Abstract
Mbd3 (Methyl-CpG binding domain protein), a core member of NuRD (nucleosome remodelling and deacetylation) is essential for embryogenesis. However, its role in reprogramming of somatic cells into induced pluripotent stem cells (iPSC) remains controversial. Some reports suggest that Mbd3 inhibits pluripotency, whilst others show that it greatly enhances reprogramming efficiency. Our study is the first to assess the role of Mbd3 on reprogramming of primary human fibroblasts using Yamanaka episomal plasmids (Reprogramming factors (RF) under feeder-free conditions. We showed that shRNA-mediated partial depletion of Mbd3 resulted in >5-fold reduction in the efficiency of reprogramming of primary human fibroblasts. Furthermore, iPSC that emerged after knock-down of Mbd3 were incapable of trilineage differentiation even though they expressed all markers of pluripotency. In contrast, over-expression of the Mbd3b isoform along with the Yamanaka episomal plasmids increased the number of fibroblast derived iPSC colonies by at least two-fold. The resulting colonies were capable of trilineage differentiation. Our results, therefore, suggest that Mbd3 appears to play an important role in reprogramming of primary human fibroblasts, which provides further insight into the biology of reprogramming but also has direct implication for translation of iPSC to clinic.
Keywords: Mbd3, Induced pluripotent stem cells, Episomal reprogramming, Fibroblasts
Introduction
The pioneering work by Takahashi and Yamanaka showed that induced pluripotent stem (iPSC) cells can be generated from somatic cells by ectopic expression of four different transcription factors, originally Oct4, Sox2, Klf4 and Myc (1–3). Understanding the molecular mechanisms that regulate the pluripotency and self-renewal of iPSC is critical for regenerative medicine in generating target cells for transplantation. Reprogramming requires major epigenetic modifications of DNA and chromatin remodelling and several groups are investigating the role of Nucleosome Remodelling and Histone Deacetylase (NuRD) complex and in particular Mbd3 (Methyl-CpG binding domain protein). Mbd3 is ubiquitously expressed in all somatic cells and is required for formation of stable NuRD complex (4). Deletion of the Mbd3 gene causes embryonic lethality in mice. Additionally, embryonic stem cells lacking Mbd3 protein fail to commit to developmental lineages (4, 5). Whilst it is clear that Mbd3 is required for early embryonic development and stem cell fate decisions, its exact role in somatic cell reprogramming is currently unclear. Two reports suggest that Mbd3 inhibits pluripotency as its reduction or deletion leads to an enhanced kinetics of reprogramming with iPSC colonies appearing within 7 days and with an almost 100% reprogramming efficiency (6, 7). In contrast, a study by dos Santos et al. (8), showed that loss of Mbd3 expression, especially, during the initiation phase of reprogramming leads to a reduction in the efficiency of reprogramming. It is unclear if these disparate results reflect the different reprograming techniques used or a function of the target cells.
Here, we investigated the role of Mbd3 in direct reprogramming of primary human fibroblasts using episomal-plasmid based method (2, 3). Our results show that over-expression of Mbd3b significantly increased the yield of iPSC colonies and reduction of Mbd3 expression not only reduced the yield of iPSC colonies but repressed subsequent trilineage differentiation. Our findings suggest that Mbd3 plays a critical role in reprogramming of primary human fibroblasts at least in the context of episomal-based method of reprogramming.
Materials and Methods
Cell culture
All human primary fibroblast cells were generated in vitro after written informed consent using protocols approved by the Royal Free research ethics committee, Royal Free Hospital, London, UK. Fibroblasts were derived from a skin biopsy and cultured as described previously (3). We used PD1 (70 year old suffering from Parkinson disease) and RDP1 (49 year old with rapid onset Parkinsonism dystonia) as described previously (3). We also used BJ fibroblast cell line (a normal neonatal fibroblast cell line from ATCC).
We employed episomal plasmid method of reprogramming as described previously (2, 3) and used the following plasmids: pCXLE-hOct3/4-shp53 (Addgene number 27077), pCXLE-hSK (Addgene number 27078), pCXLE-hUL (Addgene number 27080), (the latter three referred to as reprogramming factors-(RF)), and pCXLE-EGFP (Addgene number 27082). For inhibition of Mbd3, we used pSMP-MBD3_1 (Addgene number 36371) (9). The Mbd3 isoform that we used for over-expression experiments was Mbd3b (8) which shares >80% homology to human Mbd3 (10) and is the most abundant isoform in embryonic stem cells (11). The outline of experiments is summarized in Fig. 1A. Briefly, cells were transfected using program U-023 on a Nucleofector (I) 2b device (Amaxa Nucleofector kit R) or Pulse code – EO150 (solution P2) on AMAXA 4D Nucleofector (LONZA). GFP transfection efficiency was measured using flow cytometry after a minimum of 24 hours of transfection. Established iPSC cell lines were maintained as described previously (3) and were passaged at 1 : 1~1 : 12 using gentle cell dissociation reagent (12). At day 25~30 post transfection, cells were either fixed for whole plate alkaline phosphatase staining as per manufacturer’s instructions or iPSC colonies were picked for expansion.
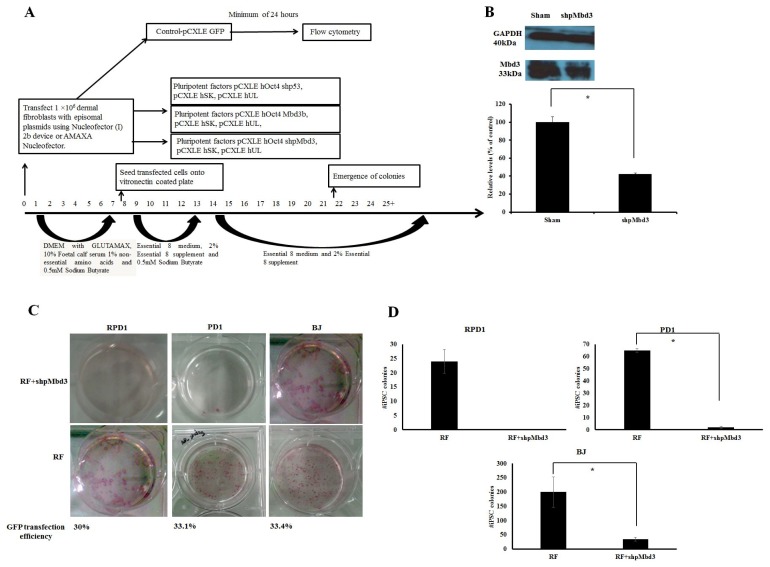
Fig. 1.
Mbd3 is required for Episomal Reprogramming. (A) Experimental design used to address the kinetics and efficiency of reprogramming in human dermal fibroblasts. Human fibroblasts were transfected with episomal plasmids encoding pCXLE-hOct3/4-shp53, pCXLE-hSK (Sox-2 and Klf4) and pCXLE-hUL (L-Myc and Lin28) (Reprogramming factors-RF) or RF+shpMbd3 or RF+Mbd3b or GFP only. After transfection, the cells were maintained in Essential 8 media supplemented with 0.5mM sodium butyrate until day 12 and thereafter maintained in Essential 8 media only. Transfection efficiency was measured by Flow Cytometry after a minimum of 24 hours of transfection with an average of 32.8±1.68%. (B) Immunoblot analysis for Mbd3 expression transfected with shpMbd3 compared to sham. Densitometry plot shows that cells transfected with shpMbd3 had 50% reduction in expression of Mbd3 compared to sham (p<0.05). (C) iPSC colony formation in fibroblasts was assessed by alkaline phosphatase (AP) staining between day 25~30 and the results showed that shpMbd3 reduced the number of iPSC colonies. (D) Quantification of AP staining further confirmed reduction of iPSC colonies formed in cells treated with RF+shpMbd3 compared with RF in patient fibroblasts RPD1 and PD1 and BJ neonatal cell line (n=3). Colony number is per 1×106 fibroblasts (p<0.05).
Immunoblotting
Immunoblotting was carried out as described previously (13). The blots were probed either with Mbd3 (ABCAM), α-tubulin (ABCAM), or GAPDH (Cell Signalling). Densitometry was performed using NIH image J software and a sampling window was used to measure the intensity of all bands on the blot and normalized against the band intensity of the internal control.
Characterisation of iPSC
Immunofluorescence was carried on established iPSC cell lines as described previously (3). Primary antibodies namely TRA-1-60 and TRA-1-80 (Santa Cruz Biotechnologies) were used at a 1 : 100 dilution followed by goat anti-mouse IgG/IgM Alexa 488 as a secondary antibody at a concentration of 1 : 400. The cells were visualised using Axiovision microscope equipped with 10× objective. For established iPSC cell lines, RNA was harvested with TRIzol and 1μg of RNA was used for subsequent reverse transcriptase reactions with Superscript III first strand synthesis system. For qRT-PCR cDNA was diluted in 1 in 20 and were set up in duplicates using SYBR Green Master Mix. Delta Ct values with GAPDH were calculated and brought to power −2. Error bars represent±SEM of technical duplicates. Oligonucleotide primers details have been described elsewhere (3). To demonstrate spontaneous in vitro differentiation, iPSC were grown to confluency and maintained in suspension in EB media. The expression of trilineage marker genes were assayed after 14 days of differentiation (3). Statistical significance was determined by non-parametric Mann-Whitney U-test and in all cases p≤0.05 was considered to be significant (Supplementary Methods).
Results
Mbd3 is required for reprogramming of primary human fibroblasts
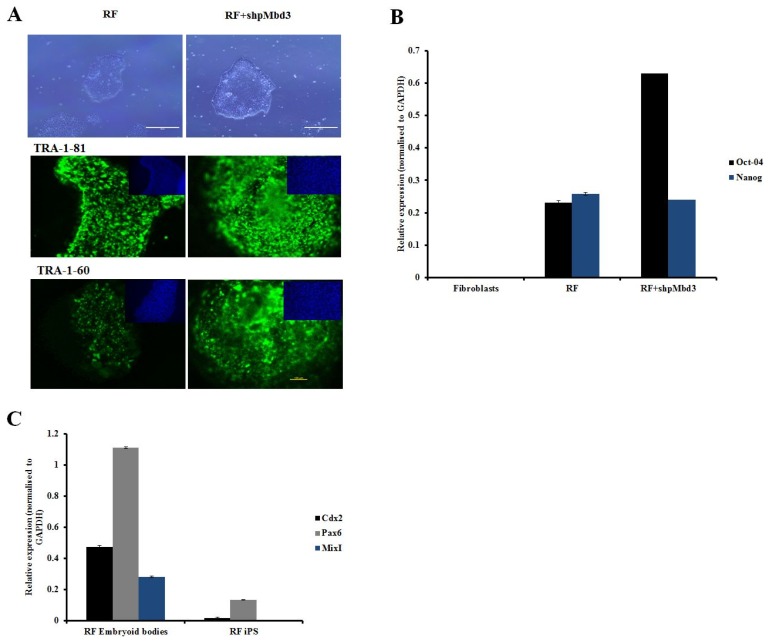
In order to assess the role of Mbd3 in establishment of pluripotency of human somatic cells, a plasmid construct expressing short-hairpin (shp) RNA targeted to human Mbd3 transcript (shpMbd3) was transfected together with Yamanaka episomal plasmids expressing human Oct3/4, shRNA against p53, Sox2, Klf4 and L-Myc/Lin28 (referred to as RF) in BJ foetal fibroblast cell line and two adult patient derived fibroblast lines; PD1 (70 years old with Parkinson’s disease) and RDP1 (49 years old with rapid onset of Parkinsonism dystonia) (Fig. 1A). GFP transfection efficiency of all three lines was comparable with mean (±SD) expression levels of 32.8±1.68%. Immunoblotting after 24~48 hours of transfection showed 50% knock-down of Mbd3 compared to sham (p<0.05) (Fig. 1B), but did not result in a difference in the proliferation rate of cells transfected with RF+shpMbd3 when compared with cells transfected with RF alone as seen morphologically. This suggests that partial knock-down of Mbd3 does not cause cell cycle arrest (8) of human fibroblasts, however, cell cycle analysis are required to further confirm this. iPSC colonies emerged at approximately the same time (days 12~20 post transfection) in the cells transfected with RF+shpMbd3 and RF alone cultures suggesting that Mbd3 does not alter the kinetics of reprogramming as reported previously (8). We used alkaline phosphatase activity as an early indicator of pluripotency prior to performing an extended assessment of pluripotent stem cell markers (14). Our results showed that the number of alkaline phosphatase positive colonies in cells transfected with RF+shpMbd3 was significantly reduced (p<0.05) by 5~33 fold in established BJ fibroblast cell line and in PD1 and RPD1 primary fibroblast cells compared to the numbers observed with cells transfected with RF alone (BJ mean (±SD): RF 200±52.9 and RF+shpMbd3 35±6.8; RPD1 mean (±SD): RF 24±4.2 and RF+shpMbd3 0±0; PD1 mean (±SD): RF 65±1.5 and RF+shpMbd3 2±1; (n=3) Fig. 1C, D). Interestingly, this effect was more profound in patient derived fibroblasts than in neonatal BJ fibroblast cell-line. The reason for this difference is unclear but could reflect the biological age of these lines and disease status of the patients. It was however, possible to expand iPSC colonies from cells treated with RF+ shpMbd3 although less efficiently and with delayed kinetics compared to colonies expanded from cells treated with RF only. Phenotypic characterisation of expanded iPSC colonies showed that they expressed TRA-1-60 and TRA-1-80 pluripotent markers with no difference in level of expression between cells treated with RF+shpMbd3 and RF only (Fig. 2A). Additionally, qRT-PCR analysis showed that iPSCs expanded from RF and RF+shpMbd3 expressed pluripotent genes such as Oct-4 and Nanog (Fig. 2B) at comparable levels. To investigate the role of NuRD in differentiation, iPSCs were maintained in suspension in EB media and under these conditions iPSCs formed aggregates called embryoid bodies which consists of a heterogeneous mix of derivatives of the three germ layers. iPSCs derived from RF+shpMbd3 failed to form EBs as previous reports (4). In contrast, EBs emerged from RF iPSCs with evidence of expression of trilineage markers such as Cdx2 (mesoderm), Pax6 (ectoderm) and MixI (endoderm) by qRT-PCR analysis (Fig. 2C).
Fig. 2.
Characterisation of iPSC colonies. (A) Representative phase images of iPSC derived from fibroblasts transfected RF and cells transfected with RF+shpMbd3. Morphologically they appear to be similar. Immunostaining with pluripotency markers (green) and counter staining with DAPI (blue) of iPSC derived from fibroblasts transfected with RF only and those transfected with RF+shpMbd3 with TRA-1-81 and TRA-1-60. Scale bar is 200μm. (B) Q-PCR showed expression of pluripotency genes (Oct-4 and Nanog respectively) in cells treated with RF and RF+ shpMbd3. All results were normalised to GAPDH. (n=3 Error bars±S.E.M). (C) Q-PCR showed up- regulation of lineage specific genes: Cdx2 (mesoderm), Pax6 (ectoderm) and MixI (endoderm) in cells treated with RF embryoid bodies relative to respective iPSC line (n=3 Error bars± S.E.M).
Over-expression of Mbd3b enhances reprogramming of human fibroblasts
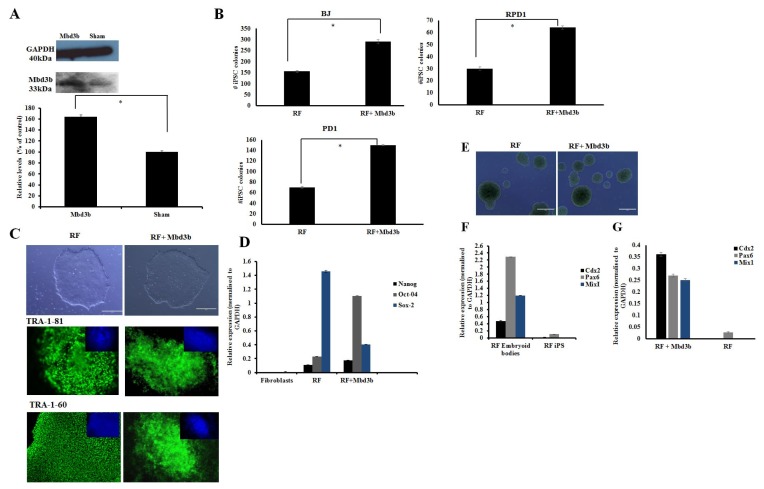
We next looked at over-expression of Mbd3b, a major isoform expressed in pluripotent stem cells (8, 11) and confirmed by immunoblotting an approximately 50% significant increase in the levels of Mbd3b (Fig. 3A). An almost two-fold significant (p<0.05) increase in the number of alkaline phosphatase colonies was observed in cultures transfected with RF+Mbd3b when compared with RF alone (Fig. 3B) (BJ: RF 155±5 and RF+Mbd3b 290±10; RPD1: RF 30±1.5 and RF+Mbd3b 64±1.3; PD1: RF 70±2.0 and RF+Mbd3b 150±1.0). iPSC colonies emerged at approximately the same time in cells treated with RF+Mbd3b and in RF alone cultures and the overall proliferation kinetics of iPSC between these two conditions were similar (data not shown). iPSC clones from both these conditions expressed pluripotent markers (TRA-1-81 and TRA-1-60) with evidence of endogenous expression of Nanog, Oct 4 and Sox 2 by qRT-PCR (Fig. 3C, D) with no difference in level of expression. Importantly, iPSCs derived from fibroblasts transfected with RF+Mbd3b formed EBs with comparable kinetics to that observed with cells treated RF alone (Fig. 3E) and expressed markers indicative of the three germ layers (Fig. 3F, G).
Fig. 3.
Over-expression of Mbd3b facilitates reprogramming. (A) Immunoblot analysis for cells over-expressing Mbd3b compared to sham. Densitometry plot showed that cells over-expressing Mbd3b increased its expression by almost 50% compared to sham (p<0.05). (B) Quantification of iPSC colonies generated from fibroblasts transfected with RF+Mbd3b compared to cells treated with RF in BJ neonatal cell line and patient fibroblasts (RPD1 and PD1). A two fold increase is seen in the number of colonies in RF+Mbd3b compared to RF only (p<0.05). (C) Representative phase images of iPSC of established cell lines transfected with RF and RF+Mbd3b. Immunostaining with pluripotency markers (green) and counter staining with DAPI (blue) of cells treated with RF and RF+Mbd3b with TRA-1-81 and TRA-1-60. Scale bar is 200μm. (Error bars±S.E.M). (D) Q-PCR showed expression of pluripotency genes (Oct-4, Nanog and Sox-2 respectively) in cells treated with RF and RF+Mbd3b. All results were normalised to GAPDH (n=3 Error bars±S.E.M). (E) Representative images of embryoid bodies (EBs) generated from RF (left), and RF+Mbd3b (right). (F) Q-PCR showed up-regulation of lineage specific genes: Cdx2 (mesoderm), Pax6 (ectoderm) and MixI (endoderm) in conditions treated with RF EBs and RF+Mbd3b EBs (G) relative to respective iPSC line.
Discussion
Our study is the first to investigate the role of Mbd3 in the induction of pluripotency in patient derived primary fibroblasts, a convenient source of cells for iPSC cell-based therapies. Therefore, the identification of factors that enhance reprogramming efficiency of primary fibroblast derived lines will have direct clinical relevance (3). Our results show that partial knock-down of Mbd3 in human fibroblasts reduced the reprogramming efficiency. The iPSCs expanded from fibroblasts transfected with RF+shpMbd3 displayed all the hallmarks of pluripotency suggesting that Mbd3 is not required for the establishment of pluripotency as reported previously (7, 8). However, the iPSCs expanded and derived from fibroblasts treated with RF+shpMbd3 failed to form EBs confirming that Mbd3 is essential for forward differentiation into trilineage progenitors (4, 8).
In contrast, we found that over-expressing Mbd3b along with Yamanaka episomal plasmids enhances reprogramming efficiency by a two-fold. Whilst both forms of Mbd3 (Mbd3a and Mbd3b) interact with NuRD components, over-expression of Mbd3b has been shown to result in more pronounced phenotype than other isoforms in reversing the effects of knock-down of Mbd3 (11). Our results showed that over-expression of Mbd3b along with RF increased the number of iPSC cell colonies formed without affecting the attributes of pluripotency including the ability generate embryoid bodies with evidence of trilineage differentiation. The in-vitro differentiation potential in this study provides with robust reproducible alternative to teratoma assay and is sensitive enough to assess the pluripotency characteristics of iPSCs (15). However, further studies in a teratoma model are nevertheless required to establish this aspect in an unequivocal manner.
Our results reported herein are in agreement with reports by Onder et al.; 2012 (9) dos Santos et al.; 2014 (8, 9) and Zhang et al.; 2016 (16) but are at odds with two other reports (6, 7). In our opinion, the latter studies are not directly comparable as they used different methods of reprogramming and target cell lines. However, future studies are required to show the effect of Mbd3 using retrovirus or lentiviral method of reprogramming fibroblasts. Interestingly, others have shown that the Mbd3-miR-134 axis plays a key role in induction of pluripotency with Mbd3 overcoming the repression of reprogramming caused by over-expression of miR-134 (16). How this happens is under investigation with recent data showing an interdependence among Mbd3/NuRD, Tet1 and 5hmC to maintain normal chromatin structure and gene regulation in pluripotent stem cells (17).
We, therefore, conclude that Mbd3 plays an important role in reprogramming of human fibroblast cells using the episomal plasmid-based system of reprogramming. Understanding detailed molecular mechanism of Mbd3 in reprogramming and their function in enhancing the reprogramming efficiency are worthy of further investigation, and will be helpful in deriving excellent quality of iPSCs to facilitate clinical application.
Supplementary Information
Acknowledgments
This work was supported by Medical Research Council MR/L022842/1. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Potential Conflict of Interest
The authors have no conflicting financial interest.
Supplementary data including supplementary methods can be found with this article online at http://pdf.medrang.co.kr/paper/pdf/IJSC/IJSC-11-s18036.pdf.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 3.Goh PA, Caxaria S, Casper C, Rosales C, Warner TT, Coffey PJ, Nathwani AC. A systematic evaluation of integration free reprogramming methods for deriving clinically relevant patient specific induced pluripotent stem (iPS) cells. PLoS One. 2013;8:e81622. doi: 10.1371/journal.pone.0081622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- 5.Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo M, Ling T, Xie W, Sun H, Zhou Y, Zhu Q, Shen M, Zong L, Lyu G, Zhao Y, Ye T, Gu J, Tao W, Lu Z, Grummt I. NuRD blocks reprogramming of mouse somatic cells into pluripotent stem cells. Stem Cells. 2013;31:1278–1286. doi: 10.1002/stem.1374. [DOI] [PubMed] [Google Scholar]
- 7.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M, Maza I, Mor N, Baran D, Weinberger L, Jaitin DA, Lara-Astiaso D, Blecher-Gonen R, Shipony Z, Mukamel Z, Hagai T, Gilad S, Amann-Zalcenstein D, Tanay A, Amit I, Novershtern N, Hanna JH. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 8.dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4:1. doi: 10.1186/1471-2164-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 12.Beers J, Gulbranson DR, George N, Siniscalchi LI, Jones J, Thomson JA, Chen G. Passaging and colony expansion of human pluripotent stem cells by enzyme-free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7:2029–2040. doi: 10.1038/nprot.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McIntosh J, Lenting PJ, Rosales C, Lee D, Rabbanian S, Raj D, Patel N, Tuddenham EG, Christophe OD, McVey JH, Waddington S, Nienhuis AW, Gray JT, Fagone P, Mingozzi F, Zhou SZ, High KA, Cancio M, Ng CY, Zhou J, Morton CL, Davidoff AM, Nathwani AC. Therapeutic levels of FVIII following a single peripheral vein administration of rAAV vector encoding a novel human factor VIII variant. Blood. 2013;121:3335–3344. doi: 10.1182/blood-2012-10-462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Štefková K, Procházkováv J, Pacherník J. Alkaline phosphatase in stem cells. Stem cells Int. 2015 doi: 10.1155/2015/628368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buta C, David R, Dressel R, Emgård M, Fuchs C, Gross U, Healy L, Hescheler J, Kolar R, Martin U, Mikkers H, Müller FJ, Schneider RK, Seiler AE, Spielmann H, Weitzer G. Reconsidering pluripotency tests: do we still need teratoma assays? Stem Cell Res. 2013;11:552–562. doi: 10.1016/j.scr.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Zheng Y, Sun Y, Zhang Y, Yan J, Chen Z, Jiang H. MiR-134-Mbd3 axis regulates the induction of pluripotency. J Cell Mol Med. 2016;20:1150–1158. doi: 10.1111/jcmm.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, Lahesmaa R, Orkin SH, Rodig SJ, Daley GQ, Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.