Abstract
High blood pressure is a highly heritable and modifiable risk factor for cardiovascular disease. We report the largest genetic association study of blood pressure traits (systolic, diastolic, pulse pressure) to date in over one million people of European ancestry. We identify 535 novel blood pressure loci that not only offer new biological insights into blood pressure regulation but also reveal shared genetic architecture between blood pressure and lifestyle exposures. Our findings identify new biological pathways for blood pressure regulation with potential for improved cardiovascular disease prevention in the future.
Introduction
High blood pressure (BP) is a leading heritable risk factor for stroke and coronary artery disease, responsible for an estimated 7.8 million deaths and 148 million disability life years lost worldwide in 2015 alone1. Blood pressure is determined by complex interactions between life-course exposures and genetic background2–4. Previous genetic association studies have identified and validated variants at 274 loci with modest effects on population BP, explaining in aggregate ~3% of the trait variance5–12.
Here, we report genome-wide discovery analyses of BP traits - systolic (SBP), diastolic (DBP) and pulse pressure (PP) - in people of European ancestry drawn from UK Biobank (UKB)13 and the International Consortium of Blood Pressure-Genome Wide Association Studies (ICBP)11,12. We adopted a combination of a one- and two-stage study design to test common and low-frequency single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) ≥ 1% associated with BP traits (Fig. 1). In all, we studied over 1 million people of European descent, including replication data from the US Million Veterans Program (MVP, N=220,520)14 and the Estonian Genome Centre, University of Tartu (EGCUT, N=28,742) Biobank15.
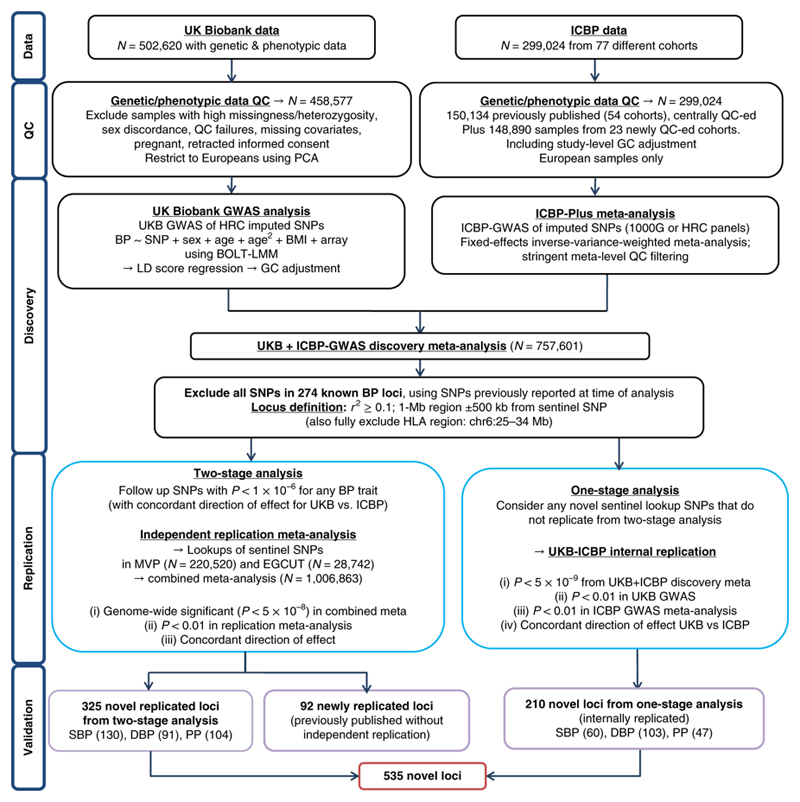
Figure 1.
Study design schematic for discovery and validation of loci. ICBP; International Consortium for Blood Pressure; N, sample size; QC, quality control; PCA, principal-component analysis; GWAS, Genome-wide Association Study; 1000G 1000 Genomes; HRC, Haplotype Reference Panel; BP: blood pressure; SNPs, single nucleotide polymorphisms; BMI, body mass index; LMM; linear mixed model; UKB, UK Biobank, MAF, minor allele frequency; HLA, Human Leukocyte Antigen; MVP, Million Veterans Program; EGCUT; Estonian Genome Center, University of Tartu; SBP, systolic blood pressure; DBP, diastolic blood pressure; PP, pulse pressure.
UKB is a prospective cohort study of ~500,000 richly phenotyped individuals, including BP measurements13, with genotyping by customized array and imputation from the Haplotype Reference Consortium (HRC) panel, yielding ~7 million SNPs (imputation quality score (INFO) ≥ 0.1 and MAF ≥ 1%)16. We performed genome-wide association studies (GWAS) of BP traits (N=458,577 Europeans) under an additive genetic model17 (Supplementary Table 1a). Following LD-score regression18, genomic control (GC) was applied to the UKB data prior to meta-analysis (Online methods).
In addition, we performed GWAS analyses for BP traits in newly extended ICBP GWAS data comprising 77 independent studies for up to 299,024 Europeans genotyped with various arrays, and imputed to either the 1,000 Genomes Reference Panel or the HRC platforms (Supplementary Table 1b). After QC we applied GC at the individual study level and obtained summary effect sizes for ~7 million SNPs with INFO ≥ 0.3 and heterogeneity Cochran’s Q statistic19 filtered at P ≥ 1 × 10-4 (Online Methods).
We then combined the UKB and ICBP GWAS results using inverse-variance weighted fixed effects meta-analysis (Online Methods), giving a total discovery sample of up to 757,601 individuals20.
In our two-stage design we attempted replication (in MVP and EGCUT, Supplementary Table 1c) of 1,062 SNPs at P < 1 × 10-6 from discovery with concordant effect direction between UKB and ICBP, using the sentinel SNP (i.e. SNP with smallest P-value at the locus) after excluding the HLA region (chr 6:25-34MB) and all SNPs in Linkage Disequilibrium (LD) (r2 ≥ 0.1) or ±500 Kb from any previously validated BP-associated SNPs at the 274 published loci. Our replication criteria were genome-wide significance (P < 5 × 10-8) in the combined meta-analysis, P < 0.01 in the replication data and concordant direction of effect between discovery and replication.
We additionally undertook a one-stage design to reduce type II error from the two-stage analysis. We used P < 5 × 10-9 as threshold from the discovery meta-analysis, i.e. an order of magnitude more stringent than genome-wide significance21, and required an internal replication P < 0.01 in each of the UKB and ICBP GWAS analyses, with concordant direction of effect, to minimize false positive findings.
We carried out conditional analyses using genome-wide complex trait analysis (GCTA)22. We then explored putative function of BP-associated signals using a range of in silico resources, and evaluated co-occurrence of BP-associated loci with lifestyle exposures and other complex traits and diseases. Finally, we developed a genetic risk score (GRS) and assessed impact of BP-associated variants on BP level, risk of hypertension (HTN), other cardiovascular diseases and in other ethnicities.
Results
We present a total of 535 novel loci (Fig.2, Supplementary Fig. 1): 325 loci claimed from the two-stage design (Supplementary Tables 2a-c) and an additional 210 claimed from our one-stage design with internal replication (Supplementary Tables 3a-c). Our two-stage design uniquely identified 121 variants, while 204 also met the one-stage criteria (Fig. 3a); large numbers of loci would not have been detected by either the one- or two-stage designs alone (Fig. 3a). For SBP, the distributions of effect sizes are similar for the one-stage (median = 0.219 mmHg per allele; Inter-Quartile Range (IQR) = 0.202-0.278) and two-stage loci (median = 0.224; IQR = 0.195-0.267) (P = 0.447) (Supplementary Fig. 2). Of the 210 loci found only in the one-stage analysis, 186 are also genome-wide significant (P < 5 × 10-8) in the combined meta-analysis, with all variants, except one, having concordant direction of effect between discovery and replication (Supplementary Tables 3a-c); of the remaining 24 SNPs, 10 still have concordant direction of effect.
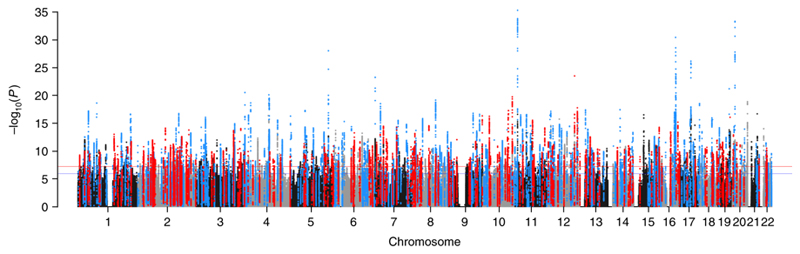
Figure 2.
Manhattan plot showing the minimum P-value for the association across all blood pressure traits in the discovery stage excluding known and previously reported variants. Manhattan plot of the discovery genome-wide association meta-analysis in 757,601 individuals excluding variants in 274 known loci. The minimum P-value, computed using inverse variance fixed effects meta-analysis, across SBP, DBP and PP is presented. The y axis shows the –log10 P values and the x axis shows their chromosomal positions. Horizontal red and blue line represents the thresholds of P = 5 x 10-8 for genome-wide significance and P = 1 x 10-6 for selecting SNPs for replication, respectively. SNPs in blue are in LD (r2 > 0.8) with the 325 novel variants independently replicated from the 2-stage design whereas SNPs in red are in LD (r2 > 0.8) with 210 SNPs identified through the 1-stage design with internal replication. Any loci in black or grey that exceed the significance thresholds were significant in the discovery meta-analysis, but did not meet the criteria of replication in the one- or two-stage designs.
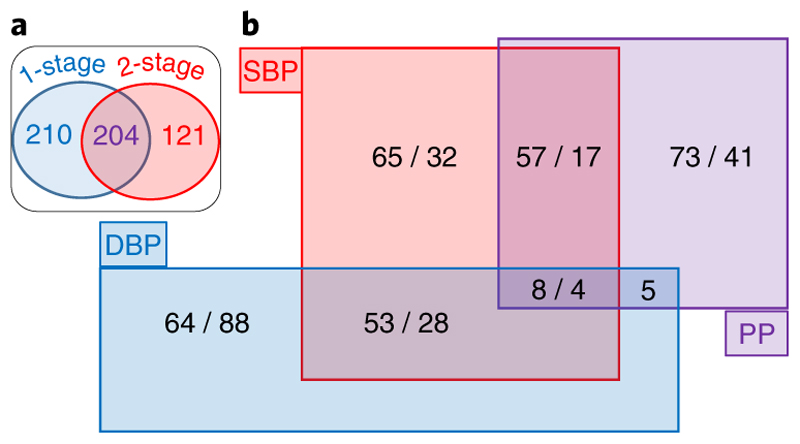
Figure 3.
Venn Diagrams of Novel Loci Results (a) “Comparison of 1-stage and 2-stage design analysis criteria”: For all 535 novel loci, we compare the results according to the association criteria used for the one-stage and the two-stage design. Two-hundred and ten loci exclusively met the one-stage analysis criteria (P <5x10-9 in the discovery meta-analysis [N=757,601], P < 0.01 in UKB [N=458,577], P < 0.01 in ICBP [N=299,024] and concordant direction of effect between UKB and ICBP). The P-values for the discovery and the ICBP meta-analyses were calculated using inverse variance fixed effects meta-analysis. The P-values in UKB were derived from linear mixed modeling using BOLT-LMM. Of the 325 novel replicated loci from the 2-stage analysis (genome-wide significance in the combined meta-analysis, P < 0.01 in the replication meta-analysis and concordant direction of effect), 204 loci would also have met the one-stage criteria, whereas 121 were only identified by the two-stage analysis. (b) “Overlap of Associations across Blood Pressure Traits”. For all 535 novel loci, we show the number of loci associated with each blood pressure trait. We present the two-stage loci first, followed by the one-stage loci. SBP: systolic blood pressure; DBP: diastolic blood pressure; PP: pulse pressure; UKB: UK Biobank; ICBP: International Consortium of Blood Pressure.
We find support in our data for all 274 previously published BP loci (Supplementary Fig. 1 & 2 and Supplementary Table 4); >95% of the previously reported SNPs covered within our data are genome-wide significant. Only 6 available SNPs did not reach Bonferroni-significance, likely because they were originally identified in non-European ancestries (e.g. rs6749447, rs10474346, rs11564022), or from a gene-age interaction analysis (rs16833934). In addition, we confirmed a further 92 previously reported, but not replicated, loci (Supplementary Table 5)9; together with 274 previously reported loci confirmed, and 535 novel loci identified here, there are 901 BP-associated loci in total.
Novel genetic loci for blood pressure
Of the 535 independent novel loci, 363 SNPs were associated with one trait, 160 with two traits and 12 with all three BP traits (Fig. 3b, Supplementary Fig. 3). Using GCTA we additionally identified 163, genome-wide significant, independent secondary signals with MAF ≥ 1% associated with BP (Supplementary Table 6), of which 19 SNPs are in LD (r2 ≥ 0.1) with previously reported secondary signals. This gives a total of 144 new secondary signals; hence we now report over 1,000 independent BP signals.
The estimated SNP-wide heritability (h2) of BP traits in our data was 0.213, 0.212 and 0.194 for SBP, DBP and PP respectively, with a gain in percentage of BP variance explained. For example, for SBP, percentage variance explained increased from 2.8 % for the 274 previously published loci to 5.7% for SNPs identified at all 901 loci (Supplementary Table 7).
Functional analyses
Our functional analyses approach is summarised in Supplementary Figure 4. First, for each of the 901 loci we annotated all SNPs (based on LD r2 ≥ 0.8) to the nearest gene within 5kb of a SNP, identifying 1333 genes for novel loci and 1272 genes for known loci. Then we investigated these loci for tissue enrichment, DNase hypersensitivity site enrichment and pathway analyses. At 66 of the 535 novel loci we identified 97 non-synonymous SNPs, including 8 predicted to be damaging (Supplementary Table 8).
We used chromatin interaction Hi-C data from endothelial cells (HUVEC)23, neural progenitor cells (NPC), mesenchymal stem cells (HVMSC) and tissue from the aorta (HAEC) and adrenal gland24 to identify distal associated genes. There were 498 novel loci that contained a potential regulatory SNP and in 484 of these we identified long-range interactions in at least one of the tissues or cell types. We found several potential long-range target genes that do not overlap with the sentinel SNPs in the LD block. For example, the TGFB2 gene forms a 1.2Mb regulatory loop with SNPs in the SLC30A10 locus, and the TGFBR1 promoter forms a 100kb loop with the COL15A1 locus (Supplementary Table 8).
Our eQTL analysis identified 60 novel loci with eQTLs in arterial and 20 in adrenal tissue (Supplementary Table 9), substantially increasing those identified in our previously published GWAS on ~140K UKB individuals10. An example is SNP rs31120122 which defines an aortic eQTL affecting expression of the MED8 gene within the SZT2 locus. In combination with Hi-C interaction data in MSC, this supports a role for MED8 in BP regulation, possibly mediated through repression of smooth muscle cell differentiation. Hi-C interactions provide supportive evidence for involvement of a further 36 arterial eGenes (genes whose expression is affected by the eQTLs) that were distal to their eQTLs (e.g PPHLN1, ERAP2, FLRT2, ACVR2A, POU4F1).
Using DeepSEA we found 198 SNPs in 121 novel loci with predicted effects on transcription factor binding or on chromatin marks in tissues relevant for BP biology, such as vascular tissue, smooth muscle and the kidney (Supplementary Table 8).
We used our genome-wide data at a false discovery rate (FDR) < 1% to robustly assess tissue enrichment of BP loci using DEPICT and identified enrichment across 50 tissues and cells. (Supplementary Fig 5a; Supplementary Table 10a). Enrichment was greatest for the cardiovascular system especially blood vessels (P = 1.5 × 10-11) and the heart (P = 2.7 × 10-5). Enrichment was high in adrenal tissue (P = 3.7 × 10-4) and, for the first time, we observed high enrichment in adipose tissues (P = 9.8 × 10-9) corroborated by eQTL enrichment analysis (P < 0.05) (Supplementary Fig. 6; Supplementary Table 9). Evaluation of enriched mouse knockout phenotype terms also points to the importance of vascular morphology (P = 6 × 10-15) and development (P = 2.1 × 10-18) in BP. With addition of our novel BP loci, we identified new findings from both the gene ontology and protein-protein interaction subnetwork enrichments, which highlight the TGFβ (P = 2.3 × 10-13) and related SMAD pathways (P = 7 × 10-15) (Supplementary Table 10b, Supplementary Fig. 5b-d).
We used FORGE25 to investigate the regulatory regions for cell type specificity from DNase I hypersensitivity sites, which showed strongest enrichment (P < 0.001) in the vasculature and highly vascularised tissues, as reported in previous BP genetic studies10 (Supplementary Fig. 7).
Potential therapeutic targets
Ingenuity pathway analysis and upstream regulator assessment showed enrichment of canonical pathways implicated in cardiovascular disease including pathways targeted by antihypertensive drugs (e.g. nitric oxide signalling) and also suggested some potential new targets, such as relaxin signalling. Notably, upstream regulator analysis identified several BP therapeutic targets such as angiotensinogen, calcium channels, progesterone, natriuretic peptide receptor, angiotensin converting enzyme, angiotensin receptors and endothelin receptors (Supplementary Fig. 8).
We developed a cumulative tally of functional evidence at each variant to assist in variant/gene prioritisation at each locus and present a summary of the vascular expressed genes contained within the 535 novel loci, including a review of their potential druggability (Supplementary Fig. 9). The overlap between BP-associated genes and those associated with antihypertensive drug targets further demonstrates new genetic support for known drug mechanisms. For example, we report five novel BP associations with targets of five antihypertensive drug classes (Supplementary Table 11), including the PKD2L1, SLC12A2, CACNA1C, CACNB4 and CA7 loci - targeted by potassium-sparing diuretics (amiloride), loop diuretics (bumetanide and furosemide), dihydropyridine, calcium channel blockers, non-dihydropyridines and thiazide-like diuretics (chlortalidone) respectively. Notably in all but the last case, functional variants in these genes are the best candidates in each locus.
Concordance of BP variants and lifestyle exposures
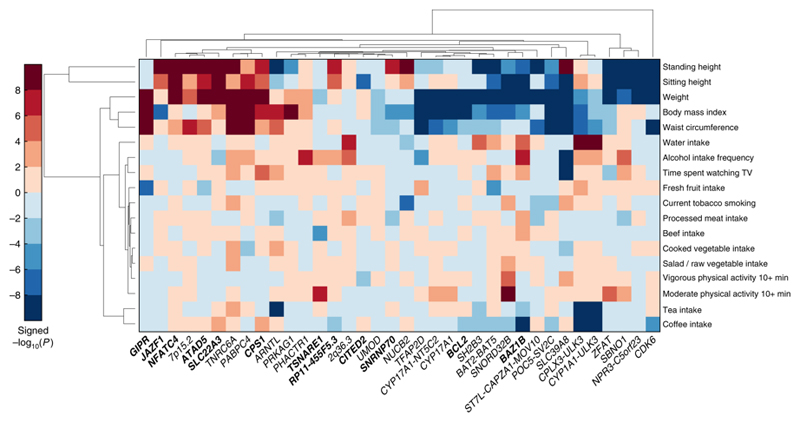
We examined association of sentinel SNPs at the 901 BP loci with BP-associated lifestyle traits14 in UKB using either the Stanford Global Biobank Engine (N=327,302) or Gene ATLAS (N=408,455). With corrected P < 1 × 10-6, we found genetic associations of BP variants with daily fruit intake, urinary sodium and creatinine concentration, body mass index (BMI), weight, waist circumference, and intakes of water, caffeine and tea (P = 1.0 × 10-7 to P = 1.3 × 10-46). Specifically, SNP rs13107325 in SLC39A8 is a novel locus for frequency of drinking alcohol (P = 3.5 × 10-15) and time spent watching TV (P = 2.3 × 10-11) as well as being associated with BMI (P = 1.6 × 10-33), weight (P = 8.8 × 10-16) and waist circumference (P = 4.7 × 10-11) (Supplementary Table 12). We used unsupervised hierarchical clustering for the 36 BP loci that showed at least one association at P < 1 × 10-6 with the lifestyle-related traits in UKB (Fig. 4). The heatmap summarises the locus-specific associations across traits and highlights heterogeneous effects with anthropometric traits across the loci examined. For example, it shows clusters of associations between BP-raising alleles and either increased or decreased adult height and weight. We note that some observed cross-trait associations are in counter-directions to those expected epidemiologically.
Figure 4.
Association of blood pressure loci with lifestyle traits. Plot shows unsupervised hierarchical clustering of BP loci based on associations with lifestyle-related factors. For the sentinel SNP at each BP locus (x-axis), we calculated the -log10(P)*sign(β) (aligned to BP-raising allele) as retrieved from the Gene Atlas catalogue (http://geneatlas.roslin.ed.ac.uk). The P-values in Gene Atlas were calculated applying linear mixed models. BP loci and traits were clustered according to the Euclidean distance amongst -log10(P)*sign(β). Red squares indicate direct associations with the trait of interest and blue squares inverse associations. Only SNPs with at least one association at P <10-6 with at least one of the traits examined are annotated in the heat-map. All 901 loci are considered, both known and novel: novel loci are printed in bold font. SNPs: Single Nucleotide Polymorphisms; BP: Blood Pressure.
Association lookups with other traits and diseases
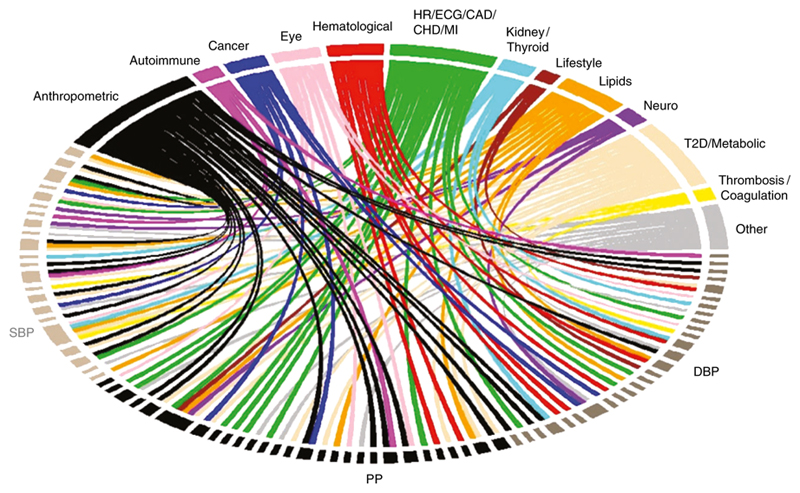
We further evaluated cross-trait and disease associations using GWAS catalog26, PhenoScanner27 and DisGeNET28,29. The GWAS catalog and PhenoScanner search of published GWAS showed that 77 of our 535 novel loci (using sentinel SNPs or proxies; r2 ≥ 0.8) are also significantly associated with other traits and diseases (Fig. 5, Supplementary Table 13). We identified APOE as a highly cross-related BP locus showing associations with lipid levels, cardiovascular-related outcomes and Alzheimer’s disease, highlighting a common link between cardiovascular risk and cognitive decline (Fig. 5). Other loci overlap with anthropometric traits, including BMI, birth weight and height (Fig. 5) and with DisGeNET terms related to lipid measurements, cardiovascular outcomes and obesity (Fig. 6).
Figure 5.
Association of blood pressure loci with other traits. Plot shows results from associations with other traits which were extracted from the GWAS catalog and PhenoScanner databases for the 535 novel sentinel SNPs including proxies in Linkage Disequilibrium (r2 ≥ 0.8) with genome-wide significant associations. SBP: Systolic Blood Pressure; DBP: Diastolic Blood Pressure; PP: Pulse Pressure; HR: Heart Rate; ECG: Electrocardiographic traits; CAD: Coronary Artery Disease CHD; Coronary Heart Disease MI; Myocardial Infraction; T2D: Type II Diabetes.
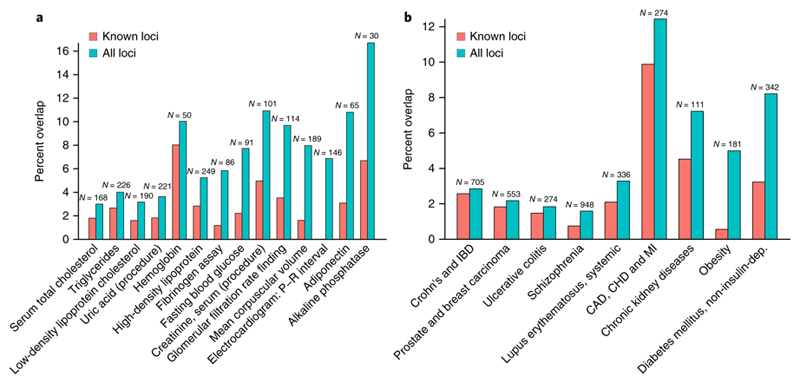
Figure 6.
Association of blood pressure loci with other traits. Plots (a) and (b) show overlap between variants associated to (a) traits and (b) diseases in the manually-curated version of the DisGeNET database, and all variants in LD r2>0.8 with the known (red bars) SNPs from the 274 published loci, and all (green bars) BP variants from all 901 loci. Numbers on top of the bars denote the number of SNPs included in DisGeNET for the specific trait or disease. Traits/diseases with an overlap of at least 5 variants in LD with all markers are shown. The Y axis shows the percentage of variants associated with the diseases that is covered by the overlap. For the sake of clarity, the DisGeNET terms for blood pressure and hypertension are not displayed, whereas the following diseases have been combined: coronary artery disease (CAD), coronary heart disease (CHD) and myocardial infarction (MI); prostate and breast carcinoma; Crohn's and inflammatory bowel diseases.
We did lookups of our sentinel SNPs in 1H NMR lipidomics data on plasma (N=2,022) and data from the Metabolon platform (N=1,941) in the Airwave Study30, and used PhenoScanner to test SNPs against published significant (P < 5 × 10-8) genome vs metabolome-wide associations in plasma and urine (Online Methods). Ten BP SNPs show association with lipid particle metabolites and a further 31 SNPs (8 also on PhenoScanner) show association with metabolites on the Metabolon platform, highlighting lipid pathways, amino acids (glycine, serine, glutamine), tri-carboxylic acid cycle intermediates (succinylcarnitine) and drug metabolites (Supplementary Tables 14 and 15). These findings suggest a close metabolic coupling of BP regulation with lipid and energy metabolism.
Genetic risk of increased blood pressure, hypertension and cardiovascular disease
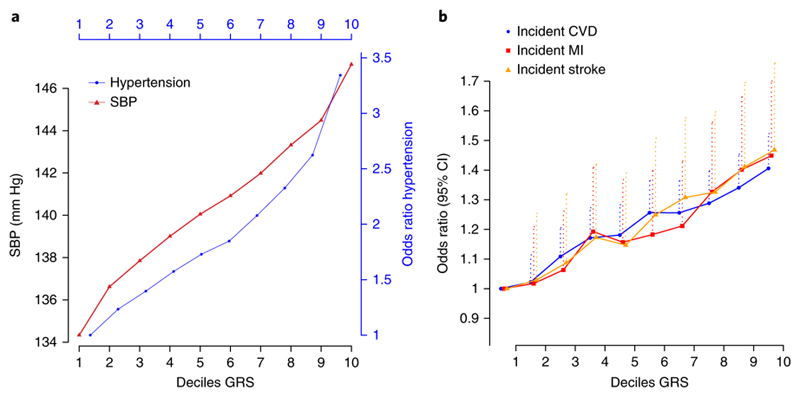
A weighted GRS for BP levels across all 901 loci was associated with a 10.4 mmHg higher, sex-adjusted mean SBP in UK Biobank comparing the upper and lower quintiles of the GRS distribution (95% CI: 10.2 to 10.6 mm Hg, P < 1 × 10-300) and with 12.9 mmHg difference in SBP (95% CI: 12.6 to 13.1, P < 1 × 10-300) comparing the upper and lower deciles (Fig. 7a, Supplementary Table 16). In addition, we observed over three-fold sex-adjusted higher risk of hypertension (OR 3.34; 95% CI: 3.24 to 3.45; P < 1 × 10-300) between the upper and lower deciles of the GRS in UK Biobank (Fig. 7a). Sensitivity analyses in the independent Airwave cohort gave similar results (Supplementary Table 17).
Figure 7.
Relationship of deciles of the genetic risk score (GRS) based on all 901 loci with blood pressure, risk of hypertension and cardiovascular disease in UK Biobank. The plots show sex-adjusted (a) mean systolic blood pressure (SBP) and odds ratios of hypertension (HTN) (N=364,520) and (b) odds ratios of incident cardiovascular disease (CVD), myocardial infarction (MI) and stroke (N=392,092), comparing each of the upper nine GRS deciles with the lowest decile; dotted lines represent the upper 95% confidence intervals.
We also show that the GRS is associated with increased, sex-adjusted risk of incident stroke, myocardial infarction and all incident cardiovascular outcomes, comparing upper and lower deciles of the GRS distribution, with odds ratios of 1.47 (95% CI: 1.35 to 1.59, P = 1.1 × 10-20), 1.50 (95% CI: 1.28 to 1.76, P = 8.0 × 10-7) and 1.52 (95% CI: 1.26 to 1.82, P = 7.7 × 10-6) respectively (Fig. 7b, Supplementary Table 16).
Extending analyses to other ancestries
We examined associations with BP of both individual SNPs and the GRS among unrelated individuals of African and South Asian descent in UKB, for the 901 known and novel loci. Compared to Europeans, 62.4%, 62.5% and 64.8% of the variants among Africans (N=7,782), and 74.2%, 72.3% and 75% South Asians (N=10,323) have concordant direction of effect for SBP, DBP and PP respectively (Supplementary Table 18; Supplementary Fig. 10). Pearson correlation coefficients with effect estimates in Europeans were r2= 0.37 and 0.78 for Africans and South Asians respectively (Supplementary Fig. 11). We then applied the European-derived GRS findings to unrelated Africans (N=6,970) and South Asians (N=8,827). BP variants in combination were associated with 6.1 mmHg (95% CI: 4.5 to 7.7; P = 4.9 × 10-14) and 7.4 mmHg (95% CI: 6.0 to 8.7; P = 1.7 × 10-26) higher, sex-adjusted mean systolic pressure among Africans and South Asians, respectively, comparing upper and lower quintiles of the GRS distribution (Supplementary Tables 19a and 19b).
Discussion
Our study of over 1 million people offers an important step forward in understanding the genetic architecture of BP. We identified over 1,000 independent signals at 901 loci for BP traits, and the 535 novel loci more than triples the number of BP loci and doubles the percentage variance explained, illustrating the benefits of large-scale biobanks. By explaining 27% of the estimated heritability for BP, we make major inroads into the missing heritability influencing BP level in the population31. The novel loci open the vista of entirely new biology and highlight gene regions in systems not previously implicated in BP regulation. This is particularly timely as global prevalence of people with SBP over 110-115 mm Hg, above which cardiovascular risk increases in a continuous graded manner, now exceeds 3.5 billion, of whom over 1 billion are within the treatment range 32,33.
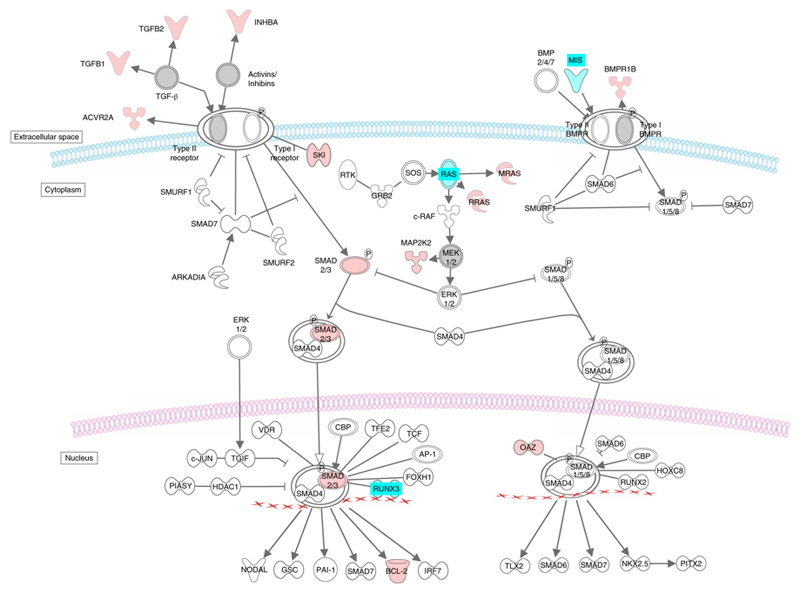
Our functional analysis highlights the role of the vasculature and associated pathways in the genetics underpinning BP traits. We show a role for several loci in the transforming growth factor beta (TGFβ) pathway including SMAD family genes and the TGFβ gene locus itself. This pathway affects sodium handling in the kidney, ventricular remodelling, while plasma levels of TGFβ have recently been correlated with hypertension (Fig. 8)34,35. The activin A receptor type 1C (ACVR1C) gene mediates the effects of the TGFβ family of signalling molecules. A BP locus contains the Bone Morphogenetic Protein 2 (BMP2) gene in the TGFβ pathway, which prevents growth suppression in pulmonary arterial smooth muscle cells and is associated with pulmonary hypertension36. Another BP locus includes the Kruppel-like family 14 (KLF14) gene of transcription factors, induced by low levels of TGFβ receptor II gene expression, and which has also been associated with type 2 diabetes, hypercholesterolaemia and atherosclerosis37.
Figure 8.
Known and novel BP associations in the TGFβ signalling pathway. Genes with known associations with BP are indicated in cyan. Genes with novel associations with BP reported in this study are indicated in red. TGFβ pathway was derived from an ingenuity canonical pathway. BP: Blood Pressure.
Our analysis shows enrichment of BP gene expression in the adrenal tissue. Autonomous aldosterone production by the adrenal glands is thought to be responsible for 5-10% of all hypertension, rising to ,20% amongst people with resistant hypertension38. Some of our novel loci are linked functionally to aldosterone secretion39,40. For example, the CTNNB1 locus encodes β-catenin, the central molecule in the canonical Wnt signalling system, required for normal adrenocortical development41,42. Somatic adrenal mutations of this gene that prevent serine/threonine phosphorylation lead to hypertension through generation of aldosterone-producing adenomas43,44.
Our novel loci also include genes involved in vascular remodelling, such as vascular endothelial growth factor A (VEGFA), the gene product of which induces proliferation, migration of vascular endothelial cells and stimulates angiogenesis. Disruption of this gene in mice resulted in abnormal embryonic blood vessel formation, while allelic variants of this gene have been associated with microvascular complications of diabetes, atherosclerosis and the antihypertensive response to enalapril45. We previously reported a fibroblast growth factor (FGF5) gene locus in association with BP10. Here, we additionally identify a new BP locus encoding FGF9, which is linked to enhanced angiogenesis and vascular smooth muscle cell differentiation by regulating VEGFA expression.
Several of our novel loci contain lipid-related genes consistent with the observed strong associations among multiple cardio-metabolic traits. For example, the apolipoprotein E gene (APOE) encodes the major apoprotein of the chylomicron. Recently, APOE serum levels have been correlated with SBP in population-based studies and in murine knockout models; disruption of this gene led to atherosclerosis and hypertension46,47. A second novel BP locus contains the low-density lipoprotein receptor-related protein 4 (LRP4) gene which may be a target for APOE and is strongly expressed in the heart in mice and humans. In addition, we identified a novel locus including the apolipoprotein L domain containing 1 gene (APOLD1) that is highly expressed in the endothelium of developing tissues (particularly heart) during angiogenesis.
Many of our novel BP loci encode proteins which may modulate vascular tone or signalling. For example, the locus containing urotensin-2 receptor (UTS2R) gene encodes a class A rhodopsin family G-protein coupled-receptor that upon activation by the neuropeptide urotensin II, produces profound vasoconstriction. One novel locus for SBP contains the relaxin gene, encoding a G-protein coupled receptor, with roles in vasorelaxation and cardiac function; it signals by phosphatidylinositol 3-kinase (PI3K)48,49, an enzyme which inhibits vascular smooth muscle cell proliferation and neo-intimal formation50. We identify the PI3K gene here as a novel BP locus. We also identify the novel RAMP2 locus which encodes an adrenomedullin receptor51; we previously identified the adrenomedullin (ADM) gene as a BP locus12. Adrenomedullin is known to exert differential effects on BP in the brain (vasopressor) and the vasculature (vasodilator). In addition, a locus containing Rho guanine nucleotide exchange factor 25 (ARHGEF25) gene generates a factor that interacts with Rho GTPases involved in contraction of vascular smooth muscle and regulation of responses to angiotensin II52.
We evaluated the 901 BP loci for extant or potentially druggable targets. Loci encoding MARK3, PDGFC, TRHR, ADORA1, GABRA2, VEGFA and PDE3A are within systems with existing drugs not currently linked to a known antihypertensive mechanism; they may offer repurposing opportunities e.g. detection of SLC5A1 as the strongest repurposing candidate in a new BP locus targeted by the type-2 diabetes drug canagliflozin. This is important as between 8-12% of patients with hypertension exhibit resistance or intolerance to current therapies and repositioning of a therapy with a known safety profile may reduce development costs.
This study strengthens our previously reported GRS analysis indicating that all BP elevating alleles combined could increase systolic BP by 10 mm Hg or more across quintiles or deciles of the population distribution, substantially increasing risk of cardiovascular events10. We previously suggested that genotyping BP elevating variants in the young may lead to targeted lifestyle intervention in early life that might attenuate the BP rise at older ages10.
We identified several BP-associated loci that are also associated with lifestyle traits, suggesting shared genetic architecture between BP and lifestyle exposures 53. We adjusted our BP GWAS analyses for BMI to control for possible confounding effects, though we acknowledge the potential for collider bias54. Nonetheless, our findings of possible genetic overlap between loci associated with BP and lifestyle exposures could support renewed focus on altering specific lifestyle measures known to affect BP55.
Despite smaller sample sizes, we observed high concordance with direction of effects on BP traits of BP variants in Africans (> 62%) and South Asians (> 72%). The GRS analyses show that, in combination, BP variants identified in European analyses are associated with BP in non-European ancestries, though effect sizes were 30-40% smaller.
Our use of a two- and one-stage GWAS design illustrates the value of this approach to minimize the effects of stochastic variation and heterogeneity. The one-stage approach included signals that had independent and concordant support (P < 0.01) from both UKB and ICBP, reducing the impact of winners’ curse on our findings. Indeed, all but two of the 210 SNPs discovered in the one-stage analysis reach P < 5 × 10-6 in either UKB or ICBP. To further minimize the risk of reporting false positive loci within our one-stage design, we set a stringent overall discovery meta-analysis P-value threshold of P < 5 × 10-9, an order of magnitude smaller than a genome-wide significance P-value, in line with thresholds recommended for whole genome sequencing22. We found high concordance in direction of effects between discovery data in the one-stage approach and the replication resources, with similar distributions of effect sizes for the two approaches. We note that 24 of the one-stage SNPs which reached P < 5 × 10-9 in discovery failed to reach genome-wide significance (P < 5 × 10-8) in the combined meta-analysis of discovery and replication resources, and hence may still require further validation in future, larger studies.
The new discoveries reported here more than triple the number of loci for BP to a total of 901 and represent a substantial advance in understanding the genetic architecture of BP. The identification of many novel genes across the genome, could partly support an omnigenic model for complex traits where genome-wide association of multiple interconnected pathways is observed. However, our strong tissue enrichment shows particular relevance to the biology of BP and cardiovascular disease56, suggesting trait-specificity, which could argue against an omnigenic model. Our confirmation of the impact of these variants on BP level and cardiovascular events, coupled with identification of shared risk variants for BP and adverse lifestyle could contribute to an early life precision medicine strategy for cardiovascular disease prevention.
Online Methods
UK Biobank (UKB) data
We performed a Genome Wide Association Study (GWAS) analysis in 458,577 UKB participants13 (Supplementary Methods). These consist of 408,951 individuals from UKB genotyped at 825,927 variants with a custom Affymetrix UK Biobank Axiom Array chip and 49,626 individuals genotyped at 807,411 variants with a custom Affymetrix UK BiLEVE Axiom Array chip from the UK BiLEVE study57, which is a subset of UKB. SNPs were imputed centrally by UKB using a reference panel that merged the UK10K and 1000 Genomes Phase 3 panel as well as the Haplotype Reference Consortium (HRC) panel58. For current analysis only SNPs imputed from the HRC panel were considered.
UKB phenotypic data
Following Quality Control (QC) (Supplementary Methods), we restricted our data to a subset of post-QC individuals of European ancestry combining information from self-reported and genetic data (Supplementary Methods) resulting in a maximum of N=458,577 individuals (Fig. 1, Supplementary Fig. 12).
Three BP traits were analysed: systolic (SBP), diastolic (DBP) and pulse pressure (PP) (difference between SBP and DBP). We calculated the mean SBP and DBP values from two automated (N=418,755) or two manual (N=25,888) BP measurements. For individuals with one manual and one automated BP measurement (N=13,521), we used the mean of these two values. For individuals with only one available BP measurement (N=413), we used this single value. After calculating BP values, we adjusted for medication use by adding 15 and 10 mmHg to SBP and DBP, respectively, for individuals reported to be taking BP-lowering medication (N=94,289)59. Descriptive summary statistics are shown in Supplementary Table 1a.
UKB analysis models
For the UKB GWAS we performed linear mixed model (LMM) association testing under an additive genetic model of the three (untransformed) continuous, medication-adjusted BP traits (SBP, DBP, PP) for all measured and imputed genetic variants in dosage format using the BOLT-LMM (v2.3) software17. We also calculated the estimated SNP-wide heritability (h2) in our data. Within the association analysis, we adjust for the following covariates: sex, age, age2, BMI and a binary indicator variable for UKB vs UK BiLEVE to account for the different genotyping chips. The analysis of all HRC-imputed SNPs was restricted to variants with MAF ≥ 1% and INFO > 0.1.
Genomic inflation and confounding
We applied the univariate LD score regression method (LDSR)18 to test for genomic inflation (expected for polygenic traits like BP, with large sample sizes, and especially also from analyses of such dense genetic data with many SNPs in high LD)60. LDSR intercepts (and standard errors) were 1.217 (0.018), 1.219 (0.020) and 1.185 (0.017) for SBP, DBP and PP respectively, and were used to adjust the UKB GWAS results for genomic inflation, prior to the meta-analysis.
International Consortium for Blood Pressure (ICBP) GWAS
ICBP GWAS is an international consortium to investigate BP genetics6. We combined previously reported post-QC GWAS data from 54 studies (N=150,134)11,12,61, with newly available GWAS data from a further 23 independent studies (N=148,890) using a fixed effects inverse variance weighted meta-analysis. The 23 studies providing new data were: ASCOT-SC, ASCOT-UK, BRIGHT, Dijon 3C, EPIC-CVD, GAPP, HCS, GS:SFHS, Lifelines, JUPITER, PREVEND, TWINSUK, GWAS-Fenland, InterAct-GWAS, OMICS-EPIC, OMICS-Fenland, UKHLS, GoDARTS-Illumina and GoDarts-Affymetrix, NEO, MDC, SardiNIA, METSIM.
All study participants were Europeans and were imputed to either the 1000 Genomes Project Phase 1 integrated release v.3 [March 2012] all ancestry reference panel62 or the HRC panel16. The final enlarged ICBP GWAS dataset included 77 cohorts (N=299,024).
Full study names, cohort information and general study methods are included in Supplementary Table 1b and in Supplementary Tables 20a-c. GC was applied at study-level. The LDSR intercepts (standard error) for the ICBP GWAS meta-analysis were 1.089 (0.012), 1.086 (0.012) and 1.066 (0.011) for SBP, DBP and PP, respectively.
Meta-analyses of discovery datasets
We performed a fixed-effects inverse variance weighted meta-analysis using METAL20,63 to obtain summary results from the UKB and ICBP GWAS, for up to N=757,601 participants and ~7.1 M SNPs with MAF ≥ 1% for variants present in both the UKB data and ICBP meta-analysis for all three traits. The LDSR intercepts (standard error), in the discovery meta-analysis of UKB and ICBP were 1.156 (0.020), 1.160 (0.021) and 1.113 (0.018) for SBP, DBP and PP respectively. The LDSR intercept (standard error), after the exclusion of all published BP variants (see below) in the discovery meta-analysis of UKB and ICBP was 1.090 (0.018), 1.097 (0.017) and 1.064 (0.015) for SBP, DBP and PP respectively, hence showing little inflation in the discovery GWAS after the exclusion of published loci (Supplementary Fig. 13). No further correction was applied to the discovery meta-analysis of UKB and ICBP GWAS.
Previously reported variants
We compiled from the peer-reviewed literature all 357 SNPs previously reported to be associated with BP at the time that our analysis was completed, that have been identified and validated as the sentinel SNP in primary analyses from previous BP genetic association studies. These 357 published SNPs correspond to 274 distinct loci, according to locus definition of: (i) SNPs within ±500kb distance of each other; (ii) SNPs in Linkage Disequilibrium (LD), using a threshold of r2 ≥ 0.1, calculated with PLINK (v2.0). We then augment this list to all SNPs present within our data, which are contained within these 274 published BP loci, i.e. all SNPs which are located ±500kb from each of the 357 published SNPs and/or in LD with any of the 357 previously validated SNPs (r2 ≥ 0.1).
Identification of novel signals: Two-stage and one-stage study designs
To identify novel signals of association with BP, two complementary study designs (which we term here “two-stage design” and “one-stage design”) were implemented in order to maximize the available data and minimize reporting of false positive associations.
Two-stage design: Overview
All of the following criteria had to be satisfied for a signal to be reported as a novel signal of association with BP using our two-stage design:
-
(i)
the sentinel SNP shows significance (P < 1 × 10-6) in the discovery meta-analysis of UKB and ICBP, with concordant direction of effect between UKB and ICBP;
-
(ii)
the sentinel SNP is genome-wide significant (P < 5 × 10-8) in the combined meta-analysis of discovery and replication (MVP and EGCUT) (replication, described below);
-
(iii)
the sentinel SNP shows support (P < 0.01) in the replication meta-analysis of MVP and EGCUT alone (Supplementary Methods);
-
(iv)
the sentinel SNP has concordant direction of effect between the discovery and the replication meta-analyses;
-
(v)
the sentinel SNP must not be located within any of the 274 previously reported loci described above.
The primary replicated trait was then defined as the BP trait with the most significant association from the combined meta-analysis of discovery and replication (in the case where a SNP was replicated for more than one BP trait.)
Two-stage design: Selection of variants from the discovery meta-analysis
We considered for follow-up SNPs in loci non-overlapping with previously reported loci according to both an LD threshold at r2 of 0.1 and a 1Mb interval region, as calculated by PLINK64. We obtained a list of such SNPs with P < 1 × 10-6 for any of the three BP traits, which also had concordant direction of effect between UKB vs ICBP (Supplementary Table 21). By ranking the SNPs by significance in order of minimum P-value across all BP traits, we performed an iterative algorithm to determine the number of novel signals (Supplementary Methods), and identify the sentinel SNP (most significant) per locus.
Two-stage design: Replication analysis
We considered SNPs with MAF ≥ 1% for an independent replication in MVP (max N=220,520)14 and in EGCUT Biobank (N=28,742)15 (Supplementary Methods). This provides a total of N=249,262 independent samples of European descent available for replication. Additional information on the analyses of the two replication datasets is provided in Supplementary Methods and in Supplementary Table 1c.
The two datasets were then combined using fixed effects inverse variance weighted meta-analysis and summary results for all traits were obtained for the replication meta-analysis dataset.
Two-stage design: Combined meta-analysis of discovery and replication meta-analyses
The meta-analyses were performed within METAL software63 using fixed effects inverse variance weighted meta-analysis (Supplementary Methods). The variants from the discovery GWAS that required proxies for replication are shown in Supplementary Table 22. The combined meta-analysis of both the discovery data (N=757,601) and replication meta-analysis (max N=249,262) provided a maximum sample size of N=1,006,863.
One-stage design: Overview
Variants that were looked-up but did not replicate according to the two-stage criteria were considered in a one-stage design. All of the following criteria had to be satisfied for a signal to be reported as a novel signal of association with BP using our one-stage criteria:
-
i)
the sentinel SNP has P < 5 × 10-9 in the discovery (UKB+ICBP) meta-analysis;
-
ii)
the sentinel SNP shows support (P < 0.01) in the UKB GWAS alone;
-
iii)
the sentinel SNP shows support (P < 0.01) in the ICBP GWAS alone;
-
iv)
the sentinel SNP has concordant direction of effect between UKB and ICBP datasets;
-
v)
The sentinel SNP must not be located within any of the 274 previously reported loci described above (Supplementary Table 4) or the recently reported non-replicated loci from Hoffman et al9 (Supplementary Table 23).
We selected the one-stage P-value threshold to be an order of magnitude more stringent than a genome-wide significance P-value, so as to ensure robust results and to minimize false positive findings. The threshold of P < 5 × 10-9 has been proposed as a more conservative statistical significance threshold, e.g. for whole-genome sequencing-based studies21.
Selection of variants from the meta-analysis of UKB and ICBP was performed as described above for the two-stage design.
Conditional Analysis
We performed conditional analyses using the GWAS discovery meta-analysis data, in order to identify any independent secondary signals in addition to the sentinel SNPs at the 901 loci. We used two different methodological approaches, each using the Genome-wide Complex Traits Analysis (GCTA) software22: (i) full “genome-wide conditional analysis” with joint multivariate analysis and stepwise model selection across all three BP traits; and (ii) “locus-specific conditional analysis” for the primary BP trait conditioning on the sentinel SNPs within each locus (Supplementary Methods). For robustness, secondary signals are only reported if obtained from both approaches. All secondary signals were selected at genome-wide significance level, with MAF ≥ 1% and confirmed to be pairwise-LD-independent (r2 < 0.1), as well as not being in LD with any of the published or sentinel SNPs at any of the 901 BP-associated loci (r2 < 0.1). In all cases the UKB data was used as the reference genetic data for LD calculation, restricted to individuals of European ancestry only.
Functional analyses: Variants
We used an integrative bioinformatics approach to collate functional annotation at both the variant level (for each sentinel SNP within all BP loci) and the gene level (using SNPs in LD r2 ≥ 0.8 with the sentinel SNPs). At the variant level, we use Variant Effect Predictor (VEP) to obtain comprehensive characterization of variants, including consequence (e.g. downstream or non-coding transcript exon), information on nearest genomic features and, where applicable, amino acid substitution functional impact, based on SIFT and PolyPhen. The biomaRt R package is used to further annotate the nearest genes.
We evaluated all SNPs in LD (r2 ≥ 0.8) with our novel sentinel SNPs for evidence of mediation of expression quantitative trait loci (eQTL) in all 44 tissues using the Genotype-Tissue Expression (GTEx) database, to highlight specific tissue types which show eQTLs for a larger than expected proportion of novel loci. We further seek to identify novel loci with the strongest evidence of eQTL associations in arterial tissue, in particular. A locus is annotated with a given eGene only if the most significant eQTL SNP for the given eGene is in high LD (r2 ≥ 0.8) with the sentinel SNP, suggesting that the eQTL signal co-localises with the sentinel SNP.
We annotated nearest genes, eGenes (genes whose expression is affected by eQTLs) and Hi-C interactors with HUVEC, HVMSC and HAEC expression from the Fantom5 project. Genes that had higher than median expression levels in the given cell types were indicated as expressed.
To identify SNPs in the novel loci that have a non-coding functional effect (influence binding of transcription factors or RNA polymerase, or influence DNase hypersensitivity sites or histone modifications), we used DeepSEA, a deep learning algorithm, that learnt the binding and modification patterns of ~900 cell/factor combinations65. A change of >0.1 in the binding score predicted by DeepSEA for the reference and alternative alleles respectively was used as cut-off to find alleles with non-coding functional effect (Supplementary Methods)
We identified potential target genes of regulatory SNPs using long-range chromatin interaction (Hi-C) data from HUVECs23, aorta, adrenal glands, neural progenitor and mesenchymal stem cell, which are tissues and cell types that are considered relevant for regulating BP24. We find the most significant promoter interactions for all potential regulatory SNPs (RegulomeDB score ≤ 5) in LD (r2 ≥ 0.8) with our novel sentinel SNPs and published SNPs, and choose the interactors with the SNPs of highest regulatory potential to annotate the loci.
We then performed overall enrichment testing across all loci. Firstly, we used DEPICT66 (Data-driven Expression Prioritized Integration for Complex Traits) to identify tissues and cells which are highly expressed at genes within the BP loci (Supplementary Methods). Secondly, we used DEPICT to test for enrichment in gene sets associated with biological annotations (manually curated and molecular pathways, phenotype data from mouse KO studies) (Supplementary Methods). We report significant enrichments with a false discovery rate <0.01. The variants tested were i) the 357 published BP associated SNPs at the time of analysis and ii) a set including all (published and novel) variants (with novel SNPs filtered by highest significance, P < 1 × 10-12).
Furthermore, to investigate cell type specific enrichment within DNase I sites, we used FORGE, which tests for enrichment of SNPs within DNase I sites in 123 cell types from the Epigenomics Roadmap Project and ENCODE25 (Supplementary Methods). Two analyses were compared (i) using published SNPs only; (ii) using sentinel SNPs at all 901 loci, in order to evaluate the overall tissue specific enrichment of BP associated variants.
Functional analyses: Genes
At the gene level, we used Ingenuity Pathway Analysis (IPA) software (IPA®, QIAGEN Redwood City) to review genes with prior links to BP, based on annotation with the “Disorder of Blood Pressure”, “Endothelial Development” and “Vascular Disease” Medline Subject Heading (MESH) terms. We used the Mouse Genome Informatics (MGI) tool to identify BP and cardiovascular relevant mouse knockout phenotypes for all genes linked to BP in our study. We also used IPA to identify genes that interact with known targets of anti-hypertensive drugs. Genes were also evaluated for evidence of small molecule druggability or known drugs based on queries of the Drug Gene Interaction database.
Lookups in non-European ancestries
As a secondary analysis, we look up all known and novel BP-associated SNPs in Africans (7,782) and South Asians (10,322) from UKB using BOLT-LMM analysis for each BP trait within each ancestry (Supplementary Methods).
Effects on other traits and diseases
We queried SNPs against GWAS catalog26 and PhenoScanner27, including genetics and metabolomics databases, to investigate cross-trait effects, extracting all association results with genome-wide significance at P < 5 × 10-8 for all SNPs in high LD (r2 ≥ 0.8) with the 535 sentinel novel SNPs, to highlight the loci with strongest evidence of association with other traits. We further evaluated these effects using DisGeNET28,29. At the gene level, overrepresentation enrichment analysis (ORA) with WebGestalt67 on the nearest genes to all BP loci was carried out. Moreover, we tested sentinel SNPs at all published and novel (N=901) loci for association with lifestyle related data including food, water and alcohol intake, anthropomorphic traits and urinary sodium, potassium and creatinine excretion using the recently developed Stanford Global Biobank Engine and the Gene ATLAS68. Both are search engines for GWAS findings for multiple phenotypes in UK Biobank. We used a Bonferroni corrected significance threshold of P < 1 × 10-6 to deem significance.
Genetic risk scores and percentage of variance explained
We calculated a weighted genetic risk score (GRS) (Supplementary Table 24) to provide an estimate of the combined effect of the BP raising variants on BP and risk of hypertension and applied this to the UKB data (Supplementary Methods). Our analysis included 423,713 unrelated individuals of European ancestry of whom 392,092 individuals were free of cardiovascular events at baseline.
We assessed the association of the continuous GRS variable on BP and with the risk of hypertension, with and without adjustment for sex. We then compared BP levels and risk of hypertension, respectively, for individuals in the top vs bottom quintiles of the GRS distribution. Similar analyses were performed for the top vs bottom deciles of the GRS distribution. All analyses were restricted to the 392,092 unrelated individuals of European ancestry from UKB. As a sensitivity analysis to assess for evidence of bias in the UKB results, we also carried out similar analyses in Airwave, an independent cohort of N=14,004 unrelated participants of European descent30 (Supplementary Methods).
We calculated the association of the GRS with cardiovascular disease in unrelated participants in UKB data, based on self-reported medical history, and linkage to hospitalization and mortality data (Supplementary Table 25). We use logistic regression with binary outcome variables for composite incident cardiovascular disease (Supplementary Methods), incident myocardial infarction and incident stroke (using the algorithmic UKB definitions) and GRS as explanatory variable (with and without sex adjustment).
We also assessed the association of this GRS with BP in unrelated individuals Africans (N=6,970) and South Asians (N=8,827) from the UKB to see whether BP-associated SNPs identified from GWAS predominantly in Europeans are also associated with BP in populations of non-European ancestry.
We calculated the percentage of variance in BP explained by genetic variants using the independent Airwave cohort (N=14,004) (Supplementary Methods). We considered three different levels of the GRS: (i) all pairwise-independent, LD-filtered (r2 < 0.1) published SNPs within the known loci; (ii) all known SNPs and sentinel SNPs at novel loci; (iii) all independent signals at all 901 known and novel loci including the 163 secondary SNPs.
Supplementary Material
Acknowledgements
H.R.W. was funded by the National Institute for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Centre at Barts and The London School of Medicine and Dentistry. D.M-A is supported by the Medical Research Council [grant number MR/L01632X.1]. B.M. holds an MRC eMedLab Medical Bioinformatics Career Development Fellowship, funded from award MR/L016311/1. H.G. was funded by the NIHR Imperial College Health Care NHS Trust and Imperial College London Biomedical Research Centre. C.P.C. was funded by the National Institute for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Center at Barts and The London School of Medicine and Dentistry. S.T. was supported by Canadian Institutes of Health Research; Université Laval (Quebec City, Canada). G.P. was supported by Canada Research Chair in Genetic and Molecular Epidemiology and CISCO Professorship in Integrated Health Biosystems. I.K. was supported by the EU PhenoMeNal project (Horizon 2020, 654241). C.P.K. is supported by grant U01DK102163 from the NIH-NIDDK, and by resources from the Memphis VA Medical Center. C.P.K. is an employee of the US Department of Veterans affairs. Opinions expressed in this paper are those of the authors’ and do not necessarily represent the opinion of the Department of Veterans Affairs. S.D. was supported for this work by grants from the European Research Council (ERC), the EU Joint Programme - Neurodegenerative Disease Research (JPND), the Agence Nationale de la Recherche (ANR). T.B., J.MART., V.V., A.F.W. and C.H. were supported by a core MRC grant to the MRCHGU QTL in Health and Disease research programme. M.BOE is supported by NIH grant R01-DK062370. H.W. and A.G. acknowledge support of the Tripartite Immunometabolism Consortium [TrIC], Novo Nordisk Foundation (grant NNF15CC0018486). N.V. was supported by Marie Sklodowska-Curie GF grant (661395) and ICIN-NHI. C.M. is funded by the MRC AimHy (MR/M016560/1) project grant. M.A.N participation is supported by a consulting contract between Data Tecnica International and the National Institute on Aging, NIH, Bethesda, MD, USA. M.BR., M.CO., I.G., P.G., G.G, A.MO., A.R., D.V., C.M.B., C.F.S., M.T., D.T. were supported by the Italian Ministry of Health RF2010 to Paolo Gasparini, RC2008 to Paolo Gasparini. D.I.B. is supported by the Royal Netherlands Academy of Science Professor Award (PAH/6635). J.C.C. is supported by the Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research Investigator (STaR) Award (NMRC/STaR/0028/2017). C.C., P.B.M and M.R.B were funded by the National Institutes for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Centre at Barts. T.F. is supported by the NIHR Biomedical Research Centre, Oxford. M.R. is recipient from China Scholarship Council (No. 2011632047). C.L. was supported by the Medical Research Council UK (G1000143; MC_UU_12015/1; MC_PC_13048; MC_U106179471), Cancer Research UK (C864/A14136), EU FP6 programme (LSHM_CT_2006_037197). G.B.E is supported by the Swiss National Foundation SPUM project FN 33CM30-124087, Geneva University, and the Fondation pour Recherches Médicales, Genève. C.M.L is supported by the Li Ka Shing Foundation, WT-SSI/John Fell funds and by the NIHR Biomedical Research Centre, Oxford, by Widenlife and NIH (CRR00070 CR00.01). R.J.F.L. is supported by the NIH (R01DK110113, U01HG007417, R01DK101855, R01DK107786). D.O.M-K. is supported by Dutch Science Organization (ZonMW-VENI Grant 916.14.023). M.M was supported by the National Institute for Health Research (NIHR) BioResource Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. H.W. and M.F. acknowledge the support of the Wellcome Trust core award (090532/Z/09/Z) and the BHF Centre of Research Excellence (RE/13/1/30181). A.G, H.W. acknowledge European Union Seventh Framework Programme FP7/2007-2013 under grant agreement no. HEALTH-F2-2013-601456 (CVGenes@Target) & and A.G, the Wellcome Trust Institutional strategic support fund. L.R. was supported by Forschungs- und Förder-Stiftung INOVA, Vaduz, Liechtenstein. M.TO. is supported by British Heart Foundation (PG/17/35/33001). P.S. is recipient of an NIHR Senior Investigator Award and is supported by the Biomedical Research Centre Award to Imperial College Healthcare NHS Trust. P.v.d.H. was supported by ICIN-NHI and Marie Sklodowska-Curie GF (call: H2020-MSCA-IF-2014, Project ID: 661395). N.J.W. was supported by the Medical Research Council UK (G1000143; MC_UU_12015/1; MC_PC_13048; MC_U106179471), Cancer Research UK (C864/A14136), EU FP6 programme (LSHM_CT_2006_037197). E.Z. was supported by the Wellcome Trust (WT098051). J.N.H. was supported by the Vanderbilt Molecular and Genetic Epidemiology of Cancer (MAGEC) training program, funded by T32CA160056 (PI: X.-O. Shu) and by VA grant 1I01CX000982. A.G. was supported by VA grant 1I01CX000982. T.L.E. and D.R.V.E. were supported by grant R21HL121429 from NIH/NHLBI. A.M.H. was supported by VA Award #I01BX003360. C.J.O. was supported by the VA Boston Healthcare, Section of Cardiology and Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School. The MRC/BHF Cardiovascular Epidemiology Unit is supported by the UK Medical Research Council [MR/L003120/1]; British Heart Foundation [RG/13/13/30194]; and UK National Institute for Health Research Cambridge Biomedical Research Centre. J.DA is a British Heart Foundation Professor and NIHR Senior Investigator. L.V.W. holds a GlaxoSmithKline/British Lung Foundation Chair in Respiratory Research. P.E. acknowledges support from the NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London, the NIHR Health Protection Research Unit in Health Impact of Environmental Hazards (HPRU-2012-10141), and the Medical Research Council (MRC) and Public Health England (PHE) Centre for Environment and Health (MR/L01341X/1). P.E. is a UK Dementia Research Institute (DRI) professor, UK DRI at Imperial College London, funded by the MRC, Alzheimer’s Society and Alzheimer’s Research UK. He is also associate director of Health Data Research-UK London funded by a consortium led by the Medical Research Council. M.J.C. was funded by the National Institute for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Center at Barts and The London School of Medicine and Dentistry. M.J.C. is a National Institute for Health Research (NIHR) senior investigator and this work is funded by the MRC eMedLab award to M.J.C. and M.R.B. and the NIHR Biomedical Research Centre at Barts.
This research has been conducted using the UK Biobank Resource under Application Numbers 236 and 10035. This research was supported by the British Heart Foundation (grant SP/13/2/30111). Large-scale comprehensive genotyping of UK Biobank for cardiometabolic traits and diseases: UK CardioMetabolic Consortium (UKCMC).
Computing: This work was enabled using the computing resources of the i) UK MEDical BIOinformatics partnership - aggregation, integration, visualisation and analysis of large, complex data (UK MED-BIO) which is supported by the Medical Research Council [grant number MR/L01632X/1] and ii) the MRC eMedLab Medical Bioinformatics Infrastructure, supported by the Medical Research Council [grant number MR/L016311/1].
URLs
FORGE: http://browser.1000genomes.org/Homo_sapiens/UserData/Forge?db=core
Fantom5 data: http://fantom.gsc.riken.jp/5/
ENCODE DNase I data: (wgEncodeAwgDnaseMasterSites; accessed using Table browser)
ENCODE cell type data: http://genome.ucsc.edu/ENCODE/cellTypes.html.
GTEx: www.gtexportal.org
DeepSEA: http://deepsea.princeton.edu/
WebGestalt: http://www.webgestalt.org
Mouse Genome Informatics (MGI): http://www.informatics.jax.org/batch
Drug Gene Interaction database: www.dgidb.org
PhenoScanner: http://www.phenoscanner.medschl.cam.ac.uk (Phenoscanner integrates results from the GWAS catalogue: https://www.ebi.ac.uk/gwas/ and GRASP: https://grasp.nhlbi.nih.gov/)
DisGeNEt: http://www.disgenet.org
GeneATLAS: http//geneatlas.roslin.ed.ac.uk
Global Biobank Engine: https://biobankengine.stanford.edu
Data availability statement
The UKB GWAS data can be assessed from the UK Biobank data repository (http://biota.osc.ox.ac.uk/). The genetic and phenotypic UKB data are available upon application to the UK Biobank (https://www.ukbiobank.ac.uk). ICBP summary data can be assessed through request to ICBP steering committee. Contact Mark Caulfield (m.j.caulfield@qmul.ac.uk) or Paul Elliott (p.elliott@imperial.ac.uk) to apply for access to the data. The UKB+ICBP summary data can be assessed through request to Paul Elliott (p.elliott@imperial.ac.uk) or Mark Caulfield (m.j.caulfield@qmul.ac.uk). All replication data generated during this study are included in the published article. For example, association results of look-up variants from our replication analyses and the subsequent combined meta-analyses are contained within the Supplementary Tables provided.
Ethics Statement
The UKB study has approval from the North West Multi-Centre Research Ethics Committee. Any participants from UKB who withdrew consent have been removed from our analysis. Each cohort within the ICBP meta-analysis as well as our independent replication cohorts of MVP and EGCUT had ethical approval locally. More information on the participating cohorts is available in Supplementary Methods.
Conflicts/Disclosures
K.W. is a Commercial partnerships manager for Genomics England, a UK Government Company
M.A.N. consults for Illumina Inc, the Michael J. Fox Foundation and University of California Healthcare among others.
A.S.B. has received grants outside of this work from Merck, Pfizer, Novartis, AstraZeneca, Biogen and Bioverativ and personal fees from Novartis
J.DA. has the following competing interests: Pfizer Population Research Advisory Panel (grant), AstraZeneca (grant), Wellcome Trust (grant), UK Medical Research Council (grant), Pfizer(grant), Novartis (grant), NHS Blood and Transplant(grant), National Institute of Health Research(grant), UK MEDICAL RESEARCH COUNCIL(grant), BRITISH HEART FOUNDATION(grant),UK NATIONAL INSTITUTE OF HEALTH RESEARCH (grant), EUROPEAN COMMISSION (grant), Merck Sharp and Dohme UK Atherosclerosis (personal fees), Novartis Cardiovascular and Metabolic Advisory Board (personal fees), British Heart Foundation (grant), European Research Council (grant), Merck (grant).
B.M.P. serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson.
M.J.C. is Chief Scientist for Genomics England, a UK Government company.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U. S. Department of Health and Human Services. This publication does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Author contributions
Central analysis: E.E., H.R.W., D.M-A., B.M., R.P., H.G., G.N., N.D., C.P.C., I.K., F.N., M.E., K.W., E.T. L.V.W.
Writing of the manuscript: E.E., H.R.W., D.M-A., B.M., R.P., H.G., I.T., M.R.B., L.V.W., P.E., M.J.C. (with group leads EE, H.R.W, L.V.W., P.E., M.J.C.)
ICBP-Discovery contributor: (3C-Dijon) S.D., M.S., P.AM., G.C., C.T.; (AGES-Reykjavik) V.GU., L.J.L., A.V.S., T.B.H.; (ARIC) D.E.A., E.B., A.CH. A.C.M., P.N.; (ASCOT) N.R.P., D.C.S., A.S., S.THO., P.B.M., P.S., M.J.C., H.R.W.; (ASPS) E.H., Y.S., R.S., H.S.; (B58C) D.P.S., BHSA.J., N.SHR.; (BioMe (formerly IPM)) E.P.B., Y.LU., R.J.F.L.; (BRIGHT) J.C., M.F., M.J.B., P.B.M., M.J.C., H.R.W. ; (CHS) J.C.B., K.R., K.D.T., B.M.P.; (Cilento study) M.C., T.NU., D.R., R.SO.; (COLAUS) M.B., Z.K., P.V.; (CROATIA_Korcula) J.MART., A.F.W.; (CROATIA_SPLIT) I.KO., O.P., T.Z.; (CROATIA_Vis) J.E.H., I.R., V.V.; (EPIC) K-T.K., R.J.F.L., N.J.W.; (EPIC-CVD) W-Y.L., P.SU., A.S.B., J.DA., J.M.M.H.; (EPIC-Norfolk, Fenland-OMICS, Fenland-GWAS) J-H.Z.; (EPIC-Norfolk, Fenland-OMICS, Fenland-GWAS, InterAct-GWAS) J.L., C.L., R.A.S., N.J.W.; (ERF) N.A., B.A.O., C.M.v.D.; (Fenland-Exome, EPIC-Norfolk-Exome) S.M.W., FHSS-J.H., D.L.; (FINRISK (COROGENE_CTRL)) P.J., K.K., M.P., A-P.S.; (FINRISK_PREDICT_CVD) A.S.H., A.P., S.R., V.S.; (FUSION) A.U.J, M.BOE., F.C., J.T., (GAPP) S.T., G.P., D.CO., L.R.; (Generation Scotland (GS:SFHS)) T.B., C.H., A.C., S.P.; (GoDARTs) N.S., A.S.F.D., A.D.M., C.N.A.P.; (GRAPHIC) P.S.B., C.P.N., N.J.SA., M.D.T.; (H2000_CTRL) A.JU., P.K., S.KO., T.N.; (HABC) Y.L., M.A.N., T.B.H.; (HCS) J.R.A., E.G.H., C.O., R.J.SC.; (HTO) K.L.A., H.J.C., B.D.K., M.TO, C.MA.; (ICBP-SC) G.A., T.F., M-R.J., A.D.J., M.LA., C.N.; (INGI-CARL) I.G., G.G., A.MO., A.R.; (INGI-FVG) M.BR., M.CO., P.G., D.V.; (INGI-VB) C.M.B., C.F.S., D.T., M.T.; (JUPITER) F.G., L.M.R., P.M.R., D.I.C.; (KORA S3) C.G., M.L., E.O., S.S.; (KORA S4) A.PE., J.S.R.; (LBC1921) S.E.H., D.C.M.L., A.PA., J.M.S.; (LBC1936) G.D., I.J.D., A.J.G., L.M.L.; (Lifelines) N.V., M.H.d.B., M.A.S., P.v.d.H.; (LOLIPOP) J.C.C., J.S.K., B.L., W.Z.; (MDC) P.A., O.M.; (MESA) X.G., W.P., J.I.R., J.Y.; (METSIM) A.U.J., M.LAA.; (MICROS) F.D.G.M., A.A.H., P.P.P.; (MIGEN) R.E., S.K., J.M., D.SI.; (NEO) R.L., R.d.M., R.N., D.O.M-K.; (NESDA) Y.M., I.M.N., B.W.J.H.P., H.SN.; (NSPHS) S.E., U.G., Å.JO.; (NTR) D.I.B., E.J.d.G., J-J.H., G.W.; (ORCADES) H.C., P.K.J., S.H.W., J.F.W.; (PIVUS) L.LI., C.M.L., J.S., A.M.; (Prevend) N.V., P.v.d.H.; (PROCARDIS) M.F., A.G., H.W.; (PROSPER) J.DE., J.W.J., D.J.S., S.TR.; (RS) O.H.F., A.HO., A.U., G.C.V.; (SardiNIA) J.D., Y.Q., F.CU., E.G.L.; (SHIP) M.D., R.R., A.T., U.V.; (STR) M.FR., A.H., R.J.S., E.I.; (TRAILS) C.A.H., A.J.O., H.R., P.J.v.d.M.; (TwinsUK) M.M., C.M., T.D.S.; (UKHLS) B.P.P., E.Z.; (ULSAM) V.G., A.P.M., A.M., E.I.; (WGHS) F.G., L.M.R., P.M.R., D.I.C.; (YFS) M.K., T.L., L-P.L., O.T.R.
Replication study contributor: (MVP) J.N.H., A.G., D.R.V.E., Y.V.S., K.C., J.M.G., P.W.F.W., P.S.T., C.P.K., A.M.H., C.J.O., T.L.E.; (EGCUT) T.E., R.M., L.M. A.ME.
Airwave Health Monitoring Study: E.E, H.G, A-C.V., R.P., I.K., I.T., P.E.
All authors critically reviewed and approved the final version of the manuscript
References
- 1.Forouzanfar MH, et al. Global Burden of Hypertension and Systolic Blood Pressure of at Least 110 to 115 mm Hg, 1990-2015. JAMA. 2017;317:165–182. doi: 10.1001/jama.2016.19043. [DOI] [PubMed] [Google Scholar]
- 2.Munoz M, et al. Evaluating the contribution of genetics and familial shared environment to common disease using the UK Biobank. Nat Genet. 2016;48:980–3. doi: 10.1038/ng.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulter NR, Prabhakaran D, Caulfield M. Hypertension. Lancet. 2015;386:801–12. doi: 10.1016/S0140-6736(14)61468-9. [DOI] [PubMed] [Google Scholar]
- 4.Feinleib M, et al. The NHLBI twin study of cardiovascular disease risk factors: methodology and summary of results. Am J Epidemiol. 1977;106:284–5. doi: 10.1093/oxfordjournals.aje.a112464. [DOI] [PubMed] [Google Scholar]
- 5.Cabrera CP, et al. Exploring hypertension genome-wide association studies findings and impact on pathophysiology, pathways, and pharmacogenetics. Wiley Interdiscip Rev Syst Biol Med. 2015;7:73–90. doi: 10.1002/wsbm.1290. [DOI] [PubMed] [Google Scholar]
- 6.Ehret GB, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surendran P, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–70. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann TJ, et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2017;49:54–64. doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren HR, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wain LV, et al. Novel Blood Pressure Locus and Gene Discovery Using Genome-Wide Association Study and Expression Data Sets From Blood and the Kidney. Hypertension. 2017 doi: 10.1161/HYPERTENSIONAHA.117.09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Consortium for Blood Pressure Genome-Wide Association Studies et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaziano JM, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–23. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 15.Leitsalu L, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 2015;44:1137–47. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14:379–89. doi: 10.1038/nrg3472. [DOI] [PubMed] [Google Scholar]
- 21.Pulit SL, de With SA, de Bakker PI. Resetting the bar: Statistical significance in whole-genome sequencing-based association studies of global populations. Genet Epidemiol. 2017;41:145–151. doi: 10.1002/gepi.22032. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt AD, et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunham IKE, Iotchkova V, Morganella S, Birney E. FORGE: A tool to discover cell specific enrichments of GWAS associated SNPs in regulatory regions. F1000Research. 2015;4 [Google Scholar]
- 26.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinero J, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015 doi: 10.1093/database/bav028. bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinero J, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott P, et al. The Airwave Health Monitoring Study of police officers and staff in Great Britain: rationale, design and methods. Environ Res. 2014;134:280–5. doi: 10.1016/j.envres.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 31.Ehret GB, Caulfield MJ. Genes for blood pressure: an opportunity to understand hypertension. Eur Heart J. 2013;34:951–61. doi: 10.1093/eurheartj/ehs455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blood Pressure Lowering Treatment Trialists C et al. Blood pressure-lowering treatment based on cardiovascular risk: a meta-analysis of individual patient data. Lancet. 2014;384:591–8. doi: 10.1016/S0140-6736(14)61212-5. [DOI] [PubMed] [Google Scholar]
- 33.GBD 2015 Risk Factors Collaborators. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1659–1724. doi: 10.1016/S0140-6736(16)31679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakao E, et al. Elevated Plasma Transforming Growth Factor beta1 Levels Predict the Development of Hypertension in Normotensives: The 14-Year Follow-Up Study. Am J Hypertens. 2017;30:808–814. doi: 10.1093/ajh/hpx053. [DOI] [PubMed] [Google Scholar]
- 35.Feng W, Dell'Italia LJ, Sanders PW. Novel Paradigms of Salt and Hypertension. J Am Soc Nephrol. 2017;28:1362–1369. doi: 10.1681/ASN.2016080927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International PPH Consortium et al. Heterozygous germline mutations in BMPR2, encoding a TGF-beta receptor, cause familial primary pulmonary hypertension. Nat Genet. 2000;26:81–4. doi: 10.1038/79226. [DOI] [PubMed] [Google Scholar]
- 37.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Douma S, et al. Prevalence of primary hyperaldosteronism in resistant hypertension: a retrospective observational study. Lancet. 2008;371:1921–6. doi: 10.1016/S0140-6736(08)60834-X. [DOI] [PubMed] [Google Scholar]
- 39.Rossi GP, et al. A prospective study of the prevalence of primary aldosteronism in 1,125 hypertensive patients. J Am Coll Cardiol. 2006;48:2293–300. doi: 10.1016/j.jacc.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 40.Calhoun DA, Nishizaka MK, Zaman MA, Thakkar RB, Weissmann P. Hyperaldosteronism among black and white subjects with resistant hypertension. Hypertension. 2002;40:892–6. doi: 10.1161/01.hyp.0000040261.30455.b6. [DOI] [PubMed] [Google Scholar]
- 41.Drelon C, Berthon A, Mathieu M, Martinez A, Val P. Adrenal cortex tissue homeostasis and zonation: A WNT perspective. Mol Cell Endocrinol. 2015;408:156–64. doi: 10.1016/j.mce.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 42.El Wakil A, Lalli E. The Wnt/beta-catenin pathway in adrenocortical development and cancer. Mol Cell Endocrinol. 2011;332:32–7. doi: 10.1016/j.mce.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 43.Teo AE, et al. Pregnancy, Primary Aldosteronism, and Adrenal CTNNB1 Mutations. N Engl J Med. 2015;373:1429–36. doi: 10.1056/NEJMoa1504869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tissier F, et al. Mutations of beta-catenin in adrenocortical tumors: activation of the Wnt signaling pathway is a frequent event in both benign and malignant adrenocortical tumors. Cancer Res. 2005;65:7622–7. doi: 10.1158/0008-5472.CAN-05-0593. [DOI] [PubMed] [Google Scholar]
- 45.Oliveira-Paula GH, et al. Polymorphisms in VEGFA gene affect the antihypertensive responses to enalapril. Eur J Clin Pharmacol. 2015;71:949–57. doi: 10.1007/s00228-015-1872-5. [DOI] [PubMed] [Google Scholar]
- 46.Yang R, et al. Hypertension and endothelial dysfunction in apolipoprotein E knockout mice. Arterioscler Thromb Vasc Biol. 1999;19:2762–8. doi: 10.1161/01.atv.19.11.2762. [DOI] [PubMed] [Google Scholar]
- 47.Sofat R, et al. Circulating Apolipoprotein E Concentration and Cardiovascular Disease Risk: Meta-analysis of Results from Three Studies. PLoS Med. 2016;13:e1002146. doi: 10.1371/journal.pmed.1002146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrad KP. Unveiling the vasodilatory actions and mechanisms of relaxin. Hypertension. 2010;56:2–9. doi: 10.1161/HYPERTENSIONAHA.109.133926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun HJ, et al. Relaxin in paraventricular nucleus contributes to sympathetic overdrive and hypertension via PI3K-Akt pathway. Neuropharmacology. 2016;103:247–56. doi: 10.1016/j.neuropharm.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto Y, et al. Phosphatidylinositol 3-kinase inhibition induces vasodilator effect of sevoflurane via reduction of Rho kinase activity. Life Sci. 2017;177:20–26. doi: 10.1016/j.lfs.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Pawlak JB, Wetzel-Strong SE, Dunn MK, Caron KM. Cardiovascular effects of exogenous adrenomedullin and CGRP in Ramp and Calcrl deficient mice. Peptides. 2017;88:1–7. doi: 10.1016/j.peptides.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohtsu H, et al. Signal-crosstalk between Rho/ROCK and c-Jun NH2-terminal kinase mediates migration of vascular smooth muscle cells stimulated by angiotensin II. Arterioscler Thromb Vasc Biol. 2005;25:1831–6. doi: 10.1161/01.ATV.0000175749.41799.9b. [DOI] [PubMed] [Google Scholar]
- 53.Tzoulaki I, Elliott P, Kontis V, Ezzati M. Worldwide Exposures to Cardiovascular Risk Factors and Associated Health Effects: Current Knowledge and Data Gaps. Circulation. 2016;133:2314–33. doi: 10.1161/CIRCULATIONAHA.115.008718. [DOI] [PubMed] [Google Scholar]
- 54.Munafo MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol. 2017;47:226–235. doi: 10.1093/ije/dyx206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pazoki R, et al. Genetic predisposition to high blood pressure and lifestyle factors: Associations with midlife blood pressure levels and cardiovascular events. Circulation. 2018;137:653–661. doi: 10.1161/CIRCULATIONAHA.117.030898. [DOI] [PubMed] [Google Scholar]
- 56.Boyle EA, Li YI, Pritchard JK. An expanded view of complex traits. From polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–81. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bycroft CFC, Petkova D, Band G, Elliot LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O'Conell J, Cortes A, Welsh S, et al. Genome-wide geentic data on 500,000 UK Biobank Participants. bioRxiv. 2017 166298. [Google Scholar]
- 59.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 60.Marouli E, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wain LV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–11. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–4. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6 doi: 10.1038/ncomms6890. 5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Canela-Xandri O, Konrad R, Tenesa Albert. An atlas of genetic associations in UK Biobank. bioRxiv. 2017 doi: 10.1038/s41588-018-0248-z. 176834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.