Animal behavior is increasingly being recorded in systematic imaging studies that generate large data sets. To maximize the usefulness of these data there is a need for improved resources for analyzing and sharing behavior data that will encourage re-analysis and methodological developments1. However, unlike genomic or protein structural data, there are no widely used standards for behavior data. It is therefore desirable to make data available in a relatively raw form so that different investigators can use their own representations and derive their own features. For computational ethology to approach the level of maturity of other areas of bioinformatics, we need to address at least three challenges: storing and accessing video files, defining flexible data formats to facilitate data sharing, and developing software to read, write, browse, and analyze the data. We have generated an open resource to begin addressing these challenges for C. elegans behavioral data.
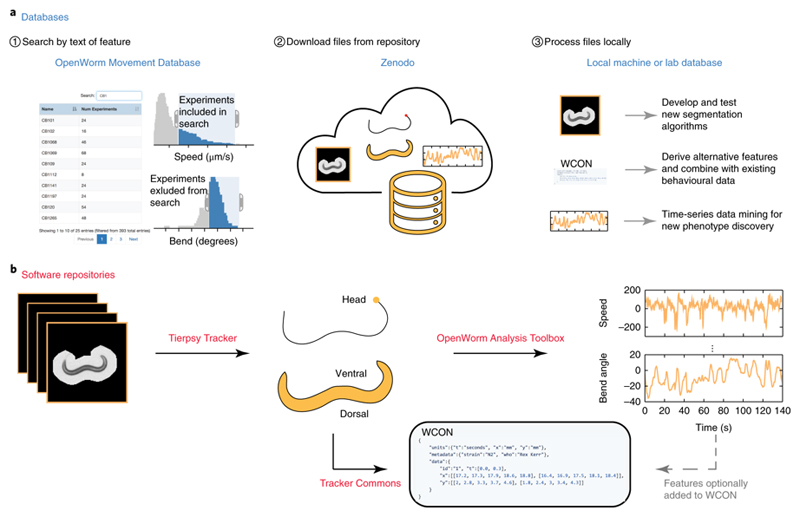
To store video files and the associated feature and metadata, we use a Zenodo.org community (an open-access repository for data) that provides durable storage and citability, and that supports contributions from other groups. We have also developed a web interface that enables filtering of the video files based on feature histograms that can return, for example, fast and curved worms in addition to more standard searches for particular strains or genotypes (Fig. 1 and http://movement.openworm.org/). The database currently consists of 14,874 single-worm tracking experiments representing 386 genotypes (building on 9,203 experiments and 305 genotypes in a previous publication2) and includes data from several larval stages as well as data from ageing experiments consisting of over 2,700 videos of animals tracked daily from the L4 stage to death (see Life Sciences Reporting Summary). Full resolution videos are available in HDF5 containers that include gzip-compressed video frames, timestamps, worm outline and midline, feature data, and experiment metadata. HDF5 files are compatible with multiple languages including MATLAB, R, Python, and C. We have also developed an HDF5 video reader that allows video playback with adjustable speed and zoom (important when reviewing high-resolution, multi-worm tracking data), as well as toggling of worm segmentation over the original video to verify segmentation accuracy during playback.
Figure 1. Schematic of searchable database and Tierpsy analysis pipeline.
(a) The OpenWorm Movement Database provides a web interface for searching by genotype, strain, and/or other discrete values, and interactive histograms with sliders to filter results based on feature values. The interface points to data stored on Zenodo. The video and feature data can be further analyzed or combined with data collected using other worm trackers through the Worm tracker Commons Object Notation (WCON), a human and machine readable JSON format. (b) Tierpsy (short for Tierpsychology, the German word for ethology) segments and tracks worms, extracting the outline and skeleton of each animal then determining the head-tail orientation. These data are saved in WCON. The Open Worm Analysis Toolbox is then used to extract behavioral features.
Secondly, we have defined an interchange format named Worm tracker Commons Object Notation (WCON), to facilitate data sharing and software reuse among groups working on worm behavior. WCON uses the widely supported JSON format to store tracking data as text that is both human and machine readable. It is compatible with single and multi-worm3 tracking data, at any resolution: from a single point representing worm position over time4, to many points representing the high-resolution skeleton of a moving worm2. It also supports custom feature additions so that individual labs can store their own specific data sets alongside the existing set of basic worm data. WCON readers are available for Python, MATLAB, Scala, and C. Detailed documentation for the file formats and software is available on the project page (https://github.com/openworm/tracker-commons).
Finally, we have complemented the database and file formats with open-source software written in Python for single and multi-worm tracking, feature extraction, review, and analysis (Supplementary Discussion; code and documentation is available as Supplementary Software, and at https://doi.org/10.5281/zenodo.1323782, where compiled versions are available in addition).
The suite of tools we have reported makes quantitative behavior (re-)analysis accessible for both experimentalists and computational scientists. It may also serve as a template for similar efforts in other model organism communities.
Supplementary Material
Acknowledgements
This work was supported by the MRC through grant MC-A658-5TY30 to AEX Brown. Q Ch’ng is supported by an ERC Starting Grant (NeuroAge 242666), Research Councils UK Fellowship, and the University of London Central Research Fund. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Data Availability Statement
Videos, skeleton (WCON) files, and feature files are available with a Creative Commons attribution (CC BY) license through the database page http://movement.openworm.org/ and Zenodo community page https://zenodo.org/communities/open-worm-movement-database/
Code Availability Statement
Tierpsy Tracker is available as Supplementary Software and at https://doi.org/10.5281/zenodo.1323782. Updated versions will be made available at http://ver228.github.io/tierpsy-tracker/.
Author contributions
A.J. wrote Tierpsy Tracker and analyzed data; M.C. wrote WCON viewer, database, and OpenWorm Analysis Toolbox; C.W.L. wrote database, web interface, and WCON viewer; J.H. wrote MATLAB WCON viewer and OpenWorm Analysis Toolbox; K.L. wrote stage alignment code; C.N.M. collected data; E.Y. wrote skeletonization algorithm and stage alignment code; L.J.G. collected data; C.L. contributed strains and planned experiments; Q.C. contributed strains and planned experiments; W.R.S. planned the study; E.A.A.N. contributed strains and planned experiments; R.K. designed WCON and wrote several readers; A.E.X.B. planned the study and wrote the manuscript.
Competing Financial Interests Statement
The authors declare that they have no competing financial or non-financial interests as defined by Nature Research.
References
- 1.Gomez-Marin A, Paton JJ, Kampff AR, Costa RM, Mainen ZF. Nat Neurosci. 2014;17:1455–1462. doi: 10.1038/nn.3812. [DOI] [PubMed] [Google Scholar]
- 2.Yemini E, Jucikas T, Grundy LJ, Brown AEX, Schafer WR. Nat Methods. 2013;10:877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swierczek NA, Giles AC, Rankin CH, Kerr RA. Nat Methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramot D, Johnson BE, Berry TL, Carnell L, Goodman MB. PLoS ONE. 2008;3:e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.