Abstract
Background
Gut microbiota can affect human behavior and mood in many ways. Several studies have shown that patients with depression were also accompanied with gut microbiota disorder, in which Firmicutes are related to the protective function of intestinal barrier. In this study, we explore the changes and effects of Firmicutes in the patients with major depressive disorder (MDD).
Method
We recruited 54 subjects, including 27 patients with MDD. Fecal samples were collected for identification by 16S rRNA sequencing and bioinformatics analysis.
Results
The study shows that the alpha diversity indices of MDD patients are lower than those of the healthy controls. Firmicutes is the most significantly decreased phylum in the MDD samples. There are totally 13 taxonomic biomarkers with P-value <0.01 from Firmicutes. There are differences in 17 KEGG pathways between the two groups.
Conclusion
This study found that there is a significant disorder of gut microbiota in the patients with depression, in which the Firmicutes decreased significantly. Defects of the Firmicutes may lead to the depression in short-chain fatty acids, which could account for the physiological basis of low-level inflammation of depression.
Limitations
This is a cross-sectional study and the sample size is comparatively small. Though several diet-related factors were controlled in the study, there is no quantified assessment of it.
Keywords: gut microbiota, brain–gut axis, depression, Firmicutes, short-chain fatty acids
Introduction
Depression is a serious disabling health problem with high incidence worldwide;1 however, the mechanism of its occurrence and development remains unclear. Recent studies suggested that the microbial–gut–brain axis may affect people’s mood and behavior in a variety of ways. By interacting with the vagus nerve, directly changing the function of the central nervous system, affecting the intestinal nervous system, changing the plasticity of the brain,2 activating the immune system, and even in more ways,3,4 these conditional pathogenic bacteria can cause the disease. Increasing evidences were found which lead to the belief that the association between gut microbiota and depression is significant.
In the case of mice depression model, the changes of gut microbiota and fecal metabolic phenotype were found to correlate with depression through 16S rRNA sequencing and research methods based on liquid chromatography–mass spectrometry metabolomics.5 Moreover, three studies have shown that germ-free mice displayed more depression-like behavior after being transplanted gut microbiota from depressed people.6–8 These animal experiments suggest that the disorder of gut microbiota may cause depression. Furthermore, growing evidences of continuous low-level immune inflammatory reaction also cannot be ignored in the pathological process of depression development,5,9 since the source of this immune inflammation reaction is probably related to the disorder of gut microbiota. First, the bacteria of Firmicutes in gut microbiota can ferment carbohydrates to a variety of short-chain fatty acids (SCFAs),10 and the lack of these SCFAs can lead to decreased intestinal barrier function.11 Then, when many conditional pathogens and their metabolites in the intestinal tract cross the barrier, and stimulate the immune response, the “gut leaky” is formed, which can affect the occurrence and development of the disease.12 This can be supported by the study of Yu et al, which showed that there was a significant decrease in Firmicutes in depressed mice.13 Another study also found a significant correlation between stress-induced behavioral changes in mice and the disorder of Firmicutes in gut microbiota.14 In patients with inflammatory bowel disease (IBD), the amount of Faecalibacterium prausnitzii in Firmicutes is minimal, and the decrease in the proportion of bacteria was associated with the decrease in intestinal mucosal protection function.15 These studies suggest that Firmicutes, as a protective factor of the intestine, deserve further exploration.
It can be observed that Firmicutes and Bacteroidetes are still two major focuses in the human studies related to the gut microbiota and depression. At different levels, certain difference in gut microbiota has been shown between patients and the healthy control (HC) group, but the results of studies on Firmicutes are inconsistent. In the study by Jiang et al,16 it was found that there was a significant decrease in Firmicutes. However, in three other studies, there was no obvious difference of Firmicutes at phylum level.6 Furthermore, some bacteria associated with Firmicutes were decreased greatly at lower levels while others displayed certain increment. The inconsistency of these findings may be due to the following factors: 1) The reference HC group was not entirely normal. 2) The individual health status of the recruited patients was different. 3) The patients’ age span varied in those studies. 4) The effects of related treatment. 5) Differences in diet between typical symptoms and atypical symptoms of depression. Although the results of various studies are inconsistent, the disorder of Firmicutes can still be considered as one of the characteristics of patients with depression.
In order to explore a more certain correlation between the disorder of Firmicutes and the occurrence and development of depression, we adjusted inclusion criteria to better limit the possible interference of the abovementioned factors on gut microbiota so as to avoid the inconsistency appeared in previous studies. We are aiming to clarify the changes of Firmicutes in patients with depression and their related effects.
Materials and methods
Participants
This study was approved by the ethics committee of Peking University Sixth Hospital and Beijing Hospital of Chinese Traditional and Western Medicine. Clinical information was collected at Beijing Hospital of Chinese Traditional and Western Medicine. All subjects signed their written informed consent before participation. The clinical information and sample collection were conducted after obtaining all subjects’ informed consent, and the whole procedures conformed to the directives of the Declaration of Helsinki.
We recruited subjects following a redesigned inclusion criteria with emendation to previous studies.6,8,16–18 Some adjustments were made according to the specific medical standards in Beijing region. From March 30 to June 30, 2018, we recruited 30 patients with depression, in which 27 met the study criteria and formed the major depressive disorder (MDD) group; then 27 healthy subjects were selected as the HC group according to the MDD group’s age and sex. Both groups are Han Chinese residents living in Beijing for a long time, with no special eating habits and their BMI range from 18 to 30 kg/m2. The MDD group met the diagnostic criteria of ICD-10 MDD; they were at the first episode and without systemic antidepressive treatment. The depressive episodes caused by organic and substance abuse and those with atypical characteristics were excluded. The HC group was assessed by two certified attending physician to exclude any other mental illness.
In addition, by carefully examining the approaches applied in former researches,19 we put stricter limitation on the exclusion criteria as well. We reviewed the previous medical data provided by the subjects and none of the following subjects were included in this study: 1) suffering from any other chronic disease that may affect the stability of the gut microbiota, such as hypertension, diabetes mellitus, metabolic syndrome, immune deficiency, autoimmune disease, cancer, IBD, diarrhea in the last 3 months; 2) drugs that affect gut microbiota have been used in the last 6 months, including antibiotics, glucocorticoids, cytokines, large doses of probiotics and biological agents, and so on; 3) gastroscopy, colonoscopy, or barium meal in the digestive tract were performed in the last 6 months; 4) people who had major gastrointestinal surgery (cholecystectomy, appendicectomy, and intestinal tract resection) in the past 5 years; 5) people with restricted movement due to a major physical or mental illness; 6) people who had experienced significant dietary changes in the last 6 months; and 7) gestating women.
Clinical information collection
We collected general information of all subjects through questionnaires. The general information includes age, gender, race, height, weight, past medical history, drug history, smoking history, and drinking history.
16S rRNA amplification and sequencing
Participants’ fecal samples were put in sterile containers by themselves and collected at the fecal center by specialist. All 54 fresh fecal samples were stored at −80°C prior to DNA extraction. DNA was extracted from 200 mg fecal sample using PowerSoil DNA Kit (Missouri Biotechnology Association, Jefferson, MO, USA) and operating as per manufacturer’s instructions. The V3–V4 region of 16S rRNA was amplified and observed under universal primer pairs 341F (5′-GGACTACHVGGGTWTCTAAT-3′) and 805R (5′-ACTCCTACGGGAGGCAGCAG-3′) by KAPA HiFi HotStart ReadyMix (KAPA Biosystems, Inc., Wilmington, MA, USA). Unique 8 nt barcodes were added to primers in different samples. PCR was conducted at cycling conditions: 95°C for 5 minutes, 20 cycles of 98°C for 20 seconds, 58°C for 30 seconds, 72°C for 30 seconds, and 72°C for 5 minutes. We added 10 pmol primers and 100 ng templates to the 50 µL PCR reactions, then the PCR was performed in triplicate and the PCR products were pooled. QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) was used to select DNA segments in proper size. All selected DNA segments were sequenced in paired-end mode using Illumina HiSeq2500 in Novogene Bioinformatics Institute, Beijing, China.
Statistical analyses
Demographic analysis
The SPSS 23.0 statistical package for Windows was used for data analysis. Demographic data and clinical characteristics were compared between groups. Continuous variables were performed by independent samples t-test. Level of significance was set at 0.05 (two-tailed).
Sequencing data analysis
Raw reads were demultiplexed using seqtk (https://github.com/lh3/seqtk). Paired-end reads were merged using FLASH and quality filtered using Trimmomatic: pairs with >15 nt overlap were merged;20,21 merged sequences were trimmed where average quality score over 4 bases window was <20, and sequences containing ambiguous bases or <400 bp were removed. All qualified sequences were pooled and operational taxonomic units (OTUs) were picked using pick_open_reference_otus.py script from QIIME 1.9.1,22 and chimeras were removed by aligning sequences to the “gold” reference database using UCHIME. OTU sequences were assigned to taxonomy using assign_taxonomy.py from QIIME. All representative sequences (OTUs) were mapped against Greengenes database23 using UCLUST algorithm with 97% identity.24 Representative sequences were aligned by mafft,25 and phylogenetic tree was generated by FastTree using QIIME.26 Singletons and OTUs only occur in one sample were removed, and OTU table was rarefied using QIIME.
ACE, Chao1, and Shannon diversity values were calculated using vegan,27 and statistical tests were performed using R.28 Faith’s phylogenetic diversity was analyzed using alpha_diversity.py and compare_alpha_diversity.py from QIIME. Unweighted and weighted Unifrac distances were calculated using beta_diversity.py from QIIME, and principal coordinates analysis (PCoA) was performed using R. Significance test of Faith’s phylogenetic diversity was performed using Monte Carlo permutation test in QIIME, and all other significance tests were performed using Wilcoxon test in R.
Taxonomic biomarkers of HC and MDD groups were analyzed using LEfSe (linear discriminant analysis [LDA] Effect Size), and the taxa with P-value <0.01 and LDA score >2.0 were picked out as biomarkers. The metagenome functional profiling was predicted using PICRUSt,29 and the de novo OTUs were removed before the functional profiling prediction according to the manual of PICRUSt. Predicted KOs (KEGG orthology) and pathways were analyzed using STAMP,30 and P-value <0.01 was used to pick out differential KOs and pathways between HC and MDD samples.
Results
Demographic data and clinical characteristics of the subjects
We totally recruited 54 subjects, including 27 patients with MDD and 27 HCs; both groups had the same male to female ratio of 7:20. The mean age of the patient group was 48.7±12.8 and HCs was 42.3±14.1. As the table shows, there was no significant difference in age, height, weight, and BMI between the two groups (Table 1).
Table 1.
Demographic and clinical characteristics
| Characteristics | MDD (mean ± SD) | HC (mean ± SD) | t-value | P-value |
|---|---|---|---|---|
|
| ||||
| Age | 48.7±12.8 | 42.3±14.1 | 1.749 | 0.086 |
| Height | 1.65±0.1 | 1.65±0.1 | 0.390 | 0.698 |
| Weight | 65.4±11.0 | 63.6±8.5 | 0.693 | 0.492 |
| Body mass index | 23.8±2.8 | 23.4±2.9 | 0.488 | 0.628 |
Abbreviations: HC, healthly control; MDD, major depressive disorder.
OTU picking
Raw sequencing read pairs of samples range from 11,015 to 1,035,838, and the length of reads is 250 bp. After merging pair-end reads, quality filtering, and OTU clustering, the available sequences of samples are between 3,505 and 662,238. The utilization rate of total reads is 52.26%. After OTUs picking and taxonomy assignment, 2,888 OTUs were picked from all sequences, and 183 known taxa were identified.
Lower gut microbiota diversity in MDD patients
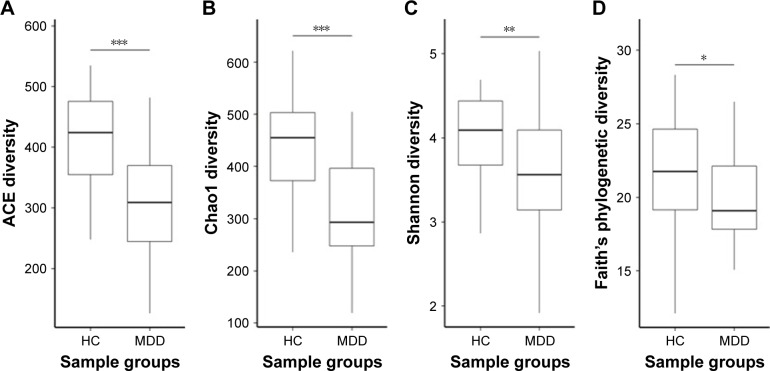
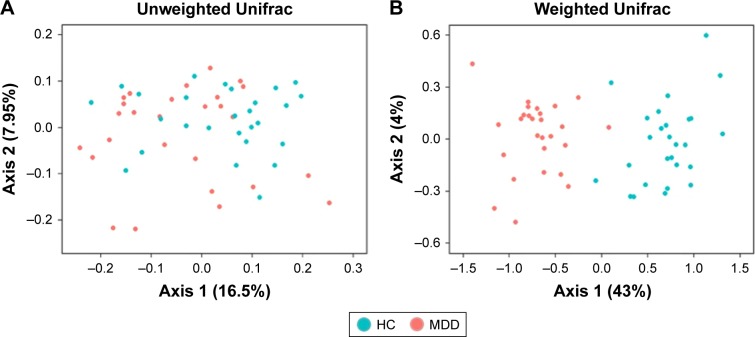
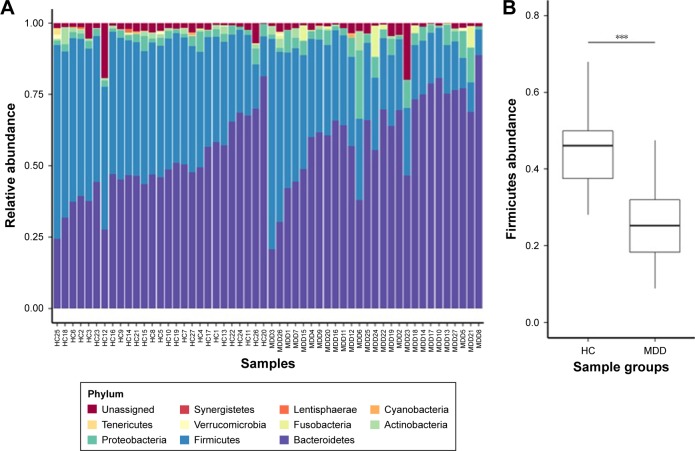
Our results show that alpha diversity indices of HC are higher than those of MDD patients (Figure 1). Chao1 and ACE diversity indices can be used to assess the species richness of samples. These two indices are both significantly higher in HC than in MDD (P<0.0008, Wilcox test), indicating that there are richer species in healthy people. The Shannon index can be used to estimate the species evenness and richness of sample, which is significantly higher in HC than in MDD samples (P=0.003, Wilcox test), indicating that healthy people have higher species diversity. The Faith’s phylogenetic diversity index can be used to estimate the phylogenetic diversity of species within a sample, and this index is also significantly higher in HC than in MDD samples (P=0.04, Monte Carlo permutation test). These all indicate that there is a significant decrease in the diversity of gut microbiota in MDD than in HC people. The PCoA plot based on weighted Unifrac distance also shows that the samples from MDD and HC are obviously different in community profile (Figure 2). After taxonomy assignment, the relative abundance of Bacteroides and Firmicutes are the highest two phyla in both HC and MDD samples, which together add up to 92% relative abundance in HC samples and 90% in MDD samples (Figure 3A). One other major difference between HC and MDD samples is the percentage of Firmicutes phylum (Figure 3B). The average relative abundance of Firmicutes in HC samples is 43.46%, while in MDD samples it is only 28.72% (P=0.00016, Wilcox test).
Figure 1.
Alpha diversity of HC and MDD samples.
Notes: (A–D) The ACE, Chao1, Shannon, and Faith’s phylogenetic diversity of HC and MDD calculated using rarefied OTU matrix. All four diversity indices in HC are significantly higher than MDD. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: HC, healthly control; MDD, major depressive disorder; OTU, operational taxonomic unit.
Figure 2.
Beta diversity of HC and MDD.
Notes: (A and B) PCoA plot of unweighted Unifrac and weighted Unifrac distance of HC and MDD samples using rarefied OTU matrix. Green dots represent HC samples and red dots represent MDD samples.
Abbreviations: HC, healthly control; MDD, major depressive disorder; OTU, operational taxonomic unit; PCoA, principal coordinates analysis.
Figure 3.
Taxa in phylum level of HC and MDD.
Notes: (A) The chart plot of taxa in HC and MDD samples. (B) The statistical test of Firmicutes relative abundance between HC and MDD samples. ***P<0.001.
Abbreviations: HC, healthly control; MDD, major depressive disorder.
Taxonomic biomarkers in HC are all from Firmicutes
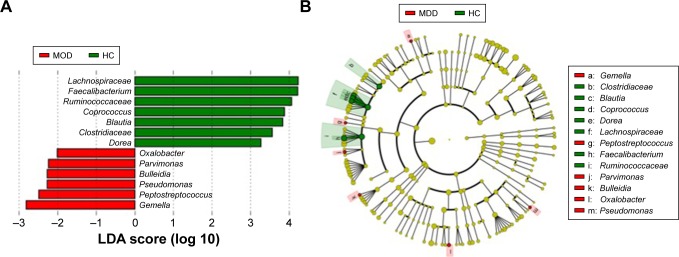
In total, there are 13 taxonomic biomarkers found with P-value <0.01 (Kruskal–Wallis test) and LDA score (log 10) >2.0, and among which seven are enriched in HC and six are enriched in MDD (Figure 4). The six biomarkers in HC are all from Firmicutes, including Lachnospiraceae, Ruminococcaceae, Coprococcus, Blautia, Clostridiaceae, and Dorea. The six biomarkers enriched in MDD are from Proteobacteria (Oxalobacter and Pseudomonas) and Firmicutes (Parvimonas, Bulleidia, Peptostreptococcus, and Gemella).1 This suggests that Firmicutes is the most important phylum that is correlated to depression.
Figure 4.
Taxonomic biomarkers in HC and MDD.
Notes: (A) Taxonomic biomarkers found by LEfSe in HC (green) and MDD (red). All taxa enriched in HC are from Firmicutes. (B) Cladogram plot of biomarkers. The size of node represents the abundance of taxa. Only taxa with P-value <0.01 and LDA score (log 10) >2 are shown.
Abbreviations: HC, healthly control; LEfSe, LDA effect size; MDD, major depressive disorder.
Function profiling prediction
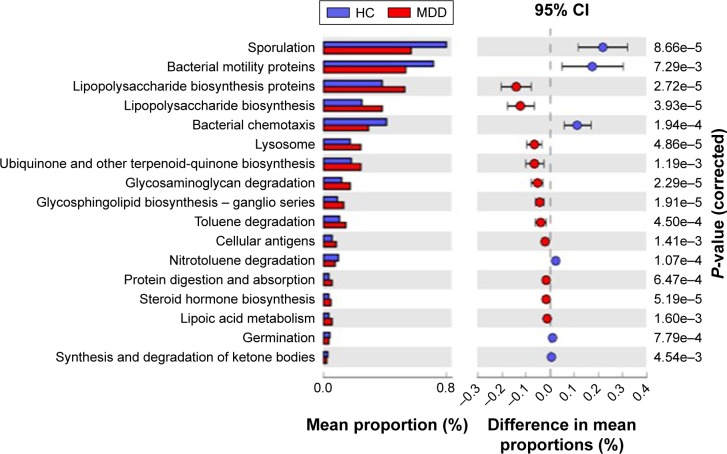
There are eleven KEGG pathways enriched in MDD (P<0.01, Welch’s t-test), including lipopolysaccharide biosynthesis, ubiquinone and other terpenoidquinone biosynthesis, glycosaminoglycan degradation, glycosphingolipid biosynthesis, toluene degradation, cellular antigens, protein digestion and absorption, steroid hormone biosynthesis, lipoic acid metabolism. Six pathways are enriched in HC, including sporulation, bacterial motility proteins, bacterial chemotaxis, nitrotoluene degradation, germination, synthesis and degradation of ketone bodies (Figure 5). These micro-biota change in MDD patients and the effects in metabolite could be further explored in future studies.
Figure 5.
Predicted differential KEGG pathways in HC and MDD.
Notes: The extended error bar plot of significantly differential KEGG pathways predicted using PICRUSt. Only P-value <0.01 are shown.
Abbreviations: HC, healthly control; MDD, major depressive disorder.
Discussion
Firmicutes account for about 40%–65% of the colon or fecal microbiota. According to the results of previous 16S rRNA sequencing, the dominant abundance flora includes three major Clostridium clusters (IV, IX, and XIV), while other clusters have lower abundance.10 In our study, it shows that the overall content of Firmicutes in patients with depression was significantly lower than that in the healthy group; this is consistent with the findings of Jiang et al.16 At the genus level, the significantly reduced genus of Firmicutes mainly fell into three families, which are the Faecalibacterium of the Ruminococcaceae and the Dorea, while Coprococcus of Lachnospiraceae have the most significant difference (P<0.001). These genera belong to Clostridium cluster IV and XIVa, respectively, and can metabolize various carbohydrate substrates to form various SCFAs such as acetate, butyrate, and lactate.10 The reduction of these fermentation-related bacteria leads to a decrease in the production of SCFA, which in turn leads to intestinal barrier dysfunction.11 This natural barrier function is weakened, multiple antigenic substances are exposed, and the weak intestinal tract becomes the source of inflammation.
Previous studies emphasized that SCFAs produced in the gut play an important role in improving chronic inflammatory diseases and promoting colonic epithelial cells. It has been reported that SCFAs can inhibit the production of proinflammatory cytokines, enhance IL-10 expression, activate regulatory T cells (Tregs), and alleviate colonic inflammation.31,32 SCFAs mainly include acetate, propionate and butyric acid, which have significant effects on intestinal epithelial cell proliferation, differentiation, and metabolism. Among them, butyrate not only can provide energy for the long epithelium but also strengthen the colon defense barrier. In addition, butyric acid can also play a role in immunoregulating cell cycle inhibition, inducing programmed cell death, and cell differentiation in various cell types. Recent evidence suggests that butyrate and propionate are keys to the regulation of Foxp3+ production of Tregs, whereas Tregs play an important role in suppressing inflammatory responses.33 Since most bacteria that produce butyric acid belong to the Firmicutes,11 with the decrease of Firmicutes, these protective factors are weakened, and the body is further exposed to the risk of inflammation.
A number of studies have indicated that cytokines and inflammation are closely related to depressive symptoms in patients with depression. It has been suggested that depression can be seen as a group of symptoms caused by peripheral inflammation and a response to inflammation.34,35 A meta-analysis suggests that the concentration of IL-6 and TNF-α in the blood is significantly elevated in depression without any physical illness.36 A large number of longitudinal studies have shown that exogenous cytokines can aggravate depressive symptoms.37–41 Similarly, injection of lipopolysaccharide endotoxin or related vaccine can increase both proinflammatory cytokine concentrations and depressive symptoms.42–44 In the mice study of Zhang et al, it was found that there was a significant decrease of Firmicutes in the social defeat stress model, while the change of proteobacteria was not significant. They also found that intravenous injection of MR16-1 induce antidepressant effects by normalizing the altered composition of the gut microbiome.45 This is consistent with our conclusion.
In addition, some cell-mediated immune processes may also be involved in the development of depression.46 Two meta-analyses have indicated that there are multiple activations of cellular immune pathways in patients with depression.36,47 Although there is currently no direct evidence that low-grade inflammation in depressive patients is derived from the intestine, there is increasing evidence that gut microbiota is important in causing this inflammatory process. Although some preclinical and clinical studies have confirmed the positive effects of probiotic supplementation on depressive symptoms, a meta-analysis showed that probiotic supplementation has an overall insignificant effect on mood.48 Therefore, it is still necessary to further clarify the changes of gut microbiota in depression, which might help the targeted supplementary to achieve a better effect.
In conclusion, our study found that there is a significant disorder of gut microbiota in patients with depression, in which the Firmicutes decreased significantly. Defects of the Firmicutes may lead to the depression in SCFA, which may be the physiological basis for low-level inflammation of depression. In the future, we can further explore the role of Firmicutes in depression through the method of multiomics.
Limitation
This study still has some limitations. First, the sample size we used was comparatively small due to financial limit. Second, although the results of this study support that gut microbiota plays a role in the development of depression, we are currently unable to investigate how exactly gut microbiota changed along with this process. In future study, the changes of gut microbiota should be further observed in high-risk groups throughout their possible development of symptom. Third, there is a lack of relevant inflammatory indicators in this study. Finally, even though we carefully selected subjects aiming to reduce the influence of related factors on gut microbiota, some confounding factors, such as diet, still need more control or detailed assessments. Moreover, atypical symptoms of depression, such as greediness and somnolence, could also have potential impact on gut microbiota, which requests a more detailed classification of depression in future research.
Acknowledgments
We thank all the subjects who took part in this study. All the costs of this study are self-financed.
Footnotes
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289(23):3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 2.Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O’Leary OF. Adult hippocampal neurogenesis is regulated by the microbiome. Biol Psychiatry. 2015;78(4):e7–e9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 3.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Maes M, Kubera M, Leunis JC. The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuroendocrinol Lett. 2008;29(1):117–124. [PubMed] [Google Scholar]
- 5.Kiecolt-Glaser JK, Derry HM, Fagundes CP. Inflammation: depression fans the flames and feasts on the heat. Am J Psychiatry. 2015;172(11):1075–1091. doi: 10.1176/appi.ajp.2015.15020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly JR, Borre Y, O’Brien C, et al. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–118. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Guo K, Zeng L, et al. Metabolite identification in fecal microbiota transplantation mouse livers and combined proteomics with chronic unpredictive mild stress mouse livers. Transl Psychiatry. 2018;8(1):34. doi: 10.1038/s41398-017-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21(6):786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 9.Lotrich FE. Inflammatory cytokine-associated depression. Brain Res. 2015;1617:113–125. doi: 10.1016/j.brainres.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan SH, Louis P, Flint HJ. Cultivable bacterial diversity from the human colon. Lett Appl Microbiol. 2007;44(4):343–350. doi: 10.1111/j.1472-765X.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- 11.Stilling RM, van de Wouw M, Clarke G, Stanton C, Dinan TG, Cryan JF. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem Int. 2016;99:110–132. doi: 10.1016/j.neuint.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494(7435):116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu M, Jia H, Zhou C, et al. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J Pharm Biomed Anal. 2017;138:231–239. doi: 10.1016/j.jpba.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Bangsgaard Bendtsen KM, Krych L, Sørensen DB, et al. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One. 2012;7(10):e46231. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Ling Z, Zhang Y, et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Lin P, Ding B, Feng C, et al. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord. 2017;207:300–304. doi: 10.1016/j.jad.2016.09.051. [DOI] [PubMed] [Google Scholar]
- 18.Naseribafrouei A, Hestad K, Avershina E, et al. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26(8):1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 19.Shen Y, Xu J, Li Z, et al. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross-sectional study. Schizophr Res. 2018;197:470–477. doi: 10.1016/j.schres.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 25.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oksanen J, Blanchet F, Kindt R, Legendre P. Vegan: Community Ecology Package Version 2.0. 2013 [Google Scholar]
- 28.R Development Core Team R . A Language and Environment for Statistical Computing. Vienna, Austria: The R Foundation for Statistical Computing; 2013. [Google Scholar]
- 29.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonnenburg ED, Zheng H, Joglekar P, et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6(12):5786–5805. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat Rev Neurosci. 2008;9(1):46–56. doi: 10.1038/nrn2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raison CL, Capuron L, Miller AH. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends Immunol. 2006;27(1):24–31. doi: 10.1016/j.it.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dowlati Y, Herrmann N, Swardfager W, et al. A meta-analysis of cytokines in major depression. Biol Psychiatry. 2010;67(5):446–457. doi: 10.1016/j.biopsych.2009.09.033. [DOI] [PubMed] [Google Scholar]
- 37.Caraceni A, Gangeri L, Martini C, et al. Neurotoxicity of interferon-alpha in melanoma therapy: results from a randomized controlled trial. Cancer. 1998;83(3):482–489. doi: 10.1002/(sici)1097-0142(19980801)83:3<482::aid-cncr17>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 38.Malaguarnera M, Di Fazio I, Restuccia S, Pistone G, Ferlito L, Rampello L. Interferon alpha-induced depression in chronic hepatitis C patients: comparison between different types of interferon alpha. Neuropsychobiology. 1998;37(2):93–97. doi: 10.1159/000026485. [DOI] [PubMed] [Google Scholar]
- 39.Pavol MA, Meyers CA, Rexer JL, Valentine AD, Mattis PJ, Talpaz M. Pattern of neurobehavioral deficits associated with interferon alfa therapy for leukemia. Neurology. 1995;45(5):947–950. doi: 10.1212/wnl.45.5.947. [DOI] [PubMed] [Google Scholar]
- 40.Raison CL, Dantzer R, Kelley KW, et al. CSF concentrations of brain tryptophan and kynurenines during immune stimulation with IFN-alpha: relationship to CNS immune responses and depression. Mol Psychiatry. 2010;15(4):393–403. doi: 10.1038/mp.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer M, Engelbrecht MA, Gut O, et al. Interferon alpha (IFN-alpha) and psychiatric syndromes: a review. Prog Neuropsychopharmacol Biol Psychiatry. 2002;26(4):731–746. doi: 10.1016/s0278-5846(01)00324-4. [DOI] [PubMed] [Google Scholar]
- 42.Brydon L, Walker C, Wawrzyniak A, et al. Synergistic effects of psychological and immune stressors on inflammatory cytokine and sickness responses in humans. Brain Behav Immun. 2009;23(2):217–224. doi: 10.1016/j.bbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geubelle F. Spectrophotometric semi-micro method for the determination of the hemoglobin-oxyhemoglobin ratio in the blood. Clin Chim Acta. 1956;1(3):225–228. doi: 10.1016/0009-8981(56)90068-7. [DOI] [PubMed] [Google Scholar]
- 44.Strike PC, Wardle J, Steptoe A. Mild acute inflammatory stimulation induces transient negative mood. J Psychosom Res. 2004;57(2):189–194. doi: 10.1016/S0022-3999(03)00569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JC, Yao W, Dong C, et al. Blockade of interleukin-6 receptor in the periphery promotes rapid and sustained antidepressant actions: a possible role of gut-microbiota-brain axis. Transl Psychiatry. 2017;7(5):e1138. doi: 10.1038/tp.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonard B, Maes M. Mechanistic explanations how cell-mediated immune activation, inflammation and oxidative and nitrosative stress pathways and their sequels and concomitants play a role in the pathophysiology of unipolar depression. Neurosci Biobehav Rev. 2012;36(2):764–785. doi: 10.1016/j.neubiorev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y, Ho RC, Mak A. Interleukin (IL)-6, tumour necrosis factor alpha (TNF-α) and soluble interleukin-2 receptors (sIL-2R) are elevated in patients with major depressive disorder: a meta-analysis and meta-regression. J Affect Disord. 2012;139(3):230–239. doi: 10.1016/j.jad.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 48.Ng QX, Peters C, Ho CYX, Lim DY, Yeo WS. A meta-analysis of the use of probiotics to alleviate depressive symptoms. J Affect Disord. 2018;228:13–19. doi: 10.1016/j.jad.2017.11.063. [DOI] [PubMed] [Google Scholar]