Abstract
CRISPR/Cas systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using crRNAs to guide the silencing of invading nucleic acids. We show here that in a subset of these systems, the mature crRNA base-paired to trans-activating tracrRNA forms a two-RNA structure that directs the CRISPR-associated protein Cas9 to introduce double-stranded (ds) breaks in target DNA. At sites complementary to the crRNA-guide sequence, the Cas9 HNH nuclease domain cleaves the complementary strand while the Cas9 RuvC-like domain cleaves the non-complementary strand. The dual-tracrRNA:crRNA, when engineered as a single RNA chimera, also directs sequence-specific Cas9 dsDNA cleavage. Our study reveals a family of endonucleases that use dual-RNAs for site-specific DNA cleavage and highlights the potential to exploit the system for RNA-programmable genome editing.
One-Sentence Summary:
A two-RNA structure directs an endonuclease to cleave target DNA.
Bacteria and archaea have evolved RNA-mediated adaptive defense systems called CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas (CRISPR-associated) that protect organisms from invading viruses and plasmids(1-3). These defense systems rely on small RNAs for sequence-specific detection and silencing of foreign nucleic acids. CRISPR/Cas systems are composed of cas genes organized in operon(s) and a CRISPR array consisting of unique genome-targeting sequences (called spacers) interspersed with identical repeats (1-3). CRISPR/Cas mediated immunity occurs in three steps. In the adaptive phase, bacteria and archaea harboring one or more CRISPR loci respond to viral and plasmid challenge by integrating short fragments of foreign sequence (protospacers) into the host chromosome at the proximal end of the CRISPR array (1-3). In the expression and interference phases, transcription of the repeat-spacer element into precursor CRISPR RNA (pre-crRNA) molecules followed by enzymatic cleavage yields the short crRNAs that can base pair with complementary protospacer sequences of invading viral or plasmid targets (4-11). Target recognition by crRNAs directs the silencing of the foreign sequences by means of Cas proteins that function in complex with the crRNAs (10, 12-20).
There are three types of CRISPR/Cas systems (21-23). The Type I and III systems share some overarching features: specialized Cas endonucleases process the pre-crRNAs, and once mature, each crRNA assembles into a large multi-Cas protein complex capable of recognizing and cleaving nucleic acids complementary to the crRNA. In contrast, Type II systems process pre-crRNAs by a different mechanism in which a trans-activating crRNA (tracrRNA) complementary to the repeat sequences in pre-crRNA triggers processing by the double-stranded RNA-specific ribonuclease RNase III in the presence of the Cas9 (formerly Csn1) protein (4, 24) (fig. S1). Cas9 is thought to be the sole protein responsible for crRNA-guided silencing of foreign DNA (25-27).
We show here that in Type II systems, Cas9 proteins constitute a family of enzymes that require a base-paired structure formed between the activating tracrRNA and the targeting crRNA to cleave target double-stranded (ds) DNA. Site-specific cleavage occurs at locations determined by both base-pairing complementarity between the crRNA and the target protospacer DNA and a short motif (referred to as the protospacer adjacent motif, or PAM) juxtaposed to the complementary region in the target DNA. Our study further demonstrates that the Cas9 endonuclease family can be programmed with single RNA molecules to cleave specific DNA sites, thereby raising the exciting possibility of developing a simple and versatile RNA-directed system to generate dsDNA breaks (DSBs) for genome targeting and editing.
Cas9 is a DNA endonuclease guided by two RNAs.
Cas9, the hallmark protein of Type II systems, has been hypothesized to be involved in both crRNA maturation and crRNA-guided DNA interference (fig. S1) (4, 25-27). Cas9 is involved in crRNA maturation (4), but its direct participation in target DNA destruction has not been investigated. To test whether and how Cas9 might be capable of target DNA cleavage, we used an overexpression system to purify Cas9 protein derived from the pathogen Streptococcus pyogenes (fig. S2) and tested its ability to cleave a plasmid DNA or an oligonucleotide duplex bearing a protospacer sequence complementary to a mature crRNA, and a bona fide PAM. We found that mature crRNA alone was incapable of directing Cas9-catalyzed plasmid DNA cleavage (Fig. 1A and fig. S3A). However, addition of tracrRNA, which can base pair with the repeat sequence of crRNA and is essential to crRNA maturation in this system, triggered Cas9 to cleave plasmid DNA (Fig. 1A and fig. S3A). The cleavage reaction required both magnesium and the presence of a crRNA sequence complementary to the DNA; a crRNA capable of tracrRNA base-pairing but containing a non-cognate target DNA-binding sequence did not support Cas9-catalyzed plasmid cleavage (Fig. 1A, fig. S3A, compare crRNA-sp2 to crRNA-sp1 and fig. S4A). Similar results were obtained with a short linear dsDNA substrate (Fig. 1B and fig. S3B, C). The trans-activating tracrRNA is thus a small non-coding RNA with two critical functions: triggering pre-crRNA processing by the enzyme RNase III (4) and subsequently activating crRNA-guided DNA cleavage by Cas9.
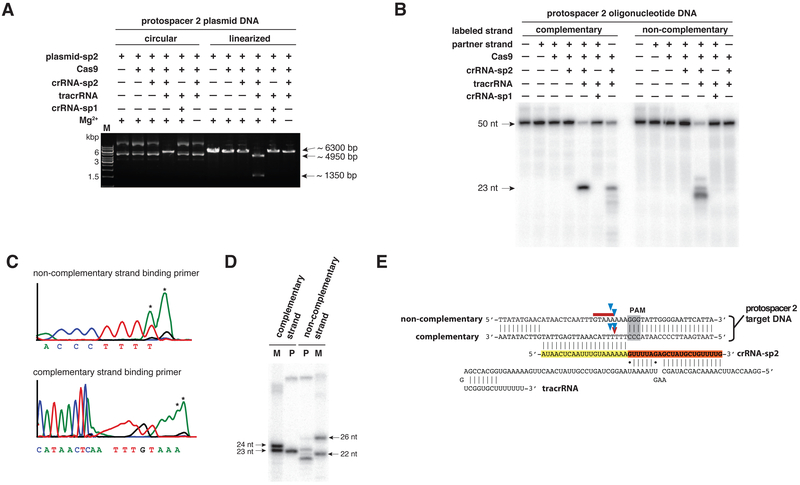
Fig. 1. Cas9 is a DNA endonuclease guided by two RNA molecules.
(A) Cas9 was programmed with a 42-nucleotide crRNA-sp2 (crRNA containing spacer 2 sequence) in the presence or absence of 75-nucleotide tracrRNA. The complex was added to circular or XhoI-linearized plasmid DNA bearing a sequence complementary to spacer 2 and a functional PAM. crRNA-sp1, specificity control; M, DNA marker; refer to fig. S3A. (B) Cas9 was programmed with crRNA-sp2 and tracrRNA (nucleotides 4-89). The complex was incubated with double- or single-stranded DNAs harboring a sequence complementary to spacer 2 and a functional PAM (4). The complementary or non-complementary strands of the DNA were 5’-radiolabeled and annealed with a non-labeled partner strand. Refer to fig. S3B, C. (C) Sequencing analysis of cleavage products from Fig. 1A. Termination of primer extension in the sequencing reaction indicates the position of the cleavage site. The 3’ terminal A overhang (asterisk) is an artifact of the sequencing reaction. Refer to fig. S5A, C. (D) The cleavage products from Fig. 1B were analyzed alongside 5’ end-labeled size markers derived from the complementary and non-complementary strands of the target DNA duplex. M, marker; P, cleavage product. Refer to fig. S5B, C. (E) Schematic representation of tracrRNA, crRNA-sp2 and protospacer 2 DNA sequences; regions of crRNA complementarity to tracrRNA (orange) and the protospacer DNA (yellow) are represented; PAM sequence, grey; cleavage sites mapped in (C) and (D) are represented by blue arrows (C), a red arrow (D, complementary strand) and a red line (D, non-complementary strand).
Cleavage of both plasmid and short linear dsDNA by tracrRNA:crRNA-guided Cas9 is site-specific (Fig. 1C-E and fig. S5A, B). Plasmid DNA cleavage produced blunt ends at a position three base pairs upstream of the PAM sequence (Fig. 1C, E and fig. S5A, C) (26). Similarly, within short dsDNA duplexes, the DNA strand that is complementary to the target-binding sequence in the crRNA (the complementary strand) is cleaved at a site three base pairs upstream of the PAM (Fig. 1D, E and fig. S5B, C). The non-complementary DNA strand is cleaved at one or more sites within 3 to 8 base pairs upstream of the PAM. Further investigation revealed that the non-complementary strand is first cleaved endonucleolytically and subsequently trimmed by a 3’-5’ exonuclease activity (fig. S4B). The cleavage rates by Cas9 under single turnover conditions ranged from 0.3 to 1 min−1, comparable to those of restriction endonucleases (fig. S6A), while incubation of wild-type Cas9-tracrRNA:crRNA complex with a 5-fold molar excess of substrate DNA provided evidence that the dual-RNA-guided Cas9 is a multiple-turnover enzyme (fig. S6B). In contrast to the CRISPR Type I Cascade complex (20), Cas9 cleaves both linearized and supercoiled plasmids (Fig. 1A, 2A). An invading plasmid can therefore in principle be cleaved multiple times by Cas9 proteins programmed with different crRNAs.
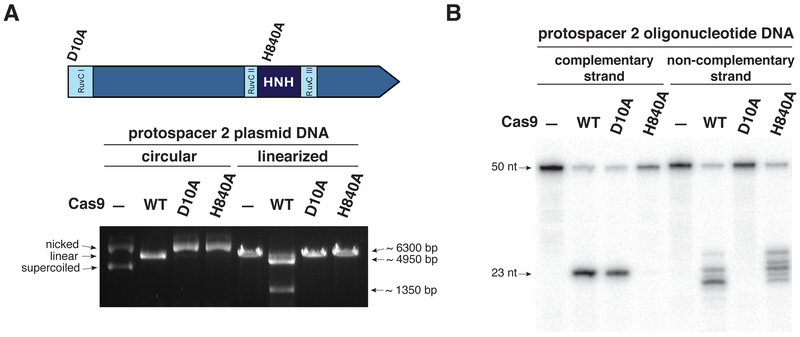
Fig. 2. Cas9 uses two nuclease domains to cleave the two strands in the target DNA.
(A) Top: Schematic representation of Cas9 domain structure showing the positions of domain mutations. Bottom: Complexes of wild-type or nuclease mutant Cas9 proteins with tracrRNA:crRNA-sp2 were assayed for endonuclease activity as in Fig. 1A. (B) Complexes of wild-type Cas9 or nuclease domain mutants with tracrRNA and crRNA-sp2 were tested for activity as in Fig. 1B.
Each Cas9 nuclease domain cleaves one DNA strand.
Cas9 contains domains homologous to both HNH and RuvC endonucleases (Fig. 2A, fig. S7) (21-23, 27, 28). We designed and purified Cas9 variants containing inactivating point mutations in the catalytic residues of either the HNH or RuvC-like domains (Fig. 2A and fig. S7) (23, 27). Incubation of these variant Cas9 proteins with native plasmid DNA showed that dual-RNA-guided mutant Cas9 proteins yielded nicked open circular plasmids, while the wild-type Cas9 protein-tracrRNA:crRNA complex produced a linear DNA product (Fig. 1A, 2A and fig. S3A, S8A). This result indicates that the Cas9 HNH and RuvC-like domains each cleave one plasmid DNA strand. To determine which strand of the target DNA is cleaved by each Cas9 catalytic domain, we incubated the mutant Cas9-tracrRNA:crRNA complexes with short dsDNA substrates in which either the complementary or the non-complementary strand was radiolabeled at its 5’ end. The resulting cleavage products indicated that the Cas9 HNH domain cleaves the complementary DNA strand, while the Cas9 RuvC-like domain cleaves the non-complementary DNA strand (Fig. 2B and fig. S8B).
Dual-RNA requirements for target DNA binding and cleavage.
tracrRNA might be required for target DNA binding and/or to stimulate the nuclease activity of Cas9 downstream of target recognition. To distinguish between these possibilities, we used an electrophoretic mobility shift assay to monitor target DNA binding by catalytically inactive Cas9 in the presence or absence of crRNA and/or tracrRNA. Addition of tracrRNA substantially enhanced target DNA binding by Cas9, whereas little specific DNA binding was observed with Cas9 alone or Cas9-crRNA (fig. S9). This indicates that tracrRNA is required for target DNA recognition, possibly by properly orienting the crRNA for interaction with the complementary strand of target DNA. The predicted tracrRNA:crRNA secondary structure includes base-pairing between the 3’-terminal 22-nucleotides of the crRNA and a segment near the 5’ end of the mature tracrRNA (Fig. 1E). This interaction creates a structure in which the 5’-terminal 20 nucleotides of the crRNA, which vary in sequence in different crRNAs, are available for target DNA binding. The bulk of the tracrRNA downstream of the crRNA base-pairing region is free to form additional RNA structure(s) and/or to interact with Cas9 or the target DNA site. To determine whether the entire length of the tracrRNA is necessary for site-specific Cas9-catalyzed DNA cleavage, we tested Cas9-tracrRNA:crRNA complexes reconstituted using full-length mature (42-nt) crRNA and various truncated forms of tracrRNA lacking sequences at their 5’ or 3’ ends. These complexes were tested for cleavage using a short target dsDNA. A substantially truncated version of the tracrRNA retaining nucleotides 23-48 of the native sequence was capable of supporting robust dual-RNA-guided Cas9-catalyzed DNA cleavage (Fig. 3A, C and fig. S10A, B). Truncation of the crRNA from either end showed that Cas9-catalyzed cleavage in the presence of tracrRNA could be triggered with crRNAs missing the 3’-terminal 10 nucleotides (Fig. 3B, C). In contrast, a 10-nucleotide deletion from the 5’ end of crRNA abolished DNA cleavage by Cas9 (Fig. 3B). We also analyzed Cas9 orthologs from various bacterial species for their ability to support S. pyogenes tracrRNA:crRNA-guided DNA cleavage. In contrast to closely related S. pyogenes Cas9 orthologs, more distantly related orthologs were not functional in the cleavage reaction (fig. S11). Similarly, S. pyogenes Cas9 guided by tracrRNA:crRNA pairs originating from more distant systems were unable to cleave DNA efficiently (fig. S11). Species specificity of dual-RNA-guided cleavage of DNA indicates co-evolution of Cas9, tracrRNA and the crRNA repeat, as well as the existence of a still unknown structure and/or sequence in the dual-RNA that is critical for the formation of the ternary complex with specific Cas9 orthologs.
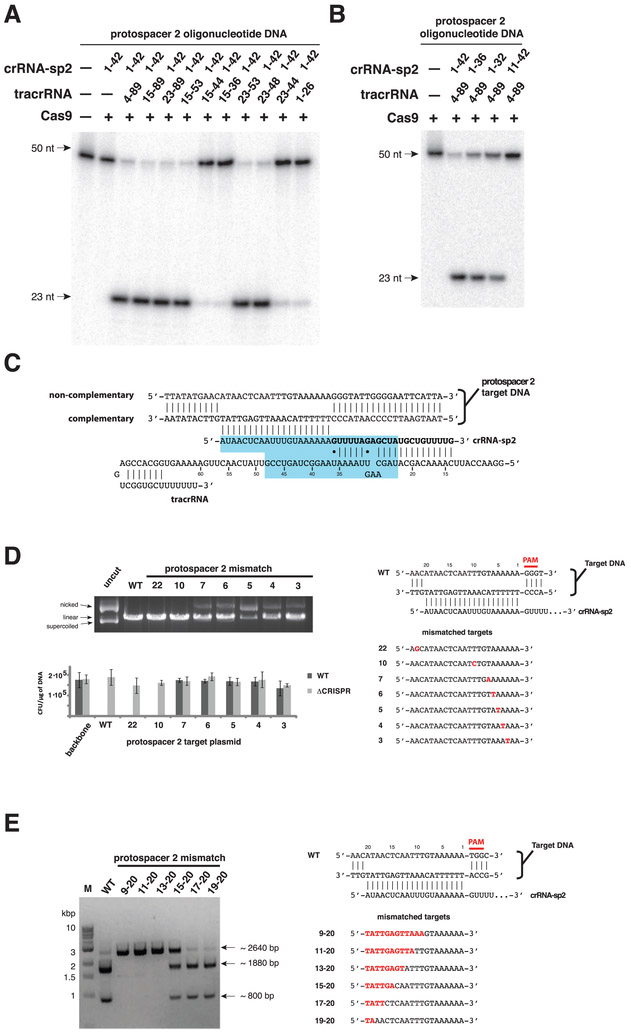
Fig. 3. Cas9-catalyzed cleavage of target DNA requires an activating domain in tracrRNA and is governed by a seed sequence in the crRNA.
(A) Cas9-tracrRNA:crRNA complexes were reconstituted using 42-nucleotide crRNA-sp2 and truncated tracrRNA constructs and assayed for cleavage activity as in Fig. 1B. (B) Cas9 programmed with full-length tracrRNA and crRNA-sp2 truncations was assayed for activity as in (A). (C) Minimal regions of tracrRNA and crRNA capable of guiding Cas9-mediated DNA cleavage (blue box). (D) Plasmids containing wild-type or mutant protospacer 2 sequences with indicated point mutations (right) were cleaved in vitro by programmed Cas9 as in Fig. 1A (left top) and used for transformation assays of wild-type or pre-crRNA-deficient S. pyogenes (left bottom). The transformation efficiency was calculated as CFU per μg of plasmid DNA; error bars represent standard deviations for three biological replicates. (E) Plasmids containing wild-type and mutant protospacer 2 inserts with varying extent of crRNA-target DNA mismatches (right) were cleaved in vitro by programmed Cas9 (left). The cleavage reactions were further digested with XmnI. The 1880 bp and 800 bp fragments are Cas9-generated cleavage products.
To investigate the protospacer sequence requirements for Type II CRISPR/Cas immunity in bacterial cells, a series of protospacer-containing plasmid DNAs harboring single-nucleotide mutations were analyzed for their maintenance following transformation in S. pyogenes and their ability to be cleaved by Cas9 in vitro. In contrast to point mutations introduced at the 5’ end of the protospacer, mutations in the region close to the PAM and the Cas9 cleavage sites were not tolerated in vivo and resulted in decreased plasmid cleavage efficiency in vitro (Fig. 3D). Our results are in agreement with a previous report of protospacer escape mutants selected in the Type II CRISPR system from S. thermophilus in vivo (27, 29). Furthermore, the plasmid maintenance and cleavage results hint at the existence of a “seed” region located at the 3’ end of the protospacer sequence that is crucial for the interaction with crRNA and subsequent cleavage by Cas9. In support of this notion, Cas9 enhanced complementary DNA strand hybridization to the crRNA and this enhancement was the strongest in the 3’-terminal region of the crRNA targeting sequence (fig. S12). Corroborating this, a contiguous stretch of at least 13 base pairs between the crRNA and the target DNA site proximal to the PAM is required for efficient target cleavage, while up to six contiguous mismatches in the 5’-terminal region of the protospacer are tolerated (Fig. 3E). These findings are reminiscent of the previously observed seed sequence requirements for target nucleic acid recognition in Argonaute proteins (30, 31) and the Cascade and Csy CRISPR complexes (13, 14).
A short sequence motif dictates R-loop formation.
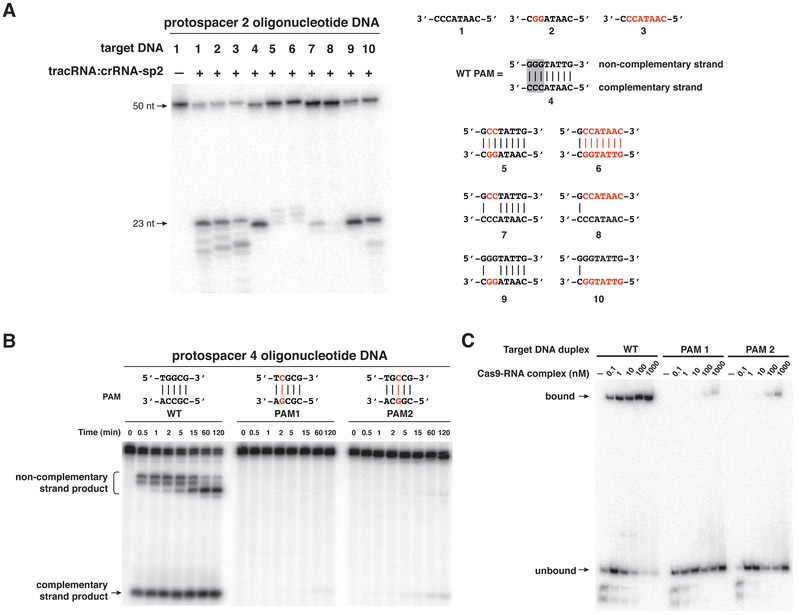
In multiple CRISPR/Cas systems, recognition of self versus non-self has been shown to involve a short sequence motif that is preserved in the foreign genome, referred to as the PAM (27, 29, 32-34). PAM motifs are only a few base pairs in length, and their precise sequence and position vary according to the CRISPR/Cas system type (32). In the S. pyogenes Type II system, the PAM conforms to an NGG consensus sequence, containing two G:C base pairs that occur one base pair downstream of the crRNA binding sequence, within the target DNA (4). Transformation assays demonstrated that the GG motif is essential for protospacer plasmid DNA elimination by CRISPR/Cas in bacterial cells (fig. S13A), consistent with previous observations in S. thermophilus (27). The motif is also essential for in vitro protospacer plasmid cleavage by tracrRNA:crRNA-guided Cas9 (fig. S13B). To determine the role of the PAM in target DNA cleavage by the Cas9-tracrRNA:crRNA complex, we tested a series of dsDNA duplexes containing mutations in the PAM sequence on the complementary or non-complementary strands, or both (Fig. 4A). Cleavage assays using these substrates showed that Cas9-catalyzed DNA cleavage was particularly sensitive to mutations in the PAM sequence on the non-complementary strand of the DNA, in contrast to complementary strand PAM recognition by Type I CRISPR/Cas systems (20, 34). Cleavage of target single-stranded DNAs was unaffected by mutations of the PAM motif. This observation suggests that the PAM motif is required only in the context of target dsDNA and may thus be required to license duplex unwinding, strand invasion, and the formation of an R-loop structure. Using a different crRNA-target DNA pair (crRNA-sp4 and protospacer 4 DNA), selected due to the presence of a canonical PAM not present in the protospacer 2 target DNA, we found that both G nucleotides of the PAM were required for efficient Cas9-catalyzed DNA cleavage (Fig. 4B and fig. S13C). To determine whether the PAM plays a direct role in recruiting the Cas9-tracrRNA:crRNA complex to the correct target DNA site, binding affinities of the complex for target DNA sequences were analyzed by native gel mobility shift assays (Fig. 4C). Mutation of either G in the PAM sequence substantially reduced the affinity of Cas9-tracrRNA:crRNA for the target DNA. This finding argues for specific recognition of the PAM sequence by Cas9 as a prerequisite for target DNA binding and possibly strand separation to allow strand invasion and R-loop formation, which would be analogous to the PAM sequence recognition by CasA/Cse1 implicated in a Type I CRISPR/Cas system (34).
Fig. 4. A PAM is required to license target DNA cleavage by the Cas9-tracrRNA:crRNA complex.
(A) Dual RNA-programmed Cas9 was tested for activity as in Fig. 1B. Wild-type and mutant PAM sequences in target DNAs are indicated (right). (B) Protospacer 4 target DNA duplexes (labeled at both 5’ ends) containing wild-type and mutant PAM motifs were incubated with Cas9 programmed with tracrRNA (nt 23-89):crRNA-sp4. At indicated time points (min), aliquots of the cleavage reaction were taken and analyzed as in Fig. 1B. (C) Electrophoretic mobility shift assays were performed using RNA-programmed Cas9 (D10A/H840A) and protospacer 4 target DNA duplexes (same as in (B)) containing wild-type and mutated PAM motifs. Cas9 (D10A/H840A)-RNA complex was titrated from 100 pM to 1 μM.
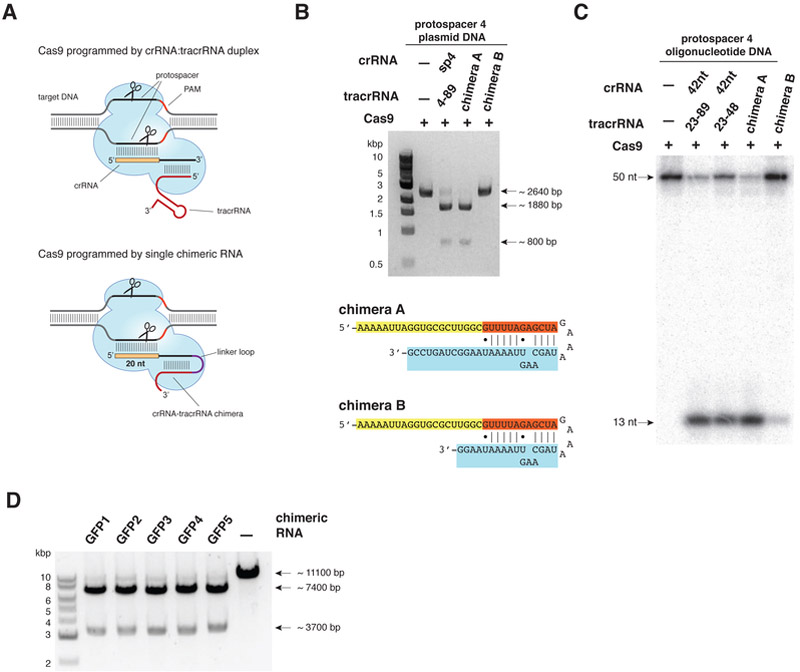
Cas9 can be programmed with a single chimeric RNA.
Examination of the likely secondary structure of the tracrRNA:crRNA duplex (Fig. 1E, 3C) suggested the possibility that the features required for site-specific Cas9-catalyzed DNA cleavage could be captured in a single chimeric RNA. Although the tracrRNA:crRNA target selection mechanism works efficiently in nature, the possibility of a single RNA-guided Cas9 is appealing due to its potential utility for programmed DNA cleavage and genome editing (Fig. 5A). We designed two versions of a chimeric RNA containing a target recognition sequence at the 5’ end followed by a hairpin structure retaining the base-pairing interactions that occur between the tracrRNA and the crRNA (Fig. 5B). This single transcript effectively fuses the 3’end of crRNA to the 5’ end of tracrRNA, thereby mimicking the dual-RNA structure required to guide site-specific DNA cleavage by Cas9. In cleavage assays using plasmid DNA, we observed that the longer chimeric RNA was able to guide Cas9-catalyzed DNA cleavage in a manner similar to that observed for the truncated tracrRNA:crRNA duplex (Fig. 5B and fig. S14A). The shorter chimeric RNA did not work efficiently in this assay, confirming that nucleotides 5-12 positions beyond the tracrRNA:crRNA base-pairing interaction are important for efficient Cas9 binding and/or target recognition. Similar results were observed in cleavage assays using short dsDNA as a substrate, which further indicate that the position of the cleavage site in target DNA is identical to that observed using the dual tracrRNA:crRNA as a guide (Fig. 5C and fig. S14B). Finally, to establish whether the design of chimeric RNA might be universally applicable, we engineered five different chimeric guide RNAs to target a portion of the gene encoding the green-fluorescent protein (GFP) (fig. S15A-C), and tested their efficacy against a plasmid carrying the GFP coding sequence in vitro. In all five cases, Cas9 programmed with these chimeric RNAs efficiently cleaved the plasmid at the correct target site (Fig. 5D, fig. S15D), indicating that rational design of chimeric RNAs is robust and could in principle enable targeting of any DNA sequence of interest with few constraints beyond the presence of a GG dinucleotide adjacent to the targeted sequence.
Fig. 5. Cas9 can be programmed using a single engineered RNA molecule combining tracrRNA and crRNA features.
(A) Top: In Type II CRISPR/Cas systems, Cas9 is guided by a two-RNA structure formed by activating tracrRNA and targeting crRNA to cleave site-specifically target dsDNA (refer to fig. S1). Bottom: A chimeric RNA generated by fusing the 3’ end of crRNA to the 5’ end of tracrRNA. (B) A plasmid harboring protospacer 4 target sequence and a wild-type PAM was subjected to cleavage by Cas9 programmed with tracrRNA(4-89):crRNA-sp4 duplex or in vitro-transcribed chimeric RNAs constructed by joining the 3’ end of crRNA to the 5’ end of tracrRNA with a GAAA tetraloop. Cleavage reactions were analyzed by restriction mapping with XmnI. Sequences of chimeric RNAs A and B are shown with DNA-targeting (yellow), crRNA repeat-derived (orange) and tracrRNA-derived (light blue) sequences. (C) Protospacer 4 DNA duplex cleavage reactions were performed as in Fig. 1B. (D) Five chimeric RNAs designed to target the GFP gene were used to program Cas9 to cleave a GFP gene-containing plasmid. Plasmid cleavage reactions were performed as in Fig. 3E, except that the plasmid DNA was restriction mapped with AvrII following Cas9 cleavage.
Conclusions.
In summary, we identify a DNA interference mechanism involving a dual-RNA structure that directs a Cas9 endonuclease to introduce site-specific double-stranded breaks in target DNA. The tracrRNA:crRNA-guided Cas9 protein utilizes distinct endonuclease domains, HNH and RuvC-like, to cleave the two strands in the target DNA. Target recognition by Cas9 requires both a seed sequence in the crRNA and a GG dinucleotide-containing PAM sequence adjacent to the crRNA-binding region in the DNA target. We further show that the Cas9 endonuclease can be programmed with guide RNA engineered as a single transcript to target and cleave any dsDNA sequence of interest. The system is efficient, versatile and programmable by changing the DNA target-binding sequence in the guide chimeric RNA. Zinc-Finger Nucleases (ZFNs) and Transcription-Activator Like Effector Nucleases (TALENs) have attracted considerable interest as artificial enzymes engineered to manipulate genomes (35-38). We propose an alternative methodology based on RNA-programmed Cas9 that could offer considerable potential for gene targeting and genome editing applications.
Supplementary Material
Acknowledgements.
We thank Kaihong Zhou, Alison Marie Smith, Rachel Haurwitz and Sam Sternberg for excellent technical assistance, and members of the Doudna and Charpentier laboratories and Jamie Cate for comments on the manuscript. We thank Barbara Meyer and Te-Wen Lo (UC Berkeley/HHMI) for providing the GFP plasmid. This work was funded by the Howard Hughes Medical Institute (M.J. and J.A.D.), the Austrian Science Fund (W1207-B09, K.C. and E.C.), the University of Vienna (K.C.), the Swedish Research Council (#K2010-57X-21436-01-3 and #621-2011-5752-LiMS, E.C.), the Kempe Foundation (E.C.) and Umeå University (K.C., E.C.). J.A.D. is an Investigator and M.J. is a Research Specialist of the Howard Hughes Medical Institute. K.C. is a fellow of the Austrian Doctoral Program in RNA Biology and co-supervised by R. Schroeder. We are grateful to A. Witte, U. Bläsi and R. Schroeder for helpful discussions, financial support to K.C and hosting K.C. in their laboratories at MFPL. M.J., K.C., J.A.D. and E.C. have filed a related patent.
Footnotes
Supplementary Materials. The Supplementary Materials contain the Supplementary Materials and Methods, Supplementary Figures S1-S15 with legends, and Supplementary Tables S1-S3, with references 39-47.
References and Notes
- 1.Wiedenheft B, Sternberg SH, Doudna JA, Nature 482, 331–338 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Bhaya D, Davison M, Barrangou R, Annu. Rev. Genet. 45, 273–297 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Terns MP, Terns RM, Curr Opin Microbiol 14, 321–327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deltcheva E et al. , Nature 471, 602–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carte J, Wang R, Li H, Terns RM, Terns MP, Genes Dev 22, 3489–3496 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haurwitz RE, Jínek M, Wiedenheft B, Zhou K, Doudna JA, Science 329, 1355–1358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R, Preamplume G, Terns MP, Terns RM, Li H, Structure 19, 257–264 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM, Nat Struct Mol Biol 18, 688–692 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Hatoum-Aslan A, Maniv I, Marraffini LA, Proc Natl Acad Sci USA 108, 21218–21222 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brouns SJJ et al. , Science 321, 960–964 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sashital DG, Jínek M, Doudna JA, Nat Struct Mol Biol 18, 680–687 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Lintner NG et al. , Journal of Biological Chemistry 286, 21643–21656 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenova E et al. , Proc Natl Acad Sci US A 108, 10098–10103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiedenheft B et al. , Proc Natl Acad Sci US A 108, 10092–10097 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiedenheft B et al. , Nature 477, 486–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale CR et al. , Cell 139, 945–956 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CR et al. , Mol Cell, 1–11 (2012). [Google Scholar]
- 18.Zhang J et al. , Mol Cell, 1–11 (2012). [Google Scholar]
- 19.Howard JAL, Delmas S, Ivančić Baće I, Bolt EL, Biochem. J. 439, 85–95 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Westra ER et al. , Mol Cell 46, 595–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova KS et al. , Nat Rev Microbiol 9, 467–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV, Biol Direct 1, 7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova KS, Aravind L, Wolf YI, Koonin EV, Biol Direct 6, 38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman S, Nature 471, 588–589 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Barrangou R et al. , Science 315, 1709–1712 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Garneau JE et al. , Nature 468, 67–71 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Sapranauskas R et al. , Nucleic Acids Res 39, 9275–9282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor GK, Heiter DF, Pietrokovski S, Stoddard BL, Nucleic Acids Res 39, 9705–9719 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deveau H et al. , J. Bacteriol. 190, 1390–1400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP, Cell 120, 15–20 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Hutvagner G, Simard MJ, Nat Rev Mol Cell Biol 9, 22–32 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C, Microbiology 155, 733–740 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Marraffini LA, Sontheimer EJ, Nature 463, 568–571 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sashital DG, Wiedenheft B, Doudna JA, Mol Cell 46, 606–615 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christian M et al. , Genetics 186, 757–761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller JC et al. , Nat Biotechnol 29, 143–148 (2010).21179091 [Google Scholar]
- 37.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, Nat Rev Genet 11, 636–646 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Carroll D, Genetics 188, 773–782 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.