Abstract
Many exciting discoveries have recently revealed the versatility of RNA and its importance in a variety of functions within the cell. Since the structural features of RNA are of major importance to their biological function, there is much interest in predicting RNA structure, either in free form or in interaction with various ligands, including proteins, metabolites and other molecules. In recent years, an increasing number of researchers have developed novel RNA algorithms for predicting RNA secondary and tertiary structures. In this review, we describe current experimental and computational advances and discuss recent ideas that are transforming the traditional view of RNA folding. To evaluate the performance of the most recent RNA 3D folding algorithms, we provide a comparative study in order to test the performance of available 3D structure prediction algorithms for an RNA data set of 43 structures of various lengths and motifs. We find that the algorithms vary widely in terms of prediction quality across different RNA lengths and topologies; most predictions have very large root mean square deviations from the experimental structure. We conclude by outlining some suggestions for future RNA folding research.
1. Introduction
It is now widely recognized that RNA is a fundamental biological macromolecule with many biological functions at all stages of cellular life. Besides the well-accepted functional properties of messenger RNA, transfer RNA and ribosomal RNA, many new non-coding RNAs are now known to perform catalytic regulatory roles that are essential to an organism’s survival and evolution. Small interference RNAs (RNAis) have a remarkable role in gene silencing [1]; transfer-messenger RNAs (tmRNAs) direct the addition of tags to peptides on stalled ribosomes, thereby affecting protein stability and transport [2]; other small non-coding RNAs (ncRNAs) regulate messenger RNA stability and translation by base pairing at various positions with their target messenger RNAs [3, 4]; and recent findings indicate that microRNAs (miRNAs) can be associated with tumorigenesis by acting either as tumor suppressors [5–7] or oncogenes [8–11]. This astonishing versatility of RNA has also been exploited for nanodesign for biomedical and technological applications [12, 13]. For example, the mechanism of RNA interference (RNAi) to silence genes in a sequence specific manner is currently being exploited as a tool to design drugs and for antiviral therapy [14, 15]. More interesting examples of RNA applications are described in table 1. Clearly, more discoveries are yet to come, given the many novel non-protein-coding transcripts identified in the human genome [16]. Many of these RNAs have yet unknown functions, and new regulatory roles continue to emerge [17, 18].
Table 1.
List of applications of natural and artificial RNAs in medicine and nanodesign.
| RNA type | Definition | Application | References |
|---|---|---|---|
| Medicine | |||
| miRNA (microRNA) | Short RNA sequences that can regulate genes in the post-transcription process | miRNA can suppress a tumor or act as an oncogene | [5–11] |
| RNAi (RNA interference) | Gene silencing mechanism | RNAi can be used as a tool for probing gene function and rational drug design | [14, 15] |
| Ribozyme | RNA molecules with catalytic properties | Ribozymes are being explored as biosensors and potential therapeutics against inflammatory disorders | [19, 20] |
| sRNA (small RNA) | Small functional RNAs that are not translated into proteins | Virulence genes are induced or repressed through sRNA regulators | [21] |
| Nanodesign | |||
| TectoRNA square | Molecular nanodesign of RNA squares | Artificial RNA building blocks can be used as jigsaw puzzle units for building larger pieces of diverse geometry and complexity | [12, 13] |
| TokenRNA aptamer biosensor | A sequence-specific, label-free fluorescent biosensor | Fluorescent signal results in the presence of its target and magnesium | [22] |
| Nanoring and nanotube | Molecular design of hexagonal ring and tube units | The helical sequences of the building blocks can include siRNAs for drug delivery | [23] |
| pRNA/siRNA nanoparticle | Design of chimeric phi29 packaging RNA (pRNA)/siRNA nanoparticles | pRNA/siRNA particles target multiple tumor cells by siRNA delivery | [24, 25] |
The structural features of RNAs are of major importance to their biological functions because sequence alone does not provide sufficient functional information. Thus, one of the goals in RNA structural biology is to provide insights into how structure and dynamics lead to specific functions of RNA, either in free form or in interaction with various molecules, in the full cellular milieu.
Our focus in this article is on reviewing recent advances in RNA structure determination and assessing current 3D prediction methods. Section 2 reviews our basic understanding of RNA structure. Section 3 discusses new discoveries in RNA dynamics that are important for understanding RNA folding. Section 4 describes current advances and limitations in experimental techniques used to determine both secondary and tertiary structures. Section 5 summarizes the recent approaches and trends in the structure determination of RNA secondary structure. Section 6 reviews computational methods for RNA 3D structure prediction, and section 7 evaluates some of the recent algorithms. We conclude with a perspective comparing performance of 3D structure prediction for a single representative RNA data set.
Developments in RNA structure prediction studies have been previously extensively reviewed. In RNA secondary-structure prediction advances, Gardner and Griegerich [26] compared secondary-structure prediction methods using multiple-sequence alignment, while Mathews and Turner [27] presented progress in free energy minimization algorithms using dynamic programming. Shapiro et al [28] provided a comprehensive review on RNA secondary-structure prediction with a focus on pseudoknots and RNA 3D structure advances with a focus on manual methods.
A more recent review by Capriotti and Marti-Renom [29] compiled current computational databases, algorithms, and computer programs that are available to the community for purposes ranging from sequence analysis to structure prediction and comparison. Schroeder [30] recently reviewed prediction progress on viral RNAs. Here we focus on exploring new ideas that could improve RNA prediction and suggest further improvements by comparing the capabilities of current 3D predictions programs.
2. RNA structure
Understanding RNA structure and function relies on our ability to identify RNA’s major structural components. RNA molecules can be studied extensively at the secondary-structure level, where building blocks include helical stems and single-stranded regions such as hairpins, internal loops, and junctions (figure 1(a)). Stems are formed by complementary canonical Watson and Crick base pairs GC and AU, along with the GU wobble base pair. A hairpin is a single-stranded region that folds back on itself via regions of complementary base pairs. The single-stranded region between two stems is known as an internal loop, while a junction can be defined as the point of connection between three or more helical stems. Single-stranded regions can form pseudoknots when base pairs intertwine (figure 1(b)). These basic secondary-structure elements are pieced together via tertiary interactions to form compact structures of active RNAs.
Figure 1.
(a) RNA secondary structure is composed of stems and loop regions. Stems are formed by canonical GC, AU and GU wobble WC base pairs. Loop regions are formed by hairpins, internal loops and junctions. (b) Loop regions can form long-range interactions as in pseudoknots, or by using non-canonical base pairs. (c) Non-canonical base pairs: A486 (blue on the left) forms a trans Watson–Watson base pair with A511 (purple on the right), a trans Hoogsteen–Sugar base pair with G482 (red on top), and a cis Sugar–Sugar base pair with C505 (green at the bottom). The base pair interactions occur in the H. marismortui 23S rRNA structure (PDB 1S72).
Structural and comparative studies have suggested that RNA structure is largely composed of repetitive modular building blocks or motifs. In particular, RNA tertiary motifs are conserved structural patterns formed by pairwise interactions between nucleotides. These include base pairing, base stacking, and base–phosphate interactions [31]. RNAs possess remarkable interaction attributes where base pair interactions can be classified into twelve geometric families in terms of pairs of interacting edges, which can be Watson and Crick (WC), Hoogsteen (H), Sugar (S), and glycosidic bond orientations cis and trans [32] (see figure 1(c)). Furthermore, base–phosphate interactions are also common and a recent classification model has been proposed [33].
Graph theory methods have also been developed to represent RNA secondary structures since the 1970s, with the pioneering work by Waterman [34], extended by others [35–37]. Recent structural or topological representations using graph representation (trees and dual graphs) of RNA secondary structures like RAG [38] constitute innovative methods from an allied field that can help RNA science [39].
3. RNA folding: proteins, dynamics, pathways and pausing
Over the past decade, many experiments have unveiled new clues into the elegant complexity and dynamics of RNA folding. Besides forming intricate long-range RNA–RNA interactions to direct and stabilize the structure, the folding is strongly affected by the speed of elongation and site-specific pausing of the RNA polymerase, as well as interactions of the RNA molecule with proteins, solvent and small metabolites [40–42].
Our current understanding of how RNA folds is that it does so by a hierarchical process: helical elements in the secondary structure are formed, and then compact structures are formed using tertiary pairwise interactions and tertiary motifs [43]. However, further studies are suggesting that RNA folding is actually quasi-hierarchical [44–46], where rearrangements of secondary-structure elements caused by tertiary interactions can in turn trigger stable native folding.
The RNA folding pathway does not always proceed uniquely. Indeed, the free energy landscape of RNA folding is highly rugged, composed of different and even parallel trajectories where RNA chains can be easily kinetically trapped into intermediate metastable states. If the RNA falls into an intermediate structure state, folding can take longer because breaking the non-native base pairs is required to reach its native state [47]. Fortunately, long-range tertiary contacts along with solvent (e.g., water, ions) interactions can guide RNA to the correct folding [48]. These tertiary interactions work cooperatively to direct folding to the native state [49].
It should be noted, however, that for some RNAs, intermediate or alternative states are functional, as for riboswitches (mentioned later) [50], and RNA regions in viruses [51–54]. For instance, there are structural domains in the hepatitis D (HDV) virus, where both branched and unbranched conformations are formed for distinct functional roles [52]. In the turnip crinkle virus (TCV), different structural configurations control the processes of virus translation and replication [53].
In contrast to in vitro folding where unfolded RNA chains fold in the presence of magnesium ions (Mg2+), in the cellular context, nascent RNA molecules start to fold before transcription is complete. Thus, achieving the correct structure can depend on the speed of transcription. This presents a problem for helices composed of long-range strands such as helix H1 shown in figure 1(a). This is because, during transcription, the 5′-strand of H1 is transcribed first; thus, it needs to wait for the 3′-strand of H1 to be fully transcribed. During this time, alternative base pairs can form at the 5′-strand. To help minimize competition from alternative folds, RNA polymerase, the molecule in charge of RNA transcription, pauses to allow the formation of non-native temporary conformations that easily unfold when the 3′-strand becomes available. Such pausing prevents formation of super-stable non-native structures and thus constitutes a general strategy for facilitating the folding of long-range helices [55, 56].
In addition to tertiary interactions, solvent molecules such as water and cations (positive ions such as Mg2+ and K+) also help initiate and guide RNA folding into the native structure. Water molecules mediate coupled molecular motions throughout a folded RNA core, and non-canonical bifurcated base pairs are formed only in the presence of water [32, 57]. In addition, the high negative charge of an RNA molecule works against its folding into its native structure. Cations promote folding by reducing the repulsion between RNA phosphates. Thus, besides helping to stabilize tertiary interactions during folding, ion–RNA interactions influence stability, pathway diversity, and transition states. It has been emphasized [42, 58] that ions of at least two types modulate the electrostatic surface potential of the negatively charged RNA molecule: chelated ions which are held in direct contact with the RNA surface by electrostatic forces, and diffuse ions which accumulate near the RNA due to the RNA electrostatic field and remain largely hydrated.
Most RNAs within the cell are parts of RNA–protein complexes. Their binding can stabilize the RNA structure, induce conformational changes, and even act as RNA chaperones to guide folding. Large RNAs such as group I intron, RNase P, and the ribosomal RNA (rRNA) structure are often stabilized by RNA-binding proteins. For instance, a recent model for rRNA folding suggested that the rRNA tertiary structure is dynamic (flexible) in the absence of proteins and that alternative structural conformations can compete with each other. Thus, ribosomal proteins may not only stabilize rRNA tertiary interactions but might also change the path of assembly, avoiding RNA misfoldings [59]. Proteins also bind at different stages in the folding process, and a hierarchical protein binding is now recognized where primary binding proteins interact at an early stage, thus marking their importance in the assembly process. Other proteins bind at the end and are related more to functional roles.
Though RNA folding is a complex process, is sensitive to the environment, and possesses a network of folding transitions and pathways, many RNAs share a common organization of their helical elements. Indeed, analyses on current solved RNA 3D structures have shown that the majority of helical elements in junctions tend to arrange roughly in parallel and perpendicular configurations. These arrangements are stabilized by both RNA–RNA interactions as well as RNA–protein interactions [60–62].
Finally, Nature exploits the potential of RNA sequences to form multiple alternative metastable structures for implementing highly sensitive molecule switches capable of controlling gene expression at the level of the mRNA. RNA riboswitches are RNAs found within some messenger RNA (mRNA) that can change conformation upon binding small metabolites and thus terminate transcription or block translation. The tertiary structure of the riboswitch binding pocket is stabilized only upon ligand binding. As mentioned earlier, RNA viruses make use of transitions between metastable structural domains for different functions. Thus, riboswitches and structural domains within viruses exemplify the dynamic properties of RNA molecules.
4. RNA structure determination
Since the ‘Era of RNA awakening’ began [63], the erroneous perception of RNA as a simple molecule has dramatically changed, due in part to advances in RNA structure determination techniques. In the early 1970s, the first full RNA structure, the transfer RNA (tRNA), was obtained by crystallization techniques [64], providing many clues to RNA’s helical organization. A decade later, the discovery of catalytic RNAs revolutionized our understanding of RNA’s complex cellular roles. Indeed, the ligand-dependent conformations of riboswitches have shown that even small RNAs are structurally and functionally complex [50]. Similarly, the initial identification of the P4–P6 fragment of the group I intron, and later the larger domain P1–P9 and the group II intron, have unraveled the intricacies of long-range RNA–RNA interaction motifs such as the tetraloop receptor, ribose zipper, A-minor and other intriguing motifs. An important milestone was reached with high resolution structures of three ribosomal RNA (rRNA) subunits, including the 23S rRNA subunit, which is the largest RNA structure solved thus far [65–67]; these structures revealed arrays of RNA–RNA and RNA–protein interactions, which in turn can form higher order interaction patterns [49]. These pioneering and far-reaching works on the ribosome structure by A E Yonath, V Ramakrishnan, T A Steitz, and others were recognized in the 2009 Nobel Prize in Chemistry. These remarkable breakthroughs, however, also demand atomic-level structural and dynamic information for understanding RNA function.
4.1. Experimental techniques
Structural information has been determined from RNA molecules using numerous experimental strategies. Many of these experimental approaches have been complemented by computational approaches, resulting in important improvements on both the computational and experimental fronts. Below, we describe some of the experimental methods for RNA structure determination. For a more detailed review, see [68].
Fluorescence resonance energy transfer (FRET) is a method often used to analyze the global structure and even dynamics of RNA elements such as junctions [69, 70]. The strength of FRET is that it allows detecting when two elements in the structure are in close proximity (10–100 Å), thus helping to determine RNA’s global helical organization.
The 2′-hydroxyl acylation RNA (SHAPE) chemistry approach is an important probing technique recently developed by the Weeks group [71, 72]. The protocol exploits the nucleophilic reactivity of the ribose-2′-hydroxyl position, which strongly correlates with the nucleotide flexibility. Nucleotides in single-stranded regions are seen as flexible, while nucleotides involved in base pairs are more rigid. The main advantage is that the method is simple and there is no limit on the size of the RNA molecule. A few secondary- and tertiary-structure prediction programs have incorporated data from RNA SHAPE to improve the prediction accuracy [73–75].
Other structure-specific probe methods such as using dimethyl sulfate (DMS) [143], which can focus on certain nucleotides (e.g., adenines), or hydroxyl radical footprinting [77], which can explore the solvent accessibility to the RNA backbone, are also used to deduce RNA structured features. Both methods can be powerful but are laborious. Another approach is the use of microarrays for chemical mapping [78]. This method has been combined with dynamic programming algorithms for predicting RNA secondary structure, replacing other labor intensive approaches such as chemical mapping and enzymatic cleavage.
NMR spectroscopy is the well-known technique that exploits the magnetic properties of certain nuclei. NMR spectroscopy is an important tool for probing the structure and dynamics of RNA [79, 80]. In addition, NMR relaxation methods can be used to obtain dynamic data on timescales ranging from picoseconds to seconds. The main limitation on NMR is molecular size (~20 kDa). However, novel methods like that reported combining small-angle scattering (SAXS) and NMR techniques [63] can be used to predict larger RNAs, as applied recently for the 100 nt TCV RNA virus [81].
Of course, x-ray crystallography analysis, based on the growth of single well-ordered crystals, remains another invaluable method for detailed structural resolution. Well-ordered single crystals are required for structural studies by x-ray diffraction methods, and obtaining them is often the most difficult step of the structure determination process [82]. Most RNAs exist naturally as protein–RNA complexes, and the crystallization of these complexes has several advantages over the crystallization of RNA alone. X-ray crystallography has been successful in determining the largest RNA molecules so far, such as the large ribosomal subunits [65–67]. Nevertheless, technical difficulties still remain, such as the availability of many conformations and transitional states for RNAs. The dynamical nature of RNA molecules also complicates matters.
Other experimental techniques such as cryo-electron microscopy (cryo-EM) have recently undergone tremendous advances, such as those reported in the study of IRES virus domains [83]. Contrary to the crystallography case, there is no size limitation and the molecule does not need to be ordered or isolated from its complex. However, due to the difficulty of accurate image reconstruction, the resolution is still limited to 10 Å at best.
4.2. Current advances in RNA structure data
Advances in sequencing technology have made available a growing amount of RNA sequence information (figure 2), but the challenge of how to interpret these data remains unmet. The RNA secondary-structure and statistics database (RNA STRAND) [144] contains, as of May 2010, 4666 secondary structures. Furthermore, Rfam, a database of sequence and secondary-structure information from several RNA families, has recorded 1446 RNA families since its last update in January 2010 [85]. Each family can contain many thousands of RNA sequences (e.g., the 5S rRNA family has 57 054 sequences).
Figure 2.
Number of RNA structures deposited in the NDB nucleic acid database (http://ndbserver.rutgers.edu/) as of December 2009.
The structures of a large majority of RNA molecules remain to be solved. Despite many advances in RNA crystallography, NMR, and chemical modification, RNA structure determination is a difficult task. This makes the need for accurate prediction using computational methods especially urgent.
5. RNA secondary-structure prediction approaches
Predicting RNA secondary structures refers to the task of determining, given a single RNA sequence or multiple RNA sequences, the set of complementary canonical Watson and Crick GC, AU, and GU wobble base pairs that define helical stems, as well as the single-stranded regions such as hairpins, internal loops, and junctions (see section 2). Many computational programs designed for predicting RNA secondary structures have been developed over the past three decades. Approaches vary widely, including free energy minimization using thermodynamic parameters [76, 86, 87], knowledge-based predictions based on known RNA structures [88–90], comparative sequence alignment algorithms [76, 91–96], combinations of these, and others. Below we describe some of the current prediction programs. More examples are listed in table 2. Other reviews on specific approaches are also available [26, 28, 97–99].
Table 2.
List of some available programs for RNA secondary-structure prediction using a single-sequence or multiple-sequence comparison.
| Program | Approach | Description | References |
|---|---|---|---|
| Prediction using a single sequence | |||
| Mfold | Free energy minimization | Determine the set of base pairs that gives the minimum free energy using dynamic programming methods | [87] |
| RNAfold | Free energy minimization | Predict the minimum free energy structure using the thermodynamic parameter approach; it can also estimate base pair probabilities | [86] |
| RNAstructure | Free energy minimization | Adapt experimental constraints such as chemical modification and SHAPE to improve prediction | [103] |
| MC-Fold | Free energy minimization | Assemble secondary structures from nucleotide cyclic motifs | [75] |
| Contrafold | Knowledge based | Predict structures using statistical data and training algorithms | [89] |
| Sfold | Knowledge based | Calculate a set of representative candidates using statistical sampling methods and Boltzmann assemble | [104] |
| RNAShapes | Knowledge based | Search the folding space using the reduced concept of RNA shape | [105, 106] |
| Kinwalker | Free energy minimization, kinetic folding | Build secondary structures using mimics of co-transcriptional folding and near optimal structures of subsequences | [107] |
| MPGAfold | Genetic algorithm | Search for possible folding pathways and functional intermediates using a massively parallel genetic algorithm | [108, 109] |
| Kinefold | Free energy minimization, stochastic folding simulations | Predict structures using a co-transcriptional folding simulation approach where RNA helices are closed and opened in a stochastic process; it can also predict pseudoknots | [112,113] |
| Pknots | Free energy minimization, parameter approximations | Predict pseudoknots using a dynamic programming algorithm and thermodynamic parameters augmented with approximated parameters for thermodynamic stability of pseudoknots | [111] |
| Prediction using multiple sequences | |||
| RNAalifold | Free energy minimization, covariation analysis | Determine a consensus secondary structure from a multiple-sequence alignment using covariation analysis | [94] |
| ILM | Free energy minimization, covariation analysis | Predict base pairs using an iterative loop matching algorithm, where base pairs are ranked using thermodynamic parameters and covariation analysis; it can also predict pseudoknots | [95] |
| CentroidFold | Knowledge based | Predict secondary structures using the centroid estimator employed by Sfold and the maximum expected accuracy estimator used by Contrafold | [90, 114] |
| Dynalign | Free energy minimization, pairwise alignment | Compute the lowest free energy sequence alignment and secondary structure common to two sequences | [76, 115,116] |
| Carnac | Free energy minimization, pairwise alignment | Predict the common structure shared by two homologous sequence using similarities, energy and covariations | [96] |
| RNAforester | Structural comparison using tree alignment models | Represent RNA secondary structures by tree graphs and compare and align secondary structures via alignment algorithms for trees and other graph objects | [93] |
| KNetfold | Machine learning | Determine pairs of aligned columns from a consensus using multiple-sequence alignment | [88] |
5.1. Secondary-structure prediction using a single sequence
It is believed that the native RNA structure corresponds to a global minimum of the relevant free energy function. Therefore prediction programs focus on determining the free energy for a secondary structure. One of the first programs developed for predicting RNA secondary structures using a single sequence, called Mfold, was developed by Zuker and Stiegler [87] in 1981. It aims to find the set of base pairs that yields the minimum free energy using dynamic programming methods. Mfold uses thermodynamic parameters obtained from experiments near 37°C, the human body temperature, to estimate different base pairing and base stacking forces [100–102]. The thermodynamic parameters are then used in a potential function that approximates the overall energy as a sum of independent terms for different loops and base pair interactions.
Because thermodynamic parameters are measured experimentally, they are incomplete or subject to inaccuracies, and thus a great number of alternative suboptimal structures can fall near the predicted global energy minimum. Therefore, Mfold and other current free energy minimization programs such as RNAfold [86], which is part of the Vienna package, consider a sample of structures near the optimal free energy conformation. In addition, RNAstructure [103] is a another program based on thermodynamic parameters which can improve prediction by incorporating constraints from experimental data using 2′-hydroxyl acylation RNA (SHAPE) chemistry (described in section 4.1).
Parameter calculation from known RNA structures is another useful approach to RNA secondary-structure prediction. Programs like Contrafold [89] use statistical data and Bayesian approaches to train the algorithm on a large set of known secondary structures using base pair probabilities. Sfold [104] uses statistical sampling methods and a Boltzmann assemble to calculate a set of representative candidates for RNA secondary structure. Similarly, RNAShapes [105, 106] performs an abstract search over all predicted RNA shapes to produce a sample of the folding space.
A recent heuristic program called Kinwalker [107] is based on a kinetic folding algorithm that simulates the dynamic properties of RNA folding during the co-transcriptional process. The algorithm constructs the secondary structure by a stepwise combination of building blocks corresponding to subsequences and their thermodynamic near optimal structures. Kinwalker can be used to fold RNA up to 1500 nucleotides long. Similarly, MPGAfold [108] is a massively parallel genetic algorithm that predicts possible folding pathways and functional intermediates using parallel computing [109].
The case for RNA structure prediction with pseudoknots has been shown to be a non-polynomial (NP) complex problem [110]. However, special cases have been explored. Programs like Pknots [111] use dynamics programs with added approximations to thermodynamic parameters needed for pseudoknot calculations. Cao and Chen [145] recently developed a predictive model for pseudoknots with inter-helix loops. The model gives conformational entropy, stabilities and free energy landscapes from RNA sequences, on the basis of a model that includes volume exclusion. Kinefold [112, 113] uses a long-time-scale stochastic folding simulation approach where RNA helices are closed and opened in a stochastic process, and pseudoknots are predicted using topological and geometrical constraints.
5.2. Multiple-sequence alignment
With the steady increase in RNA sequence data, secondary-structure prediction for RNA has also been achieved by aligning multiple sequences [91, 117–119]. Comparative sequence analysis consists of aligning many RNA sequences and looking for patterns of sequence variability between two or more nucleotides. Sequence variability (i.e., covariation) can be observed because, in contrast to nucleotides that vary randomly, base pairs that are conserved by evolution vary by compensatory changes (e.g., a GC pair in one sequence can change into an AU in another sequence). These covariations make it possible to detect the base pairs that form the helical stems, and consequently predict a secondary-structure model. Alignments of RNA sequences also allow identifying functionally important regions that are often conserved.
Several approaches for comparative sequence analysis have been developed [26]. One approach consists of aligning the sequences and then folding them on the basis of that alignment. Examples include RNAalifold [94] and ILM [95] which can predict pseudoknots. Another approach that involves simultaneous alignment, folding, and inference of structure from a set of homologous sequences is present in Dynalign [76, 115, 116] and Carnac [96] are examples of this approach. Both programs combine free energy minimization and comparative sequence analysis. If no meaningful sequence conservation is encountered during the alignment, an alternative method is used, consisting of simultaneous folding and aligning. RNAforester [93] is such an example. Alignment prediction methods for pseudoknots are more reliable on multiple sequences. KNetfold [88] is an example of a consensus prediction method based on a multiple-sequence alignment.
Successful examples include the structure models for the 5S, 16S and 23S ribosomal RNAs solved by the Gutell’s lab using alignments from hundreds of sequences [91]. These models predict base pairs in very good agreement with experimental data [92]. Similarly, the Westhof group predicted models for the group I intron [117, 118] and the ribonuclease P RNA [119] using similar methods.
Resources available to the community at large for such alignments include Gutell’s lab database (www.rna.ccbb.utexas.edu/) of aligned sequences, secondary structures, and phylogenetic information for various RNA molecules [91]. The Rfam database also contains multiple-sequence alignments and consensus secondary structures for several RNAs [85].
5.3. Advances in and limitations to RNA secondary-structure prediction
Free energy minimization methods are based on a number of thermodynamic parameters for base pairing, base stacking, loop lengths and other motifs. Although expansions and recalculations of the parameters are now available [84, 100, 101, 120], thermodynamic approaches are still limited in terms of accuracy of the parameters and the incompleteness of the thermodynamic rules used. Alternately, analyzing suboptimal states of RNA structures in addition to the free energy minimum has been valuable. However, it has been reported that about 73% of known canonical base pairs are predicted by free energy minimization for sequences with less than 700 nucleotides [76]. Similarly, parameter calculation methods are limited to the availability of structural data. While kinetic folding algorithms such as Kinwalker, Kinefold, and MPGAfold that incorporate RNA dynamics are very promising because they simulate the dynamics of co-transcriptional folding (RNA folding with sequence elongation), they are still at an early stage.
Alignments of multiple RNA sequences present a challenge for two main reasons: (1) only four letters are used on the basis of sequence similarity, and (2) sequence alignment programs such as Blast [121] or Clustal [122] do not consider information from secondary-structure conservation. Both are limitations because structure has evolved much more slowly than sequences, and compensatory mutations such as G–C by A–U, not considered in current sequence alignment approaches, frequently occur since they conserve the secondary structure.
Current rigorous alignments of distantly related RNA sequences typically require consideration of both sequence and secondary structure and are best performed manually. However, efforts are under way, with the new formation of the RNA Structure Alignment Ontology [123]. These new approaches intend to generalize sequence alignments as a set of ‘correspondence’ relations between whole regions, rather than between individual nucleotides.
Despite great advances in RNA secondary-structure prediction, many limitations remain. Most prediction programs cannot predict pseudoknots, nor the multiple configurations accessible to one sequence (e.g., for a riboswitch or domains within viruses). Pseudoknots are important, because their probability of occurrence increases sharply with RNA size [124]. Prediction also becomes less accurate as the RNA size increases, and there is currently no approach for quantifying the likelihood or error of a particular prediction. Nonetheless, as mentioned in section 4.1, many secondary-structure prediction programs are beginning to incorporate additional experimental data to improve the prediction accuracy. Such hybrid approaches like RNAstructure will indeed make advances on both experimental and computational fronts.
6. RNA tertiary-structure prediction programs
Even though determining the secondary structure provides a blueprint of the RNA molecule, it is the knowledge of the three-dimensional structure that allows us to understand its function, as well as the possible interactions with other molecules. However, in contrast to the advances achieved in protein folding programs, RNA structure prediction is still at an early stage. Current 3D RNA folding algorithms require either manual manipulation or are generally limited to simple structures. Below we describe some of the most recently developed programs. Table 3 provides more details.
Table 3.
Some available programs for RNA tertiary-structure prediction and interactive manipulation.
| Program | Input | Model | Simulation method | Description/Web page | References |
|---|---|---|---|---|---|
| Automatic prediction | |||||
| iFoldRNA | Sequence | Coarse-grained three-bead model | Replica exchange, molecular dynamics | Uses discrete molecular dynamics and force fields to simulate RNA folding dynamics; http://troll.med.unc.edu/ifoldrna/ | [132, 133] |
| FARNA (Rosetta) | Sequence, secondary structure | Coarse-grained one-bead model | Fragment assembly, Monte Carlo | Uses 3-nt fragment library, Monte Carlo simulations and a potential function to predict the structure; http://www.rosettacommons.org/manuals/archive/rosetta3.0_user_guide/index.html | [125, 127] |
| NAST | Secondary structure, tertiary contacts | Coarse-grained one-bead model | Molecular dynamics | Performs molecular dynamics simulations guided by a knowledge-based statistical potential function; https://simtk.org/home/nast | [131] |
| MC-Fold/MC-Sym | Sequence, secondary structure | Nucleotide cyclic motif | Fragment assembly, Las Vegas algorithm | Predicts RNA secondary structures using free energy minimization with structure assembled using the fragment insertion Las Vegas algorithm; http://www.major.iric.ca/MC-Pipeline/ | [75] |
| Interactive manipulation | |||||
| RNA2D3D | Secondary structure | All-atom model | Interactive manipulation | Performs molecular mechanics and dynamics, and permits insertion of coaxial stacking, and manipulation of helical elements; http://www.ccrnp.ncifcrf.gov/~bshapiro/software.html | [136] |
| Assemble | Database of known fragments and motifs | All-atom model | Interactive manipulation | Constructs a 3D structure using the insertion of tertiary motifs, and permits manipulation of torsion angles; http://www.bioinformatics.org/assemble/ | No reference |
FARNA is an energy-based program developed by Das and Baker [125] that predicts RNA 3D structures from a sequence. It was inspired by the Rosetta low resolution protein structure prediction method [126]. To represent each base, FARNA’s model consists of a one bead of a coarse-grained model, using each base’s centroid as the bead origin. To capture local conformational correlations observed in solved RNAs, FARNA builds a 3D structure library consisting of 3-nt fragments taken from a large rRNA subunit, from which torsion angles and sugar puckering parameters are stored. Then a simulation using Monte Carlo methods is used to assemble fragments into native-like structures. The folding simulation is guided by a knowledge-based energy function that takes into account both backbone conformations and side-chain interaction preferences observed in solved structures. This function includes a term for the radius of gyration, a function to penalize steric clashes, and functions that favor base stacking and the planarity of both canonical and non-canonical base pairs. Knowledge from base pairs can also be incorporated. Constraints using structural inference of native RNAs have been used more recently through an experimental method called multiplexed hydroxyl radical (–OH) cleavage analysis (MOHCA) [127], to enable detection of tertiary contacts and improve FARNA’s prediction. For instance, when the 158-nucleotide P4–P6 domain of the group I intron (PDB 1GID) is compared to the structure predicted from sequence alone, the root mean square deviation (RMSD) value is 35 Å. In contrast, prediction using secondary structure and data from MOHCA gives an RMSD value of 13 Å [127].
Parisien and Major [75] developed a method for modeling RNA 3D structures using energy minimization that builds upon earlier work using predicted cyclic building blocks [128, 129]. The approach consists of a pipeline implementation of two programs: MC-Fold and MC-Sym. MC-Fold predicts RNA secondary structure using a free energy minimization function, and the fragment insertion of nucleotide cyclic motifs formed by base pair and base stacking interactions. MC-Sym builds full-atom models of RNA structures using the 3D version of the nucleotide cyclic motif fragments. The nucleotide cyclic fragment library was built from a list of 531 RNA 3D structures. The fragment insertion simulation is performed using the Las Vegas algorithm [130], a Monte Carlo algorithm that either produces a correct result or reports that such an answer cannot be found. In the case of the MC-Sym fragment insertion simulation, the algorithm explores as many corresponding 3D fragments as possible in a given time, and all RNA 3D structures generated are consistent with the base pair and base stacking input constraints. MC-Fold can also incorporate experimental data in order to restrict the conformational space: input can be from RNA SHAPE chemistry, or dimethyl sulfate (DMS) data (mentioned in section 2).
The nucleic acid simulation tool or NAST [131] is a coarse-grained molecular dynamics simulation tool consisting of a knowledge-based statistical potential function. Each residue is represented as a single pseudo-atom centered at the C3′ atom. NAST requires secondary-structure information and, if available, tertiary contacts to direct the folding. In addition, the knowledge-based function uses geometric distances, angles and dihedrals from the available ribosomal structures using C3′ atoms between two, three, and four sequential residues respectively. A repulsive term for non-bonded interactions based on the Lennard-Jones potential is also applied.
iFoldRNA [132] is a Web-based program designed by Dokholyan’s group for predicting RNA structures from sequence. It is based on discrete molecular dynamics (DMD) [133] and a tailored force field [134] algorithms for simulating RNA folding dynamics. The coarse-grained model is composed of three beads for each nucleotide positioned at the center of mass of the phosphate group, sugar ring and six-atom ring in the base. The structure’s angles, dihedrals, and bonds are used to construct a stepwise potential function that accounts for base stacking, short-range phosphate–phosphate repulsion, and hydrophobic interactions. To explore the potential energy landscape of the molecular system, iFoldRNA uses multiple simulations or replicas of the same system performed in parallel at different temperatures. The discrete molecular dynamics algorithm is based on a stepwise potential function [133, 135]. Like NAST and MC-Sym, iFoldRNA incorporates data from tertiary contacts based on experimental contacts from SHAPE chemistry [74] to overcome size limitations.
RNA2D3D [136] is a manual-input program that uses data from sequence and secondary structure to build a first-order approximation RNA 3D model. The program allows the user to manually add or remove base pairs and incorporate coaxial stacking interactions, useful for exploring diverse RNA conformations. ASSEMBLE is a similar tool created by Jossinet and Westhof (http://www.bioinformatics.org/assemble/). The program uses either secondary-structure information or tertiary information from homologous RNAs to construct a 3D model manually. ASSEMBLE allows the manual insertion of base pairs and motifs, as well as torsion angle modifications, rotations, and translations of modular elements. While these user-input tools are useful, they rely on manual application of expert knowledge. Unfortunately, there are only a few of these experts.
7. Comparison of automated RNA 3D folding algorithms
To evaluate the performance of some of the most recent 3D prediction programs, we provide an independent comparative study for RNAs in a high resolution data set represented by various RNA lengths and structural features. Because the programs we consider are recent, they are still under development and could be improved in the future. Therefore, our data reflect the state of the art in automatic prediction programs at the outset of 2010.
7.1. Methods
We consider the tertiary-structure prediction programs FARNA, iFoldRNA, and MC-Fold/MC-Sym (MC for short) for prediction using sequence information only. In addition, a second comparison among FARNA, MC and NAST is considered for prediction based on secondary-structure data. Although NAST can accept input information from tertiary contacts, which can dramatically improve prediction accuracy, for the purpose of equal comparison, we only produce NAST structures using secondary-structure data. We did not explicitly evaluate computational efficiency; however, the general trend was that NAST is less CPU time intensive than iFoldRNA, FARNA and MC. Structures generated using NAST and FARNA were computed using a Linux Intel Xeon 3.0 GHz processor. Structures generated using iFoldRNA and MC were computed using their own Web servers. Most programs return results in less than a few hours. For instance, the calculation for predicting RNA 49-nt long takes about 4 min using NAST, 27 min using iFoldRNA, 31 min using MC-Sym, and 37 min using FARNA.
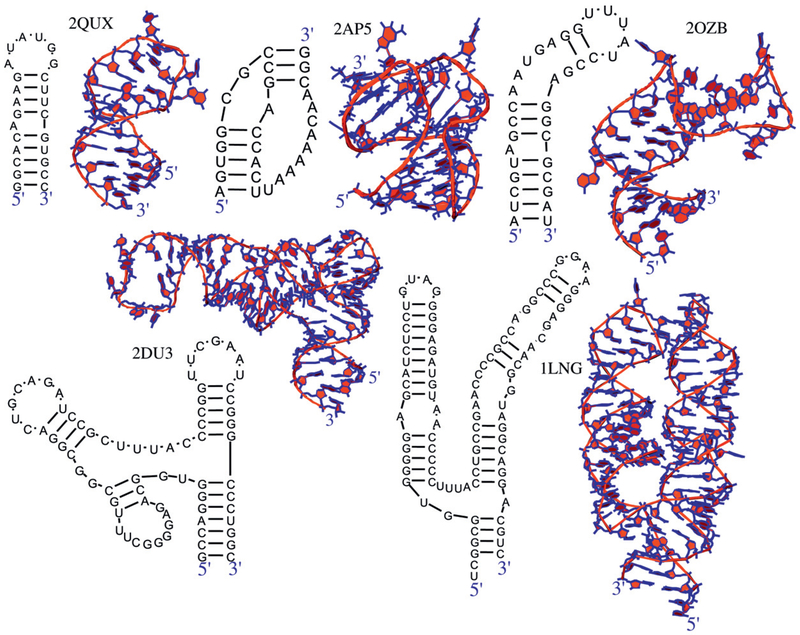
Our RNA data set consists of 43 high resolution (3.4 Å or better) structures of diverse sizes and motifs (see table 4). Both secondary and tertiary structures have been experimentally determined for these RNAs. The length varies from 16 to 128 nucleotides, and the topologies range from hairpins to complex RNAs such as riboswitches, pseudoknots, and RNAs containing junctions. Figure 3 shows representative examples of our data set. As part of the pseudoknot category, we include RNA structures with Kissing hairpins, which are loop–loop interactions between two hairpins forming WC base pairs.
Table 4.
List of RNA 3D structures predicted using several computer programs. The RNA structures are selected with diversity in size (NTs), canonical base pairs (BPs), non-canonical base pairs (NC BPs) and structural complexity such as formation of hairpins, internal loops, junctions and pseudoknots. The symbols ‘*’ and ‘#’ in the first column next to the PDB code denote the RNA structures that MC and NAST failed to predict respectively.
| PDB | NTs | BPs | NC BPs | Resolution | Organism | Structure |
|---|---|---|---|---|---|---|
| 2F8K # | 16 | 6 | 0 | 2 | S. cerevisiae | Hairpin |
| 2AB4 # | 20 | 7 | 1 | 2.4 | T. maritima | Hairpin |
| 361D | 20 | 10 | 1 | 3 | Synthetic | Hairpin |
| 2ANN * | 23 | 6 | 3 | 2.3 | H. sapiens | Hairpin |
| 1RLG * | 25 | 9 | 9 | 2.7 | A. fulgidus | Hairpin, internal loop |
| 2QUX | 25 | 9 | 0 | 2.4 | P. phage PP7 | Hairpin |
| 387D *# | 26 | 5 | 1 | 3.1 | Synthetic | Hairpin |
| 1MSY * | 27 | 10 | 4 | 1.4 | Synthetic | Hairpin |
| 1L2X * | 28 | 14 | 6 | 1.3 | Synthetic | Pseudoknot |
| 2AP5 *# | 28 | 8 | 3 | N/A | YLV virus | Pseudoknot |
| 1JID # | 29 | 12 | 2 | 1.8 | H. sapiens | Hairpin, internal loop |
| 1OOA * | 29 | 18 | 1 | 2.5 | M. musculus | Hairpin, internal loop (aptamer) |
| 430D * | 29 | 12 | 4 | 2.1 | Synthetic | Hairpin, internal loop |
| 2IPY | 30 | 12 | 0 | 2.8 | O. cuniculus | Hairpin, internal loop |
| 2OZB | 33 | 12 | 3 | 2.6 | H. sapiens | Hairpin, internal loop |
| 1MJI *# | 34 | 8 | 4 | 2.5 | T. thermophilus | Hairpin, internal loop |
| 1ET4 * | 35 | 50 | 10 | 2.3 | Synthetic | Pseudoknot |
| 2HW8 | 36 | 15 | 5 | 2.1 | T. thermophilus | Hairpin, internal loop |
| 1I6U | 37 | 34 | 4 | 2.6 | M. jannaschii | Hairpin, internal loop |
| 1F1T | 38 | 14 | 5 | 2.8 | Synthetic | Hairpin, internal loops (aptamer) |
| 1ZHO | 38 | 15 | 4 | 2.6 | T. thermophilus | Hairpin, internal loops |
| 1S03 | 47 | 13 | 3 | 2.7 | E. coli | Hairpin, internal loops |
| 1XJR | 47 | 19 | 3 | 2.7 | Synthetic | Hairpin, internal loops |
| 1U63 # | 49 | 19 | 3 | 3.4 | M. jannaschii | Hairpin, internal loop |
| 2PXB | 49 | 21 | 6 | 2 | E. coli | Hairpin, internal loops |
| 2FK6 # | 53 | 20 | 3 | 2.9 | B. subtilis | Pseudoknot, three-way junction |
| 3E5C | 53 | 21 | 5 | 2.3 | Synthetic | Three-way junction (riboswitch) |
| 1MZP * | 55 | 17 | 12 | 2.7 | S. acidocaldarius | Hairpin, internal loops |
| 1DK1 | 57 | 24 | 4 | 2.8 | T. thermophilus | Three-way junction |
| 1MMS * | 58 | 20 | 13 | 2.6 | T. maritima | Three-way junction |
| 3EGZ * | 65 | 23 | 5 | 2.2 | H. sapiens | Three-way junction (riboswitch) |
| 2QUS | 69 | 26 | 2 | 2.4 | Synthetic | Pseudoknot, three-way junction |
| 1KXK | 70 | 28 | 5 | 3 | Synthetic | Hairpin, internal loops |
| 2DU3 * | 71 | 27 | 5 | 2.6 | A. fulgidus | Four-way junction (tRNA) |
| 2OIU # | 71 | 29 | 3 | 2.6 | Synthetic | Three-way junction (ribozyme) |
| 1SJ4 *# | 73 | 19 | 8 | 2.7 | HDV virus | Pseudoknot, four-way junction |
| 1P5O * | 77 | 29 | 5 | N/A | HCV virus | Hairpin, internal loops |
| 3D2G *# | 77 | 28 | 15 | 2.3 | A. thaliana | Three-way junction (riboswitch) |
| 2HOJ *# | 79 | 27 | 12 | 2.5 | Synthetic | Three-way junction (riboswitch) |
| 2GDI * | 80 | 32 | 12 | 2 | Synthetic | Three-way junction (riboswitch) |
| 2GIS *# | 94 | 36 | 13 | 2.9 | Synthetic | Pseudoknot, four-way junction (riboswitch) |
| 1LNG * | 97 | 38 | 14 | 2.3 | M. jannaschii | Three-way junction (SRP) |
| 1MFQ * | 128 | 49 | 16 | 3.1 | H. Sapiens | Three-way junction (SRP) |
Figure 3.
Examples of RNA structures considered for prediction. These RNAs vary in length and structural complexity, including hairpins (PDB: 2QUS, 2OZB), pseudoknots (PDB: 2AP5), and junctions (PDB: 2DU3, 1LNG).
To evaluate prediction performance, we use the root mean square deviations (RMSD) from the reference known structure, as well as the deviation index (DI), which is a measure that accounts for base pair and base stacking interactions, and is defined as the quotient between the RMSD and the squared root of the specificity (PPV) times the sensitivity (STY) of base pair and base stacking interactions as defined in [137]. In brief, PPV represents the percentage of correctly predicted interactions that are in the crystal structure, while STY represents the percentage of interactions in the crystal structure predicted, without accounting for the false positives2. The smaller the DI value, the better the prediction. RMSD values were computed for the backbone using VMD [138]. Base pair and base stacking interactions were determined using FR3D [139]. An average of the RMSD and DI values for structures that have the same number of nucleotides was considered. The accuracy measured with the PPV and STY values is presented in figure 4.
Figure 4.
RNA structure prediction analysis. Specificity (PPV) and sensitivity (STY) values are sorted by length and compared among different prediction programs on the basis of sequence (upper chart) or secondary-structure information (lower chart). The curves are presented using the moving average trend lines based on two preceding values for each point.
The MC and NAST programs failed to produce a folded structure in a few cases (see table 4). MC was unable to predict some structures both from the sequence and the secondary structure, possibly due to the lack of a sufficient number of cyclic motif fragments to insert. Similarly, NAST failed in some cases, possibly due to the design of the fragment assembly algorithm. Both iFoldRNA and FARNA predicted a structure for every element in the data set.
To ensure a reliable predicted model in FARNA, we used 50 000 iteration cycles. Similarly, in NAST we consider one million steps3. In the MC-Fold/MC-Sym pipeline, when we predict the structure solely from the sequence, we consider the secondary structure that is ranked first by MC-Fold. For the 3D structure selection, we use the first-ranked structure according to the scoring function available on the MC-Sym analysis tool. Although in some programs better predictions might be possible by tuning many parameters, we used only the recommended values to make an unbiased comparison.
7.2. Tertiary-structure prediction results
Specificity (PPV) and sensitivity (STY) values describing the prediction accuracy for each program and system are shown in figure 4. Both PPV and STY take values between 0 and 1, where better predictions approach 1. Figure 4 shows that both PPV and STY values for MC are comparable and higher than those for the rest of the programs. This is due mostly to the library that MC uses that assembles nucleotide cyclic fragments by stacking base pairs. In contrast, FARNA, iFoldRNA, and NAST all have their PPV values higher than STY values. Thus, a smaller number of false interaction predictions are produced at the expense of a smaller number of correct predictions. An improvement in FARNA’s prediction is observed when the secondary-structure (base pair and base stacking) information is included, particularly in STY values where their average value increases from 0.37 to 0.63.
A different representation of the accuracy of each program is given using the RMSD and DI values as shown in figure S1 (available at stacks.iop.org/JPhysCM/22/283101/mmedia). Dashed lines represent the linear approximation to the RMSD values, and solid lines approximate the DI values. As expected, both RMSD and DI values increase rapidly as the length of the molecules increases. The best values start around 6Å RMSD, considered poor for protein predictions. We see that the performance improves dramatically when secondary-structure information is added (lower graph) as opposed to the case for prediction using the sequence only (upper graph), with the best predictions now starting around 4Å RMSD. Except for FARNA and iFoldRNA, the slopes of the lines (when scaling the RMSD to DI values) do not change drastically, reinforcing the independence of these values from RNA size.
The accuracy of each program varies from structure to structure. In general terms, MC performs better in both prediction experiments (figure 4). Program iFoldRNA performs better than FARNA for small molecules, but FARNA improves as the length increases. FARNA performs slightly better than NAST, but the difference in DI values shows that FARNA makes better use of secondary-structure information. A difference is also noted in the slope between the lines that describe the RMSD and DI accuracy values for iFoldRNA (blue lines, upper graph of figure S1, available at stacks.iop.org/JPhysCM/22/283101/mmedia). This implies that iFoldRNA’s accuracy in predicting local features such as base pair and base stacking interactions is poorer than that for global structure prediction.
For further analysis, we next separate the data by structural context (hairpins, pseudoknots and junctions). Hairpins are easiest to predict (top row of figure S2, available at stacks.iop.org/JPhysCM/22/283101/mmedia), with lowest DI values observed. Prediction using sequence show that, except for a few structures, iFoldRNA and MC perform similarly. In particular, the peak at length 23 corresponds to a hairpin with 15-nt in the single-stranded region, and because alternative secondary structures with more canonical base pairs are possible, predictions are more difficult. Similarly, peaks at length 26, 33 and 36 correspond to structures that are difficult to predict. The main reason is that for these cases (PDB 387D, 2OZB, and 2HW8) the native structure includes proteins that have kinked internal loops, thus distorting the RNA structure (e.g., see 2OZB in figure 3). Prediction using secondary structure shows that NAST and FARNA have similar accuracies. In general, MC performs better due to the nature of its cyclic fragments model that includes hairpin loops (figure 5(a)); however, many non-canonical base pair interactions in the loop regions are not predicted.
Figure 5.
Structural alignments show the accuracy of diverse prediction programs. (a) Hairpin structures predicted from sequences using FARNA (green middle), iFoldRNA (blue left) and MC-Fold/MC-Sym (magenta right) against the known structure (orange with yellow background). ((b), (c)) Pseudoknot structures are predicted both from the sequence (b) and from secondary structure for iFoldRNA, FARNA and MC, respectively in (b), and FARNA and NAST, respectively. NAST (cyan) in (c) is also included. (d) Three-way junction structure predictions using secondary structures are aligned against the solved structure (orange with yellow background), for MC, FARNA, and NAST.
The performance for pseudoknots (middle row of figure S2, available at stacks.iop.org/JPhysCM/22/283101/mmedia) shows that the accuracy of prediction of FARNA and iFoldRNA, and of FARNA and NAST are overall similar. MC performs better in both experiments but fails to produce a model for most of the pseudoknot structures predicted from secondary structure. Figures 5(b) and (c) shows the backbone conformation of the solved pseudoknot structure (PDB 1L2X) in orange with a yellow background, against the backbone of the predicted structures. Predictions from the sequence alone fail to produce a more compact structure (figure 5(b)). This is expected since in general prediction from the sequence of RNA structures with pseudoknots is difficult. In contrast, a great improvement occurs when the secondary-structure information is given (figure 5(c)).
Prediction results of structures containing a junction are shown in the last row of figure S2 (available at stacks.iop.org/JPhysCM/22/283101/mmedia). Although MC often outperforms other programs, it could not predict a structure for most of the RNA junctions even when secondary-structure information is given. DI values for FARNA and iFoldRNA, and for FARNA and NAST are comparable. Like for pseudoknot prediction, if secondary-structure data are given, then the prediction performance improves considerably. However, the long-range interactions that often stabilize helical elements are required to produce an accurate model, as observed in figure 5(d) for the models produced by NAST (cyan curve) and FARNA (green curve). Also, the peaks in DI values at lengths 65 and 79 shown in figure S2 (available at stacks.iop.org/JPhysCM/22/283101/mmedia) (last row left) correspond to two riboswitches that are structurally complex.
Structure prediction using MC regularly gives the most accurate model. However, this program often fails to predict a structure (see table 4), and while helical regions are well modeled, non-canonical base pairs occurring at the loop region are rarely predicted. Although iFoldRNA often outperforms FARNA on predicting hairpins, their performances are similar for predicting pseudoknots and junctions. Similarly, the NAST and FARNA accuracies of prediction of hairpins using secondary-structure data are comparable.
Both FARNA and MC allow prediction of multiple folds or suboptimal folds. Although in a true prediction context, one does not have knowledge of the correct structure, the study of such multiple folds helps determine the robustness in the prediction programs. We considered a group of four representative structures consisting of two hairpins and two junctions (see table 5), and predicted up to five multiple folds for each structure. In some cases, MC predicted fewer suboptimal folds. FARNA produced all five folds for each case. MC produces overall similar results for the 2D structure, since the best structures having PPV and STY values (0.8–0.9) are similar to the average. The RMSD values from the best performances are below 9.5 for hairpins, and below 18.5Å for junctions. Thus, as the degree of topological complexity increases, MC’s performance worsens. In FARNA, the STY values from the multiple folds are better for hairpins, but worse for junctions. The fact that similar 2D structures yield very different 3D folds underscores the central difficulty in tertiary RNA prediction.
Table 5.
List of performance values from suboptimal structures predicted using MC-Fold/MC-Sym (left) and FARNA (right) using secondary-structure information. The structures consist of two hairpins and two junctions. Values in bold font correspond to the ones presented in figures 4, S1, and S2 (available at stacks.iop.org/JPhysCM/22/283101/mmedia). The average and best performance values are also shown in bold.
| MC-Fold/MC-Sym | FARNA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure | Name | NTs | PPV | STY | RMSD | DI | Name | NTs | PPV | STY | RMSD | DI |
 |
1KXKm1 | 70 | 0.91 | 0.82 | 9.05 | 10.51 | lKXKrl | 70 | 0.84 | 0.70 | 20.83 | 27.29 |
| 1KXKm2 | 70 | 0.89 | 0.84 | 8.71 | 10.03 | 1KXKr2 | 70 | 0.84 | 0.76 | 14.75 | 18.46 | |
| 1KXKm3 | 70 | 0.91 | 0.83 | 9.40 | 10.79 | 1KXKr3 | 70 | 0.88 | 0.75 | 12.65 | 15.53 | |
| 1KXKm4 | 70 | 0.87 | 0.79 | 9.21 | 11.12 | 1KXKr4 | 70 | 0.86 | 0.76 | 17.36 | 21.51 | |
| 1KXKm5 | 70 | 0.87 | 0.79 | 11.06 | 13.36 | 1KXKr5 | 70 | 0.85 | 0.75 | 20.56 | 25.64 | |
| 1KXKm | 70 | 0.91 | 0.82 | 9.06 | 10.52 | 1KXKr | 70 | 0.87 | 0.66 | 14.58 | 19.26 | |
| Average | 0.89 | 0.81 | 9.49 | 11.16 | Average | 0.85 | 0.74 | 17.23 | 21.69 | |||
| Best | 0.91 | 0.84 | 8.71 | 10.03 | Best | 0.88 | 0.76 | 12.65 | 15.53 | |||
 |
1XJRml | 47 | 0.89 | 0.74 | 12.24 | 15.11 | lXJRrl | 47 | 0.84 | 0.78 | 8.88 | 10.97 |
| 1XJRm2 | 47 | 0.90 | 0.82 | 9.33 | 10.91 | lXJRr2 | 47 | 0.84 | 0.63 | 11.50 | 15.83 | |
| 1XJRm3 | 47 | 0.85 | 0.77 | 5.72 | 7.08 | lXJRr3 | 47 | 0.87 | 0.71 | 14.05 | 17.93 | |
| 1XJRm4 | 47 | 0.86 | 0.74 | 7.72 | 9.70 | lXJRr4 | 47 | 0.78 | 0.78 | 11.66 | 14.87 | |
| 1XJRm5 | 47 | 0.86 | 0.74 | 8.68 | 10.91 | lXJRr5 | 47 | 0.81 | 0.66 | 12.06 | 16.47 | |
| 1XJRm | 47 | 0.92 | 0.82 | 10.80 | 12.48 | lXJRr | 47 | 0.86 | 0.65 | 10.31 | 13.78 | |
| Average | 0.87 | 0.76 | 8.74 | 10.74 | Average | 0.83 | 0.71 | 11.63 | 15.21 | |||
| Best | 0.90 | 0.82 | 5.72 | 7.08 | Best | 0.87 | 0.78 | 8.88 | 10.97 | |||
 |
2OIUml | 71 | 0.92 | 0.74 | 19.66 | 23.86 | 2OIIM | 71 | 0.84 | 0.47 | 15.98 | 25.34 |
| 2OIUm2 | 71 | 0.92 | 0.76 | 14.02 | 16.78 | 2OIUr2 | 71 | 0.91 | 0.64 | 19.51 | 25.54 | |
| 2OIUm3 | 71 | 0.92 | 0.75 | 18.19 | 21.92 | 2OIUr3 | 71 | 0.88 | 0.56 | 19.84 | 28.35 | |
| 2OIUm4 | 71 | 0.93 | 0.80 | 15.53 | 18.00 | 2OIUr4 | 71 | 0.85 | 0.70 | 16.66 | 21.58 | |
| 2OIUr5 | 71 | 0.88 | 0.75 | 18.48 | 22.80 | |||||||
| 2OIUm | 71 | 0.91 | 0.83 | 14.02 | 16.20 | 2OIUr | 71 | 0.91 | 0.83 | 16.24 | 18.77 | |
| Average | 0.92 | 0.76 | 16.85 | 20.14 | Average | 0.87 | 0.63 | 18.10 | 24.72 | |||
| Best | 0.93 | 0.80 | 14.02 | 16.78 | Best | 0.91 | 0.75 | 15.98 | 21.58 | |||
 |
2QUSml | 69 | 0.87 | 0.79 | 15.92 | 19.19 | 2QUSrl | 69 | 0.87 | 0.52 | 16.80 | 24.88 |
| 2QUSm2 | 69 | 0.86 | 0.77 | 20.90 | 25.69 | 2QUSr2 | 69 | 0.88 | 0.73 | 12.07 | 15.03 | |
| 2QUSr3 | 69 | 0.84 | 0.46 | 17.41 | 28.21 | |||||||
| 2QUSr4 | 69 | 0.90 | 0.61 | 13.20 | 17.78 | |||||||
| 2QUSr5 | 69 | 0.80 | 0.58 | 19.18 | 28.11 | |||||||
| 2QUSm | 69 | 0.82 | 0.79 | 15.92 | 19.85 | 2QUSr | 69 | 0.92 | 0.75 | 14.06 | 16.98 | |
| Average | 0.86 | 0.78 | 18.41 | 22.44 | Average | 0.86 | 0.58 | 15.73 | 22.80 | |||
| Best | 0.87 | 0.79 | 15.92 | 19.19 | Best | 0.90 | 0.73 | 12.07 | 15.03 | |||
Furthermore, we analyzed the prediction accuracy of MC and FARNA from multiple folds using only sequence information on a hairpin (PDB 1KXK) and a junction (PDB 2QUS) structure (see table S1 in the supplementary material, available at stacks.iop.org/JPhysCM/22/283101/mmedia). MC predictions show a drop in PPV average values from 0.9 to 0.8. Interestingly, the average RMSD values are similar to the predictions using secondary-structure information shown above; this suggests that information from secondary structure affects more local features in these cases. Improving tertiary interactions, however, depends on the accurate determination of long-range contacts to position the RNA’s helical elements. Prediction accuracy with FARNA using only sequence information reduces considerably both local (PPV and STY) as well as global (RMSD) features, showing that both secondary- and tertiary-contact information is required for accurate prediction in these cases.
8. Discussion
Significant advances have been made over the last decade in RNA modeling, along with increasing computational resources and technologies for RNA investigations. It is encouraging to see that many of these advances have been achieved with relatively simple models, which combine energy or statistical potentials and fragment assembly techniques. These likely are effective due to RNA’s modular architecture and structural hierarchy. Still, further developments and refinements of the existing models are needed.
One of the major challenges in RNA tertiary-structure prediction is the determination of long-range interactions. One possible avenue for determining tertiary contacts may come from studies using multiple-sequence analysis. By comparing instances of each recurrent base pair or a larger structural element such as an RNA motif, one can identify neutral mutations that preserve its structure and function [32]. Programs like SHEVEK [140] and ISFOLD [141] that use multiple-sequence alignment approaches are a step in that direction.
Current algorithms have shown how the use of experimental data can dramatically improve the structure prediction [63, 73–75]. But data from experimental techniques such as RNA SHAPES chemistry can potentially provide more information than just canonical WC base pairs. To determine non-canonical base pairs and tertiary contacts, further refinements in order to better exploit this information should be considered. Similarly, a program for automatically restricting the structural conformation space of a model on the basis of low resolution cryo-EM data might be valuable.
Several programs have exploited the hierarchical properties of RNA molecules. For instance, part of MC-Sym protocol’s success rests on the use of cyclic motifs. Yet current automatic prediction methods cannot exploit the modular properties of the more complex tertiary motifs as is done in ASSEMBLE and RNA2D3D. Similarly, recent studies on base–phosphate interactions [33] have shown that such interactions are sufficiently frequent to be considered in an energy function similar to FARNA’s base pairing and base stacking interaction functions.
The Rosetta method has proven successful for protein prediction. A successful companion method for RNA requires changes in both the energy function and fragment elements. For instance, in contrast to the compact globular shapes of proteins, rather compact prolate ellipsoidal shapes are favored by RNA molecules [65, 142]. Functions like the radius of gyration that reward globular molecular conformations might not suit RNA. In addition, studies have shown that junctions arrange their helical arms in parallel and perpendicular configurations [60, 61]. A new scoring function that encourages these conformations can be helpful.
While the use of scoring functions that describe RNA–ion interactions will certainly improve RNA structure prediction, the computational effort of such a task might not be feasible initially. Assembly from structure fragments already in the presence of cations can circumvent this limitation, as is done in NAST, FARNA, and MC. Similarly, the use of RNA–protein interaction prediction programs can help improve predictions of RNAs in the presence of proteins.
In general, the prediction accuracy improves with added knowledge from the secondary structure and tertiary contacts, but these interactions can be greatly affected by the presence of proteins. The lack of correct functions that favor compact RNA-like structure and the failure to detect the presence of long-range interaction contacts remain challenges.
However, with the increasing interest and creation of a community devoted to RNA bioinformatics [39], we can anticipate many exciting developments in automated RNA structure prediction approaches in the coming decade. The growing appreciation for the biological importance of RNA makes such efforts more essential than ever.
Supplementary Material
Acknowledgments
The work was supported by the Human Frontier Science Program (HFSP), by a joint NSF/NIGMS initiative in Mathematical Biology (DMS-0201160), and by NSF EMT award no. CF-0727001. Partial support by NIH (grant no. R01-GM055164), NIH (grant no. 1 R01 ES 012692), NSF (grant no. CCF-0727001), and Philip Morris USA, Inc., and Philip Morris International is also gratefully acknowledged. The authors thank Abdul Iqbal, Yoon Ha, and Segun Jung for their help with data preparation, and Bruce Shapiro for a critical reading of this manuscript. We also thank Magdalena Jonikas for helping with NAST implementation and Feng Ding for helping with the iFoldRNA applications.
Footnotes
Online supplementary data available from stacks.iop.org/JPhysCM/22/283101/mmedia
PPV = TP/(TP + FP), STY = TP/(TP + FN), where TP = number of correctly predicted interactions found in the solved structure, FP = number of predicted interactions not found in the solved structure, and FN = number of interactions in the solved structure that are not predicted by the algorithm.
The PDB structure 1N32 (16S rRNA) was used for fragment samples during the all-atom model formation step.
References
- [1].Hannon GJ 2002. RNA interference Nature 418 244–51 [DOI] [PubMed] [Google Scholar]
- [2].Gillet R and Felden B 2001. Emerging views on tmRNA-mediated protein tagging and ribosome rescue Mol. Microbiol 42 879–85 [DOI] [PubMed] [Google Scholar]
- [3].Masse E and Gottesman S 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli Proc. Natl Acad. Sci. USA 99 4620–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ruvkun G 2001. Molecular biology. Glimpses of a tiny RNA world Science 294 797–9 [DOI] [PubMed] [Google Scholar]
- [5].Calin GA et al. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia Proc. Natl Acad. Sci. USA 99 15524–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Michael MZ et al. 2003. Reduced accumulation of specific microRNAs in colorectal neoplasia Mol. Cancer Res 1 882–91 [PubMed] [Google Scholar]
- [7].Takamizawa J et al. 2004. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival Cancer Res 64 3753–6 [DOI] [PubMed] [Google Scholar]
- [8].Hayashita Y et al. 2005. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation Cancer Res 65 9628–32 [DOI] [PubMed] [Google Scholar]
- [9].He L et al. 2005. A microRNA polycistron as a potential human oncogene Nature 435 828–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Metzler M et al. 2004. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma Genes Chromosom. Cancer 39 167–9 [DOI] [PubMed] [Google Scholar]
- [11].Tagawa H and Seto M 2005. A microRNA cluster as a target of genomic amplification in malignant lymphoma Leukemia 19 2013–6 [DOI] [PubMed] [Google Scholar]
- [12].Chworos A et al. 2004. Building programmable jigsaw puzzles with RNA Science 306 2068–72 [DOI] [PubMed] [Google Scholar]
- [13].Jaeger L and Chworos A 2006. The architectonics of programmable RNA and DNA nanostructures Curr. Opin. Struct. Biol 16 531–43 [DOI] [PubMed] [Google Scholar]
- [14].Haasnoot J and Berkhout B 2006. RNA interference: its use as antiviral therapy Handb. Exp. Pharmacol 173 117–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haasnoot J, Westerhout EM and Berkhout B 2007. RNA interference against viruses: strike and counterstrike Nat. Biotechnol 25 1435–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Birney E et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project Nature 447 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Amaral PP et al. 2008. The eukaryotic genome as an RNA machine Science 319 1787–9 [DOI] [PubMed] [Google Scholar]
- [18].Sharp PA 2009. The centrality of RNA Cell 136 577–80 [DOI] [PubMed] [Google Scholar]
- [19].Breaker RR 2002. Engineered allosteric ribozymes as biosensor components Curr. Opin. Biotechnol 13 31–9 [DOI] [PubMed] [Google Scholar]
- [20].Sioud M 2006. Ribozymes and siRNAs: from structure to preclinical applications Handb. Exp. Pharmacol 173 223–42 [DOI] [PubMed] [Google Scholar]
- [21].Romby P, Vandenesch F and Wagner EG 2006. The role of RNAs in the regulation of virulence-gene expression Curr. Opin. Microbiol 9 229–36 [DOI] [PubMed] [Google Scholar]
- [22].Afonin KA et al. 2008. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules Chembiochem 9 1902–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yingling YG and Shapiro BA 2007. Computational design of an RNA hexagonal nanoring and an RNA nanotube Nano Lett 7 2328–34 [DOI] [PubMed] [Google Scholar]
- [24].Khaled A et al. 2005. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology Nano Lett 5 1797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li L et al. 2009. Evaluation of specific delivery of chimeric phi29 pRNA/siRNA nanoparticles to multiple tumor cells Mol. Biosyst 5 1361–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gardner PP and Giegerich R 2004. A comprehensive comparison of comparative RNA structure prediction approaches BMC Bioinformatics 5 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mathews DH and Turner DH 2006. Prediction of RNA secondary structure by free energy minimization Curr. Opin. Struct. Biol 16 270–8 [DOI] [PubMed] [Google Scholar]
- [28].Shapiro BA et al. 2007. Bridging the gap in RNA structure prediction Curr. Opin. Struct. Biol 17 157–65 [DOI] [PubMed] [Google Scholar]
- [29].Marti-Renom MA and Capriotti E 2008. Computational RNA Structure Prediction Curr. Bioinform 3 32–45 [Google Scholar]
- [30].Schroeder SJ 2009. Advances in RNA structure prediction from sequence: new tools for generating hypotheses about viral RNA structure–function relationships J. Virol 83 6326–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leontis NB, Lescoute A and Westhof E 2006. The building blocks and motifs of RNA architecture Curr. Opin. Struct. Biol 16 279–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Leontis NB, Stombaugh J and Westhof E 2002. The non-Watson–Crick base pairs and their associated isostericity matrices Nucleic Acids Res 30 3497–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zirbel CL et al. 2009. Classification and energetics of the base–phosphate interactions in RNA Nucleic Acids Res 37 4898–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Waterman MS and Smith TF 1978. RNA secondary structure: a complete mathematical analysis Math. Biosci 42 257–66 [Google Scholar]
- [35].Benedetti G and Morosetti S 1996. A graph-topological approach to recognition of pattern and similarity in RNA secondary structures Biophys. Chem 59 179–84 [DOI] [PubMed] [Google Scholar]
- [36].Le SY, Nussinov R and Maizel JV 1989. Tree graphs of RNA secondary structures and their comparisons Comput. Biomed. Res 22 461–73 [DOI] [PubMed] [Google Scholar]
- [37].Shapiro BA and Zhang KZ 1990. Comparing multiple RNA secondary structures using tree comparisons Comput. Appl. Biosci 6 309–18 [DOI] [PubMed] [Google Scholar]
- [38].Gan HH et al. 2004. RAG: RNA-As-graphs database—concepts, analysis, and features Bioinformatics 20 1285–91 [DOI] [PubMed] [Google Scholar]
- [39].Schlick T. Mathematical and biological scientists assess the state-of-the-art in RNA science at an IMA workshop ‘RNA in biology, bioengineering and biotechnology’. Int. J. Multiscale Comput. Eng. 2009 at press. [Google Scholar]
- [40].Al-Hashimi HM and Walter NG 2008. RNA dynamics: it is about time Curr. Opin. Struct. Biol 18 321–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cruz JA and Westhof E 2009. The dynamic landscapes of RNA architecture Cell 136 604–9 [DOI] [PubMed] [Google Scholar]
- [42].Draper DE 2008. RNA folding: thermodynamic and molecular descriptions of the roles of ions Biophys. J 95 5489–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tinoco I Jr and Bustamante C 1999. How RNA folds J. Mol. Biol 293 271–81 [DOI] [PubMed] [Google Scholar]
- [44].Greenleaf WJ et al. 2008. Direct observation of hierarchical folding in single riboswitch aptamers Science 319 630–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Noeske J et al. 2007. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch Nucleic Acids Res 35 572–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wu M Jr and Tinoco I 1998. RNA folding causes secondary structure rearrangement Proc. Natl Acad. Sci. USA 95 11555–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pan J, Thirumalai D and Woodson SA 1997. Folding of RNA involves parallel pathways J. Mol. Biol 273 7–13 [DOI] [PubMed] [Google Scholar]
- [48].Chauhan S and Woodson SA 2008. Tertiary interactions determine the accuracy of RNA folding J. Am. Chem. Soc 130 1296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xin Y et al. 2008. Annotation of tertiary interactions in RNA structures reveals variations and correlations RNA 14 2465–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Montange RK and Batey RT 2008. Riboswitches: emerging themes in RNA structure and function Annu. Rev. Biophys 37 117–33 [DOI] [PubMed] [Google Scholar]
- [51].Koev G et al. 2002. The 3′or minute-terminal structure required for replication of Barley yellow dwarf virus RNA contains an embedded 3′ or minute end Virology 292 114–26 [DOI] [PubMed] [Google Scholar]
- [52].Linnstaedt SD et al. 2006. The role of a metastable RNA secondary structure in hepatitis delta virus genotype III RNA editing RNA 12 1521–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McCormack JC et al. 2008. Structural domains within the 3′ untranslated region of Turnip crinkle virus J. Virol 82 8706–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Olsthoorn RC et al. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication EmboJ 18 4856–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pan T and Sosnick T 2006. RNA folding during transcription Annu. Rev. Biophys. Biomol. Struct 35 161–75 [DOI] [PubMed] [Google Scholar]
- [56].Wong TN, Sosnick TR and Pan T 2007. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures Proc. Natl Acad. Sci. USA 104 17995–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Leontis NB and Westhof E 2001. Geometric nomenclature and classification of RNA base pairs RNA 7 499–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Draper DE 2004. A guide to ions and RNA structure RNA 10 335–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ramaswamy P and Woodson SA 2009. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20 J. Mol. Biol 392 666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Laing C et al. 2009. Tertiary motifs revealed in analyses of higher-order RNA junctions J. Mol. Biol 393 67–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Laing C and Schlick T 2009. Analysis of four-way junctions in RNA structures J. Mol. Biol 390 547–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lescoute A and Westhof E 2006. Topology of three-way junctions in folded RNAs RNA 12 83–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang J et al. 2009. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements J. Mol. Biol 393 717–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kim SH et al. 1974. The general structure of transfer RNA molecules Proc. Natl Acad. Sci. USA 71 4970–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ban N et al. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution Science 289 905–20 [DOI] [PubMed] [Google Scholar]
- [66].Schluenzen F et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution Cell 102 615–23 [DOI] [PubMed] [Google Scholar]
- [67].Wimberly BT et al. 2000. Structure of the 30S ribosomal subunit Nature 407 327–39 [DOI] [PubMed] [Google Scholar]
- [68].Felden B 2007. RNA structure: experimental analysis Curr. Opin. Microbiol 10 286–91 [DOI] [PubMed] [Google Scholar]
- [69].Hohng S et al. 2004. Conformational flexibility of four-way junctions in RNA J. Mol. Biol 336 69–79 [DOI] [PubMed] [Google Scholar]
- [70].Walter F et al. 1998. Global structure of four-way RNA junctions studied using fluorescence resonance energy transfer RNA 4 719–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Merino EJ et al. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc 127 4223–31 [DOI] [PubMed] [Google Scholar]
- [72].Mortimer SA and Weeks KM 2009. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution Nat. Protoc 4 1413–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Deigan KE et al. 2009. Accurate SHAPE-directed RNA structure determination Proc. Natl Acad. Sci. USA 106 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Gherghe CM et al. 2009. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics J. Am. Chem. Soc 131 2541–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Parisien M and Major F 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data Nature 452 51–5 [DOI] [PubMed] [Google Scholar]
- [76].Mathews D. Predicting the secondary structure common to two RNA sequences with Dynalign. Curr. Protoc. Bioinform. 2004 doi: 10.1002/0471250953.bi1204s08. chapter 12, p unit 12.4. [DOI] [PubMed] [Google Scholar]
- [77].Sclavi B et al. 1998. Following the folding of RNA with time-resolved synchrotron x-ray footprinting Methods Enzymol 295 379–402 [DOI] [PubMed] [Google Scholar]
- [78].Duan S, Mathews DH and Turner DH 2006. Interpreting oligonucleotide microarray data to determine RNA secondary structure: application to the 3′ end of Bombyx mori R2 RNA Biochemistry 45 9819–32 [DOI] [PubMed] [Google Scholar]
- [79].Latham MP et al. 2005. NMR methods for studying the structure and dynamics of RNA Chembiochem 6 1492–505 [DOI] [PubMed] [Google Scholar]
- [80].Zidek L, Stefl R and Sklenar V 2001. NMR methodology for the study of nucleic acids Curr. Opin. Struct. Biol 11 275–81 [DOI] [PubMed] [Google Scholar]
- [81].Zuo X et al. 2010. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus Proc. Natl Acad. Sci. USA 107 1385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Holbrook SR, Holbrook EL and Walukiewicz HE 2001. Crystallization of RNA Cell. Mol. Life Sci 58 234–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Spahn CM et al. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor Cell 118 465–75 [DOI] [PubMed] [Google Scholar]
- [84].Andronescu M et al. 2007. Efficient parameter estimation for RNA secondary structure prediction Bioinformatics 23 i19–28 [DOI] [PubMed] [Google Scholar]
- [85].Griffiths-Jones S et al. 2003. Rfam: an RNA family database Nucleic Acids Res 31 439–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hofacker IL and Stadler PF 2006. Memory efficient folding algorithms for circular RNA secondary structures Bioinformatics 22 1172–6 [DOI] [PubMed] [Google Scholar]
- [87].Zuker M and Stiegler P 1981. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information Nucleic Acids Res 9 133–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bindewald E and Shapiro BA 2006. RNA secondary structure prediction from sequence alignments using a network of k-nearest neighbor classifiers RNA 12 342–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Do CB, Woods DA and Batzoglou S 2006. CONTRAfold: RNA secondary structure prediction without physics-based models Bioinformatics 23 e90–8 [DOI] [PubMed] [Google Scholar]
- [90].Hamada M et al. 2009. Prediction of RNA secondary structure using generalized centroid estimators Bioinformatics 25 465–73 [DOI] [PubMed] [Google Scholar]
- [91].Cannone JJ. et al. The comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gutell RR, Lee JC and Cannone JJ 2002. The accuracy of ribosomal RNA comparative structure models Curr. Opin. Struct. Biol 12 301–10 [DOI] [PubMed] [Google Scholar]
- [93].Hochsmann M, Voss B and Giegerich R 2004. Pure multiple RNA secondary structure alignments: a progressive profile approach IEEE/ACM Trans. Comput. Biol. Bioinform 1 53–62 [DOI] [PubMed] [Google Scholar]
- [94].Hofacker IL, Fekete M and Stadler PF 2002. Secondary structure prediction for aligned RNA sequences J. Mol. Biol 319 1059–66 [DOI] [PubMed] [Google Scholar]
- [95].Ruan J, Stormo GD and Zhang W 2004. An iterated loop matching approach to the prediction of RNA secondary structures with pseudoknots Bioinformatics 20 58–66 [DOI] [PubMed] [Google Scholar]
- [96].Touzet H and Perriquet O 2004. CARNAC: folding families of related RNAs Nucleic Acids Res 32 w142–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Jossinet F, Ludwig TE and Westhof E 2007. RNA structure: bioinformatic analysis Curr. Opin. Microbiol 10 279–85 [DOI] [PubMed] [Google Scholar]
- [98].Machado-Lima A, del Portillo HA and Durham AM 2008. Computational methods in noncoding RNA research J. Math. Biol 56 15–49 [DOI] [PubMed] [Google Scholar]
- [99].Mathews DH 2006. Revolutions in RNA secondary structure prediction J. Mol. Biol 359 526–32 [DOI] [PubMed] [Google Scholar]
- [100].Mathews DH et al. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure J. Mol. Biol 288 911–40 [DOI] [PubMed] [Google Scholar]
- [101].Xia T et al. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs Biochemistry 37 14719–35 [DOI] [PubMed] [Google Scholar]
- [102].Znosko BM et al. 2002. Thermodynamic parameters for an expanded nearest-neighbor model for the formation of RNA duplexes with single nucleotide bulges Biochemistry 41 10406–17 [DOI] [PubMed] [Google Scholar]
- [103].Mathews DH. RNA secondary structure analysis using RNAstructure. Curr. Protoc. Bioinform. 2006 doi: 10.1002/0471250953.bi1206s13. chapter 12, p unit12.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ding Y and Lawrence CE 2003. A statistical sampling algorithm for RNA secondary structure prediction Nucleic Acids Res 31 7280–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Giegerich R, Voss B and Rehmsmeier M 2004. Abstract shapes of RNA Nucleic Acids Res 32 4843–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Voss B, Giegerich R and Rehmsmeier M 2006. Complete probabilistic analysis of RNA shapes BMC Biol 4 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Geis M et al. 2008. Folding kinetics of large RNAs J. Mol. Biol 379 160–73 [DOI] [PubMed] [Google Scholar]
- [108].Shapiro BA et al. 2001. The massively parallel genetic algorithm for RNA folding: MIMD implementation and population variation Bioinformatics 17 137–48 [DOI] [PubMed] [Google Scholar]
- [109].Shapiro BA et al. 2001. RNA folding pathway functional intermediates: their prediction and analysis J. Mol. Biol 312 27–44 [DOI] [PubMed] [Google Scholar]
- [110].Lyngso RB and Pedersen CN 2000. RNA pseudoknot prediction in energy-based models J. Comput. Biol 7 409–27 [DOI] [PubMed] [Google Scholar]
- [111].Rivas E and Eddy SR 1999. A dynamic programming algorithm for RNA structure prediction including pseudoknots J. Mol. Biol 285 2053–68 [DOI] [PubMed] [Google Scholar]
- [112].Xayaphoummine A, Bucher T and Isambert H 2005. Kinefold Web server for RNA/DNA folding path and structure prediction including pseudoknots and knots Nucleic Acids Res 33 w605–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Xayaphoummine A et al. 2003. Prediction and statistics of pseudoknots in RNA structures using exactly clustered stochastic simulations Proc. Natl Acad. Sci. USA 100 15310–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sato K et al. 2009. CENTROIDFOLD: a Web server for RNA secondary structure prediction Nucleic Acids Res 37 w277–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Harmanci AO, Sharma G and Mathews DH 2007. Efficient pairwise RNA structure prediction using probabilistic alignment constraints in Dynalign BMC Bioinform 8 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Mathews DH and Turner DH 2002. Dynalign: an algorithm for finding the secondary structure common to two RNA sequences J. Mol. Biol 317 191–203 [DOI] [PubMed] [Google Scholar]
- [117].Jaeger L, Westhof E and Michel F 1993. Monitoring of the cooperative unfolding of the sunY group I intron of bacteriophage T4. The active form of the sunY ribozyme is stabilized by multiple interactions with 3 terminal intron components J. Mol. Biol 234 331–46 [DOI] [PubMed] [Google Scholar]
- [118].Lehnert V et al. 1996. New loop–loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme Chem. Biol 3 993–1009 [DOI] [PubMed] [Google Scholar]
- [119].Massire C, Jaeger L and Westhof E 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis J. Mol. Biol 279 773–93 [DOI] [PubMed] [Google Scholar]
- [120].Dima RI, Hyeon C and Thirumalai D 2005. Extracting stacking interaction parameters for RNA from the data set of native structures J. Mol. Biol 347 53–69 [DOI] [PubMed] [Google Scholar]
- [121].Altschul SF et al. 1990. Basic local alignment search tool J. Mol. Biol 215 403–10 [DOI] [PubMed] [Google Scholar]
- [122].Larkin MA et al. 2007. Clustal W and Clustal X version 2.0 Bioinformatics 23 2947–8 [DOI] [PubMed] [Google Scholar]
- [123].Brown JW et al. 2009. The RNA structure alignment ontology RNA 15 1623–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gan HH, Pasquali S and Schlick T 2003. Exploring the repertoire of RNA secondary motifs using graph theory; implications for RNA design Nucleic Acids Res 31 2926–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Das R and Baker D 2007. Automated de novo prediction of native-like RNA tertiary structures Proc. Natl Acad. Sci. USA 104 14664–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Rohl CA et al. 2004. Protein structure prediction using Rosetta Methods Enzymol 383 66–93 [DOI] [PubMed] [Google Scholar]
- [127].Das R et al. 2008. Structural inference of native and partially folded RNA by high-throughput contact mapping Proc. Natl Acad. Sci. USA 105 4144–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Lemieux S and Major F 2006. Automated extraction and classification of RNA tertiary structure cyclic motifs Nucleic Acids Res 34 2340–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].St-Onge K et al. 2007. Modeling RNA tertiary structure motifs by graph-grammars Nucleic Acids Res 35 1726–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Lepage GP 1978. A new algorithm for adaptive multidimensional integration J. Comput. Phys 27 192–203 [Google Scholar]
- [131].Jonikas MA et al. 2009. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters RNA 15 189–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sharma S, Ding F and Dokholyan NV 2008 iFoldRNA: three-dimensional RNA structure prediction and folding Bioinformatics 24 1951–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ding F et al. 2008. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms RNA 14 1164–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ding F and Dokholyan NV 2006. Emergence of proteinfold families through rational design PLoS Comput. Biol 2 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dokholyan NV et al. 1998. Discrete molecular dynamics studies of the folding of a protein-like model Fold Des 3 577–87 [DOI] [PubMed] [Google Scholar]
- [136].Martinez HM, Maizel JV Jr and Shapiro BA 2008. RNA2D3D: a program for generating, viewing, and comparing 3-dimensional models of RNA J. Biomol. Struct. Dyn 25 669–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Parisien M et al. 2009. New metrics for comparing and assessing discrepancies between RNA 3D structures and models RNA 15 1875–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Hsin J. et al. Using VMD: an introductory tutorial. Curr. Protoc. Bioinform. 2008 doi: 10.1002/0471250953.bi0507s24. chapter 5, p unit 5.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sarver M et al. 2008. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures J. Math. Biol 56 215–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Pang PS et al. 2005. Prediction of functional tertiary interactions and intermolecular interfaces from primary sequence data J. Exp. Zool. B 304 50–63 [DOI] [PubMed] [Google Scholar]
- [141].Mokdad A and Frankel AD 2008. ISFOLD: structure prediction of base pairs in non-helical RNA motifs from isostericity signatures in their sequence alignments J. Biomol. Struct. Dyn 25 467–72 [DOI] [PubMed] [Google Scholar]
- [142].Tirumalai D and Hyeon C 2009. Theory of RNA Folding: From Hairpins to Ribozymes (Springer Series in Biophysicsvol 13) pp 27–47 [Google Scholar]
- [143].Wells SE et al. 2000. Use of dimethyl sulfate to probe RNA structure in vivo Methods Enzymol 318 479–93 [DOI] [PubMed] [Google Scholar]
- [144].Andronescu M et al. 2008. RNA STRAND: the RNA secondary structure and statistical analysis database BMC Bioinformatics 9 340–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Cao S and Chen SJ 2009. Predicting structures and stabilites for H-type pseudoknots with interhelix loops RNA 15 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.