Figure 2.
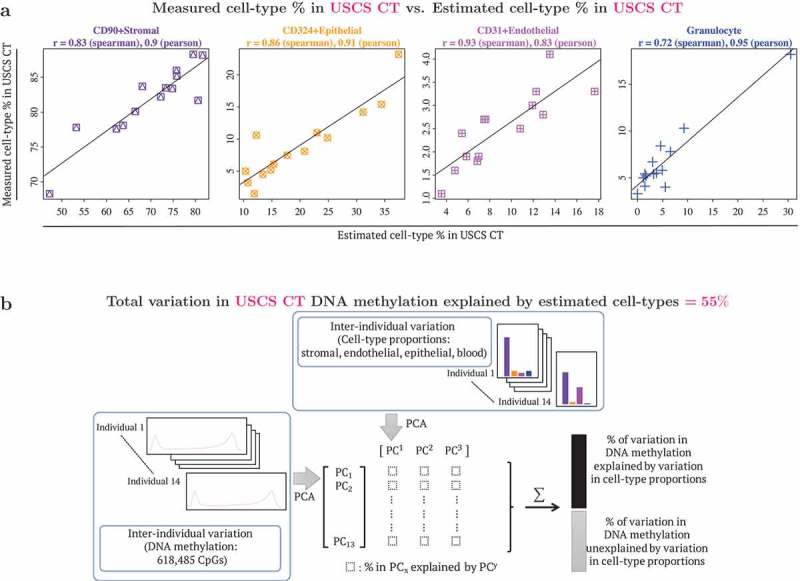
Benchmarking DNA methylation reference panel: use of reference panel to capture cell type composition in unsorted single cell suspension (USCS) cord tissue (CT). USCS CT refers to a sample of the single cell suspension of CT, that was obtained prior to the sequential isolation of stromal (CD90+), endothelial (CD31+) and epithelial (CD324+) cell types. (a) Scatterplots of cell type % measured in USCS infant CT (vertical axis) vs. estimated cell type % in USCS CT (horizontal axis). Cellular proportions were estimated using the reference panel in the current study following the method described by Houseman et al. (2012), where pairwise t-tests were used to identify 1000 cell type specific CpGs, by both P values and directionality of effect sizes (500 CpGs each). Granulocytes isolated from CB were included in the reference panel to capture CB contamination in CT. (b) Quantification of contribution of interindividual variation in cell type composition to interindividual variation in DNA methylation in CT. Additional details provided in the methods section.
