Abstract
Background/Aims
To investigate the etiology of esophageal cancer (EC) related with human papillomavirus (HPV) infection.
Materials and Methods
Fresh surgically resected tissue samples and clinical information were obtained from 189 patients. Genomic DNA was extracted, and HPV was detected using polymerase chain reaction (PCR) with HPV L1 gene primers of MY09/11; HPV16 was detected using HPV16 E6 type-specific primer sets. Copies of HPV16 E2, E6, and the human housekeeping gene β-actin were tested using quantitative PCR to analyze the relationship between HPV16 integration and esophageal squamous cell carcinoma and the relationship between the HPV16 integration status and clinical information of patients.
Results
Of the 189 samples, 168 HPV-positive samples were detected, of which 76 were HPV16 positive. Among the HPV16 positive samples, 2 cases (E2/E6 ratio>1) were 2.6% (2/76) purely episomal, 65 (E2/E6 ratio between 0 and 1) were 85.6% (65/76) mixture of integrated and episomal, and 9 (E2/E6 ratio=0) were 11.8% (9/76) purely integrated. The results indicate that integration of HPV16 was more common in the host genome than in the episome genome. The prevalence rate of HPV16 integration is increasing with the pathological stage progression of esophageal carcinoma (EC).
Conclusion
A high prevalence of HPV16 suggested that HPV16 has an etiological effect on the progress of EC. Integration of HPV16 is more common than episome genome in the host cells, indicating that continuous HPV infection is the key to esophageal epithelial cell malignant conversion and canceration.
Keywords: Esophageal carcinoma, human papillomavirus, infection, integration status
INTRODUCTION
Esophageal cancer (EC) is one of the dominating carcinomas and is about half of all cases occurring in China, which contributes to natural circumstance, living habits, and nutrient deficiency (1–3). In 1982, Syrjanen (4) assumed that the incidence of esophageal squamous cell cancer was associated with human papillomavirus (HPV) infection. Previously, studies have reported that HPV was present in almost 99% cervical carcinoma (CC) cases. Notably, HPV is regarded as the most significant etiological factor in CC (5,6). However, many studies in the field supported the hypothesis that the occurrence of EC is related to HPV infection, the causal role of HPV infection in EC development remains controversial (7–13). HPV integrated into the host cell chromosome may play a crucial role in the malignant transformation and canceration of CC. E2 is a down regulator of the oncogenes E6 and E7; this course results in the disruption of the viral E2 open-reading frames (ORF) and thereby promotes E6 and E7 expression (14–16). Down regulate their anti-tumor functions for E6 and E7 viral oncoproteins and may play a role in cellular transformation or canceration (17–19). A study by Si et al. (20) showed that the HPV16 viral load was very low, approximately <1–157 copies per genome, and <10 copies per genome in most positive cancers.
In this study, HPV16 infection and HPV16 integration status were evaluated in EC cells. Briefly, esophageal carcinoma specimens were obtained and genomic DNA was extracted from each sample. For each specimen, the HPV DNA was examined using polymerase chain reaction (PCR) amplification using MY09/11 primers, and HPV16 was detected using HPV16 type-specific primers (21). The viral load and integration state for HPV16 positive specimens were determined using quantitative (q)PCR with HPV16 E2, E6, and β-actin primers; the number of HPV16 copies was calculated using the formula: (E6 copy/β-actin copy)× 2, and the HPV16 integration status was verified using the ratio of E2 to E6 copy number. The relationship between HPV16 integrated status and patient characteristics was analyzed using a statistical method.
MATERIALS AND METHODS
Ethical approval
This study was approved by the Ethics Committee of North China University of Science and Technology.
Sample collection
Fresh surgically resected tissue samples and clinical information was collected from 189 patients between March 2013 and December 2015. A consent form was obtained from each specimen donor. All donors were diagnosed with EC and were treated at the pathology department of Tangshan People’s Hospital of Hebei Province. The average patient age was 58 (range, 40–76) years; the study included 136 male and 53 female participants. All samples were esophageal squamous cell cancers consisting 30 well differentiated, 104 moderately differentiated, and 55 poorly differentiated cancers. These specimens were categorized as 98 early, 63 middle, and 28 late stage according to the clinical and pathological stages of EC; all fresh samples were stored at −80°C prior to analysis.
Plasmids
The plasmids HPV16/pBR322 and HPV18/pBR322 DNA (containing the whole HPV genome), β-actin human housekeeping genome, and the HEK293 cell DNA were obtained from the National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, China. The DNA samples were stored at −20°C.
DNA extraction
Nucleic acids were extracted from each specimen using the QIAamp DNA mini kit (QIAGEN, Germany) according to the manufacturer’s instructions. The DNA was finally eluted in approximately 50 μL sterile distilled water and was stored at −20°C.
Primer sets design
Human papillomavirus E2 breakage was deemed to a label of HPV integrated. To explain the probability that a breakage might occur in the E2 gene, three pairs of nonoverlapping quantitative PCR primers were synthesized to contain the entire E2 ORF; three regions of the E2 ORF were designed in HPV16 gene sequence 2755–2942, 3355–3526, and 3602–3786, respectively; and one region of the E6 ORF was designed in 214–533. The HPV genome in an HPV-positive sample is supposed to “complete integration status” if the E2 gene copy number is not detectable using quantitative PCR analysis. When the ratio of the E2 to E6 gene copy number of the E6 gene is ≥1, it indicates “complete episomal status” for the HPV genome. When the ratio of the E2 to E6 gene copy number is <0 and >1, it indicates a mix (including episomal and integrated) status for HPV genome. Real-time PCR primers were designed based on the GenBank-provided HPV16 nucleotide (http://www.ncbi.nlm.nih.gov/nuccore/K02718). The primers made by Sangon biotech of Shanghai (Table 1).
Table 1.
PCR primers used in the study
| Primer | Sequences (5′-3′) | Position | Amplicon size(bp) |
|---|---|---|---|
| HPV L1 Primers | |||
| MY09 | CGT CCM ARR GGA WAC TGA TC | 450 | |
| MY11 | GCM CAG GGW CAT AAY AAT GG | ||
| HPV16 E2 Primers | |||
| HPV16 E2-1 | ATGGAGACTCTTTGCCAACG | 2755–2774 | 188 |
| HPV16 E2-2 | GCCAGTGTTGGCACCACTTGGTGG | 2919–2942 | |
| HPV16 E2-3 | AGCAGCAACGAAGTATCCTCTC | 3355–3376 | 172 |
| HPV16 E2-4 | GCAACAACTTAGTGGTGTGGC | 3506–3526 | |
| HPV16 E2-5 | GTAACACTACACCCATAGTAC | 3602–3622 | 185 |
| HPV16 E2-6 | GTCACGTTGCCATTCACTATC | 3766–3786 | |
| HPV16 E6 Primers | |||
| HPV16 E6-3 | ACTGCGACGTGAGGTATATGAC | 214–235 | 320 |
| HPV16 E6-4 | TTGATGATCTGCAACAAGACATAC | 510–533 | |
| β-actin Primers | |||
| Forward | TCACCCACACTGTGCCCATCT | 549–560 | 290 |
| Reverse | GAACCGCTCATTGCCAATGG | 819–838 | |
HPV: human papillomavirus; PCR: polymerase chain reaction
Detection of specimen quality and HPV DNA
The specimen DNA was extracted from each sample, and the DNA quality was detected by PCR using β-actin primers and 293 cell DNA as positive template control to meet the requirement of further experiments.
Human papillomavirus DNA for each specimen was tested using PCR as described by Karlsen et al. (21) HPV L1 primer MY09/11. HPV 16 was detected using HPV16 E6 type-specific primer sets; all primer sets are presented in Table 1.
PCR amplification was conducted using Takara Ex Taq kit (Takara Bio, China) in a 20-μL reaction mixture, which contained 1×Ex Buffer (MgCl2 free), 2.5 mM MgCl2, 0.2 mM mixture deoxynucleoside triphosphate (dNTPs), 5 pmol forward and reverse primers (Table 1), and 1 U Ex Taq DNA polymerase; 100 ng extracted DNA or control DNA was added in the reaction as template. The thermal cycling profile was as follows: 95°C/5 min following 31 cycles at 95°C/30 sec, 55°C/30 sec, and 72°C/30 sec. The final extension was performed at 72°C/5 min, and the products were finally stored at 4°C.
The PCR products were observed on a 1.0% agarose gel using Goldview II nuclear staining dyes (BioTeke, China).
Sequence analysis products of type-specific HPV16
To confirm the accuracy of type-specific PCR for HPV16, HPV16 E6 PCR products were depurated using the Qiaquick Gel Extraction Kit according to the manufacturer’s instructions. These depurated products were subjected to sequence analysis by Beijing Rui Bo Xing ke Biological Technology company. The results were sequence aligned using the DNAman software Lynnon Biosoft, USA (http://www.shinegene.org.cn/q2.html).
Integration status of HPV16 and viral load detected by qPCR
The integration status and viral load were determined by qPCR using HPV16 E2/E6/β-actin primers. Briefly, the number of HPV16 copies was calculated using the formula: (E6 copy/β-actin copy)×2, and the HPV16 integration status was confirmed using the rate of E2 to E6 copy number. The standard curves were obtained using quantified DNA (10-fold dilution series of full length HPV16/pBR322 and β-actin plasmids). The PCR reactions were performed in a 25-μL volume using the AmpliTaq Gold DNA polymerase kit (AB Applied Biosystems, USA) containing 1× AmpliTaq gold buffer (MgCl2-free), dNTPs (0.2 mmol/L), MgCl2 (2.5 mmol/L), primers (2 pmol/L), 20× EvaGreen (Biotium, USA; 1.25 μL), 0.25 μL RX (stabilizer), 2 μL AmpliTaq Gold DNA polymerase, and 0.4 μL DNA template (80 ng/μl). The initial denaturation was performed at 95°C/5 min, followed by 5 cycles at 95°C/1 min, 55°C/1 min, and 72°C/1 min, followed by 35 cycles at 95°C/30 sec, 55°C/50 sec, and 72°C/50 sec; final extension was performed at 72°C/5 min. The threshold cycle (22,23) was determined using the CFX96 Touch Real-time PCR System (Bio-rad, USA), with the baseline set automatically to standard deviations above the background fluorescence generated in the first five cycles. For each sample, equivalent DNA quantity was analyzed for E2 and E6 and compared with the expression of the internal reference gene β-actin. The specificity of each amplified response was verified by checking the disaggregation curve against the anticipative fusion temperature of qPCR production.
Statistical analysis
A database with Excel was established. Statistical analysis was conducted using the Statistical Package for the Social Sciences version 13.0 software (SPSS Inc.; Chicago, IL, USA). p<0.05 was considered statistically significant.
RESULTS
Detection of specimen quality and HPV DNA
To evaluate the quality of the DNA samples, a total of 189 samples were investigated for β-actin. A 290-bp band representing β-actin was detected in each sample (data not shown). This result suggested that the DNA of each specimen was of high quality. Overall, 168 specimens were detected as HPV DNA positive from 189 samples using MY09/11 primers (Figure 1a), and 76 were HPV16 positive using HPV16 E6-specific primer sets (Figure 1b).
Figure 1. a, b.
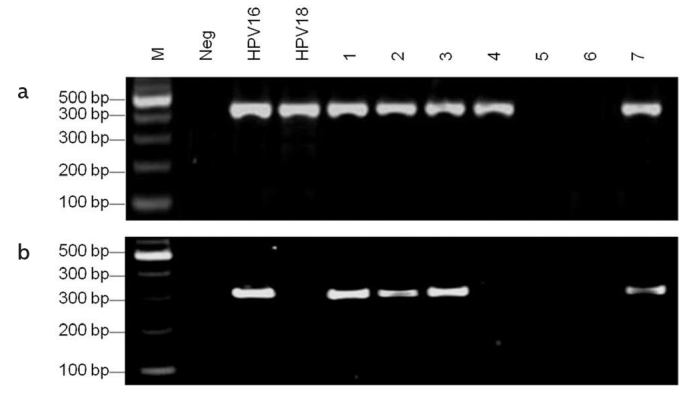
Detection of HPV DNA using MY09/11 primers
M, 100 bp DNA ladder; Neg, negative control with sterilized water as template; HPV16 and HPV18, positive controls with HPV16/pBR322 and HPV18/pBR322 templates; lanes 1–7 were the results of HPV DNA detected from different esophageal carcinoma samples
B. Detection of HPV 16 using HPV16 E6 specific primers
M, 100 bp DNA ladder; Neg, negative control with sterilized water as template; HPV16, positive control with HPV16/pBR322 template; HPV18, specific control for HPV16 using HPV18/pBR322 DNA template; lanes 1–7 were the results of HPV16 detected from different esophageal carcinoma samples
Sequence analysis for HPV16 E6
The sequencing results of positive amplicons for HPV16 were compared with the reported corresponding sequence K02718 (https://www.ncbi.nlm.nih.gov/nuccore/K02718). The analysis results verified two HPV16 E6 samples positive with two mutations, and three samples positive with one mutation in the viral DNA (Figure 2).
Figure 2.
Sequencing results of positive amplicons for HPV16 E6
Sequences 1–5 represented 5 cases of HPV16 positive amplicons with some mutations from freshly surgically resected esophageal carcinoma samples
Relationship between patient background with HPV16 integration and viral load
In this study, very low viral load was detected using highly sensitive quantitative real-time PCR. The HPV16 integration status was confirmed using the rate of E2 to E6 copy number, and the results indicated that 2.6% (2/76) with E2/E6 ratios >1 in the HPV16 positive samples uniquely belonged to the episome status, 85.6% (65/76) with 0<E2/E6 ratios<1 belonged to the mix status (including episome and integration), and 11.8% (9/76) with E2/E6 ratios=0 exclusively belonged to the integrated status. These results indicate the predominance of integrated viral genome in the host chromosome. The number of HPV16 copies was calculated using the formula: (E6copy/β-actincopy)×2; the viral load of HPV16 E6 was 1.8–89.2 copies/cell in 76 HPV16 E6-positive samples. Information regarding clinicopathological features, HPV16 integration, and viral load are shown in Table 2. Statistical analysis suggested that the HPV16 integration status was not significantly correlated with age, sex, and tumor differentiation of patients.
Table 2.
Information on clinicopathological features, HPV16 integration, and viral load
| Category | HPV16 viral load (Copy/cell) |
HPV16 Positive |
HPV16 integration status | ||
|---|---|---|---|---|---|
|
| |||||
| Pure episomal (%) Mixture # | Integration (%) | χ2 p* | |||
| Patient age, years | 2.73>0.05 | ||||
| ≤45 | 9.2–33.6 | 18 | 1(5.6%) 16(88.8%) | 1(5.6%) | |
| 46–64 | 1.8–89.2 | 28 | 1(3.6%) 24(85.7%) | 3(10.7%) | |
| ≥65 | 10.9–75.5 | 30 | 0 25(83.3%) | 5(16.7%) | |
| Gender | 0.98>0.05 | ||||
| Male | 6.8–89.2 | 55 | 2 46 | 7 | |
| Female | 1.8–67.9 | 21 | 0 19 | 2 | |
| Differentiation degree | 1.3>0.05 | ||||
| Well differentiated | 10.9–55.6 | 11 | 0 10 | 1 | |
| Moderately differentiated | 1.8–78.6 | 49 | 1 42 | 6 | |
| Poorly differentiated | 23.9–89.2 | 16 | 1 13 | 2 | |
| Pathological stage | 4.76>0.05 | ||||
| Early stage | 1.8–66.5 | 25 | 0 23 | 2 | |
| Middle stage | 23.9–55.6 | 40 | 2 34 | 4 | |
| Late stage | 10.9–89.2 | 11 | 0 8 | 3 | |
Note: There was no statistically significant difference p>0.05
Mixture of episomal and integrated virus
HPV: human papillomavirus
DISCUSSION
Pathogenesis of EC involved many factors, including geographic environment, smoking, alcohol abuse, depletion of nitrosamines or moldy food, special nutrition deficiencies, poor oral hygiene, and hereditary predisposition (24–28). To date, the etiology of EC occurrence has not been inconclusive. The present study was conducted to investigate the relationship between HPV and the occurrence of EC. Si et al. (20) reported that the viral load of HPV in the EC samples might be very low. Therefore, in this study, HPV16 was detected using PCR and the ratios of E2 to E6 were tested using qPCR. In the present study, 76 HPV16-positive cases in 189 samples were detected with a positive rate of 40.2%. The sequencing results showed 5 samples with mutations based on the reported corresponding sequence K02718 in the database from positive amplicons of HPV16.
The HPV16 physical status was determined on the assumption that the E2 gene is disrupted in integrated viral genome, and confirmed using the rate of E2 to E6 copy number: if E2/E6 ratios >1, HPV16 belonged to the episome status, if 0<E2/E6 ratios<1, HPV16 belonged to the mix status (including episome and integration), and if E2/E6 ratios=0, HPV16 belonged to the integrated status (29–31). Therefore, three pairs of quantitative real-time PCR primers were synthesized to cover the complete E2 ORF. According to the GenBank-provided HPV16 gene sequences of K02718, three regions of the E2 ORF were designed in 2755–2942, 3355–3526, and 3602–3786, respectively. Thus, the results of the quantitative real-time PCR were accurate for the HPV16 integration status of HPV16. The real-time quantitative PCR results showed more integrated than episomal status, which suggests that persistent HPV16 infection is a key factor for the carcinogenesis of the esophagus. The HPV16 viral load of 76 samples was approximately 1.8–89.2 copies/cell. The reversely low viral load (<10 copies each cell) in many positive samples was comparable with the previous results of Si et al. (20). However, considering that some CC cell lines exhibited very low HPV load, for instance, Theelen et al. (32) reported that there are approximately 2–600 HPV copies in the CaSki, SiHa, and HeLacell lines because of the low HPV load in some EC cells. Theelen et al. (32) demonstrated that particularly when the viral genome integrates to the host chromosome, low HPV load is enough to generate or accelerate the occurrence of cancer and lead to other genetic changes in the host genome. In this study, the HPV positivity rate of 168/189 is high for ECs. Among these, 40.2% samples were HPV16 positive. These results suggest that high prevalence of HPV16 acts as a pathogenic factor in the occurrence of esophageal carcinoma for the Tangshan City of Hebei province. The HPV gene integrated into the human genome indicates that continuous HPV infection is the key for carcinogenesis. However, HPV infection may be one of the important pathogenic factors for EC. Further studies are needed to illuminate HPV integrated sites in the human genome and to understand the molecular mechanism of EC.
Limitations, drawbacks, or shortcomings for this study
Constandinou-Williams et al. (33) reported that a sole measurement of viral load could not predict the risk of cervical pre-cancer and carcinoma lesions. Table 2 indicates that there is no significant for HPV16 integration and viral load correlation with the clinical features of EC patients; hence, the viral load could not be deemed a useful marker of EC progression. The results of statistical analyses revealed that the HPV16 integration status was not significantly correlated with age, sex, differentiation, and pathological stage of the cancer patients; however, nine patient samples is extremely small to indicate that the HPV16 integration status is associated with age, sex, degree of differentiation, and the pathological stage of EC.
Of the total 189 samples, 76 specimens were HPV16 positive, indicating that HPV16 infection was prevalent and that HPV16 has an etiological effect on the progress of EC. The integration of HPV16 was more common than episome genome in the host cells, indicating that continuous HPV infection is the key for esophageal epithelial cell malignant conversion and canceration.
Acknowledgements
The authors would like to thank department of Tangshan people’s hospital in Hebei Province, China for their support and assistance in this study.
Footnotes
Ethics Committee Approval: Ethics committee approval was received for this study from the Ethics Committee of North China University of Science and Technology.
Informed Consent: Informed consent was obtained from each specimen donor.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - S.L., K.Z., Jintao L.; Design - S.L., K.Z., Jintao L.; Supervision - S.L., K.Z.; Resource - S.L.; Materials - K.Z., Jintao L.; Data Collection and/or Processing - H.S., J.L., X.H.; Analysis and/or Interpretation - S.L., K.Z., Jintao L.; Literature Search - H.S., J.L., X.H.; Writing - S.L., K.Z., Jintao L.; Critical Reviews - S.L., K.Z., Jintao L.
Conflict of Interest: No conflict of interest was declared by the authors.
Financial Disclosure: The project of science and technology activities for overseas student (No: CY201620), an research fund from North China University of Science and Technology (No:28656002), Health and Family Planning Commission of Hebei (No: 20170895).
REFERENCES
- 1.Zhao J, He YT, Zheng RS, et al. Analysis of esophageal cancertime trends in China, 1989- 2008. Asian Pac J Cancer Prev. 2012;13:4613–7. doi: 10.7314/apjcp.2012.13.9.4613. https://doi.org/10.7314/APJCP.2012.13.9.4613 [DOI] [PubMed] [Google Scholar]
- 2.Sun X, Chen W, Chen Z, et al. Population-based casecontrol study on risk factors for esophageal cancer in five high-risk areas in China. Asian Pac J Cancer Prev. 2010;11:1631–6. [PubMed] [Google Scholar]
- 3.Li M, Wan X, Wang Y, Sun Y, Yang G, Wang L. Time trends of esophageal and gastric cancer mortality in China, 1991–2009: an age-period-cohort analysis. Sci Rep. 2017;7:6797. doi: 10.1038/s41598-017-07071-5. https://doi.org/10.1038/s41598-017-07071-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syrjanen KJ. Histological changes identical to those of condylomatous lesions found in esophageal squamous cell carcinomas. Arch Geschwulstforsch. 1982;52:283–92. [PubMed] [Google Scholar]
- 5.zur Hausen H. Immortalization of human cells and their malignant conversion by high risk human papillomavirus genotypes. Semin Cancer Biol. 1999;9:405–11. doi: 10.1006/scbi.1999.0144. https://doi.org/10.1006/scbi.1999.0144 [DOI] [PubMed] [Google Scholar]
- 6.Bosch XF, lorincz A, Munoz N, et al. The causal relation between Human Papillomavirus and cervical cancer. J Clin Pathol. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. https://doi.org/10.1136/jcp.55.4.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benamouzig R, Pigot F, Quiroga G, et al. Human papillomavirusinfection in esophageal squamous cell carcinoma in western countries. Int J Cancer. 1992;50:549–52. doi: 10.1002/ijc.2910500409. https://doi.org/10.1002/ijc.2910500409 [DOI] [PubMed] [Google Scholar]
- 8.Chang F, Syrjänen S, Shen Q, et al. Human papillomavirus (HPV)DNA in esophageal precancer lesions and squamous cell carcinoma from China. Int J Cancer. 1990;45:21–5. doi: 10.1002/ijc.2910450106. https://doi.org/10.1002/ijc.2910450106 [DOI] [PubMed] [Google Scholar]
- 9.Lavergno D, de Villier EM. Papillomavirus in esophageal papillomas and carcinomas. Int J Cancer. 1999;80:681–4. doi: 10.1002/(sici)1097-0215(19990301)80:5<681::aid-ijc8>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Lu ZM, Chen KN, et al. Human papillomavirus type 16 is animportant infectious factor in the high incidence of esophageal cancer in Anyang area of China. Carcinogenesis. 2001;22:929–34. doi: 10.1093/carcin/22.6.929. https://doi.org/10.1093/carcin/22.6.929 [DOI] [PubMed] [Google Scholar]
- 11.Suzuk L, Noffsinger AE, Hui YZ, et al. Detection of human papil lomavirus in esophageal squamous cell carcinoma. Cancer. 1996;78:704–10. doi: 10.1002/(SICI)1097-0142(19960815)78:4<704::AID-CNCR2>3.0.CO;2-E. https://doi.org/10.1002/(SICI)1097-0142(19960815)78:4<704::AID-CNCR2>3.0.CO;2-E [DOI] [PubMed] [Google Scholar]
- 12.Togawa K, Jaskiewicz K, Takahashi H, et al. Human papillomavirus DNA sequences in esophagus squmous cell carcinoma. Gastroenterology. 1994;107:128–36. doi: 10.1016/0016-5085(94)90070-1. https://doi.org/10.1016/0016-5085(94)90070-1 [DOI] [PubMed] [Google Scholar]
- 13.Toh Y, Kuwano H, Tanaka S, et al. Detection of HPV DNA in esophageal carcinoma in Japan by polymerase chain reaction. Can-cer. 1992;70:2234–8. doi: 10.1002/1097-0142(19921101)70:9<2234::aid-cncr2820700903>3.0.co;2-3. https://doi.org/10.1002/1097-0142(19921101)70:9<2234::AID-CNCR2820700903>3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 14.Dreer M, van de Poel S, Stubenrauch F. Control of viral replication and transcription by the papillomavirus E8E2 protein. Virus Res. 2017;231:96–102. doi: 10.1016/j.virusres.2016.11.005. https://doi.org/10.1016/j.virusres.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 15.Ramírez-Salazar E, Centeno F, Nieto K, et al. HPV16 E2 could act as down-regulator in cellular genes implicated in apoptosis, proliferation and cell differentiation. Virol J. 2011;8:247. doi: 10.1186/1743-422X-8-247. https://doi.org/10.1186/1743-422X-8-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romanczuk H, Howley PM. Disruption of either the E1 or the E2regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159–63. doi: 10.1073/pnas.89.7.3159. https://doi.org/10.1073/pnas.89.7.3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choo KB, Pan CC, Han SH. Integration of human papillomavirustype 16 into cellular DNA of cervical carcinoma: preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology. 1987;161:259–61. doi: 10.1016/0042-6822(87)90195-4. https://doi.org/10.1016/0042-6822(87)90195-4 [DOI] [PubMed] [Google Scholar]
- 18.Dyson N, Howley PM, Munger K, et al. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–7. doi: 10.1126/science.2537532. https://doi.org/10.1126/science.2537532 [DOI] [PubMed] [Google Scholar]
- 19.Scheffner M, Werness BA, Huibregtse JM, et al. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. https://doi.org/10.1016/0092-8674(90)90409-8 [DOI] [PubMed] [Google Scholar]
- 20.Si HX, Tsao SW, Poon CS, et al. Viral load of HPV in esophageal squamous cell carcinoma. Int J Cancer. 2003;103:496–500. doi: 10.1002/ijc.10865. https://doi.org/10.1002/ijc.10865 [DOI] [PubMed] [Google Scholar]
- 21.Karlsen F, Kalantari M, Jenkins A, et al. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol. 1996;34:2095–100. doi: 10.1128/jcm.34.9.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ST, Chun JW, Park G, et al. Comparative quantification ofplasma TDRD7 mRNA in cataract patients by real-time polymerase chain reaction. Korean J Ophthalmol. 2014;28:343–50. doi: 10.3341/kjo.2014.28.4.343. https://doi.org/10.3341/kjo.2014.28.4.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expressiondata using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. https://doi.org/10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Han-Ze, Jin Guang-Fu, Shen Hong-Bing. Epidemiologic differences in esophageal cancer between Asian and Western populations. Chin J Cancer. 2012;31:281–6. doi: 10.5732/cjc.011.10390. https://doi.org/10.5732/cjc.011.10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esophageal cancer: epidemiology, pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol. 2008;5:517–26. doi: 10.1038/ncpgasthep1223. https://doi.org/10.1038/ncpgasthep1223 [DOI] [PubMed] [Google Scholar]
- 26.Castro C, Peleteiro B, Lunet N. Modifiable factors and esophageal cancer: a systematic review of published meta-analyses. J Gastroenterol. 2018;1 doi: 10.1007/s00535-017-1375-5. https://doi.org/10.1007/s00535-017-1375-5 [DOI] [PubMed] [Google Scholar]
- 27.Jain S, Dhingra S. Pathology of esophageal cancer and Barrett’sesophagus. Ann Cardiothorac Surg. 2017;6:99–109. doi: 10.21037/acs.2017.03.06. https://doi.org/10.21037/acs.2017.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. https://doi.org/10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugo AP, Peyton CL, Joste NE, et al. Human papillomavirus type16 integration in cervical carcinoma in situ and invasive cervical cancer. J Clin Microbiol. 2006;44:1755–62. doi: 10.1128/JCM.44.5.1755-1762.2006. https://doi.org/10.1128/JCM.44.5.1755-1762.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badaracco G, Venuti A, Sedati A, et al. HPV16 and HPV18 in genitaltumors: significantly different levels of viral integration and correlation to tumor invasiveness. J Med Virol. 2002;67:574–82. doi: 10.1002/jmv.10141. https://doi.org/10.1002/jmv.10141 [DOI] [PubMed] [Google Scholar]
- 31.Si HX, Tsao SW, Poon CS, et al. Physical status of HPV16 inesophageal squamous cell carcinoma. J Clin Virol. 2005;32:19–23. doi: 10.1016/j.jcv.2004.04.004. https://doi.org/10.1016/j.jcv.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 32.Theelen W, Reijans M, Simons G, et al. A new multiparameter assay to assess HPV 16/18, viral load and physical status together with gain of telomerase genes in HPV-related cancers. Int J Cancer. 2010;126:959–75. doi: 10.1002/ijc.24844. [DOI] [PubMed] [Google Scholar]
- 33.Constandinou-Williams C, Collins SI, Roberts S, et al. Is human papillomavirus viral load a clinically useful predictive marker: a longitudinal study. Cancer Epidemiol Biomarkers Prev. 2010;19:832–7. doi: 10.1158/1055-9965.EPI-09-0838. https://doi.org/10.1158/1055-9965.EPI-09-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]



