acta Med Indones. 2011;43:105–11. [PubMed] [Google Scholar]
- 4.Jafarzadeh A, Esmaeeli-Nadimi A, Nemati M, Tahmasbi M, Ahmadi P. Serum concentrations of Helicobacter pylori IgG and the virulence factor CagA in patients with ischaemic heart disease. East Mediterr Health J. 2010;16:1039–44. https://doi.org/10.26719/2010.16.10.1039. [PubMed] [Google Scholar]
- 5.Tan HJ, Goh KL. Extragastrointestinal manifestations of Helicobacter pylori infection: facts or myth? A critical review. J Dig Dis. 2012;13:342–9. doi: 10.1111/j.1751-2980.2012.00599.x. https://doi.org/10.1111/j.1751-2980.2012.00599.x [DOI] [PubMed] [Google Scholar]
- 6.Jafarzadeh A, Ahmedi-Kahanali J, Bahrami M, Taghipour Z. Seroprevalence of anti-Helicobacter pylori and anti-CagA antibodies among healthy children according to age, sex, ABO blood groups and Rh status in south-east of Iran. Turk J Gastroenterol. 2007;18:165–71. [PubMed] [Google Scholar]
- 7.Berge C, Terradot L. Structural Insights into Helicobacter pyloriCag Protein Interactions with Host Cell Factors. Curr Top Microbiol Immunol. 2017;400:129–47. doi: 10.1007/978-3-319-50520-6_6. https://doi.org/10.1007/978-3-319-50520-6_6 [DOI] [PubMed] [Google Scholar]
- 8.Jafarzadeh A, Mirzaee V, Ahmad-Beygi H, Nemati M, Rezayati MT. Association of the CagA status of Helicobacter pylori and serum lev-els of interleukin (IL)-17 and IL-23 in duodenal ulcer patients. J Dig Dis. 2009;10:107–12. doi: 10.1111/j.1751-2980.2009.00371.x. [DOI] [PubMed] [Google Scholar]
- 9.Jafarzadeh A, Hassanshahi GH, Nemati M. Serum levels ofhigh-sensitivity C-reactive protein (hs-CRP)in Helicobacter pylori-infected peptic ulcer patients and its association with bacterial CagA virulence factor. Dig Dis Sci. 2009;54:2612–6. doi: 10.1007/s10620-008-0686-z. https://doi.org/10.1007/s10620-008-0686-z [DOI] [PubMed] [Google Scholar]
- 10.Pachathundikandi SK, Lind J, Tegtmeyer N, El-Omar EM, Backert S. Interplay of the Gastric Pathogen Helicobacter pylori with Toll-Like Receptors. Biomed Res Int. 2015;2015:192420. doi: 10.1155/2015/192420. https://doi.org/10.1155/2015/192420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Cao M, Song L, Qi P, Chen C, Wang X, et al. The contribution of toll-like receptor 2 on Helicobacter pylori activation of the nuclear factor-kappa B signaling pathway in gastric epithelial cells. Microb Pathogen. 2016;98:63–8. doi: 10.1016/j.micpath.2016.06.028. https://doi.org/10.1016/j.micpath.2016.06.028 [DOI] [PubMed] [Google Scholar]
- 12.Nemati M, Larussa T, Khorramdelazad H, Mahmoodi M, Jafarzadeh A. Toll-like receptor 2: An important immunomodulatory molecule during Helicobacter pylori infection. Life Sci. 2017;178:17–29. doi: 10.1016/j.lfs.2017.04.006. https://doi.org/10.1016/j.lfs.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 13.Abdollahi H, Shams S, Zahedi MJ, Darvish Moghadam S, Hayatbakhsh MM, Jafarzadeh A. IL-10, TNF-alpha and IFN-gamma levels in serum and stomach mucosa of Helicobacter pylori-infected patients. Iran J Allergy Asthma Immunol. 2011;10:267–71. [PubMed] [Google Scholar]
- 14.Hu Y, Liu JP, Zhu Y, Lu NH. The Importance of Toll-like Receptors in NF-kappaB Signaling Pathway Activation by Helicobacter pylori Infection and the Regulators of this Response. Helicobacter. 2016;21:428–40. doi: 10.1111/hel.12292. https://doi.org/10.1111/hel.12292 [DOI] [PubMed] [Google Scholar]
- 15.Tongtawee T, Bartpho T, Kaewpitoon S, Kaewpitoon N, Dechsukhum C, Leeanansaksiri W, et al. Genetic polymorphisms in TLR1, TLR2, TLR4, and TLR10 of Helicobacter pylori-associated gastritis: a prospective cross-sectional study in Thailand. Eur J Cancer Prev. 2018;27:118–23. doi: 10.1097/CEJ.0000000000000347. https://doi.org/10.1097/CEJ.0000000000000347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uno K, Kato K, Shimosegawa T. Novel role of toll-like receptors in Helicobacter pylori - induced gastric malignancy. World J Gastroenterol. 2014;20:5244–51. doi: 10.3748/wjg.v20.i18.5244. https://doi.org/10.3748/wjg.v20.i18.5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Zhang N, Liu L, Zheng K, Zhu L, Zhu J, et al. Polymorphisms of toll-like receptors 2 and 9 and severity and prognosis of bacterial meningitis in Chinese children. Sci Rep. 2017;7:42796. doi: 10.1038/srep42796. https://doi.org/10.1038/srep42796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naderi M, Hashemi M, Hazire-Yazdi L, Taheri M, Moazeni-Roodi A, Eskandari-Nasab E, et al. Association between toll-like receptor2 Arg677Trp and 597T/C gene polymorphisms and pulmonary tuberculosis in Zahedan, Southeast Iran. Biomed Environ Sci. 2013;17:516–20. doi: 10.1016/j.bjid.2012.12.009. https://doi.org/10.1016/j.bjid.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Oliveira JG, Silva AE. Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J Gastroenterol. 2012;18:1235–42. doi: 10.3748/wjg.v18.i11.1235. https://doi.org/10.3748/wjg.v18.i11.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziakas PD, Prodromou ML, El Khoury J, Zintzaras E, Mylonakis E. The role of TLR4 896 A>G and 1196 C>T in susceptibility to infections: a review and meta-analysis of genetic association studies. PloS one. 2013;8:e81047. doi: 10.1371/journal.pone.0081047. https://doi.org/10.1371/journal.pone.0081047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moura SB, Almeida LR, Guerra JB, Rocha GA, Camargos Rocha AM, Melo FF, et al. Toll-like receptor (TLR2, TLR4 and TLR5) gene polymorphisms and Helicobacter pylori infection in children with and without duodenal ulcer. Microbes Infect. 2008;10:1477–83. doi: 10.1016/j.micinf.2008.08.009. https://doi.org/10.1016/j.micinf.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 22.Achyut BR, Ghoshal UC, Moorchung N, Mittal B. Association of Toll-like receptor-4 (Asp299Gly and Thr399Ileu) gene polymorphisms with gastritis and precancerous lesions. Hum Immunol. 2007;68:901–7. doi: 10.1016/j.humimm.2007.10.006. https://doi.org/10.1016/j.humimm.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 23.Loganathan R, Nazeer M, Goda V, Devaraju P, Ali M, Karunakaran P, et al. Genetic variants of TLR4 and TLR9 are risk factors for chronic Helicobacter pylori infection in South Indian Tamils. Hum Immunol. 2017;78:216–20. doi: 10.1016/j.humimm.2016.12.002. https://doi.org/10.1016/j.humimm.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 24.Pohjanen VM, Koivurova OP, Huhta H, Helminen O, Makinen JM, Karhukorpi JM, et al. Toll-Like Receptor 4 Wild Type Homozygozity of Polymorphisms +896 and +1196 Is Associated with High Gastrin Serum Levels and Peptic Ulcer Risk. PloS on. 2015;10:e0131553. doi: 10.1371/journal.pone.0131553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafiei A, Abedini M, Hosseini SH, Hosseini-Khah Z, Bazrafshan B, Tehrani M. Toll like receptor-4 896A/G gene variation, a risk factor for migraine headaches. Iran J Immunol. 2012;9:159–67. [PubMed] [Google Scholar]
- 26.Zhang F, Gao XD, Wu WW, Gao Y, Zhang YW, Wang SP. Polymorphisms in toll-like receptors 2, 4 and 5 are associated with Legionella pneumophila infection. Infection. 2013;41:941–8. doi: 10.1007/s15010-013-0444-9. https://doi.org/10.1007/s15010-013-0444-9 [DOI] [PubMed] [Google Scholar]
- 27.Chen KH, Gu W, Zeng L, Jiang DP, Zhang LY, Zhou J, et al. Identification of haplotype tag SNPs within the entire TLR2 gene and their clinical relevance in patients with major trauma. Shock. 2011;35:35–41. doi: 10.1097/SHK.0b013e3181eb45b3. https://doi.org/10.1097/SHK.0b013e3181eb45b3 [DOI] [PubMed] [Google Scholar]
- 28.Fang J, Hu R, Hou S, Ye Z, Xiang Q, Qi J, et al. Association of TLR2 gene polymorphisms with ocular Behcet’s disease in a Chinese Han population. Invest Ophthalmol Vis Sci. 2013;54:8384–92. doi: 10.1167/iovs.13-12878. https://doi.org/10.1167/iovs.13-12878 [DOI] [PubMed] [Google Scholar]
- 29.Pellicano A, Imeneo M, Leone I, Larussa T, Luzza F. Enhanced activation of cyclooxygenase-2 downregulates Th1 signaling pathway in Helicobacter pylori-infected human gastric mucosa. Helicobacter. 2007;12:193–9. doi: 10.1111/j.1523-5378.2007.00498.x. https://doi.org/10.1111/j.1523-5378.2007.00498.x [DOI] [PubMed] [Google Scholar]
- 30.Tran CT, Garcia M, Garnier M, Burucoa C, Bodet C. Inflammatory signaling pathways induced by Helicobacter pylori in primary human gastric epithelial cells. Innate Immun. 2017;23:165–74. doi: 10.1177/1753425916681077. https://doi.org/10.1177/1753425916681077 [DOI] [PubMed] [Google Scholar]