ABSTRACT
Prenatal arsenic exposure is associated with adverse birth outcomes and disease risk later in life, which could be mediated through epigenetic dysregulation. We evaluated the association between arsenic and gestational age (GA) that was mediated through DNA methylation (DNAm) using data from a Bangladeshi birth cohort. Arsenic exposure was measured in maternal drinking water at ≤16 weeks GA and maternal toenails collected ≤1 month postpartum. Cord blood DNAm was measured using Infinium HumanMethylation450 arrays (n = 44, discovery phase). Top loci identified in the discovery phase were then pyrosequenced in a second group (n = 569, validation phase). Structural equation models (SEM) evaluated the direct and indirect effects of arsenic and DNAm on GA. In the discovery phase, arsenic was associated with differential DNAm of 139 loci that were associated with GA (P < 1.10X10−6; |β regression|>0.10). Each doubling in water arsenic concentration decreased GA by 2 days, which was fully mediated through the main principal component of the top-ten CpGs (P < 0.001). In the validation phase, there were direct and indirect effects of miR214-3 and MCC DNAm on GA. In an adjusted SEM model, mediation of the association between arsenic and GA by miR124-3 was borderline significant (P = 0.061). This study therefore identified DNAm at specific loci in cord blood that mediated the effect of arsenic exposure on GA. Specifically, prenatal arsenic exposure was associated with lower methylation of miR124-3 that mediated the exposure-response of arsenic on GA. Future research should evaluate if these epigenetic changes are persistent and associated with disease risk.
KEYWORDS: Arsenic, DNA methylation, epigenetics, Illumina 450K, in utero exposure, environmental epigenetics, fetal programming, mediation
Introduction
Arsenic-contaminated drinking water is a global public health problem. Worldwide, it has been estimated that >200 million people are chronically exposed to drinking water that contains arsenic levels that surpass the World Health Organization and the US Environmental Protection Agency standard of 10 µg/L [1]. Bangladesh has been particularly affected by arsenic-contaminated drinking water, where national surveys estimate that 20 million people rely on drinking water supplies that exceed the Bangladesh national standard of 50 µg/L [1]. This is a pressing environmental health problem because arsenic is classified as a known Group 1 human carcinogen by the International Agency for Research on Cancer [2]. Additionally, arsenic readily crosses the placenta and maternal exposures are highly correlated with foetal concentrations [3]. Many studies have linked maternal arsenic exposure with adverse reproductive outcomes and adverse health effects in early childhood. For instance, prenatal arsenic exposure has been associated with increased risk of spontaneous abortions, still birth, reduced birth weight, and both neonatal and infant mortality [4]. Prenatal arsenic exposure has also been associated with increased susceptibility to infections during early childhood [5,6] and adverse neurological and cognitive development [4,6–10]. Additionally, studies have also observed that arsenic exposure early in life, particularly during gestation, increases the risk of disease and susceptibility to adverse health conditions later in life [11,12].
The disruption of foetal programming events through epigenetic mechanisms has been postulated to mediate the association between environmental toxicants and health effects later in life [13–15]. Epidemiological studies in adults and children report that arsenic acts as an epigenetic toxicant and alters DNA methylation (DNAm) in cord or adult whole blood [16–25]. These studies provide convincing data that arsenic exposure in the environment can alter DNAm in leukocytes, although there is less information linking these DNAm alterations to health outcomes.
In this study, our goal was to examine the association between arsenic exposure, DNAm in cord blood leukocytes, and reproductive outcomes in an established prospective birth cohort recruited in Bangladesh. We utilized a two-stage approach to test the hypothesis that DNAm at specific loci would mediate the association between prenatal arsenic exposure and reproductive health outcomes. This two-stage approach was economical and potentially reduced the possibility of false discoveries by coupling an agnostic epigenome-wide association study in a small discovery set with a larger candidate gene association study using pyrosequencing as a second technology.
Methods
Study population
This analysis was nested in a prospective birth cohort recruited by Dhaka Community Hospital (DCH) Trust in Bangladesh (N = 1,458). The details describing this cohort have been previously described [22]. Briefly, pregnant women of ≤16 weeks of gestational age (GA) were recruited into a prospective birth cohort by DCH Trust in Bangladesh. Participants were eligible for the cohort if they had a single pregnancy, used a tube well as the main source of drinking water, and planned to live in their current residence for the duration of the pregnancy. As part of the study protocol, women received monthly prenatal vitamins and gave birth at a local clinic or at home with DCH trained medical personnel.
This analysis was nested within the larger birth cohort. It was designed to examine the potential mediating effect of arsenic-induced DNAm changes on reproductive outcomes using a more economical two-stage approach. We randomly selected 44 newborns to span a range of drinking water arsenic concentrations (<1–510 µg/L) for the discovery phase, which measured DNAm in whole cord blood with Infinium HumanMethylation450 (450K) BeadChip technology. For the validation phase, we randomly selected 569 newborns from the cohort, which measured DNAm in whole cord blood using pyrosequencing. Twenty-five samples from the discovery phase were also included to assess DNAm replication across the two technology platforms.
This study was approved by the Human Research Committees at the Harvard School of Public Health, Oregon State University, and DCH Trust.
Maternal drinking water arsenic
Arsenic was measured in tube-wells identified by participants as their main source of drinking water at the time of enrollment, as previously described [22]. Briefly, water samples were collected, preserved with nitric acid to a pH <2, and stored at room temperature prior to analysis by inductively coupled plasma-mass spectrometry (ICP-MS) using US EPA method 200.8 (Environmental Laboratory Services, North Syracuse, NY) [26]. The average percent recovery for arsenic from plasmaCal multi-element QC standard #1 solution (SCP Science) was 102 ± 7%. The limit of detection (LOD) for arsenic was 1 µg/L. Thirty samples were below the LOD and were assigned LOD/2.
Maternal toenail arsenic and quality control
Toenail arsenic concentrations represent exposure during several months to year prior to collection [27]. Maternal toenail clippings were collected at enrollment and ≤1 month after delivery. To remove contamination, toenail clippings were sonicated in 1% Triton X-100 solution (Sigma-Aldrich, Inc., St. Louis, MO) and rinsed in Milli-Q water (Millipore Corporation, Billerica, MA). Samples were digested using Trace Select Ultra Pure nitric acid (HNO3; Sigma-Aldrich, Inc.) and diluted with Milli-Q water. Total arsenic was measured using an inductively coupled plasma mass spectrometer (Perkin-Elmer Model DRC-II 6100, Norwalk, CT). Toenail references are not available and therefore measured arsenic concentrations were corrected for method error using blank correction and normalization based on arsenic concentrations of batch-specific human hair references (CRM Hair; Shanghai Institute of Nuclear Research, Academia Sinica, China). Samples with a mass < 5 mg (n = 6) or relative standard deviation > 25% (n = 5) were excluded from analyses. Additionally, only one remaining toenail sample was below the respective batch LOD ranging 0.004–0.85 µg As/g and subsequently excluded from the analyses.
Cord blood DNAm and quality control
Illumina Infinium HumanMethylation450 BeadChip: A sample of umbilical cord blood was collected after delivery into an EDTA-coated vacutainer tube (B.D. Scientific). DNA was extracted from whole blood using the Purgene DNA isolation solutions (Qiagen/Gentra Systems), following manufacturer’s instructions. DNA samples were analyzed for DNAm at the University of Minnesota Biomedical Genomic Center using the Illumina Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA), which simultaneously profiles the methylation status for > 485,000 CpG loci at a single nucleotide resolution covering 99% of the RefSeq genes.
Samples were analyzed in one plate and randomly allocated to 16 chips. DNAm image files were normalized using the functional normalization method with two principal components to account for technical variation between samples using the minfi package of R [28,29]. Methylation measurements at CpG loci on X and Y chromosomes were excluded from the analysis to avoid gender-specific methylation bias. Previously identified non-specific and cross-reactive probes within the array along with polymorphic CpG loci (≥ 5% of the minor allele frequency) were excluded from the analysis [30]. Additionally, detection P values were computed for all CpGs and probes with non-significant detection (P > 0.01) in greater than 10% of the samples were also excluded from the analysis. Lastly, a beta-mixture quantile intra sample normalization procedure (BMIQ) was further applied to reduce the potential bias that can arise from type-2 probes, as previously described [31]. The total number of autosomal loci left for analysis after quality control procedures was 383,940. Methylation values were logit-transformed to M-values to evaluate the sex adjusted linear association between CpG methylation and prenatal maternal water arsenic exposure.
Bisulfite pyrosequencing: In the discovery phase, the top 10 CpG sites were highly correlated. Therefore, we opted to only conduct gene specific DNAm for miR124-3, GNAL, and MCC (n = 569). Custom pyrosequencing was performed by EpigenDx (Hopkington, MA). Briefly, 500 ng of whole cord blood DNA was bisulfite treated using the EZ DNA Methylation kit (Zymo Research, Inc., CA). Bisulfite-treated DNA was purified following manufacturer’s protocols and eluted to a final volume of 46 µL. Custom PCR assays were performed using 1 µL of bisulfite-treated DNA and 0.2 µM of each primer. One primer was biotin-labeled and HPLC purified in order to purify the final PCR product using sepharose beads. PCR product was bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences) after which the immobilized PCR products were purified, washed, denatured with a 0.2 µM NAOH solution, and rewashed using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Qiagen) following manufacturer’s protocol. Next, 0.5 μM of sequencing primer was annealed to the purified single stranded PCR products, and 10 μL of the PCR products were sequenced by Pyrosequencing on the PSQ96 HS System (Pyrosequencing, Qiagen) following the manufacturer’s instructions.
The methylation status of each CpG site was determined individually as an artificial C/T SNP using QCpG software (Pyrosequencing, Qiagen). The methylation level at each CpG site was calculated as the percentage of the methylated alleles divided by the sum of all methylated and unmethylated alleles. The mean methylation level was calculated using methylation levels of all measured CpG sites within the targeted region of each gene. Each experiment included non-CpG cytosines as internal controls to detect incomplete bisulfite conversion of the input DNA. In addition, a series of unmethylated and DNAm are included as controls in each PCR. The average quantity of DNAm and standard deviation (SD) measured at these 3 controls [e.g., low methylation (0%), medium methylation (50%), and high methylation (100%)] were estimated for each site. For miR124-3, the average and standard deviation (SD) of DNAm for the low control was 0.0 ± 0.0, medium control was 48.1 ± 22.8, and high control was 89.4 ± 10.0. For GNAL, the average and SD of DNAm for the low control was 0.07 ± 1.4, medium control was 53.2 ± 5.7, and the high control was 93.2 ± 4.7. For MCC, the average and SD of DNAm for the low control was 0.1 ± 0.7, the medium control was 64.4 ± 11.5, and the high control was 88.1 ± 7.2. Furthermore, PCR bias testing was performed by mixing unmethylated control DNA with in vitro methylated DNA at different ratios (0%, 5%, 10%, 25%, 50%, 75%, and 100%), followed by bisulfite modification, PCR, and pyrosequencing analysis. The linearity of the assays were assessed using a 7 sample serial dilution (R2 = 0.91 for miRNA124-3, 0.95 for GNAL, and 0.96 for MCC).
Statistical analysis: discovery approach
Epigenome-Wide Association (Step 1): A three stage filtering method was implemented to identify candidate loci that could mediate the association between prenatal arsenic exposure and birth outcomes. The first step was to conduct an Epigenome-Wide Association Study (EWAS) in cord blood (Step 1, Figure 1). The EWAS in cord blood has been previously published for this sample, but we re-analyzed the data to implement the latest technical processing steps for 450K data described in the quality control section [22]. After quality control, the sex adjusted linear association between maternal drinking water arsenic and individual CpG methylation levels were evaluated using the limma function (linear models for microarray analysis) from the minfi package of R. A selection criteria was set a priori for both significance (P < 1x10−6) and effect size (|β|> 0.10) to identify differentially methylated CpGs associated with arsenic exposure in utero on the M-value scale. We did not adjust for cell type composition because it would not be feasible to adjust for nucleated cell type composition in the rest of the archived samples that were subsequently pyrosequenced in the validation phase due to the absence of epigenome-wide information.
Figure 1.
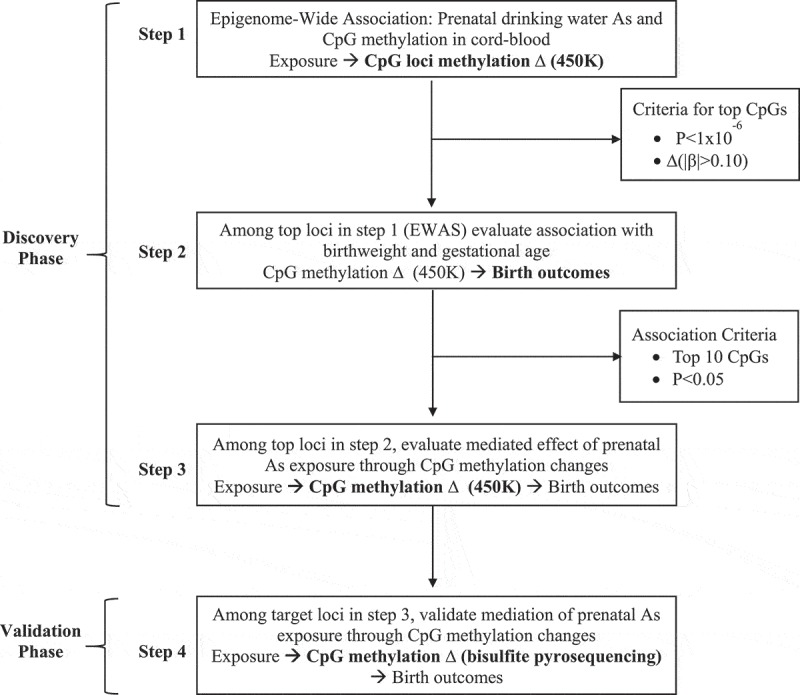
Experimental approach for the discovery of DNA methylation disruption induced by prenatal arsenic exposure and subsequent mediation of birth outcomes.
Phenotype Association (Step 2): All loci selected from the EWAS using the a priori criteria were then evaluated for their sex adjusted association with both birth GA and birth weight using multivariate linear regression models on a CpG-by-CpG basis (Step 2, Figure 1). Two selection criteria were used to test for association between individual CpG methylation levels and birth outcomes. Specifically, CpGs were considered to be significantly associated with birth gestational age and birth weight if: i) they reached an uncorrected level of significance of P < 0.05 and ii) if multiple loci were associated only the top-ten loci ranked on lowest P value would be selected for subsequent validation using pyrosequencing.
Mediation Analysis (Step 3): Finally, CpGs identified to be differentially methylated relative to prenatal arsenic exposure in Step 1 and found to be significantly associated with either birth GA or birth weight in Step 2 were evaluated for their potential to mediate the effect of exposure on these birth outcomes using structural equation models (SEMs) (Step 3, Figure1). First, we checked conditions previously postulated for a variable to be considered a potential mediator [32]. Specifically, mediation requires a significant association between the exposure and the outcome, a significant association between the mediator and the exposure, and a significant association of the mediator to the outcome while controlling for the exposure.
We proposed a conceptual model for mediation based on the a priori assumption that a mediated effect through CpG DNAm is biologically plausible (Figure 2). First, we tested the independent effects of exposure on birth outcomes, exposure on CpG methylation and CpG methylation on birth outcomes while adjusting for the exposure. We then conceptualized a model in which the direct effects between prenatal arsenic exposure, CpG methylation and birth outcomes (a, b, c) was evaluated while also testing the indirect effect of exposure on birth outcomes mediated through CpG methylation levels (a b) while adjusting for infant sex (Figure 2). Two SEMs were used to evaluate the direct effect of log2-transformed maternal drinking water arsenic on both birth gestational age and birth weight. Bias corrected standard errors and 95% bootstrap Confidence Intervals (CIs) were calculated from 10,000 replicates as the sample size available was relatively small in the discovery phase.
Figure 2.
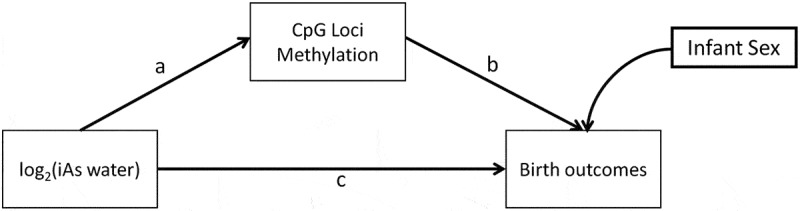
Conceptual Structural Equation Model (SEM) for the direct and indirect effect of exposure (maternal drinking water arsenic ≤ 16 weeks of gestation) and infant birth outcomes, birth gestational age, and birth weight in the discovery phase.
Histograms and scatter plots along with regression lines and locally weighted smoothing lines were plotted for bivariate association between exposure, methylation, and birth outcomes. All pairwise Pearson correlation coefficients were evaluated among the top candidate CpGs considered for mediation analyses. Due to high correlation among all top-ten CpGs found to be associated with birth GA, a post-hoc Principal Component Analysis (PCA) was implemented to deconvolute the major source of variability into a single factor. The scores from the first principal component that accounted for the maximum amount of variability of the methylation levels of all top-ten CpGs were then evaluated as a mediator of the exposure and GA relationship into the conceptualized SEM model.
Statistical analysis: validation approach
Within each gene, the relationships between CpGs were assessed using Pearson correlations with the Benjamini-Hochberg false discovery rate (FDR) adjustment [33]. SEMs were built to assess mediation between arsenic exposure and GA using three steps. First, CpGs were used as indicator variables to construct a latent variable for each gene. A SNP located within MCC was found to be significantly associated with DNAm of nearby CpGs, and therefore dummy variables for the variant genotypes were included as predictors of the latent variable. Second, mediation of the association between log2-transformed maternal drinking water arsenic concentration and birth GA by each latent variable (i.e., DNAm of each gene) was assessed. Third, a single SEM was constructed with genes that significantly mediated the association between exposure and GA in individual models, and adjusted for the potential confounders of sex, maternal weight gain between enrollment and delivery, maternal education (> primary vs. ≤ primary education), and birth type (cesarean section vs. vaginal birth). Due to skew in DNAm variables, robust estimates of model fit were used [34,35]. At each step, model indices greater than 35 were used to identify residual correlations that would improve model fit. All analyses were carried out using the R statistical package, version 3.4.0 or Stata, version 12.1. SEMs in R were conducted using the lavaan package [36].
Results
Discovery stage
A total of 44 infants (29 male and 15 female) had cord blood DNAm measurements along with maternal drinking water arsenic concentrations available for analysis. The mean maternal drinking water arsenic concentration at ≤ 16 weeks of gestation was 63.7 µg/L (range: < 1–510 µg/L). The mean gestational age at delivery was 37.6 weeks (range: 33–41 weeks) and the average birth weight was 2923 grams (range: 2080–4050 grams). Selected sample characteristics are summarized in Table 1.
Table 1.
Selected sample characteristics of the study population.
| Discovery set (N = 44) |
Validation set (n = 569) |
||||
|---|---|---|---|---|---|
| Sample characteristics | Mean ± SD | Range | Mean ± SD | Range | P a |
| Drinking water As at recruitment (µg/L) | 63.7 ± 116.5 | < 1 – 510 | 56.0 ± 99.8b | < 1 – 629 | 0.898 |
| Maternal toenail As at delivery (ng/µg) | 7.0 ± 10.2 | 0.3 – 46.6 | 2.9 ± 3.8c | 0.04 – 34.8 | 0.008 |
| Gestational age at recruitment (weeks) | 12.2 ± 2.5 | 6 – 16 | 11.4 ± 2.5 | 4 – 16 | |
| Gestational age at delivery (weeks) | 37.6 ± 2.1 | 33 – 41 | 37.7 ± 2.2d | 22 – 42 | 0.753 |
| Birth weight (grams) | 2923 ± 372 | 2080 – 4050 | 2824 ± 444b | 1400 – 4600 | 0.141 |
| Gender | n (%) | n (%)b | |||
| Male | 29 (65.9%) | 291 (51.2) | 0.060 | ||
| Female | 15 (34.1%) | 277 (48.8) | |||
| a. Wilcoxon rank sum test for continuous variables and chi-squared test for categorical variables. | |||||
| b. n = 568 | |||||
| c. n = 533 | |||||
| c. n = 566 | |||||
Using our a priori selection criteria for significance of P < 1.0x10−6 and effect size of |β regression|> 0.10, a total of 380 loci were selected to evaluate their association with birth weight and GA (Figure 3). Among the selected 380 CpG loci identified to be differentially methylated relative to prenatal arsenic exposure, none overlapped with the top 600 Houseman-probes used in differentiating white blood cell composition of whole blood samples when compared to the adult reference methylome or among the 700 CpGs used in the new cord blood reference set [37,38]. In sensitivity analyses, we adjusted this initial EWAS for cell type composition and, while the results were attenuated in magnitude and significance, they remained consistent (Supplemental Table 1). We also evaluated associations with log2(postpartum maternal toenail arsenic concentration), which are considered a very good biomarker of internal dose. The regression coefficients for the top CpGs generated from the models that used toenail arsenic as the exposure metric were similar to models using maternal drinking water arsenic as the measure of exposure, although significance was lower (Supplemental Table 1).
Figure 3.
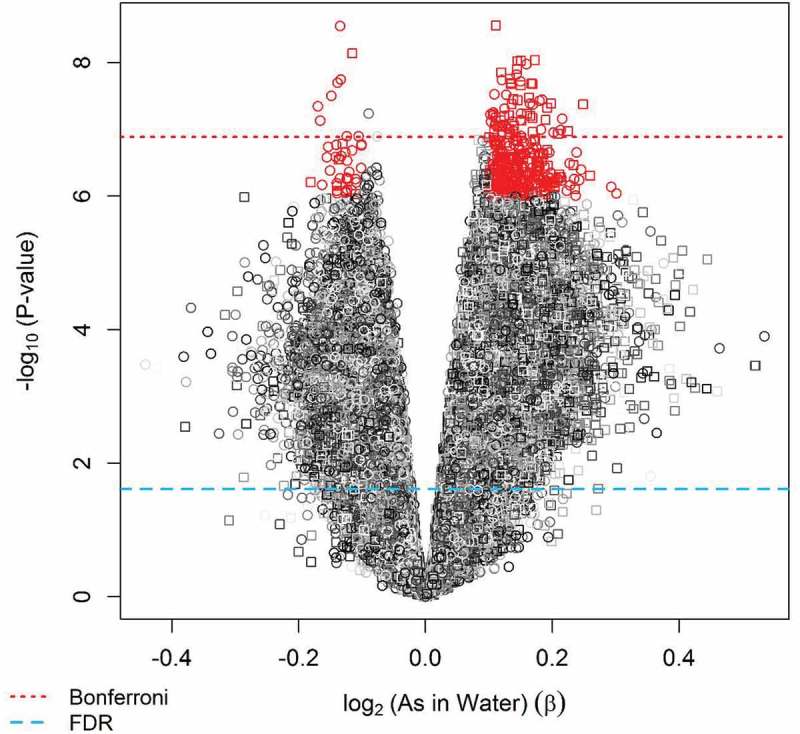
Selected differentially methylated CpG loci found in cord blood relative to prenatal arsenic exposure using |β|> 0.10 and nominal P < 1x10−6 criteria for association (Step 1 of discovery phase) .
CpG methylation and birth gestational age
Multivariate linear regression models adjusted for sex revealed that methylation levels of 139 CpGs (35.1%) from the 380 candidate loci were significantly associated with birth gestational age (P < 0.05) (Figure 4(a)). Among these loci, the top 10 CpGs ranked on lowest P value were selected to be evaluated as mediators of the exposure and birth outcome relationship. Six of the top-ten loci were located in CpG islands and the other four in shore regions of CpG islands among unique genes and chromosomes (Table 2). Nine of the top-ten CpGs had higher methylation relative to prenatal arsenic exposure and only one was observed to have lower methylation (Figure 5). The nine CpGs observed to be positively associated with log2-transformed arsenic exposure (higher methylation) were inversely associated with GA at birth, while the single CpG loci with lower methylation was positively associated with birth GA (Figure 6).
Figure 4.
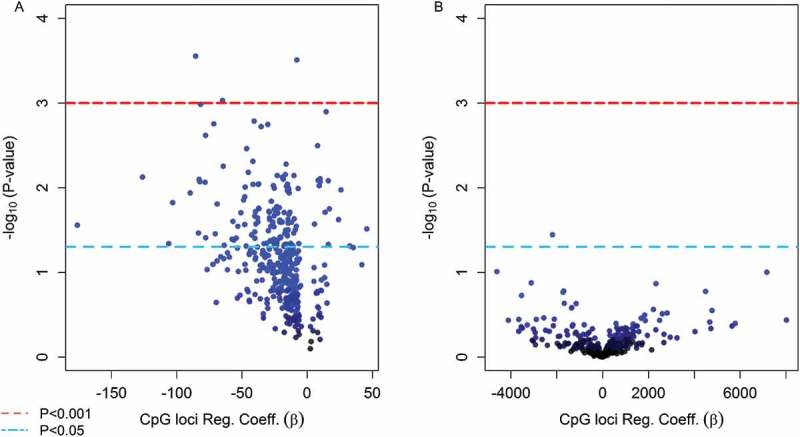
(Step 2 of discovery phase) Volcano plots for the association between the top 380 CpG loci in cord blood found to be significantly associated with As exposure in utero and infant health outcomes (a) gestational age and (b) birth weight.
Table 2.
Top-ten CpG loci that were significantly associated with birth gestational age in the EWAS for the discovery phase.
| CpG ID | P value | β Coeff. | Relation to CpG Island | Chromosome | Gene | Gene Group |
|---|---|---|---|---|---|---|
| cg01163597 | 2.9x10−4 | −62.3 | N_Shore | Chr6 | SLC22A23 | Body |
| cg16081457 | 3.1x10−4 | −9.0 | S_Shore | Chr12 | ||
| cg06522054 | 3.8x10−4 | −68.1 | Island | Chr18 | GNAL;GNAL;GNAL | 1stExon;Body;Body |
| cg20382695 | 5.5x10−4 | −52.8 | Island | Chr10 | ATRNL1 | Body |
| cg24937280 | 1.1x10−3 | −56.7 | Island | Chr5 | MCC | Body |
| cg01910639 | 1.3x10−3 | 14.7 | N_Shore | Chr1 | S100A6 | Body |
| cg18115406 | 1.4x10−3 | −35.9 | Island | Chr9 | LMX1B | TSS200 |
| cg04874129 | 1.5x10−3 | −31.1 | Island | Chr16 | SLC6A2 | 1stExon |
| cg20277905 | 1.7x10−3 | −39.9 | Island | Chr20 | miR124-3 | TSS200 |
| cg00398764 | 1.7x10−3 | −28.0 | N_Shore | Chr15 |
Figure 5.
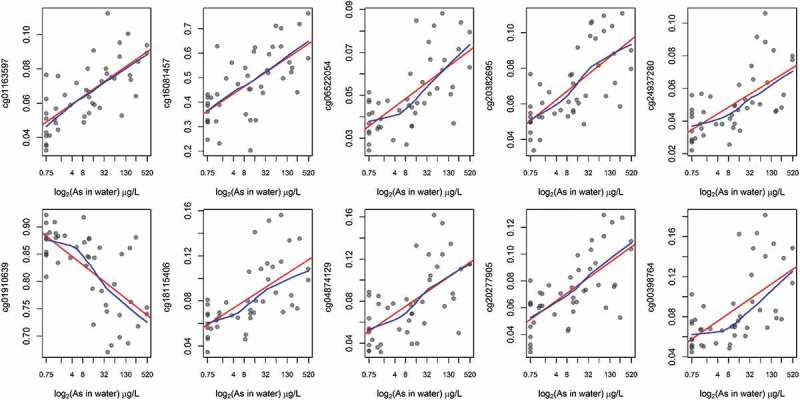
Unadjusted associations between prenatal As exposure and CpG methylation among the top-ten CpGs associated with gestational age.
red: linear regression line; blue: locally weighted scatter plot smoothing
Figure 6.
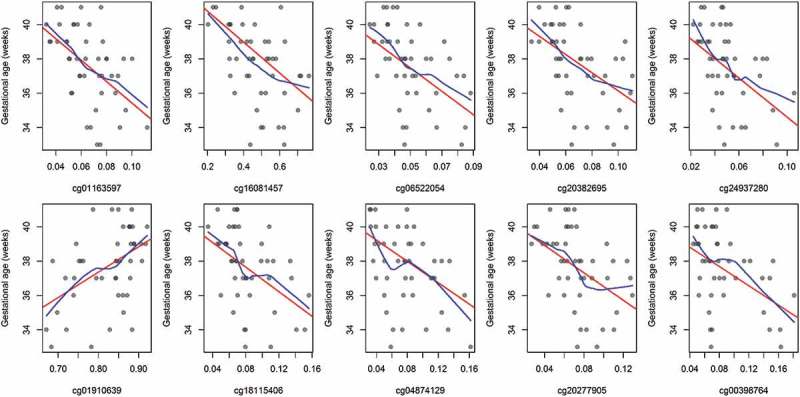
Unadjusted associations between gestational age and CpG methylation among the top-ten CpGs.
red: linear regression line
blue: locally weighted scatter plot smoothing
Nine of the top-ten CpG loci were observed to have higher methylation relative to prenatal arsenic exposure and were positively correlated (Pearson’s ρ range: 0.61 to 0.90), while the single loci with lower methylation was negatively correlated with the other nine (Pearson’s ρ range: −0.85 to −0.68) (Figure 7). The Principal Component Analysis (PCA) of the top-ten loci selected demonstrated that 80% of the variance was accounted in the first and main principal component (Figure 8(a)) and that the cumulative variance explained by four principal components was 93% (Figure 8(b)). The relative loadings of the top-ten CpG loci on the first principal component (PC1) are presented in Supplemental Table 2. Thus, we chose to use the first principal component due to the high level of correlation among CpGs.
Figure 7.
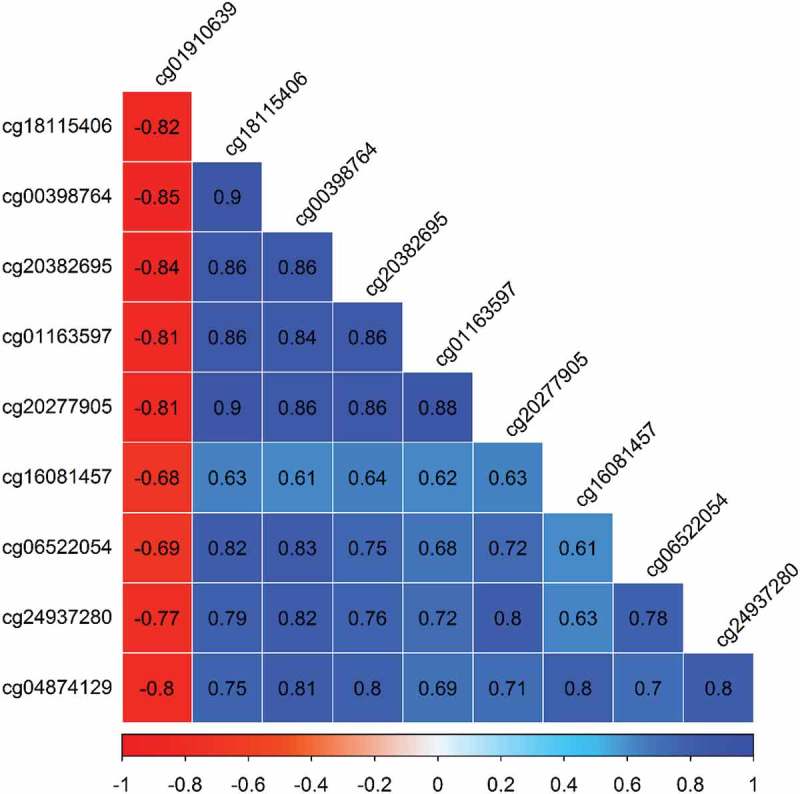
Correlation among the top-ten CpGs associated with As exposure in utero and gestational age at birth.
Figure 8.
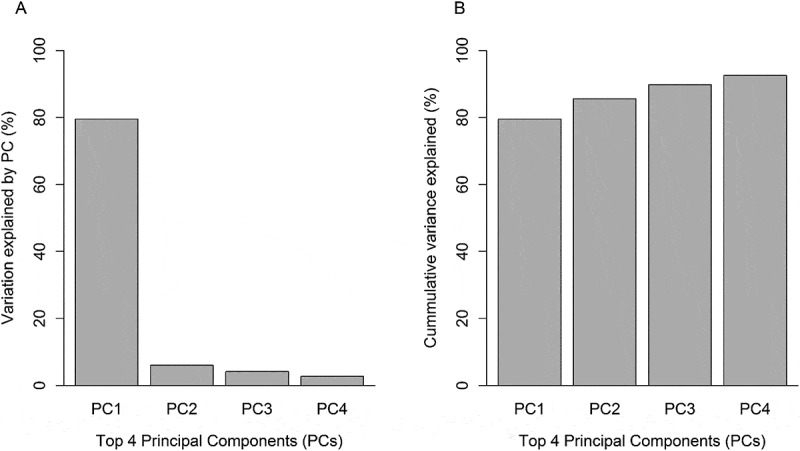
Proportion of variance explained by the first four principal components (PCs). (a) Proportion of variance explained by each PC. (b) Cumulative proportion of variance explained by all 4 PCs.
Before implementing the SEM we evaluated assumptions for mediation analysis. Namely, in this subsample, log2-transformed arsenic was significantly associated with birth gestational age (β = -0.25, 95% CI: −0.47, −0.05; P = 0.017) and with the scores for PC1 capturing the maximum amount of variation (80%) for the DNAm levels of the top-ten loci (β = 0.70, 95% CI: 0.49, 0.88; P < 0.001). In turn, the scores for the first principal component were significantly associated with birth GA while also including log2-transformed arsenic exposure in the model (β = -0.47, 95% CI: −0.77, −0.17; P = 0.003), meeting the postulated conditions for mediation.
In the sex adjusted conceptual SEM log2-tranformed maternal drinking water arsenic was positively associated with PC1 scores of methylation levels for the top-ten CpGs (β = 0.69, 95% CI: 0.50, 0.87; P < 0.001). The principal component scores of PC1 were negatively associated with birth GA (β = -0.42, 95% CI: −0.59, −0.25; P < 0.001) (Figure 9). The effect of prenatal arsenic exposure on birth GA was completely mediated through PC1. Specifically, each doubling in prenatal maternal drinking water arsenic decreased birth GA by 0.29 weeks or approximately two days and this was fully mediated through the PC1 scores for the methylation levels of the selected top-ten CpGs (β = -0.29, 95% CI: −0.42, −0.15; P < 0.001). The direct effect of maternal drinking water arsenic on birth GA was non-significant after accounting for the mediation pathway and therefore not included in the final mediation model (β = 0.06, 95% CI: −0.18, 0.30; P = 0.62). The direct and indirect results for the conceptual model are summarized in Table 3. This final SEM conformed to all model fit indices for good fit, summarized in Table 4.
Figure 9.
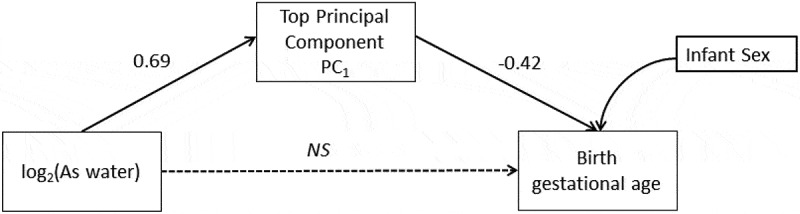
Structural Equation Model (SEM) conceptualized for the mediated association of the main principal component (PC1) that explained 80% of the variance for the top-10 CpG loci and birth gestational age in the discovery phase.
NS=non-significant
Table 3.
Structural equation model for the mediated effect of arsenic exposure on birth gestational age through variation of the top-ten CpGs captured in the main principal component (PC1) in the discovery phase.
| Pathway | Effect | β Coefficient (95% CIs) | P value |
|---|---|---|---|
| log2 (As Water) → PC1 | Direct | 0.69 (0.50, 0.87) | <0.001 |
| PC1→ Birth gestational age | Direct | −0.42 (−0.59, −0.25) | <0.001 |
| Sex→ Birth gestational age | Direct | −0.05 (−1.17, 1.07) | 0.93 |
| log2 (As Water)→ Birth gestational age | Direct | 0.06 (−0.18, 0.30) | 0.62 |
| log2 (As Water)→ PC1→ Birth gestational age | Indirect | −0.29 (−0.42, −0.15) | <0.001 |
Table 4.
Fit indices for the final the structural equation model that describe the indirect effect of prenatal arsenic exposure on birth gestational age that is mediated through the main principal component of CpG methylation levels (top-ten CpGs) in the discovery phase (n = 44) .
| Index | Criterion for Good Fit | Model Fit |
|---|---|---|
| χ2 p-value | >0.05 | 0.81 |
| Root Mean Square Error of Approximation (RMSEA) | <0.05 | <0.001 |
| Comparative Fit Index (CFI) | >0.95 | 1 |
| Tucker-Lewisnon-normed Fit Index | >0.90 | 1 |
| Standardized Root Mean Squared Residual | >0.05 | 0.02 |
| Coefficient of Determination | NA | 0.54 |
CpG methylation and birth weight
From the 380 candidate CpGs, only one loci (cg24484905), located in an open sea region of the DAB1 gene and observed to have higher methylation relative to prenatal arsenic exposure, was also associated with birth weight (P = 0.035, FDR = 0.99) (Figure 4(b)). No direct significant association was observed between maternal drinking water arsenic and birth weight (β = 0.977, 95% CI: −27.40, 46.93; P = 0.59), and the direct effect of methylation levels of the DAB1 loci (cg24484905) on birth weight was significant after controlling for arsenic exposure (β = -2717, 95% CI: −5186, −248; P = 0.032). However, no significant mediation for the effect of prenatal arsenic exposure on birth weight was observed through methylation levels of this single loci (β = 0.02, 95% CI: −0.01, 0.05; P = 0.15).
Validation stage
Bisulfite pyrosequencing was performed for target CpGs located in miR124-3, GNAL, and MCC on cord blood DNA from 569 infants. Among the 25 samples with Infinium 450K array and pyrosequencing data, there were strong correlations for DNAm measured at cg20277905 (miR124-3; Pearson r = 0.728, P < 0.001) and cg24937280 (MCC; Pearson r = 0.760; P < 0.001); however, correlation between platforms for cg06522054 was not significant (MCC; Pearson r = 0.138; P < 0.500). Approximately half the infants were male (51.2% male, 48.8% female) (Table 1). The mean maternal drinking water arsenic concentration at ≤ 16 weeks GA was 56.0 µg/L (range: < 1–629 µg/L) and the mean maternal toenail arsenic concentration < 1 month postpartum was 2.9 ng/µg (range: 0.04–34.8 µg). The mean GA was 37.7 weeks (range: 22–42 weeks) and the mean birth weight was 2824 grams (range: 1400–4600 grams). There were no significant differences in drinking water arsenic concentration, birthweight, GA, or sex between participants included in the discovery and validation sets. Median postpartum maternal toenail arsenic concentration was significantly higher among participants in the discovery set than the validation set (Wilcoxon rank sum P = 0.008).
In each gene, CpG loci were located within 69–81 base pairs and had mean methylation ranging 0.26–5.76% (Supplemental Table 3). Within genes, all CpGs were significantly and positively correlated (FDR adjusted P < 0.05) with the exception of nine CpG pairs located on miR124-3 (Supplemental Table 4–6). For each gene, SEMs were used to evaluate mediation between log2-transformed maternal drinking water arsenic and GA by DNAm (Supplemental Tables 7–9; Supplemental Figure 1–3). The latent variables representing DNAm of miR124-3 and MCC were found to significantly mediate the association between prenatal arsenic exposure and GA (miR124-3 indirect effect: β = -0.02; 95% CI: −0.04, 0.00; P = 0.030; MCC indirect effect: β = -0.03; 95% CI:-0.05, −0.01; P = 0.004); however, GNAL was not a significant mediator (GNAL indirect effect: β = -0.01; 95% CI:-0.02, 0.00; P = 0.174).
Table 6.
Fit indices for the final the structural equation model that describe the indirect effect of prenatal arsenic exposure on birth gestational age that is mediated through variation of DNAm of CpGs in miR124-3 and MCC in the validation phase (n = 569) .
| Index | Criterion for Good Fit | Model Fit |
|---|---|---|
| χ2 P value | >0.05 | <0.001 |
| Robust root Mean Square Error of Approximation (RMSEA) | <0.05 | 0.038 |
| Robust Comparative Fit Index (CFI) | >0.95 | 0.893 |
| Robust Tucker-Lewisnon-normed Fit Index | >0.90 | 0.884 |
| Standardized Root Mean Squared Residual | >0.05 | 0.049 |
Mediation between log2-transformed maternal drinking water arsenic and GA by DNAm of miR124-3 and MCC were assessed in a single SEM. In an unadjusted model, log2-transformed maternal drinking water arsenic concentration was significantly associated with DNAm of miR124-3 and MCC. Furthermore, DNAm of miR124-3, but not MCC, was significantly associated with GA (P < 0.05) (Supplemental Table 10; Supplemental Figure 4). Mediation of the association between prenatal drinking water arsenic exposure and GA by miR124-3 DNAm achieved borderline significance (indirect effect: β = -0.02; 95% CI:-0.03, 0.00; P = 0.051), whereas mediation by MCC was not significant (indirect effect: β = -0.01; 95% CI:-0.03, 0.01; P = 0.224).
Results from a SEM adjusted for infant sex, maternal weight gain, maternal education, and birth type were consistent. There were significant direct effects of log2-transformed maternal drinking water arsenic concentration on DNAm of miR124-3 and MCC, and of miR124-3 DNAm on gestational age (P < 0.05) (Figure 10 and Table 5). Mediation by DNAm of miR124-3 was borderline significant (indirect effect: β = -0.02; 95% CI:-0.03, 0.00; P = 0.061). DNAm of MCC did not act as a mediator (indirect effect: β = -0.01; 95% CI: −0.03, 0.01; P = 0.276).
Figure 10.
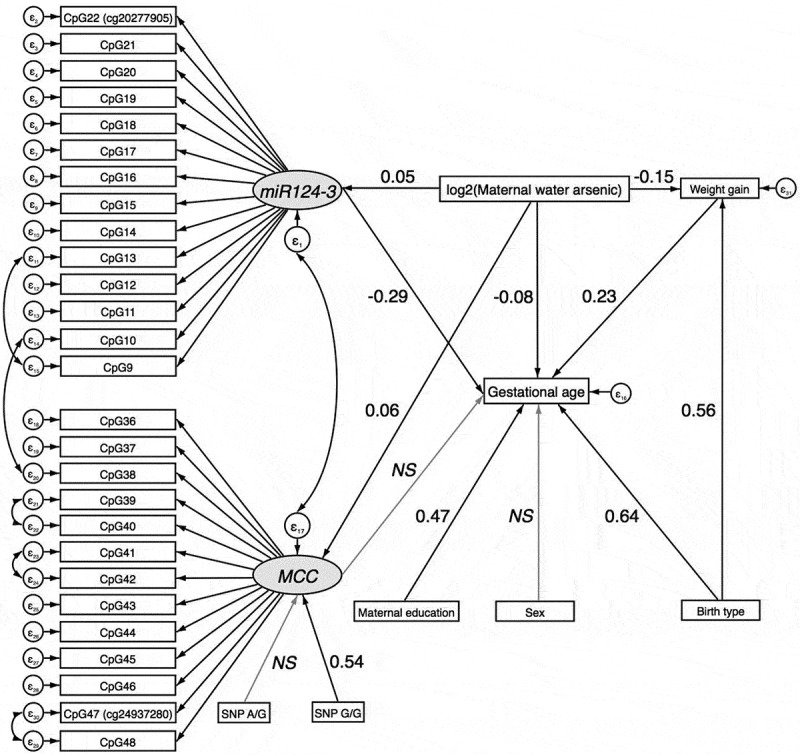
Structural Equation Model (SEM) for the mediated effect of arsenic exposure on birth gestational age through DNAm of CpGs in miR124-3 and MCC in the validation phase.
NS=non-significant
Table 5.
Adjusted structural equation model for the mediated effect of arsenic exposure on birth gestational age through variation of DNAm of CpGs in miR124-3 and MCC in the validation phase (n = 569).
| Pathway | Effect | β Coefficient |
P |
|---|---|---|---|
| (95% CIs) | |||
| miR124-3 | |||
| log2(water arsenic) → miR124-3 | Direct | 0.05 (0.01, 0.10) | 0.028 |
| MCC | |||
| log2(water arsenic) → MCC | Direct | 0.06 (0.03, 0.09) | <0.001 |
| SNP rs1057827 → MCC (reference: AA) | |||
| G/G | Direct | 0.53 (0.12, 0.94) | 0.011 |
| A/G | Direct | 0.11 (−0.06, 0.27) | 0.198 |
| Maternal weight gain | |||
| log2(water arsenic) → Weight gain | Direct | −0.15 (−0.23, −0.07) | <0.001 |
| Birth type (caesarean vs. vaginal) → Weight gain | Direct | 0.57 (0.09, 1.09) | 0.029 |
| Gestational age | |||
| log2(water arsenic) → Gestational age | Direct | −0.08 (−0.14, −0.02) | 0.011 |
| miR124-3 → Gestational age | Direct | −0.29 (−0.50, −0.09) | 0.005 |
| MCC → Gestational age | Direct | −0.17 (−0.44, 0.11) | 0.242 |
| Sex (male vs. female) → Gestational age | Direct | 0.08 (−0.23, 0.40) | 0.600 |
| Weight gain → Gestational age | Direct | 0.23 (0.17, 0.30) | <0.001 |
| Birth type (caesarean section vs. vaginal) → Gestational age | Direct | 0.64 (0.32, 0.95) | <0.001 |
| Maternal education (>primary vs. ≤primary) → Gestational age | Direct | 0.47 (0.15, 0.80) | 0.004 |
| log2(water arsenic) → Weight gain → Gestational age | Indirect | −0.04 (−0.06, −0.02) | 0.001 |
| log2(water arsenic) → miR124-3 → Gestational age | Indirect | −0.02 (−0.03, 0.00) | 0.061 |
| log2(water arsenic) → MCC → Gestational age | Indirect | −0.01 (−0.03, 0.01) | 0.276 |
Sensitivity analyses were performed using maternal toenail arsenic concentration collected at enrollment and ≤ 1 month postpartum as measures of exposure. Maternal drinking water arsenic concentrations were significantly correlated with maternal toenail arsenic concentrations collected at enrollment (Spearman r = 0.53, P < 0.001) and postpartum (Spearman r = 0.58, P < 0.001). Results from an adjusted SEM assessing mediation between log2-transformed postpartum maternal toenail arsenic and GA by DNAm of miR124-3 and MCC were consistent (Supplemental Table 11; Supplemental Figure 5). DNAm of miR124-3, but not MCC, mediated with association between arsenic exposure and GA (miR124-3 indirect effect: β = -0.04; 95% CI: −0.08, −0.01; P = 0.023; MCC indirect effect: β = -0.02; 95% CI: −0.04, 0.01; P = 0.215). Likewise, in an adjusted SEM with log2-transformed maternal toenail arsenic concentration collected at enrollment, DNAm of miR124-3 significantly mediated the association between prenatal arsenic exposure and GA (miR124-3 indirect effect: β = -0.05; 95% CI:-0.09, −0.01; P = 0.021; MCC indirect effect: β = -0.02; 95% CI:-0.05, 0.01; P = 0.210) (Supplemental Table 12; Supplemental Figure 6).
Discussion
We introduced an experimental approach for the discovery, evaluation, and validation of candidate CpG loci as mediators of the association between prenatal exposures and birth outcomes. Using this approach, we show that prenatal arsenic exposure decreased birth GA and the association was mediated through DNAm levels of selected CpG loci, namely miR124-3 and MCC. However, no significant mediation or direct association was observed between prenatal arsenic exposure, DNAm levels at CpG loci, and birth weight.
One previous study reported an epigenome-wide association of prenatal arsenic exposure with birth outcomes [25]. Namely, the authors observed 7 unique loci significantly associated with prenatal arsenic exposure that also correlated with birth GA, head circumference, or placental weight. None of the CpGs found by this group were within the top-ten differentially methylated loci found in our study that were identified in the discovery phase. This could be potentially attributed to differences in the timing of the exposure assessment as well as the type of exposure assessment. For instance, Rojas et al. used urinary arsenic measurements at the time of delivery, whereas we used drinking water arsenic collected at the time of enrollment and maternal nails collected < 1 month postpartum. Furthermore, we observed that most loci were associated with GA and none with birth weight. This is consistent with our previously reported finding in which we show that the effect of arsenic on birth weight is mediated through birth GA and to a lesser extent with maternal weight gain during pregnancy within this birth cohort [39]. Another study of arsenic exposure in utero using a candidate gene approach found that the expression of AQP9, which encodes a cell membrane channel, has the ability to mediate the effect of arsenic on birth weight. Even though it is unknown if the AQP9 has an epigenomic control mechanism, this observation raises the possibility that prenatal exposure can influence size at birth [40].
In our discovery phase, two CpGs (cg01163597; cg04874129) located in two genes of the solute carrier (SLC) superfamily were observed to mediate the association between prenatal arsenic exposure and gestational age at birth (SLC22A23; SLC6A2). The SLC6A2 is an neurotransmitter transporter across the cell membrane and has been shown to be upregulated by exposure to arsenic in animal models [41]. Furthermore, higher methylation of this gene has been associated with esophageal carcinogenesis and non-small cell lung cancer [42,43]. The SLC22A23 gene is a novel solute carrier protein transporter and its function has not been well characterized but abundant expression in the brain and liver has been observed [44]. It has been proposed that even though these transporters exist for endogenous substances, drugs, non-essential metals, and environmental toxins could potentially permeate. However, the physiological purpose in more than half of these transporters remains to be characterized [45].
The only CpG observed to have higher methylation relative to prenatal arsenic exposure and positively associated with GA was located in a north shore region of a CpG island in the body of the S100A6 gene, involved in a Ca+2-dependent insulin release. Downregulation of this specific gene has been associated with intrauterine growth restrictions [46]. In addition, high expression levels of this protein has been observed in the human heart and, in experimental models, increased cardiac expression has been shown to be anti-hypertrophic [47].
In our validation phase, candidate loci in the miR124-3 gene were observed to have lower DNAm by prenatal arsenic exposure and to mediate the association with GA. This specific microRNA has been correlated with tumor size and disease recurrence of non-small cell lung cancer and renal cell carcinoma [48,49] and has been also shown to affect neuron growth and differentiation in vitro [50]. The effect of prenatal exposure to arsenic in drinking water on miR124-3 expression has been studied in a mouse model. Tyler et al. (2014) demonstrated that exposure to 50 µg/L arsenic decreased miR124-3 expression in male embryonic brain tissue [8]. Although miR124-3 is predominantly expressed in the nervous system, miR124-3 expression has been observed to regulate hematopoiesis in human cord blood cells [51]. In addition, miR124-3 may be involved in mammalian growth; fertilized mouse eggs injected with mir124-3 microRNA resulted in increased weight at birth and in adults [52]. There is no current evidence to suggest that miR124-3 is imprinted. However, future work should address if miR124-3 is a metastable epiallele potentially playing a role in the development of adult disease [53].
GA as an outcome is a biologically significant parameter. The clinical phenotype for early GA is prematurity, defined as < 37 weeks of gestation, and preterm infants have higher rates of mortality and increased neonatal morbidity. However, recent findings also suggest a risk gradient for GA beyond 37 weeks compared to full term infants [54]. Reduced GA at birth is associated with many adverse long-term health outcomes hypothesized to be mediated by DNAm [55,56]. Our mediation approach showed that selected CpGs had the potential to mediate the effects of prenatal exposure on GA. Recently few studies have started to use mediation approaches to understand the effect of environmental exposures on relevant phenotypes. For example, the effect of smoking on birth weight has been shown to be mediated by DNAm as well as the association between air pollution and blood pressure [57,58]. We further proposed that GA is an intermediate phenotype of disease risk later in life, potentially mediated by DNAm of metastable epialleles. Future prospective studies should evaluate if these epigenetic perturbations are persistent or malleable as children grow and also test if certain birth outcomes are an intermediate phenotype to for a clinical disease stage. Malleability or persistence of epigenomic modifications could yield important information on the contribution of prenatal environmental exposures to disease risk [59,60].
Although no direct or mediated associations were observed between prenatal arsenic exposure and birth weight, the CpG in the DAB1 gene was associated with both prenatal arsenic exposure and birth weight. DAB1 expression has been shown to play an important role in brain ontogenesis and shown to be highly methylated in placentas of different species [61].
The present study has many strengths. First, the prospective measurements of the exposure, DNAm, and subsequent birth outcome present the possibility of testing for mediation that is chronologically possible. We also used objective personal exposure measures during early pregnancy in which many of the fetal programming events take place. However, separate exposure measures were available in the discovery and validation phases. The discovery phase relied on a single personal water sample, which might be reasonable for the population studied. Additionally, we were able to conduct sensitivity analyses in the validation phase using postpartum maternal toenail arsenic concentration, reflecting exposures that occurred during the gestational period, and using maternal toenail arsenic concentration at enrollment, both of which produced similar results as personal water samples. Other studies in Bangladesh have shown that drinking water arsenic exposures are relative constant and correlate with biomarkers of internal dose [27,62]. Additionally, we utilized a second technology (pyrosequencing) to validate loci-specific DNAm discovered on the Illumina Infinium HumanMethylation450 BeadChip. This provided considerable cost-savings, as well as validation across different assays used to quantify DNAm in a larger set of participants. However, it should be noted that each technology has a different sensitivity for measuring DNAm. Women also received prenatal vitamins as part of this study and reported high compliance with taking the vitamins. Thus, micronutrient deficiencies related to vitamin B or folate are less likely to be confounders in this analysis.
There are also some important limitations to be considered. Namely, functional gene expression was not evaluated and the observed epigenetic disruption might not lead to physiological changes in expression. Although the unadjusted EWAS was used for the identification of differentially methylated loci, the potential of confounding by shifts in white blood cell composition was minimized by ensuring that the probes selected did not differentiate cell types when using the Houseman method and available reference panels. Therefore, results should be interpreted in light of this and could indeed reflect cell type distribution. In addition, in the discovery phase, our relatively small sample size is an important limitation as it does not allow us to adjust for other potential confounders. However, use of a larger set of the birth cohort in the validation phase allowed for adjustment of multiple confounders. It should also be noted that maternal toenail arsenic concentrations, which can serve as a more accurate measure of exposure than drinking water arsenic concentrations, were not available when the discovery phase was completed. However, we performed sensitivity analyses in the validation phase using arsenic concentrations from maternal toenail samples collected at enrollment and postpartum. Overall, the results of these analyses were consistent. The significance of mediation of the association between prenatal arsenic exposure and GA by DNAm of miR124-3 increased with use of toenail arsenic as the measure of exposure.
Conclusions
In summary, in a two-stage experimental approach using discovery and validation phases we show that prenatal arsenic exposure is inversely associated with birth GA and the association mediated by DNAm of miR124-3 and MCC. However, no direct or mediated association was observed for birth weight. Our results support the hypothesis that arsenic exposure in utero can disrupt fetal programming leading to phenotypic consequences that may play a role in the developmental origins of health and disease. Furthermore, this experimental framework for the discovery and validation of candidate CpG loci as mediators of exposures and health outcomes could be extended to other exposures and health outcomes.
Funding Statement
This work was supported by the US National Institute of Environmental Health Sciences (NIEHS) grants R01 ES015533, R01 ES023441, P42 ES010349, and F31ES029019, and the National Center for Advancing Translational Sciences (NCATS) grant TL1 TR001875.
Acknowledgments
We would like to thank Dr John Geldhof from Oregon State University College of Public Health and Human Sciences for his expertise in structural equation modeling. We are also very grateful to Dr Andres Houseman for his epigenetic expertise and insights into how to approach this analysis. This work was supported by the US National Institute of Environmental Health Sciences (NIEHS) grants R01 ES015533, R01 ES023441, and P42 ES010349, and the National Center for Advancing Translational Sciences (NCATS) grant TL1 TR001875.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
supplemental data for this article can be accessed here
References
- 1.Naujokas MF, Anderson B, Ahsan H, et al. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121(3):295–302. Epub 2013/ 03/06 PubMed PMID: 23458756; PubMed Central PMCID: PMCPMC3621177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer A review of human carcinogens: arsenic, metals, fibres, and dusts. Lyon, France: international agency for research on cancer. 2012. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans.100. [PMC free article] [PubMed]
- 3.Concha G, Vogler G, Lezcano D, et al. Exposure to inorganic arsenic metabolites during early human development. Toxicological Sci. 1998;44(2):185–190. [DOI] [PubMed] [Google Scholar]
- 4.Quansah R, Armah FA, Essumang DK, et al. Association of arsenic with adverse pregnancy outcomes/infant mortality: a systematic review and meta-analysis. Environ Health Perspect. 2015;123(5):412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahman A, Vahter M, Ekstrom EC, et al. Arsenic exposure in pregnancy increases the risk of lower respiratory tract infection and diarrhea during infancy in Bangladesh. Environ Health Perspect. 2011;119(5):719–724. Epub 2010/ 12/15 PubMed PMID: 21147604; PubMed Central PMCID: PMCPMC3094427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farzan SF, Korrick S, Li Z, et al. In utero arsenic exposure and infant infection in a United States cohort: a prospective study. Environ Res. 2013;126:24–30. Epub 2013/ 06/19 PubMed PMID: 23769261; PubMed Central PMCID: PMCPMC3808159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dangleben NL, Skibola CF, Smith MT.. Arsenic immunotoxicity: a review. Environ Health. 2013;12(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyler CR, Allan AM. The effects of arsenic exposure on neurological and cognitive dysfunction in human and rodent studies: a review. Curr Environ Health Rep. 2014;1(2):132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardenas A, Smit E, Houseman EA, et al. and prevalence of the varicella zoster virus in the United States: NHANES (2003-2004 and 2009-2010). Environ Health Perspect. 2015;123:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardenas A, Smit E, Bethel J, et al. Arsenic exposure and the seroprevalence of total hepatitis A antibodies in the US population: NHANES, 2003–2012. Epidemiol Infect. 2016;144(8):1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farzan SF, Karagas MR, Chen Y. In utero and early life arsenic exposure in relation to long-term health and disease. Toxicol Appl Pharmacol. 2013;272(2):384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey KA, Smith AH, Tokar EJ, et al. Mechanisms underlying latent disease risk associated with early-life arsenic exposure: current research trends and scientific gaps. Environ Health Perspect. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saffery R, Novakovic B. Epigenetics as the mediator of fetal programming of adult onset disease: what is the evidence? Acta Obstet Gynecol Scand. 2014;93(11):1090–1098. [DOI] [PubMed] [Google Scholar]
- 14.Marsit CJ. Influence of environmental exposure on human epigenetic regulation. J Exp Biol. 2015;218(1):71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera F, Herbstman J. Prenatal environmental exposures, epigenetics, and disease. Reprod toxicol. 2011;31(3):363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argos M. Arsenic exposure and epigenetic alterations: recent findings based on the illumina 450K DNA methylation array. Curr Environ Health Rep. 2015;2(2):137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardenas A, Houseman EA, Baccarelli AA, et al. In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics. 2015;10(11):1054–1063. Epub 2015/12/10 PubMed PMID: 26646901; PubMed Central PMCID: PMCPMC4844206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardenas A, Koestler DC, Houseman EA, et al. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics. 2015;10(6):508–515. Epub 2015/04/30 PubMed PMID: 25923418; PubMed Central PMCID: PMCPMC4622995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broberg K, Ahmed S, Engstrom K, et al. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Orig Health Dis. 2014;5(4):288–298. Epub 2014/06/27 PubMed PMID: 24965135; PubMed Central PMCID: PMCPMC4283288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gribble MO, Tang WY, Shang Y, et al. Differential methylation of the arsenic (III) methyltransferase promoter according to arsenic exposure. Arch Toxicol. 2014;88(2):275–282. Epub 2013/ 10/25 PubMed PMID: 24154821; PubMed Central PMCID: PMCPMC3946764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kile ML, Baccarelli A, Hoffman E, et al. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect. 2012;120(7):1061–1066. Epub 2012/04/03 PubMed PMID: 22466225; PubMed Central PMCID: PMCPMC3404653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kile ML, Houseman EA, Baccarelli AA, et al. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9(5):774–782. Epub 2014/02/15 PubMed PMID: 24525453; PubMed Central PMCID: PMCPMC4063836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushal A, Zhang H, Karmaus WJJ, et al. Genome-wide DNA methylation at birth in relation to in utero arsenic exposure and the associated health in later life. Environ Health. 2017;16(1):50 Epub 2017/ 06/01 PubMed PMID: 28558807; PubMed Central PMCID: PMCPMC5450181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phookphan P, Navasumrit P, Waraprasit S, et al. Hypomethylation of inflammatory genes (COX2, EGR1, and SOCS3) and increased urinary 8-nitroguanine in arsenic-exposed newborns and children. Toxicol Appl Pharmacol. 2017;316:36–47. Epub 2016/ 12/28 PubMed PMID: 28025110. [DOI] [PubMed] [Google Scholar]
- 25.Rojas D, Rager JE, Smeester L, et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci. 2015;143(1):97–106. Epub 2014/ 10/12 PubMed PMID: 25304211; PubMed Central PMCID: PMCPMC4274382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creed J, Brockhoff C, Martin T.. Method 200.8: determination of trace elements in waters and wastes by inductively-coupled plasma-mass spectrometry. Environmental Monitoring Systems Laboratory, Office of Research and Development, US Environmental Protection Agency, Cincinnati, OH, Rev. 1994;5. [Google Scholar]
- 27.Kile ML, Houseman EA, Rodrigues E, et al. Toenail arsenic concentrations, GSTT1 gene polymorphisms, and arsenic exposure from drinking water. Cancer Epidemiol Prev Biomarkers. 2005;14(10):2419–2426. [DOI] [PubMed] [Google Scholar]
- 28.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fortin J-P, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y-A, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium human methylation450 microarray. Epigenetics. 2013;8(2):203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baron RM, Kenny DA. The moderator–mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51(6):1173. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 34.Brosseau-Liard PE, Savalei V, Li L. An investigation of the sample performance of two nonnormality corrections for RMSEA. Multivariate Behav Res. 2012;47(6):904–930. [DOI] [PubMed] [Google Scholar]
- 35.Brosseau-Liard PE, Savalei V. Adjusting incremental fit indices for nonnormality. Multivariate Behav Res. 2014;49(5):460–470. [DOI] [PubMed] [Google Scholar]
- 36.Oberski D. lavaan. survey: an R package for complex survey analysis of structural equation models. J Stat Software. 2014;57(1):1–27. [Google Scholar]
- 37.Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PloS one. 2012;7(7):e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kile ML, Cardenas A, Rodrigues E, et al. Estimating direct and indirect effects of arsenic exposure during pregnancy on maternal weight gain, gestational age, and birth weight in a Bangladeshi birth cohort. Epidemiology (Accepted in press) 2015.
- 40.Fei DL, Koestler DC, Li Z, et al. Association between In Utero arsenic exposure, placental gene expression, and infant birth weight: a US birth cohort study. Environ Health. 2013;12(58):10.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu P PIAOF-Y, Y-Y WANG, Hong Y. Study on expression of neurotransmitter transmission-related genes in cerebellum of mice exposed to arsenic by gene chip technology. J Dalian Med Univ. 2008;5:002. [Google Scholar]
- 42.Xu E, Gu J, Hawk ET, et al. Genome-wide methylation analysis shows similar patterns in Barrett’s esophagus and esophageal adenocarcinoma. Carcinogenesis. 2013;34(12):2750–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carvalho RH, Haberle V, Hou J, et al. Genome-wide DNA methylation profiling of non-small cell lung carcinomas. Epigenetics Chromatin. 2012;5(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett KM, Liu J, Hoelting C, et al. Expression and analysis of two novel rat organic cation transporter homologs, SLC22A17 and SLC22A23. Mol Cell Biochem. 2011;352(1–2):143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, Vasiliou K, Nebert DW. Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics. 2009;3(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sitras V, Paulssen R, Leirvik J, et al. Placental gene expression profile in intrauterine growth restriction due to placental insufficiency. Reprod Sci. 2009;16(7):701–711. [DOI] [PubMed] [Google Scholar]
- 47.Tsoporis J, Marks A, Haddad A, et al. S100A6 is a negative regulator of the induction of cardiac genes by trophic stimuli in cultured rat myocytes. Exp Cell Res. 2005;303(2):471–481. [DOI] [PubMed] [Google Scholar]
- 48.Kitano K, Watanabe K, Emoto N, et al. CpG island methylation of microRNAs is associated with tumor size and recurrence of non‐small‐cell lung cancer. Cancer Sci. 2011;102(12):2126–2131. [DOI] [PubMed] [Google Scholar]
- 49.Gebauer K, Peters I, Dubrowinskaja N, et al. Hsa-mir-124-3 CpG island methylation is associated with advanced tumours and disease recurrence of patients with clear cell renal cell carcinoma. Br J Cancer. 2013;108(1):131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J-Y, Chung K-H, Deo M, et al. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314(14):2618–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Huang X, Timani KA, et al. microRNA-124 targets Tip110 expression and regulates hematopoiesis. Stem Cells Dev. 2015;24(17):2009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grandjean V, Gounon P, Wagner N, et al. The miR-124-Sox9 paramutation: RNA-mediated epigenetic control of embryonic and adult growth. Development. 2009;136(21):3647–3655. [DOI] [PubMed] [Google Scholar]
- 53.Dolinoy DC, R D, Weidman JR, et al. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr Res. 2007;61(5 Part 2):30R. [DOI] [PubMed] [Google Scholar]
- 54.Boyle EM, Poulsen G, Field DJ, et al. Effects of gestational age at birth on health outcomes at 3 and 5 years of age: population based cohort study. Bmj. 2012;344:e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Platt M. Outcomes in preterm infants. Public Health. 2014;128(5):399–403. [DOI] [PubMed] [Google Scholar]
- 56.Schroeder JW, Conneely KN, Cubells JF, et al. Neonatal DNA methylation patterns associate with gestational age. Epigenetics. 2011;6(12):1498–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Küpers LK, Xu X, Jankipersadsing SA, et al. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int J Epidemiol. 2015;44(4):1224–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bellavia A, Urch B, Speck M, et al. DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. 2013;2(3):e000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cardenas A, Rifas-Shiman SL, Agha G, et al. Persistent DNA methylation changes associated with prenatal mercury exposure and cognitive performance during childhood. Sci Rep. 2017;7(1). Article No. 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cardenas A, Rifas-Shiman SL, Godderis L, et al. Prenatal exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ Health Perspect. 2017;87022:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder DI, Jayashankar K, Douglas KC, et al. Early developmental and evolutionary origins of gene body DNA methylation patterns in mammalian placentas. PLoS Genet. 2015;11(8):e1005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kile ML, Hoffman E, Hsueh Y-M, et al. Variability in biomarkers of arsenic exposure and metabolism in adults over time. Environ Health Perspect. 2009;117(3):455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


