Abstract
There is an urgent need of sensitive bioanalytical platforms for sensitive and precise quantification of low-abundance microRNA targets in complex biological samples, including liquid biopsies of tumors. Many of current miRNA biosensing methods require laborious sample pretreatment procedures, including extraction of total RNA, which largely limits their biomedical and clinical applications. Herein we developed an integrated Microfluidic Exponential Rolling Circle Amplification (MERCA) platform for sensitive and specific detection of microRNAs directly in minimally processed samples. The MERCA system integrates and streamlines solid-phase miRNA isolation, miRNA-adapter ligation, and a dualphase exponential rolling circle amplification (eRCA) assay in one analytical workflow. By marrying the advantages of microfluidics in leveraging bioassay performance with the high sensitivity of eRCA, our method affords a remarkably low limit of detection at <10 zeptomole levels, with the ability to discriminate single-nucleotide difference. Using the MERCA chip, we demonstrated quantitative detection of miRNAs in total RNA, raw cell lysate, and cellderived exosomes. Comparing with the parallel TaqMan RT-qPCR measurements verified the adaptability of the MERCA system for detection of miRNA biomarkers in complex biological materials. In particular, high sensitivity of our method enables direct detection of low-level exosomal miRNAs in as few as 2 × 106 exosomes. Such analytical capability immediately addresses the unmet challenge in sample consumption, a key setback in clinical development of exosome-based liquid biopsies. Therefore, the MERCA would provide a useful platform to facilitate miRNA analysis in broad biological and clinical applications.
Keywords: Microfluidic Exponential Rolling Circle Amplification (MERCA), microRNA detection, raw cell lysate, cell-derived exosomes, complex biological samples
1. Introduction
MicroRNAs (miRNAs) are small, endogenous, noncoding RNAs that have recently emerged as promising diagnostic markers for many diseases including cancer, Alzheimer’s, and diabetes because of their unique dysregulation patterns and great stability [1–4]. However, it is still challenging to accurately and specifically quantify this increasingly important biomarkers in complex biological samples due to the high level of sequence homology, small size, low abundance in total RNA samples, and common secondary structures of miRNAs [57]. Standard techniques, such as northern blotting [8, 9] and reverse transcription polymerase chain reaction (RT-PCR) [11,12], require large sample size, time-consuming sample processing, and sophisticated analytical procedures [13]. Recently, RCA has attracted wide attention as a robust isothermal amplification method with unique advantages in simplicity, specificity, robustness and sensitivity [14–16]. Since the Kjems group first reported RCA-based padlock probes for miRNA detection [17], several variants have been developed to enhance the sensitivity and specificity of this method, which include a target-primed branched rolling circle amplification (BRCA) reaction using a second primer [18], exponential RCA based on the padlock probes designed with nicking sites [19], and a dumbbell DNA probe-mediated RCA method [20]. However, these methods mostly demand sample preparation steps, e.g., total RNA extraction, which limit their applicability to complex biological samples.
Increasing efforts have recently been invested on developing new methods for quantitative sensing of miRNA biomarkers in complex biological samples without the need for manual, time-consuming sample treatment [21–23]. Because of its unique advantages for bioanalysis, microfluidic technology has been adapted to leverage various miRNA assays, such as digital RT-PCR for miRNA expression analysis and single cell analysis [24], sandwich hybridization assay coupled with dendritic amplification [25], encoded hydrogel particles or on-chip gel microreactors for multiplexed miRNA detection [26–28], and electrochemical miRNA sensing [29–31]. It is obvious that the majority of these microfluidic methods integrate miRNA capture by hybridization which affords sensitive and specific detection of miRNAs in complex biological samples, including crude cell lysates [28], human plasma/ serum [27], and human cerebrospinal fluid [29]. Despite the promising strength of microfluidics, to our knowledge, very few studies have been reported to adapt it to transform RCA. The Doyle group developed encoded gel microparticles modified with probes for miRNA capture and in-gel RCA amplification [27]. Kitamori et.al reported a microbead-based microchip for solid-phase capture and padlock probe-triggered RCA detection of DNA [32].
Herein, we report on an integrated Microfluidic Exponential Rolling Circle Amplification (MERCA) platform capable of highly sensitive and specific miRNA analysis with complex biological samples. As a proof-of-concept, the MERCA system was assessed for quantitative miRNA detection directly from various biological samples: purified total RNA, crude whole cell lysate, and cell-derived exosomes. Exosomes are 40–150 nm membranous extracellular vesicles secreted by cells into the circulation and hold great clinical potential as non-invasive liquid biopsy owing to their roles in many biological processes, including tumor progression, metastasis, and drug resistance [33–36]. Exosomes were found to provide enriched and protective source of miRNAs in body fluids [37–39]. Standard RT-PCR analysis of exosomal RNAs suffers from variations in RNA extraction and amplification (e.g., sequence bias) which result in limited specificity and reproducibility of reported miRNA markers [40, 41]. A variety of sensitive approaches have been adapted to detect exosomal miRNAs, including surfaceenhanced Raman scattering (SERS) [42], electrochemical biosensors [43, 44], and droplet digital PCR (dPCR) [45]. However, all of these existing methods still rely on RNA extraction which results in variable RNA yield, sequence bias, sample contaminations, and RNA degradation [39].
Compared to the existing methods, our MERCA system presents distinct technological innovation, the advantages in analytical performance, and potential impact on translational research in liquid biopsy. First, nicking enzyme-based exponential RCA (eRCA) has been established as a solution-phase assay for miRNA detection [19]. Based on this approach, we developed and optimized a new dual-phase eRCA modality that employs the surface-bound RCA reaction to trigger solution-phase eRCA (Fig. 1). Second, the previous eRCA has been only reported for analysis of purified RNA samples, reflecting its limited analytical capability. Our MERCA represents a total analytical system that combines sample processing (i.e., miRNA isolation and labeling with the RCA adapter by ligation) and miRNA quantification by dual-phase eRCA in one analytical workflow (Fig. 1), enabling direct mRNA analysis with complex biological samples without the need of RNA purification. Such ability not only enhances the analytical metrics (sensitivity, specificity, reproducibility, and speed), but also profoundly expand the adaptability to broad clinical applications. Third, distinct from the gel particles [27] and the bead-packed chip [32], the MERCA system integrates programmable microvalves and pumps for precise nanoliter-scale flow controlling to streamline and automate the analytical pipeline. We demonstrated the application of the MERCA to sensitive and specific miRNA detection directly from the crude lysates of cells and exosomes with minute sample consumption. Lastly, exosome analysis is a highly promising, yet very challenging area in the development of liquid biopsy biomarkers, as summarized above. Our work immediately addresses the technical challenges in this emerging field, and thus would have potential impact on a broad spectrum of applications across miRNA biology, biomarker development, and clinical diagnostics.
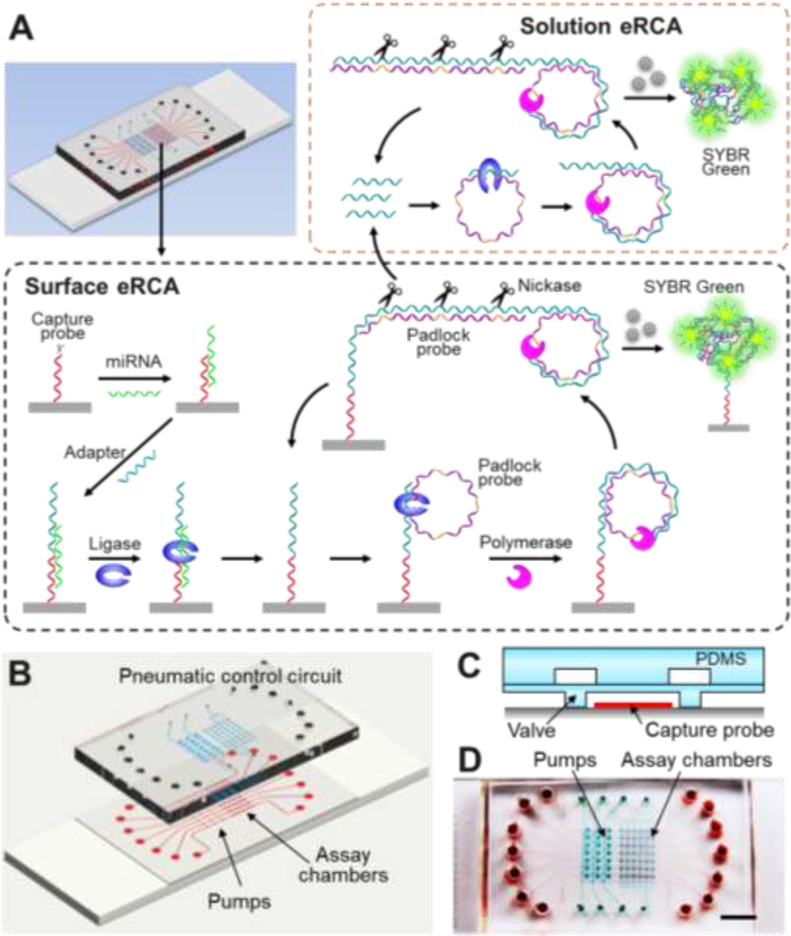
Figure 1. The MERCA system for miRNA detection.
(A) Schematic illustration of the MERCA principle. The on-chip chamber is patterned with probes to capture target miRNAs. An adapter hybridizes with the miRNA and is ligated with the capture probe by T4 DNA ligase. The adapter contains a template that binds with a padlock probe containing three template hybridization sequences linked by Nb.BbvCI nicking sites. After ligation, isothermal RCA produces a long concatenated copy which can hybridize with free padlock probes in solution. Nicking enzyme recognizes and cleaves the dsDNA at specific sites to release multiple replicates of the RCA template to trigger liquid-phase eRCA. Meanwhile, the surface-bound adapter sequence can be recycled to initiate successive rounds of solid-phase RCA reaction. The double-stranded amplification product on both surface and in the solution is detected by SYBR Green dye added to the reaction. (B) Design of the MERCA chip. The three-layer PDMS/glass chip integrates seven parallel units each consisting of a three-valve pump and an assay chamber. The assay chambers aligned vertically across an array of capture probe patterned on the substrate. (C) Schematic of an assay chamber gated by two normally-closed valves. (D) Digital photo of an assembled chip filled with red food dye in the flow channels and green dye in the pneumatic control channels.
2. Experimental section
2.1. Chemicals and materials.
Synthetic oligonucleotides and miRNAs were synthesized and purified with polyacrylamide gel electrophoresis (PAGE) by Integrated DNA Technologies (IDT, Coralville, IA). The sequences of the oligonucleotides and miRNAs were listed in Table 1. Phi29 DNA polymerase, T4 DNA ligase, Nb.BbvCI nicking endonuclease and deoxynucleotide solution mixture (dNTPs) were purchased from New England Biolabs. 1×phosphate-buffered saline solution (PBS) (Mediatech, Inc.), ethanol (100%, Decon Laboratories Inc.), diethypyrocarbonate (DEPC) treated water and TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) were purchased from Thermo Fisher scientific. SYBR Green I, 2propanol (IPA, 99.5%), bovine serum albumin (BSA), aminopropyl-trimethoxysilane (APTMS), glutaraldehyde (GL, 50%), trichloro (1H, 1H, 2H, 2H-perfluorooctyl) silane (97%), 3-glycidoxypropyltrimethoxysilane (GOPS, 97%) and triethylamine (TEA) were purchased from Sigma-Aldrich. DEPC treated deionized water was used in all experiments. TaqMan MicroRNA Reverse Transcription kit and TaqMan MicroRNA Assays, total RNA extracted from MCF-7 cells, and mirVana™ miRNA Isolation Kit were purchased from Thermo Fisher Scientific. A549 whole cell lysate (4.6 mg/mL) was obtained from Novus Biologicals. Lyophilized exosome standards isolated from human U87MG and A549 cell culture medium were purchased from HansaBioMed Life Sciences and reconstituted to prepare the stock solutions (1 × 1010 particles/mL for U87MG exosomes; 2 × 1011 particles/mL for A549 exosomes), as recommended by the manufacturer. Exosome stock solutions were aliquoted, stored at -80°C, and diluted to prepare samples immediately before use. All other chemicals were analytical grade and used without pretreatment, unless mentioned otherwise.
Table 1.
Sequences of oligonucleotides and miRNAs
| Name | Sequences |
|---|---|
| Padlock probea | 5′-phosphate-TCA CTA TGG TCC CTC AGC AGC ACA ATA AAT CAC TAT GGT CCC TCA GCA GCA CAA TAA ATC ACT ATG GTC CCT CAG CAG CAC AAT AAA-3′ |
| let-7a capture probe | 5′-NH2-C12-AAA AAA AAA AAA AAC TAT ACA AC-3′ |
| miR-21 capture probe | 5′-NH2-C12-AAA AAA AAA AAA TCA ACA TCA GT-3′ |
| let-7a RCA adapterb | 5′-phosphate-CTA CTA CCT CA A AAA AAA AAA AAG ACC ATA GTG ATT TAT TGT GCT-3′ |
| miR-21 RCA adapterb | 5′-phosphate-CTG ATA AGC TA AA AA AAA AAA AAG ACC ATA GTG ATT TAT TGT GCT-3′ |
| let-7a | 5′-UGA GGU AGU AGG UUG UAU AGU U-3′ |
| let-7bc | 5′-UGA GGU AGU AGG UUG UGU GGU U-3′ |
| let-7cc | 5′-UGA GGU AGU AGG UUG UAU GGU U-3′ |
| miR-21 | 5′-UAG CUU AUC AGA CUG AUG UUG A-3′ |
| miR-200a | 5′-UAA CAC UGU CUG GUA ACG AUG U-3′ |
| miR-200b | 5′-UAA UAC UGC CUG GUA AUG AUG A-3′ |
The bold letters show recognition sequences of nicking enzyme.
The italic letters are the sequences to hybridize with miRNAs.
The underlined bases show the different bases between let-7b or let-7c and let-7a.
2.2. Microfabrication.
The chips were fabricated according to the multilayer soft lithography approach. The integrated MERCA chip is constructed by a two-layer PDMS chip assembly and a glass substrate patterned with the miRNA capture probe, as illustrated in Fig. 1. SU-8 molds on silicon (Si) wafers were fabricated by standard photolithography for the flow channel layer, pneumatic control layer and patterning chip, respectively. The molds of pneumatic and patterning chips were made of SU-8 2025 (MicroChem) with a final thickness of ~40 μm and ~30 μm, respectively. The mold of microfluidic channel layer was made of SU-8 2010 with a final thickness of ~25 μm (i.e., a volume of ~8 nL for each reaction chamber), following the procedure recommended by the manufacturer. All Si molds were treated with trichloro (1H, 1H, 2H, 2H-perfluorooctyl) silane using gas-phase silanization under vacuum for overnight. To fabricate the pneumatic and patterning PDMS chips, a total 30 g mixture at a 10: 1 (base: curing agent) ratio was degassed and poured on the molds, followed by curing in the oven at 70 °C for 4 h. The cured PDMS slabs were peeled off from molds, cut into small pieces, and punched with biopsy punch to make access holes. For the fluidic channel layer, a 6.4 g mixture at a ratio of 15:1 (base: curing agent) was spin-coated over the mold at 1000 rpm for 30 s and cured for 40 min to make the PDMS membrane. After surface cleaning with UV Ozone, the PDMS chip was assembled by manually aligning the pneumatic layer over the fluidic layer under a stereomicroscope, followed by curing at 70 °C overnight to form the permanently bonded chips.
2.3. Surface patterning and device assembly.
The glass substrate was modified with two silanes for surface patterning of the miRNA capture probe. Firstly, glass slides were cleaned in piranha solution (3:1 concentrated sulfuric acid and 30% hydrogen peroxide) at room temperature overnight, rinsed with deionized water, and dried under nitrogen atmosphere. Precleaned glass slides were functionalized by the APTMS-GL method according to the literature with slight modification [46]. Briefly, substrates were shaken in a 95% ethanol/water solution containing 2% APTMS for 30 min at room temperature, followed by washing in ethanol and deionized water three times, respectively. The treated glasses were dried on a 110 °C hotplate for 1.5 h to crosslink the APTMS coating. APTMS-treated glass substrates were then incubated in 1× PBS (pH 7.4) containing 2.5% glutaraldehyde solution with shaking for 2 h at room temperature, followed by rinsing with deionized water and dried in a flow of nitrogen. For GOPS modification, cleaned substrates were shaken in anhydrous toluene with 2% GOPS and 0.2% TEA for 1 hour at room temperature, thoroughly rinsed with fresh toluene and isopropanol, and dried at 80°C for 2 h to crosslink the monolayer [46]. The GOPS-modified surface possesses the epoxide groups that can form a secondary amine bond upon reaction with amine groups.
Aminated capture probe was covalently immobilized onto an APTMS-GL or GOPS functionalized slide using the patterning chip, as described below. 2 μL 1× PBS containing the capture probe (50 μM) was injected into individual patterning microchannels. The chip was incubated at room temperature in a humid container to prevent drying during surface conjugation. After overnight, the capture probe solution was flushed out from the patterning channels by rinsing with deionized water twice and then 1× PBS to remove non-covalently absorbed probe. Finally, the patterning chip was peeled off and the patterned slide was rinsed by water briefly and dried by pure nitrogen. To construct the MERCA chip, an assembled PDMS chip was briefly cleaned by deionized water and ethanol, treated with UV ozone for 2 min, and sealed onto a patterned slide with the assay chambers aligned vertically with the capture probe patterns. To deactivate excessive aldehyde sites and block non-specific absorption on the channel surface, the entire chip was incubated with 5% BSA at room temperature for 1h.
2.4. Standard eRCA.
Optimization of the nicking enzyme-assisted eRCA for miRNA detection was first performed in the standard microplate form. The padlock probe with a wide concentration range of 10−11 to 10−7 M was mixed with the adapter sequence (50 nM), denatured at 65 °C for 3 min, and then slowly cooled to room temperature. After annealing, the ligation reaction was conducted by adding the adapter/padlock probe mixture to the reaction buffer to prepare a 10 μL reaction solution, which was composed of 1× ligation buffer (40 mM TrisHCl, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 500 mM ATP, pH 7.8), 10 U of T4 DNA ligase, 2 μL different concentration padlock probe, and 2 μL adapter (50 nM, final concentration). The ligation reaction was conducted with various concentrations of padlock probes at 30 °C for 2 h. The RCA reaction was then carried out at 30 °C in a 20 μL reaction mixture containing 10 μL ligation reaction products, 20 mM Tris-HCl buffer (pH 7.5), 1μL (6× SYBR Green I), 200 μg/ml BSA, 10 mM (NH4)2SO4, 10 mM MgCl2, 400 μeach dNTP, 1 U Nb.BbvCI, and 1 U Phi29 DNA polymerase. Finally, eRCA amplification products were measured using a CYTATION 5 imaging reader (BioTek).
2.5. Microfluidic eRCA.
For the method development, synthetic miRNAs diluted to various concentrations were used. To test the MERCA for complex biological samples, total RNA extracted from the breast adenocarcinoma MCF-7 cell line (1 mg/mL, Thermo Fisher Scientific) was diluted to 0.1 and 1 μg/mL with RNase-free TE buffer. A549 whole cell lysate was diluted to 46 and 460 μg/mL with the lysis buffer (1× TE buffer with 2% SDS and 200 μg/mL proteinase K) [28]. A549 and U87 exosome standards were lysed with the lysis/binding solution from the mirVana™ miRNA Isolation Kit at the 1:1 volume ratio for 10 min on ice and then diluted with TET buffer (1× TE with 0.05% Tween-20) containing 200 μg/mL proteinase K for direct miRNA detection on MERCA chips. The chips were operated using a LabVIEW program to actuate the monolithic pneumatic valves and assay chambers through the homemade solenoid controllers [34]. The air pressure and vacuum were set to 30 kPa and -87 kPa, respectively. On-chip pumping was conducted with a five-step stop-flow sequence that we developed: four consecutive valve actuation steps and a pulse step. The averaged flow rate can be conveniently adjusted by tuning the time for each valve actuation and pulse step. A mini digital incubator with 0.5 °C accuracy was used for temperature control during the assay procedure involving miRNA capture, ligation, and eRCA reactions (Fig. 1A). 10 μL samples were injected into the chips to capture miRNAs on the channel surface at the optimized flow rate and temperature (see the main text). The channels were flushed with 15 μL TET with 50 mM NaCl for 30 min at 0.5 μL min−1 to sufficiently remove excess sample and non-specifically bound species. After target capture, 10 μL ligation mixture containing 5 μL adapter (50 nM), 10 U T4 DNA ligase and 1× ligation buffer was prepared and pumped into the channels, followed by incubation for 30 min at 30 °C with stopped flow and washing with TET with 50 mM NaCl for 30 min to sufficiently remove excess adapter oligoes. Prior to MERCA, 10 μL ligation mixture containing 5 μL padlock probe (10−8 M), 10 U T4 DNA ligase and 1× ligation buffer was prepared and pumped into the channels. Padlock probe binding and ligation was carried out for 2h at 30 °C 10 μL eRCA cocktail composed of 20 mM Tris−HCl buffer (pH 7.5), 1μL (6× SYBR green I), 200 μg/ml BSA, 10 mM (NH4)2SO4, 10 mM MgCl2, 400 Μm each dNTP, 10 U T4 ligase, 1 U Phi29 DNA polymerase, 1 U Nb.BbvCI and 3 μL padlock probe (10−8 M) was pumped into the channels at a flow rate of 0.5 μL/min; the assay chambers were closed to prevent diffusion of the eRCA products; finally MERCA was carried out at 30 °C for the desired extension time. Fluorescence detection of the MERCA product was performed using a Zeiss Axiovert A1 inverted fluorescence microscope equipped with a sCMOS camera (OptiMOS, QImaging). Acquired images were analyzed with ImageJ (NIH, http://rsbweb.nih.gov/ij/) to quantify signal intensity.
2.6. RT-qPCR analysis.
TaqMan RT-qPCR of synthetic miRNAs, MCF-7 total RNA, A549 whole cell lysate, and cell-derived exosomes was conducted in parallel to the MERCA. A549 whole cell lysate was diluted with the lysis buffer by 10 and 100 folds prior to RNA extraction. Lyophilized exosome standards (100 μg) were disrupted by adding 300 μL lysis buffer. Total RNA was then extracted from the cell and exosome lysates with the mirVana™ miRNA Isolation Kit, following the manufacturer’s protocol with slight change. The RNA extract was stored at -80 ºC until use. Purified total RNA was reverse-transcribed into cDNAs using the TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific) following the manufacturer’s instruction. Each 15-μL RT reaction consists of 7 μL master mix, 3 μL of 5× RT primer, and 5 μL RNA sample. 1.33 μl of 15-fold diluted RT reaction product was mixed with 1 μl pre-formulated primer and probe set for let-7a (Applied Biosystems™ TaqMan™ MicroRNA Assays, 20×), 10 μl TaqMan® Universal PCR Master Mix II (2×), 7.67 μl nuclease-free water to prepare 20 μl PCR reactions. qPCR was conducted on a Mastercycler EP Gradient S thermocycler (Eppendorf) using a hot-start protocol: 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All reactions were run in triplicate.
3. Results and Discussion
3.1. MERCA assay and chip design.
The principle of the MERCA for miRNA detection is illustrated in Fig. 1A. The glass surface in the assay chamber is patterned with oligonucleotide probes to capture target miRNAs by hybridization with the half miRNA sequence at the 3′ end. The adapter incorporates a sequence complementary to the 5′-end miRNA sequence and another universal sequence as the RCA template. The probe and adapter that specifically hybridize with the miRNA can be ligated by T4 DNA ligase, which enhances the specificity of the MERCA. The universal template binds with a padlock probe for RCA regardless of miRNA targets, eliminating the possibility of sequence-biased miRNA amplification. The padlock probe designed here contains three template hybridization sequences linked by Nb.BbvCI nicking sites. The padlock probe hybridized onto the surface-immobilized adapter with the two termini which are then ligated to form the circular padlock probe. Isothermal linear rolling circle amplification (LRCA) with Phi29 DNA polymerase produces a long, concatenated DNA copy which will hybridize with free padlock probes in solution. Nicking enzymes present in the reaction recognize and cleave the double-stranded DNA at specific sites to release multiple replicates of the RCA template. Each replicate provides a new template for RCA reaction, leading to liquid-phase exponential amplification of the template. Meanwhile, since the circular padlock complex is cleaved off, the surface-bound adapter sequence can be recycled to initiate successive rounds of solid-phase RCA reaction. Therefore, our method presents a new dualphase eRCA modality that employs the surface-bound RCA reaction to trigger solution-phase eRCA, which enables integration of miRNA capture and enrichment to construct a total analytical platform for direct mRNA analysis with complex biological samples without the need of RNA extraction. Our dual-phase eRCA assay is distinctive from the previously reported eRCA assay that does not have ability to capture and enrich the targeted sequences and performs the RCA and nicking reactions only in the solution [19]. This homogeneous eRCA method was only demonstrated for miRNA analysis using total RNA purified from cells.
Fig. 1B shows the prototype MERCA chip that we have devised. The three-layer device integrates seven parallel units to demonstrate the potential scalability of our platform. Each unit combines a three-valve pump and the assay chambers that are pneumatically addressable.
Flow channels (25 μm high) were fabricated on a ~120 μm thick PDMS membrane and assembled with a thick PDMS slab patterned with the pneumatic control circuit. The PDMS assembly is sealed onto a glass slide patterned with miRNA capture probe. The assay chambers are gated with the normally-closed valves (Fig. 1C) which will be opened for flow-through miRNA capture and closed for isothermal amplification and fluorescence detection. A complete MERCA chip is displayed in Fig. 1D, in which the flow channels and the pneumatic network visualized by filling with red and green dyes, respectively. A microbead-based RCA microchip has been reported, in which a sample was driven by syringe pump and DNA targets were captured and amplified by linear RCA on beads trapped inside the channel [32]. For comparison, our MERCA platform integrates microvalves and pumps for precise flow control to automate the assay workflow and is amenable to scaling for multiplexed analysis. Moreover, our method implements ligation-assisted capture and exponential RCA to afford improved sensitivity and specificity for miRNA detection.
3.2. Optimization of eRCA.
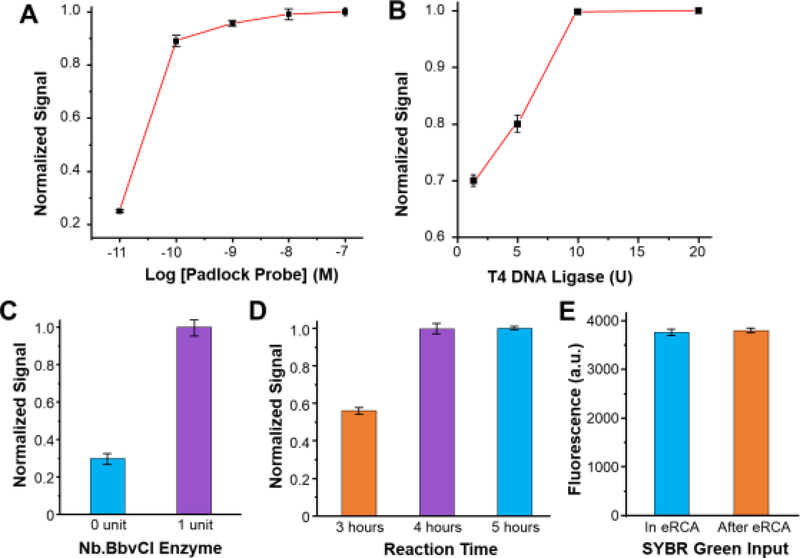
As seen in Fig. 1, the MERCA integrates a multi-step workflow for miRNA capture and eRCA detection that are affected by many variables. To facilitate our method development, we first investigated off-chip eRCA to optimize critical reaction variables, including the padlock probe, adapter, T4 DNA ligase, nicking enzyme Nb.BbvCI, and reaction time, using let-7a as the model target. The optimized sequences of capture probe, padlock probe, and adapter were listed in Table 1. The padlock probe concentration affects the efficiency of binding with the RCA template and thus was assessed across a wide range of 1011 to 10−7 M. As shown in Fig. 2A, the assay signal increases along with the concentration of padlock probe and saturates at 10−8 M, which was then used in the subsequent experiments. The effect of T4 DNA ligase was also assessed and an optimal concentration of 10 U T4 DNA ligase per 10 μL reaction was identified to maximize the efficiency of enzymatic ligation (Fig. 2B). In our approach, a nicking enzyme Nb.BbvCI is used to cleave the DNA synthesized by linear RCA to trigger exponential amplification. Fig. 2C displays that compared to the conventional LRCA, the addition of Nb.BbvCI (0.1 units/μL) resulted in a substantial signal increase, in line with the previous report [19]. This result verifies that Nb.BbvCI can recognize and cleave the DNA product of LRCA at the nicking sites to drive exponential amplification. We tested eRCA reactions over a period of time of 3–5 hours and observed that the assay reached the saturated signal in 4 hours (Fig. 2D). While microfluidic reaction is usually more efficient than in the bulk scale, we still adapted 4-hour reaction time for the on-chip eRCA to maximize detection sensitivity. In the bulk eRCA assay reported before [19], an intercalating dye, SYGR Green I, was added for detection after the reaction was completed, which dilutes the DNA product concentration and thus reduces the detection sensitivity. Therefore, we tested the eRCA in the presence of SYGR Green I, which didn’t show inhibitory effects on the enzymatic eRCA reaction (Fig. 2E). This negates extra steps of reagent mixing for on-chip signal readout, which simplifies the microchip design and operation.
Figure 2. Optimization of eRCA conditions using the adapter sequence.
(A, B) Plots of fluorescence signals normalized to the maximum signal obtained under different concentrations of (A) the padlock probe and (B) T4 DNA ligase. (C) Comparison of the fluorescent signals of RCA with and without adding Nb.BbvCI (1 U per 20 μL reaction). (D, E) Bar graphs showing the effects of (D) reaction time and (E) the SYBR Green on the assay response. The 20 μL reaction solution contained 10−8 M padlock probe and 50 nM let-7a adapter. The ligation reactions were performed at 37 °C for 2 h, and RCA reactions were performed at 30 °C. Error bars represent one standard deviation from three replicates.
3.3. MERCA for miRNA detection.
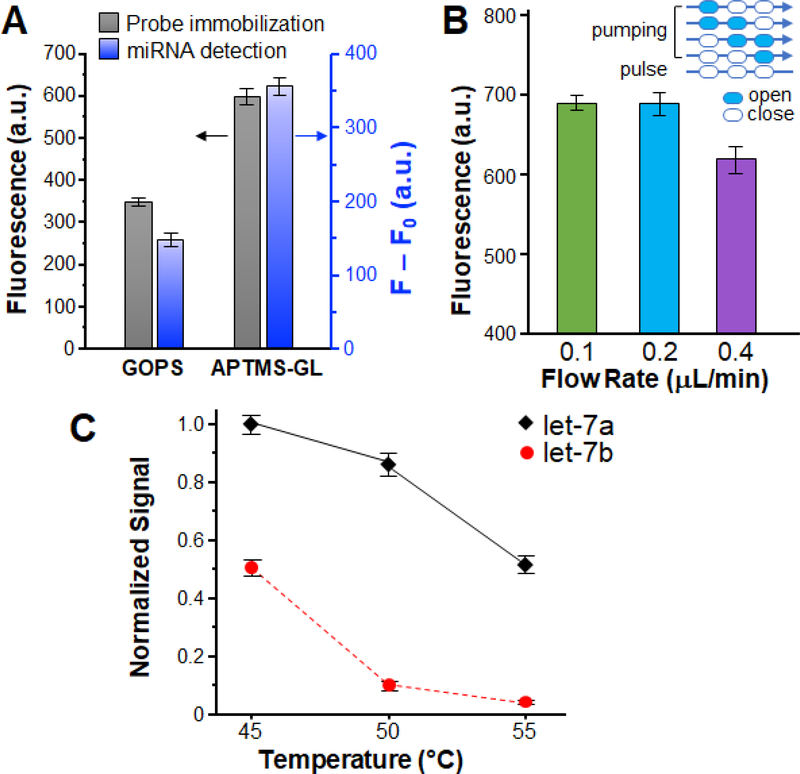
The optimized eRCA was implemented on chip and integrated with the miRNA capture step to establish the complete MERCA, as described below. The surface functionalization chemistry was first investigated as it affects surface properties, binding kinetics, and processing simplicity [46]. To this end, we evaluated the effects of two commonly used covalent silanization chemistries, APTMS-GL and GOPS, on probe immobilization and miRNA detection performance. As seen in Fig. 3A, surface immobilization by the APTMS-GL method confers significantly higher probe density on the surface than the GOPS method, which can be attributed to the greater density of reactive sites created by polymerization of glutaraldehyde [46]. The APTMS-GL modified chips was also seen to greatly augment the detection signal for detecting let-7a miRNA at 5.0 × 10−10 M (Fig. 3A), which can be mainly attributed to the better efficiency of probe immobilization. Flow rate is another critical parameter that affects surface-based affinity capture inside microchannels. While fast flow can enhance mass transfer of analyte towards the binding surface, it will also reduce the time for analyte molecules to interact with the immobilized capture probe. To promote the capture efficiency, we optimized the flow rate by conveniently tuning the frequency of microfluidic pumping. On-chip pumping was conducted in a five-step stop-flow sequence that we implemented: four valve actuation steps and a pulse step, with the pressure and vacuum set to 30 kPa and -87 kPa, respectively. Based on our prior flow optimization for surface affinity capture [47, 48], here we examined a range of flow rate from 0.1–0.4 μL/min by decreasing the time for each valve actuation step from 0.5 to 0.2 s with a fixed pulse step of 2 s. Fig. 3B shows that the maximum detection signal was reached at an averaged flow rate of ∼0.2 μL/min obtained with 0.3 s valve actuation steps and a 2 s pulse. Temperature is an essential variable in the thermodynamics of DNA/RNA hybridization that affects the balance between the efficiency and specificity of miRNA capture. Here we investigated the effects of capture temperature on specific detection of let-7a against a homologous let-7 family member, let-7b, which differ by only two nucleotides (Table 1). As seen in Fig. 3C, the detection signal for let-7a decreases as the capture temperature increases from 45 ℃ to 55 ℃, owing to enhanced dissociation of complementarily paired DNA/RNA strands. Meanwhile, increasing temperature also suppresses the non-specific signal from partial binding of let-7b, especially from 45 ℃ to 50 ℃. An optimal temperature of 50 ℃ was determined for miRNA capture, above which no improvement in the signal-to-noise ratio was observed.
Figure 3. Optimization of the MERCA assay with let-7a miRNA.
(A) Effects of conjugation chemistries on surface immobilization of capture probes (grey) and miRNA detection (blue). To assess probe immobilization, FAM labeled capture probe (50 μM) is covalently immobilized on glass slides by the APTMS-GL and GOPS methods, respectively. For RCA detection, let7a (5.0 × 10−10 M) was measured as a model sample using chip surfaces modified by the two methods. F: fluorescence intensity for miRNA detection; F0: background signal obtained with buffer. (B) Comparison of the detection of let-7a (5.0×10−10 M) using different averaged flow rates for solid-phase miRNA capture. A five-step pumping sequence was used with the actuation time for each step varied from 200 to 500 ms and the pulse time fixed at 2 s (inset). (C) Effects of capture temperature on the specificity of miRNA detection. The concentrations of let-7a and let-7b are 5.0×10−10 M, SYBR Green as fluorescence reagent, respectively. Error bars indicate one standard deviation of three replicates.
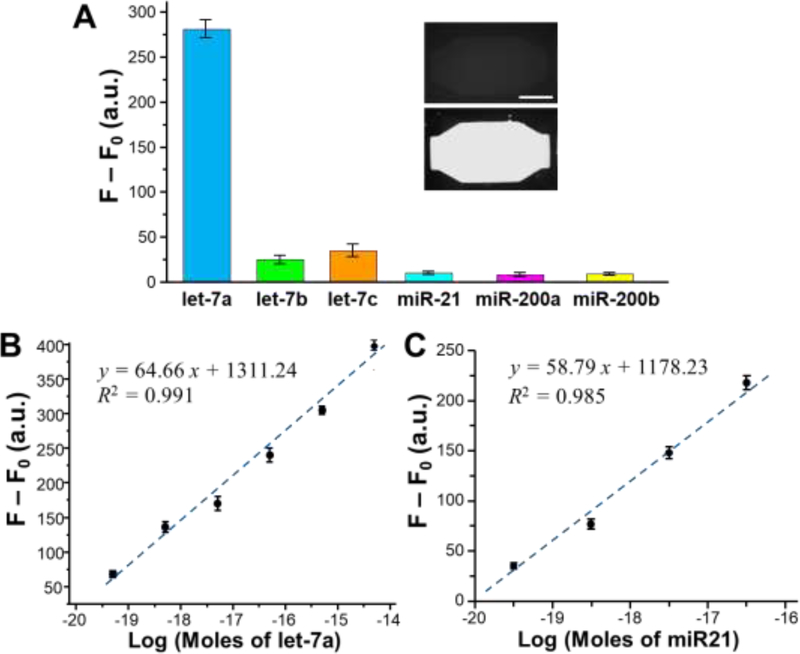
High detection specificity is required for quantitative analysis of miRNAs in complex matrices for biomarker development and clinical diagnostics. Under the optimal conditions, we systematically characterized the specificity of the MERCA platform by expanding the test to a number of miRNAs, including the let-7 family members, miR21, miR200a, and miR200b. MERCA detection of let-7a was successfully demonstrated with a low background measured from a buffer blank, as shown by the bar plot and the representative fluorescence images in Fig. 4A. Very low fluorescence signal above the background level was detected from three random miRNAs, miR21, miR200a, and miR200b, under the same conditions. In the let-7 family, let7b and let-7c differ from let-7a by only two and one nucleotide, respectively. Despite the high sequence similarity, only small fluorescence signals were observed for let-7b and let-7c, which account for 1/10 and 1/8 that of let-7a, respectively. These findings clearly demonstrate that the MERCA method affords high specificity for miRNA detection that can discriminate singlebase difference. Such high specificity can be attributed to the combined effects of miRNA capture at a relative high temperature (50 ℃), enzymatic ligation between the miRNA and adapter both complementarily hybridized with the probe, and specific eRCA reaction triggered by the universal adapter and the nicking enzyme.
Figure 4. Characterization of the MERCA for miRNA detection.
(A) Specificity tests using let-7a, let-7b, let-7c, miR21, miR200a and miR200b. The concentration of each miRNA was 5.0×10−11 M. F: fluorescence intensity for miRNA detection; F0: background signal obtained with buffer. Inset: representative fluorescence images obtained with the buffer blank (top) and the let-7a sample (bottom), respectively. Scale bar: 250 μm. (B) Semi-log calibration plot of the MERCA assay for detection of a serial dilutions of let-7a in the range from 5.0×10−20 to 5.0×10−15 moles per reaction. (C) Semi-log calibration plot of the MERCA assay for detection of a serial dilutions of miR21 in the range of 5.0×10−20 to 5.0×10−17 moles per reaction. The linear curves were fitted by the least-squares fitting. The reactions were conducted with 10−8 M padlock probe under the optimal conditions. Error bars are the standard deviation from three replicates.
We further calibrated the MERCA system for quantitative miRNA detection using a series of 10-fold dilutions of synthetic let-7a and miR-21 miRNAs. The multi-channel design allowed parallel measurements of a blank and different standards to expedite the measurements. The measured fluorescence signals were subtracted by the background and plotted as a logarithmic function of the moles of miRNAs. It was demonstrated in Fig. 4B that our MERCA system enables quantitative detection of let-7a (y = 64.66x + 1311.24, R2 = 0.991) across an extensive dynamic range of 50 zmol to 5 fmol, presumably due to the use of universal adapter ligation and dual-phase eRCA that enhance the efficiency and capacity of RCA reaction [19]. A remarkably low LOD of 5 zmol let-7a was observed, which is calculated from the value of blank signal plus three standard deviations. Quantitative detection of miR-21 was also achieved (y = 58.79x + 1178.23, R2 = 0.985), with a LOD of 8 zmol (Fig. 4C). Such low LODs are comparable with or better than that of the ultrasensitive miRNA assays reported previously, [18–20, 27] as summarized in Table 2. To assess the precision of the MERCA, a series of eight repetitive measurements of 5 × 10−15 mol let-7a was conducted on the multi-channel chip, which yielded a relative standard deviation (RSD) of 3.7%, indicating the good reproducibility of this miRNA assay. Overall, the results discussed above demonstrate that by integrating efficient and specific miRNA capture with highly sensitive dual-phase eRCA reaction, the MERCA system holds the potential for ultrasensitive and accurate quantitation of miRNA at low concentrations in complex biological samples.
Table 2.
Comparison with existing RCA-based miRNA detection methods
| Methods | LOD | Dynamic range | Sample analyzed | Reference |
|---|---|---|---|---|
| B-RCAa | 6 amol | 0.025 to 1 pM | Total RNA | 18 |
| P-ERCAb | 0.24 zmol | 0.3 zmol to 18 zmol | Total RNA | 19 |
| D-RCAc | 20 zmol | 1 fM to 100 nM | Total RNA | 20 |
| Gel particle-based RCA | 15 zmol | 300 aM to 40 pM | Synthetic miRNA spiked serum | 27 |
| Paper-based Electrochemical biosensor | 0.35fM | 1.0 fM to10 nM | Diluted human serum | 31 |
| Stem-loop RT-PCR | 7 copies | 0.013 fM to 13 pM | total RNA, cell lysate and cells | 12 |
| MERCA | 5–8 zmol | 50 zmol to 5 fmol | Total RNA; cell lysate; exosomes | This work |
branched rolling circle amplification
padlock probe-based exponential rolling circle amplification
dumbbell rolling circle amplification
3.4. miRNA analysis directly from biological samples.
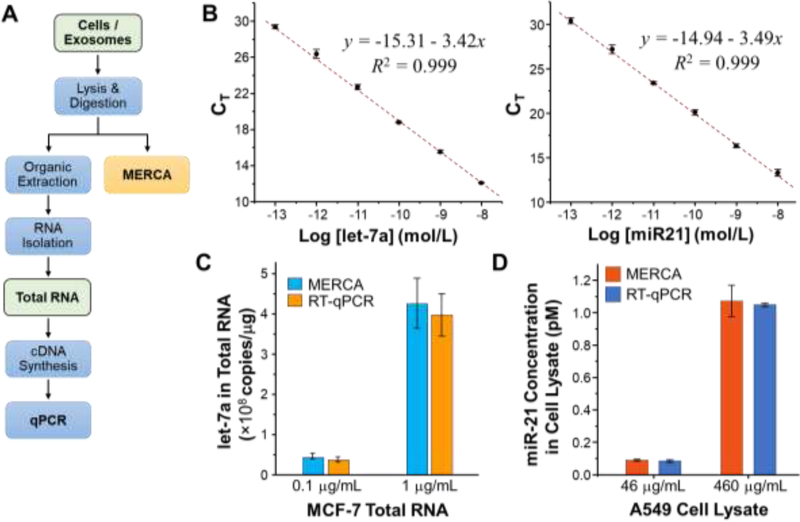
Standard RT-qPCR and the majority of existing sensing methods (Table 2) for miRNA analysis require multi-step RNA extraction from complex biological samples, as depicted in Fig. 5A. Not only are these sample preprocessing steps labor intensive and time consuming, but also prone to RNA degradation and cross-contamination. More importantly, extensive studies have shown that different RNA purification kits/protocols and starting biological materials (e.g., cells, tissue, and plasma) can lead to variable yield and quality of purified RNAs and even introduce isolation bias depending on RNA sizes and sequences [49–54]. These issues could result in significant variation and biased results in miRNA analysis and thus has become a methodological bottleneck in the advance of miRNA biology and the development of miRNA-based biomarkers and therapeutics [55, 56]. To address these challenges, the MERCA platform is designed to directly detect miRNAs in complex biological samples without the need of RNA extraction, which greatly simplifies the analysis procedure in comparison with RT-qPCR (Fig. 5A). The entire on-chip assay can be completed within 8 hours, shorter than RT-qPCR that usually needs at least one day to complete.
Figure 5. Detection of miRNAs in total RNA and crude cell lysate using the MERCA and standard RT-qPCR.
(A) Comparison between the standard RT-qPCR assay that requires multi-step sample preparation procedures and the MERCA for direct miRNA analysis with minimal sample treatment. (B) Calibration curves for the TaqMan RT-qPCR detection of synthetic let-7a and miR21 miRNAs, respectively. The linear curves were fitted by the leastsquares fitting. (C) Comparison of the concentrations of let-7a in the 0.1 μg/mL and 1 μg/mL MCF-7 total RNA samples measured by the RT-qPCR and the MERCA, respectively. (D)Comparing the MERCA and RT-qPCR analysis of miR-21 in the diluted samples of crude A549 whole cell lysate. 4.6 mg/mL A549 cell lysate was first diluted with the lysis buffer by 10 and 100 folds, respectively, prior to analysis. For RT-qPCR, total RNA was first extracted from the diluted A549 cell lysate samples, followed by RT and qPCR analysis. The concentrations of let-7a and miR-21 were calculated using the calibration curves in (B) for RTqPCR and in Fig. 4 for the MERCA. Error bars are one standard deviation (n = 3).
To demonstrate its broad applicability, we assessed the MERCA method with various biological samples, starting with purified total RNA to more complex crude cell lysate and cellderived exosomes. Commercial TaqMan RT-qPCR miRNA assays with pre-designed primers for let-7a and miR-21 were used to benchmark the performance of our method. Using this gold standard method, the calibration curves were established for let-7a and miR-21 to determine of their quantities in real samples (Fig. 5B). To assess the adaptability of our assay to total RNA, we attempted to detect let-7a in a commercially available total RNA extracted from a breast adenocarcinoma cell line MCF-7. 0.1 and 1 μg/mL MCF-7 total RNA samples were prepared from the original stock (1 mg/mL) and assayed with both MERCA system and RT-qPCR to determine the let-7a concentration, based on the established calibration curves. As shown in Fig. 5C, the concentrations of let-7a in the 0.1 and 1 μg/mL total RNA samples measured by the MERCA were (4.56 ± 0.64) × 107 and (4.26 ± 0.74) × 108 copies/μg, respectively. These results match closely with the values of (3.78 ± 0.68) × 107 and (3.97 ± 0.53) × 108 copies/μg measured by RT-qPCR, indicating the ability of our method for accurate quantification of miRNAs in total RNA samples.
In addition to purified total RNA, we further evaluated the MERCA for analyzing more complex crude cell lysate with minimal sample pre-treatment. To this end, a commercially available whole cell lysate of human lung adenocarcinoma epithelial cell line A549 (4.6 mg/mL) was diluted by 10 folds and 100 folds with the lysis buffer containing 2% SDS and 200 μg/mL proteinase K to release miRNAs from their carriers [28]. Direct analysis of miR-21 in these two cell lysate samples by the MERCA yielded a concentration of 0.089 ± 0.006 pM and 1.071 ± 0.098 pM, respectively. For comparison, RT-qPCR analysis of total RNA isolated from the same samples measured the concentrations to be 0.083 ± 0.008 pM and 1.047 ± 0.012 pM. Good agreement between the two methods (within 7%) demonstrates the high specificity of the MERCA to enable sensitive and accurate quantification of miRNA targets in a vast background of crude whole cell lysate.
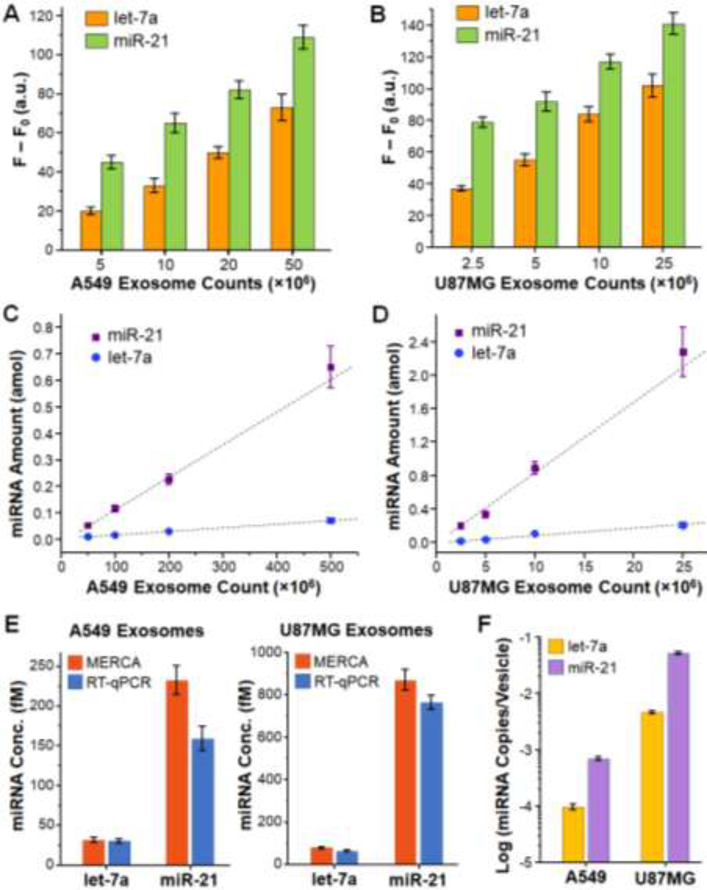
Lastly, we adapted the MERCA assay to measure miRNA contents in exosomes derived from tumor cells. Exosome is one of the major carriers of miRNA in human biofluids and exosomes secreted by tumor cells offer a promising route to explore disease-specific miRNA signatures [57–59]. However, recent studies have shown that the averaged copy number of given miRNAs can be as low as 10−5 copies per vesicle [45, 59], which poses daunting challenges to sample preparation and miRNA analysis in biomedical and clinical exosome studies. Current RNA isolation methods demand large amount of exosome samples and cause extensive variation in exosomal RNA yield and size patterns, which profoundly impacts downstream exosomal RNA analysis [41]. The MERCA system can overcome these problems by taking the advantages of microfluidic integration to eliminate preparative RNA extraction and to substantially improve the analytical performance, while greatly reducing the sample consumption. As a proof-of-concept, we measured let-7a and miR-21 in commercially purchased exosome standards purified from human lung cancer A549 cells and glioblastoma U87MG cells. Prior to miRNA detection, the only sample pretreatment was lysing the exosome standards with the lysis solution and diluting the lysate to desired concentrations by the TET buffer containing proteinase K to completely release exosomal miRNAs. Fig. 6A and B display the background-corrected fluorescence readouts for let-7a and miR-21 measured in 10 μL of 40×, 20×, 10×, and 4× dilutions of A549 exosomes (2 × 1011 mL−1) and U87MG exosomes (1 × 1010 mL−1), respectively. Based on these data, the moles of let-7a and miR-21 were calculated from the established calibration curves and plotted as a function of the counts of A549 and U87MG exosomes in Fig. 6C and D, respectively. Our results show that the amount of let-7a expressed in both types of exosomes was only a fraction of that of miR-21 (~13.8% for A549 exosomes and ~9.0 % for U87MG exosomes). The MERCA device is able to quantitatively measure both miRNAs in raw lysates of exosomes, as manifested by the linear relationship between the quantities of miRNAs and exosomes. The LOD for exosomal miRNA depends on the abundance of miRNA in exosomes. It was calculated that the theoretical LOD of our method for detection of low-abundance let-7a miRNA in U87MG exosomes is as low as 2 × 106 (Fig. 6D), which much lower than standard RT-PCR that typically requires 109-1011 exosomes isolated from >1 mL patient plasma or hundreds of mL of cell culture media.
Figure 6. MERCA analysis of miRNAs in exosomes.
(A, B) Background-subtracted fluorescence readouts for the detection of let-7a and miR-21 in a serial dilution of exosome standards derived from A549 (A) and U87MG cells (B). The concentrations of A549 and U87MG exosome standards were 2 × 1011 and 1 × 1010 vesicles/mL, respectively. Exosomes were lysed and then diluted by the TET buffer. 10 μL of each diluted sample was injected into the chip for analysis. Error bars are one standard deviation (n = 4). (C, D) Plots of the let-7a and miR-21 amount in A549 exosomes (C) and U87MG exosomes (D) calculated from the fluorescence readouts in (A, B) using the calibration curves in Figures 4 and 5, respectively. Error bars indicate standard error. (E) Bar graphs comparing the concentrations of let-7a and miR-21 in the original A549 and U87MG exosome standards measured by the MERCA and RT-qPCR. Error bars are one standard deviation (n ≥ 3). (F) Averaged miRNA copy number per exosome for let-7a and miR-21 in the A549 and U87MG exosomes, respectively, calculated from the MERCA results in (E).
To validate the MERCA, we compared our results with that measured by the commercial TaqMan RT-qPCR miRNA assays (Fig. 6E). Based on the microfluidic results presented above, the concentration of let-7a and miR-21 in the original A549 exosome standard (2 × 1011 mL−1) were determined to be 32.0 ± 3.6 fM and 232.3 ± 18.2 fM, respectively. For comparison, the let-7a and miR-21 concentrations measured by commercial TaqMan RT-qPCR miRNA assays were 30.9 ± 2.7 fM and 159.2 ± 15.2 fM, respectively. Similarly, in the original U87MG exosome standard (1 × 1010 mL−1), the concentrations of let-7a and miR-21 were determined to be 77.6 ± 4.3 fM and 867.2 ± 49.3 fM by the MERCA, which are also higher than those measured by the RT-qPCR assays (62.3 ± 1.7 fM for let-7a and 761.2 ± 34.8 fM for miR-21). Such discrepancy between the two methods can be presumably attributed to the sample loss caused by manual RNA extraction involved in the RT-qPCR analysis. These comparative studies should verify that the MERCA confer unique capability for sensitive and accurate quantification of exosomal miRNA targets directly from exosome samples, as opposed to RTqPCR and other existing methods that require extensive RNA sample preparation [42–45].
We further assessed the miRNA distribution in exosomes based on the exosomal miRNA concentrations measured by the MERCA in Fig. 6E. In A549 exosomes, the averaged exosomal let-7a level was calculated to be (9.6 ± 1.1) × 10−5 copies/exosomes, which is lower than the average level of (69.9 ± 5.5) × 10−5 copies/exosomes for miR-21, as presented in Fig. 6F. The levels of let-7a and miR-21 harbored in U87MG exosomes were found to be (4.67 ± 0.26) × 10−3 and (52.2 ± 3.0) × 10−3 copies/exosomes, respectively, which are much higher than in A549 exosomes. Our results agree with the previous stoichiometric analysis of exosomal miRNAs that reported a range of 10−5 to 10−2 copies of miRNA per exosome in average [45, 59]. We observed significantly lower abundance of let-7a than miR-21 in both cell line-derived exosomes. This trend is consistent with the well-documented evidences showing that the expression of let-7 family miRNAs is downregulated in many cancer types, including lung cancer [60], while miR-21 is overexpressed in glioblastoma and other tumors [59, 61]. It is worth noting that our method affords high sensitivity and specificity to permit quantitative detection of such low-level let-7a miRNA in as low as 2 × 106 U87MG exosomes (Fig. 6D). Collectively, our results (Figs 5 and 6) suggest that the MERCA provide a useful platform to promote miRNA detection in complex samples for broad biological and clinical applications. The performance of our prototype chip can be further improved from different aspects, such as using LNA probes to enhance miRNA capture efficiency [62] and expanding to multiplexed assays for high throughput miRNA profiling.
4. Conclusions
In summary, an integrated microfluidic exponential rolling circle amplification platform was developed here for isothermal detection of miRNAs with superior sensitivity and specificity. The MERCA system integrates a multi-step miRNA assay, including solid-phase miRNA hybridization capture, enzymatic miRNA-adapter ligation, and the dual-phase eRCA, in one analytical workflow. The MERCA system demonstrated quantitative miRNA detection with a remarkably low LOD of 5–8 zmol and single-nucleotide specificity. As a proof-of-concept, we successfully demonstrated the applications of the MERCA chip to accurate and quantitative miRNA detection directly from total RNA, raw cell lysate, and exosome samples, without the need of RNA extraction. The microfluidic analysis results were verified with the parallel measurements using standard TaqMan RT-qPCR. Overall, these studies suggest the potential of our method to facilitate quantitative miRNA analysis in complex biological materials, which is crucial to the development of miRNA-based biomarkers and molecular therapeutics.
Highlights.
We developed a pneumatically actuated Microfluidic Exponential Rolling Circle Amplification (MERCA) system for sensitive and specific detection of microRNAs directly in minimally processed samples.
The MERCA presents a total analysis lab-on-a-chip system that integrates and streamlines solid-phase miRNA isolation, miRNA-adapter ligation, and a dual-phase exponential rolling circle amplification (eRCA) assay in one analytical workflow.
By marrying the advantages of microfluidics in leveraging bioassay performance with the high sensitivity of eRCA, our method affords a remarkably low limit of detection at <10 zeptomole levels, with the ability to discriminate single-nucleotide difference.
Using the MERCA chip, we demonstrated quantitative miRNA detection directly from raw cell lysate and cell-derived exosomes with superior sensitivity than the existing microsystems.
The MERCA would provide a useful platform to facilitate miRNA analysis in broad biological and clinical applications.
Acknowledgments
We would like to acknowledge the Microfabrication and Microfluidics Core Facility at the KU COBRE Center for Molecular Analysis of Disease Pathways (CMADP) for device fabrication. This study was supported by 1R21EB024101, 1R21CA186846, 1R33CA214333, 1R21CA207816, and P20GM103638 from the NIH. H. C. was partially supported by the postdoc fellowship from the Kansas IDeA Network of Biomedical Research Excellence (KINBRE) under the grant P20GM103418 from NIH/NIGMS.
Footnotes
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH and Ferrando AA, MicroRNA expression profiles classify human cancers, Nature 7043 (2005) 834–838. [DOI] [PubMed] [Google Scholar]
- [2].Volinia S, Calin GA, Liu C, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C and Ferracin M, A microRNA expression signature of human solid tumors defines cancer gene targets, Proc. Natl. Acad. Sci. U. S. A, 7 (2006) 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Esquela-Kerscher A and Slack FJ, Oncomirs-microRNAs with a role in cancer, Nat. Rev. Cancer, 4 (2006) 259–269. [DOI] [PubMed] [Google Scholar]
- [4].Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G and Liu Y, miR2Disease: a manually curated database for microRNA deregulation in human disease, Nucleic Acids Res,1 (2008) D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cheng Y, Zhang X, Li Z, Jiao X, Wang Y and Zhang Y, Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification, Angew. Chem. Int. Ed, 18 (2009) 3318–3322. [DOI] [PubMed] [Google Scholar]
- [6].Jia H, Li Z, Liu C and Cheng Y, Ultrasensitive detection of microRNAs by exponential isothermal amplification, Angew. Chem. Int. Ed, 32 (2010) 5498–5501. [DOI] [PubMed] [Google Scholar]
- [7].Baker M, MicroRNA profiling: separating signal from noise, Nat. Methods, 9 (2010) 687–692. [DOI] [PubMed] [Google Scholar]
- [8].Várallyay É, Burgyán J and Havelda Z, MicroRNA detection by northern blotting using locked nucleic acid probes, Nat. Protoc, 2 (2008) 190–196. [DOI] [PubMed] [Google Scholar]
- [9].Pall GS and Hamilton AJ, Improved northern blot method for enhanced detection of small RNA, Nat. Protoc., 6 (2008) 1077–1084. [DOI] [PubMed] [Google Scholar]
- [10].Koshiol J, Wang E, Zhao Y, Marincola F and Landi MT, Strengths and limitations of laboratory procedures for microRNA detection, Cancer Epidem. Biomar, 4 (2010) 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V and Lianidou ES, Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non–small cell lung cancer by quantitative real-time RT-PCR, Clin. Chem, 10 (2008) 1696–1704. [DOI] [PubMed] [Google Scholar]
- [12].Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR and Andersen MR, Real-time quantification of microRNAs by stem-loop RT-PCR, Nucleic Acids Res, 20 (2005) e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moltzahn F, Olshen AB, Baehner L, Peek A, Fong L, Stöppler H, Simko J, Hilton JF, Carroll P and Blelloch R, Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res, 71(2) (2011) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miao P, Wang B, Meng F, Yin J and Tang Y, Ultrasensitive detection of microRNA through rolling circle amplification on a DNA tetrahedron decorated electrode. Bioconjugate Chem, 26 (2015) 602–607. [DOI] [PubMed] [Google Scholar]
- [15].Yao B, Liu Y, Tabata M, Zhu H and Miyahara Y, Sensitive detection of microRNA by chronocoulometry and rolling circle amplification on a gold electrode. Chem. Commun, 50 (2014) 9704–9706. [DOI] [PubMed] [Google Scholar]
- [16].JamesáYang C, A T7 exonuclease-assisted cyclic enzymatic amplification method coupled with rolling circle amplification: a dual-amplification strategy for sensitive and selective microRNA detection, Chem. Commun, 50 (2014) 1576–1578. [DOI] [PubMed] [Google Scholar]
- [17].Jonstrup SP, Koch J and Kjems J, A focus on microfluidics and nanotechnology approaches for the ultra sensitive detection of microRNA, RNA, 12 (2006) 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cheng Y, Zhang X, Li Z, Jiao X, Wang Y and Zhang Y, Highly sensitive determination of microRNA using target‐primed and branched rolling‐circle amplification, Angew. Chem. Int. Ed, 121 (2009) 33183322. [DOI] [PubMed] [Google Scholar]
- [19].Liu H, Li L, Duan L, Wang X, Xie Y, Tong L, Wang Q and Tang B, High specific and ultrasensitive isothermal detection of microRNA by padlock probe-based exponential rolling circle amplification Anal. Chem, 85 (2013)7941–7947. [DOI] [PubMed] [Google Scholar]
- [20].Zhou Y, Huang Q, Gao J, Lu J, Shen X and Fan C, A dumbbell probe-mediated rolling circle amplification strategy for highly sensitive microRNA detection, Nucleic Acids Res, 38 (2010) e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lingam S, Beta M, Dendukuri D and Krishnakumar S, A focus on microfluidics and nanotechnology approaches for the ultra sensitive detection of microRNA, MicroRNA, 3(2014) 18–28. [DOI] [PubMed] [Google Scholar]
- [22].Ahrberg CD, Manz A and Chung BG, Polymerase chain reaction in microfluidic devices, Lab Chip, 16 (2016) 3866–3884. [DOI] [PubMed] [Google Scholar]
- [23].Cui L, Lin X, Lin N, Song Y, Zhu Z, Chen X and Yang CJ, Graphene oxide-protected DNA probes for multiplex microRNA analysis in complex biological samples based on a cyclic enzymatic amplification method. Chem. Commun, 48 (2012) 194–196. [DOI] [PubMed] [Google Scholar]
- [24].White AK, VanInsberghe M, Petriv I, Hamidi M, Sikorski D, Marra MA, Piret J, Aparicio S and Hansen CL, High-throughput microfluidic single-cell RT-qPCR, Proc. Natl. Acad. Sci. U. S. A, 108 (2011) 13999–14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Arata H, Komatsu H, Han A, Hosokawa K and Maeda M, Rapid microRNA detection using power-free microfluidic chip: coaxial stacking effect enhances the sandwich hybridization, Analyst, 137 (2012) 32343237. [DOI] [PubMed] [Google Scholar]
- [26].Chapin SC, Appleyard DC, Pregibon DC and Doyle PS, Rapid microRNA profiling on encoded gel microparticles, Angew. Chem. Int. Ed, 50 (2011) 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chapin SC and Doyle PS, Ultrasensitive multiplexed microRNA quantification on encoded gel microparticles using rolling circle amplification Anal. Chem, 83 (2011) 7179–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee H, Shapiro SJ, Chapin SC and Doyle PS, Encoded hydrogel microparticles for sensitive and multiplex microRNA detection directly from raw cell lysates, Anal. Chem, 88 (2016) 3075–3081. [DOI] [PubMed] [Google Scholar]
- [29].McArdle H, Jimenez-Mateos EM, Raoof R, Carthy E, Boyle D, ElNaggar H, Delanty N, Hamer H, Dogan M and Huchtemann T, “TORNADO” – Theranostic One-Step RNA Detector; microfluidic disc for the direct detection of microRNA-134 in plasma and cerebrospinal fluid, Sci. Rep, 7 (2017) 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sun XL, He WG, Jian YN, Lan FF, Zhang LN, Liu HY, Ge SG, Yu JH, Ultrasensitive microfluidic paper-based electrochemical/visual biosensor based on spherical-like cerium dioxide catalyst for miR-21 detection, Biosens. Bioelectron, 105 (2018) 218–225. [DOI] [PubMed] [Google Scholar]
- [31].Wang H, Jian YN, Kong QK, Liu HY, Lan FF, Liang LL, Ge SG and Yu JH, Ultrasensitive electrochemical paper-based biosensor for microRNA via strand displacement reaction and metal-organic frameworks, Sensors and Actuators B, 257 (2018) 561–569 [Google Scholar]
- [32].Sato K, Tachihara A, Renberg B, Mawatari K, Sato K, Tanaka Y, Jarvius J, Nilsson M and Kitamori T, Microbead-based rolling circle amplification in a microchip for sensitive DNA detection, Lab Chip, 10 (2010) 1262–1266. [DOI] [PubMed] [Google Scholar]
- [33].O Driscoll L, Expanding on exosomes and ectosomes in cancer, New Engl. J. Med, 372 (2015) 2359–2362. [DOI] [PubMed] [Google Scholar]
- [34].Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang W, Li P, Li M, Wang X, and Zhang C, Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth, Nature, 527 (2015) 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hoshino A, Costa-Silva B, Shen T, Rodrigues G, Hashimoto A, Mark MT, Molina H, Kohsaka S, Di Giannatale A and Ceder S, Tumour exosome integrins determine organotropic metastasis, Nature, 527 (2015) 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yeung CLA, Tsuruga T, Yeung T, Kwan S, Leung CS, Li Y, Lu ES, Kwan K, Wong K and Schmandt R, Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1, Nat. Commun 7 (2016) 11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Skog J, Würdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT Jr, Carter BS, Krichevsky AM and Breakefield XO, Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers, Nat. Cell Biol, 10 (2008) 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y and Liang M, Characterization of human plasma-derived exosomal RNAs by deep sequencing, BMC Genomics, 14 (2013) 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cheng L, Sharples RA, Scicluna BJ and Hill AF, Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood,J. Extracell. Vesicles, 3 (2014) 23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Witwer KW, Circulating microRNA biomarker studies: pitfalls and potential solutions, Clin. Chem, 61 (2015) 56–63. [DOI] [PubMed] [Google Scholar]
- [41].Eldh M, Lötvall J, Malmhäll C and Ekström K, Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods, Mol. Immunol, 50 (2012) 278–286. [DOI] [PubMed] [Google Scholar]
- [42].Ma D, Huang C, Zheng J, Tang J, Li J, Yang J and Yang R, Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering,Biosens. Bioelectron, 101 (2018) 167–173. [DOI] [PubMed] [Google Scholar]
- [43].Boriachek K, Umer M, Islam MN, Gopalan V, Lam AK, Nguyen N and Shiddiky MJ, An amplification-free electrochemical detection of exosomal miRNA-21 in serum samples, Analyst, 143 (2018) 1662–1669. [DOI] [PubMed] [Google Scholar]
- [44].Wei F, Yang J and Wong DT, Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM), Biosens. Bioelectron, 44 (2013) 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK and Gallichotte EN, Quantitative and stoichiometric analysis of the microRNA content of exosomes, Proc. Natl. Acad. Sci. U.S.A, 111 (2014) 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goddard JM and Erickson D, Bioconjugation techniques for microfluidic biosensors,Anal. Bioanal. Chem, 394 (2009) 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang T, Zhang M, Dreher DD and Zeng Y, Ultrasensitive microfluidic solid-phase ELISA using an actuatable microwell-patterned PDMS chip, Lab Chip, 13 (2013) 4190–4197 [DOI] [PubMed] [Google Scholar]
- [48].Shang Y, Zeng Y and Zeng Y, Integrated microfluidic lectin barcode platform for high-performance focused Glycomic profiling, Sci. Rep, 6 (2016) 20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].El-Khoury V, Pierson S, Kaoma T, Bernardin F and Berchem G, Assessing cellular and circulating miRNA recovery: the impact of the RNA isolation method and the quantity of input material, Sci. Rep, 6 (2016) 19529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tan GW, Khoo ASB and Tan LP, Evaluation of extraction kits and RT-qPCR systems adapted to high-throughput platform for circulating miRNAs, Sci. Rep, 5 (2015) 9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sourvinou IS, Markou A and Lianidou ES, Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability, J. Mol. Diagn, 15 (2013) 827–834 [DOI] [PubMed] [Google Scholar]
- [52].Podolska A, Kaczkowski B, Litman T, Fredholm M and Cirera S, How the RNA isolation method can affect microRNA microarray results, Acta Biochim Pol, 58 (2011) 535–40. [PubMed] [Google Scholar]
- [53].Monleau M, Bonnel S, Gostan T, Blanchard D, Courgnaud V and Lecellier C, Comparison of different extraction techniques to profile microRNAs from human sera and peripheral blood mononuclear cells, BMC genomics, 15 (2014) 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gaarz A, Debey-Pascher S, Classen S, Eggle D, Gathof B, Chen J, Fan J, Voss T, Schultze JL and Staratschek-Jox A, Schultze and A. Staratschek-Jox, Bead array–based microRNA expression profiling of peripheral blood and the impact of different RNA isolation approaches, J. Mol. Diagn, 12 (2010) 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Witwer KW, Circulating microRNA biomarker studies: pitfalls and potential solutions Clin. Chem, 61(2015) 56–63. [DOI] [PubMed] [Google Scholar]
- [56].Moldovan L, Batte KE, Trgovcich J, Wisler J, Marsh CB and Piper M, Methodological challenges in utilizing miRNAs as circulating biomarkers,J. Cell. Mol. Med, 18 (2014) 371–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Saadatpour L, Fadaee E, Fadaei S, Mansour RN, Mohammadi M, Mousavi SM, Goodarzi M, Verdi J and Mirzaei H, Glioblastoma: exosome and microRNA as novel diagnosis biomarkers, Cancer Gene Ther, 23 (2016) 415–418. [DOI] [PubMed] [Google Scholar]
- [58].Melo SA, Sugimoto H, O Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT and Melo CA, Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis, Cancer Cell, 26 (2014) 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, Kalinina J, Hua W, Kesari S and Mao Y, MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development, Plos One, 8 (2013) e78115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Boyerinas B, Park S, Hau A, Murmann AE and Peter ME, The role of let-7 in cell differentiation and cancer, Endoc-Relat. Cancer, 17 (2010) F19–F36. [DOI] [PubMed] [Google Scholar]
- [61].Shi J, Considering exosomal miR-21 as a biomarker for cancer, J. Clin. Med, 5 (2016) 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Østergaard ME and Hrdlicka PJ, Pyrene-functionalized oligonucleotides and locked nucleic acids (LNAs): Tools for fundamental research, diagnostics, and nanotechnology, Chem. Soc. Rev, 40 (2011) 5771–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]