Abstract
Purpose
To determine the potential for detection of incidental germline cancer predisposition mutations through cell-free DNA (cfDNA) analyses in patients who underwent solid tumor somatic mutation evaluation.
Patients and Methods
Data were evaluated from 10,888 unselected patients with advanced (stage III/IV) cancer who underwent Guardant360 testing between November 2015 and December 2016. The main outcome was prevalence of putative germline mutations identified among 16 actionable hereditary cancer predisposition genes.
Results
More than 50 cancer types were studied, including lung (41%), breast (19%), colorectal (8%), prostate (6%), pancreatic (3%), and ovarian (2%). Average patient age was 63.5 years (range, 18 to 95 years); 43% were male. One hundred and fifty-six individuals (1.4%) had suspected hereditary cancer mutations in 11 genes. Putative germline mutations were more frequent in individuals younger than 50 years versus those 50 years and older (3.0% v 1.2%, respectively; P < .001). Highest yields of putative germline findings were in patients with ovarian (8.13%), prostate (3.46%), pancreatic (3.34%), and breast (2.2%) cancer. Putative germline mutation identification was consistent among 12 individuals with multiple samples. Patients with circulating tumor DNA copy number variation and/or reversion mutations suggestive of functional loss of the wild-type allele in the tumor DNA also are described.
Conclusion
Detection of putative germline mutations from cfDNA is feasible across multiple genes and cancer types without prior mutation knowledge. Many mutations were found in cancers without clear guidelines for hereditary cancer genetic counseling/testing. Given the clinical significance of identifying hereditary cancer predisposition for patients and their families as well as targetable germline alterations such as in BRCA1 or BRCA2, research on the best way to validate and return potential germline results from cfDNA analysis to clinicians and patients is needed.
INTRODUCTION
Identification of individuals and families at high risk for cancer can lead to opportunities for cancer prevention and early detection and can influence cancer treatment and management decisions.1-3 However, current guideline-directed genetic testing for cancer risk misses a significant portion of hereditary cancer predisposition gene mutation carriers.4,5
Germline mutation detection in the context of tumor genomic sequencing has been reported in single-institution and retrospective consortia (eg, The Cancer Genome Atlas) settings,6,7 although established mechanisms for return of results are lacking. Novel methods to identify patients with germline mutations could serve as useful adjuncts to conventional genetic cancer risk assessment.2,3 In addition, mutations in germline homologous recombination repair genes (eg, BRCA1, BRCA2) identify candidates for poly (ADP-ribose) polymerase inhibitor treatment and other targeted therapies in clinical trials.8
Cell-free DNA (cfDNA) studies in cancer have shown promise for tumor detection, molecular heterogeneity assessments, identification of therapeutically relevant genomic alterations, and monitoring of tumor dynamics and response to therapy.9 Incidental maternal germline cancer risk gene mutations have been reported in noninvasive prenatal cfDNA testing for fetal aneuploidies.10 Similarly, because circulating tumor cfDNA (ctDNA) sequencing is from mostly leukocyte-derived DNA, cfDNA next-generation sequencing (NGS) for somatic tumor mutations may identify heterozygous germline mutations at approximately 50% mutant allele fraction (MAF), which generally are distinguishable from somatic mutations that typically occur at lower MAFs.11
Few studies have evaluated germline cancer predisposition through cfDNA,12-14 and no publications have reported the utility of ctDNA analyses in hereditary predisposition mutation detection across multiple genes or cancer types when the germline mutation status is unknown. The purpose of this study was to describe putative (ie, probable) incidental germline mutations identified through commercial cfDNA testing of patients who underwent solid tumor somatic genotyping. Furthermore, we define clinical factors associated with putative germline mutation detection.
PATIENTS AND METHODS
Participants and Sequencing
This observational, noninterventional case series (completed under Quorum Review Institutional Review Board protocol 30-001) included coded data from 11,681 de-identified cfDNA samples from 10,888 consecutive patients with advanced (all stage III/IV) solid tumors who underwent Guardant360 (Guardant Health, Redwood City, CA) testing (70 to 73 genes) as part of clinical care between November 2015 and December 2016. No individuals with sequencing data available were excluded from the analysis. cfDNA was extracted from plasma, quantified, and used to prepare sequencing libraries, which were sequenced to approximately 15,000 times the average read depth using methods previously described.15,16 The Guardant360 assay detects single nucleotide variants, small (fewer than 50 bases) indels, MET exon 14 large indels, copy number amplifications, and fusion events.
Variant Curation
Variants in 16 clinically actionable genes with defined hereditary cancer associations were analyzed, including APC, ATM, BRCA1, BRCA2, CDKN2A, KIT, MLH1, NF1, PTEN, RB1, RET, SMAD4, STK11, TP53, TSC1, and VHL (Table 1). Of this list, full exon sequencing was performed for BRCA1, BRCA2, CDKN2A, KIT, NF1, PTEN, RB1, TP53, and VHL; select exons (those with previously reported somatic mutations) were sequenced for the remaining genes.
Table 1.
Putative Germline Mutation Occurrences by Cancer Type
All variants suspected to be of germline origin (allele fraction, 40% to 60%) were included in the screen for pathogenic germline alterations. The allele fraction cutoff was selected as a conservative approach on the basis of expected 50% MAFs with a standard deviation of 2.3% for germline heterozygous alterations. Our decision rule to evaluate variants of 40% or more allele fraction was selected a priori without additional adjustments. Variants with an allele fraction less than 40% are more likely to represent somatic alterations related to ctDNA or clonal hematopoiesis.17,18 In addition, variants with allele fractions greater than 60% were included if a concomitant copy number change was identified, concordant with deviations in allele fractions for neighboring common germline single nucleotide polymorphisms (SNPs) from their expected allele fractions. Generally, such deviations may occur as a result of either copy number gain of the variant allele or copy number loss of the wild-type (nonvariant containing) allele. Equivocal cases with high ctDNA load (defined as more than one circulating tumor mutation present at an MAF greater than 30%), were manually reviewed for relevant gene copy number status and allele frequencies of common neighboring germline SNPs to characterize variants as germline or somatic. If manual review did not adjudicate status, the equivocal variant was not included in the putative germline set.
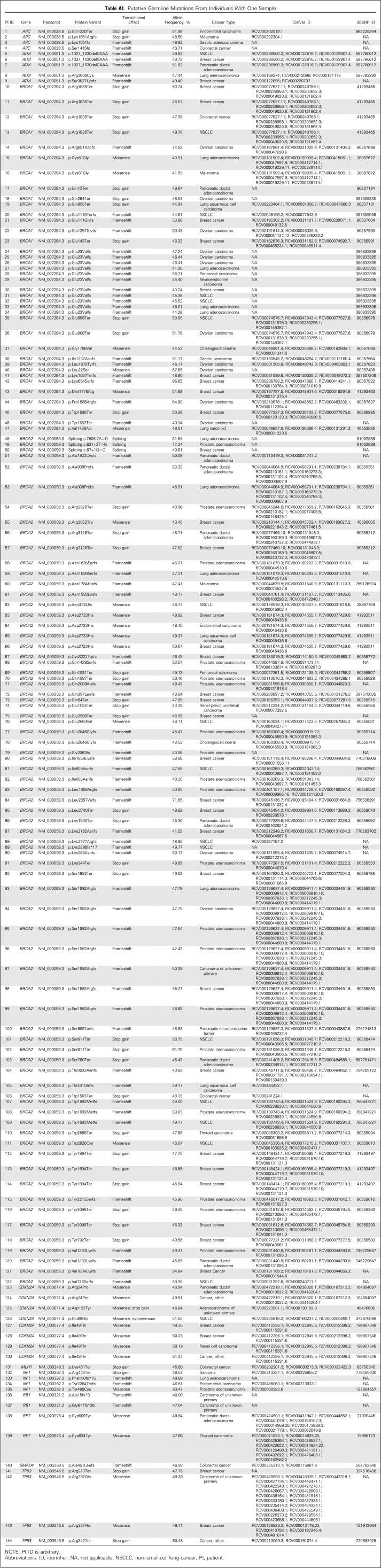
Using Ingenuity Variant Analysis (QIAGEN, Redwood City, CA), variants observed with an allele frequency of 2.0% or greater in the 1000 Genomes Project,19 National Heart, Lung, and Blood Institute Exome Sequencing Project,20 Exome Aggregation Consortium,21 or Genome Aggregation Database21 databases were excluded. Remaining single nucleotide variants and indels suspicious for germline origin were classified by Ingenuity Variant Analysis as pathogenic, likely pathogenic, unknown significance, likely benign, or benign according to American College of Medical Genetics and Genomics guidelines.22 All pathogenic and likely pathogenic variants and other frameshifts, start losses or gains, and ± 1, 2 splice site variants were manually reviewed by a board-certified individual in molecular diagnostics by the American Board of Clinical Chemistry (T.P.S.) and a genetic counselor (K.C.B.) to establish a final set of putative mutations (our summary term for likely pathogenic and pathogenic variants22). During manual review, ClinVar (National Center for Biotechnology Information, Bethesda, MD) was used to confirm final determinations when available for a particular variant. For variants that lacked consensus among ClinVar commercial laboratories (Online Mendelian Inheritance in Man excluded), Ingenuity Variant Analysis classification was considered confirmed if consistent with at least one of the following hereditary gene testing laboratories: Ambry Genetics (Aliso Viejo, CA), Invitae (San Francisco, CA), GeneDx (Gaithersburg, MD), or the Sharing Clinical Reports Project (Myriad Genetics, Salt Lake City, UT). Last exon protein-truncating variants were called as likely pathogenic or pathogenic unless located after a known benign polymorphic stop codon in ClinVar. Appendix Table A1 (online only) includes a final list of the putative mutations by individual.
Statistical Analysis
Data are presented as means or proportions. Descriptive statistics were used to analyze clinical, demographic, and genetic test result characteristics; χ2 tests were used to compare groups. A two-sided P < .05 was considered statistically significant.
RESULTS
More than 50 cancer types were studied, including lung (41%), breast (19%), colorectal (8%), prostate (6%), pancreas (3%), and ovarian (2%). Average patient age was 63.5 years (range, 18 to 95 years), and 43% were male. Overall, 239 potential germline mutations in 227 unique patients were identified. Of these, 83 (34.7%) in 74 patients were excluded because of high ctDNA load, which made germline-somatic origin unclear (see Variant Curation in Patients and Methods).
Of the 83 excluded mutations, 44 (53%) were in TP53, 20 (24.1%) in APC, nine (10.8%) in RB1, four (4.8%) in PTEN, and two (2.4%) each in CDKN2A, NF1, and STK11. Excluded variants generally were congruent with the known somatic mutation spectrum of their respective tumor types: 77% of excluded TP53 alterations (n = 34) were in lung, breast, or colorectal cancer (CRC); 75% of excluded APC alterations (n = 15) were in CRC; 33% of excluded RB1 alterations (n = 3) were in non–small-cell lung cancer (which potentially indicates small-cell transformation), and one was in small cell lung cancer; and 50% of excluded PTEN mutations (n = 2) were in breast cancer, one in melanoma and one in non–small-cell lung cancer. No mutations in ATM, BRCA1, BRCA2, RET, MLH1, or SMAD4 were excluded because of high ctDNA fraction.
In total, 156 mutations in 156 patients (1.4%) were considered likely germline (Table 1; Appendix Tables A1 and A2, online only), with BRCA1 and BRCA2 mutations being the most common (78% combined [n = 122]). Eighty-eight (82.2%) of 107 unique mutations were previously identified in ClinVar as likely pathogenic or pathogenic. Only one putative germline mutation implicated in Lynch syndrome was identified most likely because only a single Lynch syndrome gene exon (exon 12 of MLH1) was included in Guardant360.
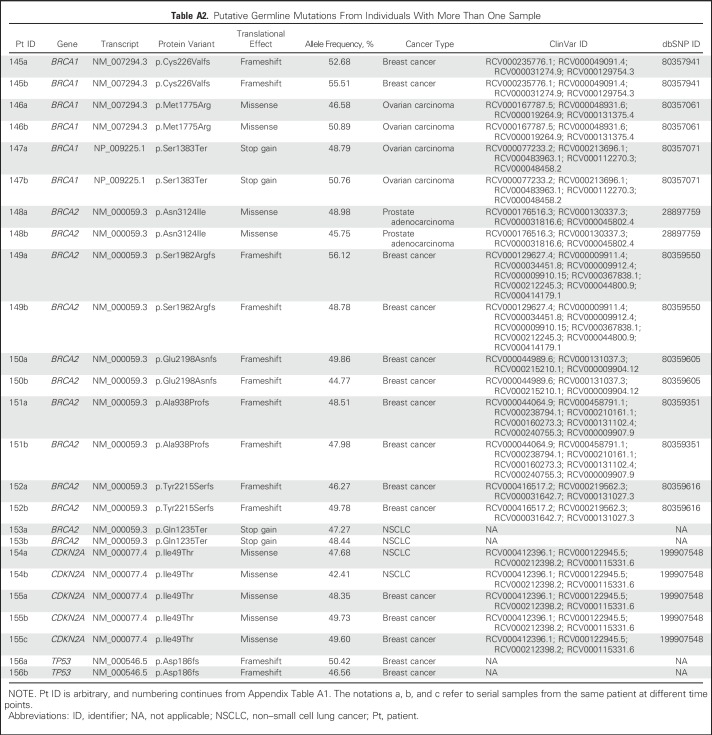
Putative germline mutation identification was consistent among 12 individuals with multiple samples evaluated. Serial sampling MAFs ranged from 42.4% to 56.1%, with the largest intrapatient variation being 7.3% (Appendix Table A2).
Overall, the highest prevalence of putative germline mutations was in patients with ovarian (8.1%), prostate (3.5%), pancreatic (3.3%), and breast (2.2%) cancer (Table 1). Putative mutation rates were higher in patients younger than 50 years of age across all tumor types (3.0% v 1.2% [P < .001]; breast cancer excluded, 2.1% v 1.2% [P = .017]) and only in patients with breast cancer (4.7% v 1.4%; P < .001).
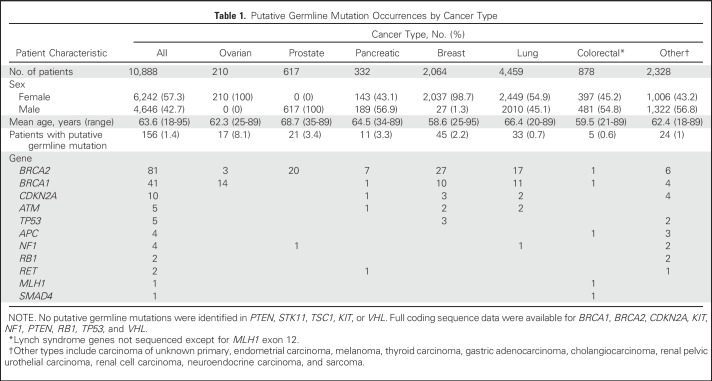
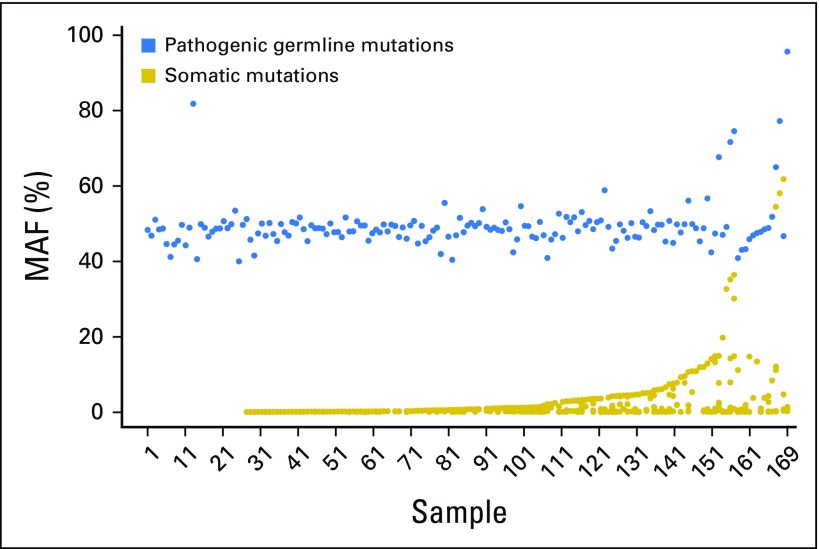
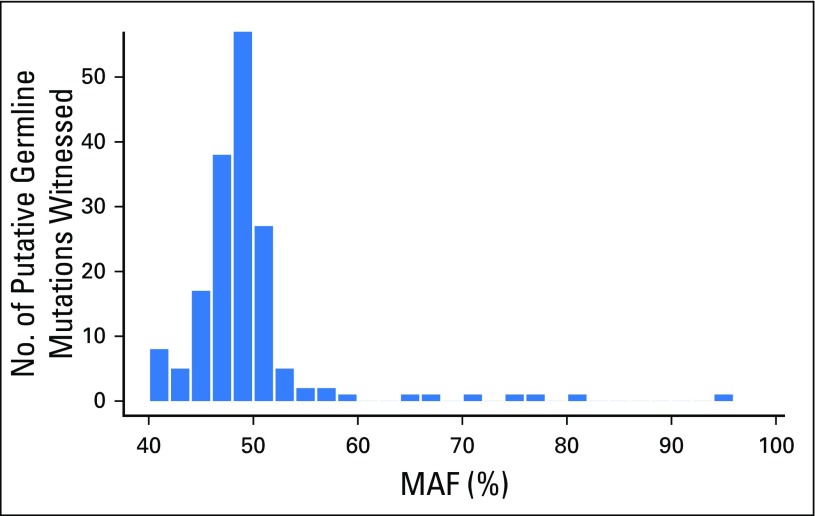
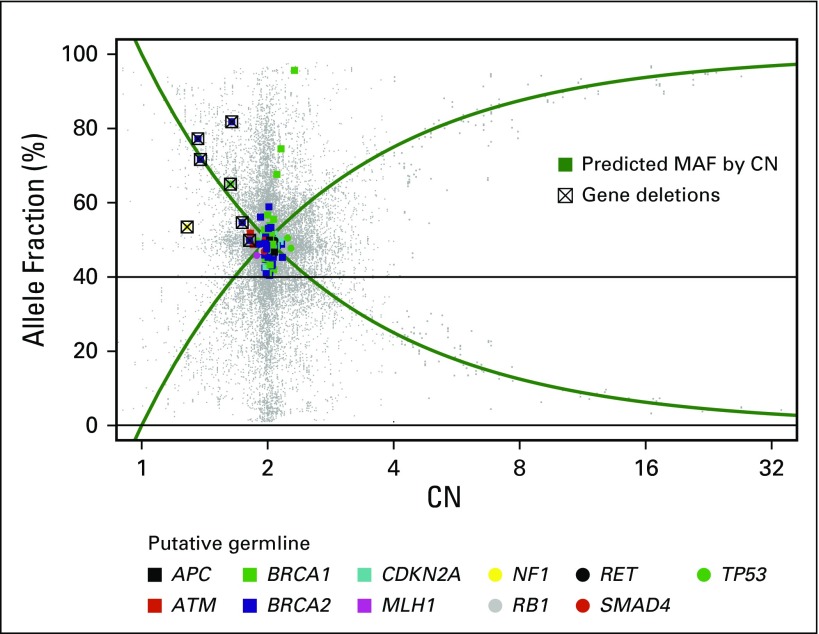
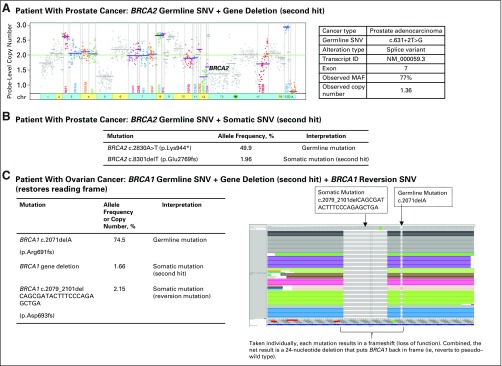
The mean MAF of putative germline findings was 48.7%, and MAFs ranged from the cohort limit of 40% to 95.6% (Fig 1). Examination of the relationship between germline MAF estimates and copy number variation established that deviation from expected germline allelic frequency (50%) often is concordant with copy number estimates (Figs 2 and 3). Putative germline mutations in tumor suppressor genes were observed with ctDNA copy number losses in the same gene (presumably loss of heterozygosity of the wild-type allele). Distinct mechanisms explain secondary hits evident in the data, including gene deletions (ie, loss of heterozygosity) and somatic truncating mutations (Fig 4). In addition, somatic reversion mutations that restore expression of functional proteins were identified, which likely evolved as a resistance mechanism to prior therapy23 (Fig 4).
Fig 1.
Somatic mutation and putative pathogenic germline mutant allele fractions (MAFs) for all 156 individuals with putative germline mutations arranged by lowest to highest somatic MAF. This included 169 samples in total; 144 individuals had one sample evaluated, and 12 had more than one sample evaluated (25 samples total; Appendix Tables A1 and A2). Putative germline mutations (pathogenic germline) had MAFs close to 50%, whereas somatic mutations had lower MAFs, as expected. Copy number variation corrections account for MAFs greater than 60%. Some samples did not have somatic mutations.
Fig 2.
Distribution of mutant allele fractions (MAFs) of putative germline mutations. The majority of mutations had MAFs close to 50%, as expected. Mutations with MAFs less than 40% were not evaluated; those with MAFs greater than 60% were included if concomitant locus copy number loss was identified (see Figs 3 and 4).
Fig 3.
Relationship between mutant allele fractions (MAFs) of identified putative germline mutations and common single nucleotide polymorphisms. The putative germline MAFs are plotted against, and in agreement with, genomic copy number (CN) measured using corresponding single nucleotide polymorphisms at the same allele. Mutations under 40% MAF were not evaluated.
Fig 4.
Three example genetic profiles of patients with secondary tumor mutations. The putative germline mutations identified are presented with any respective BRCA1 or BRCA2 circulating tumor findings. (A) Patient with a BRCA2 gene deletion and concomitant putative germline single nucleotide variant (SNV). (B) Patient with both germline and somatic BRCA2 SNVs. (C) Patient with a reversion of the germline SNV in the circulating tumor DNA with an allele frequency of 2.15%, potentially explaining the patient’s reported resistance to platinum-based chemotherapy (the wild-type allele presumed lost because of a BRCA1 gene deletion). chr, chromosome; ID, identifier; MAF, mutant allele fractions.
Germline testing of leukocyte DNA confirmed germline mutation status in five patients (one with ATM, one with BRCA1, three with BRCA2) for which ancillary germline testing results were available (100% concordance), and six additional patients (five with BRCA2, one with MLH1) had congruent germline-positive results by clinician report (no primary germline laboratory report available). In addition, 19 BRCA Ashkenazi Jewish founder mutations were identified24; most (58% [n = 11 of 19]) were in patients with breast, ovarian, or prostate cancer, consistent with hereditary breast and ovarian cancer syndrome,3 whereas another six were in patients with lung cancer and one each in patients with neuroendocrine carcinoma and carcinoma of unknown primary. A Brazilian TP53 p.R337H founder mutation associated with Li-Fraumeni syndrome also was identified in a 38-year-old patient with breast cancer.25
DISCUSSION
We leveraged cfDNA NGS analysis to identify and differentiate germline mutations from somatic mutations without a priori knowledge of germline mutation status to identify a spectrum of putative germline findings in a large cohort of patients with advanced solid tumors who underwent somatic genomic testing as part of routine clinical care. This patient cohort primarily represented the four most common cancers in the United States in 2018—breast, lung, prostate, and colorectal—as well as pancreatic cancer, the 11th most common cancer type.26 Of note, in addition to diseases where genetic counseling and testing recommendations are supported by evidence-based guidelines, we identified putative germline mutations in common tumor types, including lung, pancreatic, and prostate, where routine germline testing is not common, which suggests that incidental putative germline mutation reporting in these cancer types could significantly affect clinical care.
Although we observed a lower prevalence of cancer predisposition germline mutations than previously reported among other advanced cancer cohorts,7,27-29 we used a conservative approach to establish the final putative germline set, which likely underestimates of the true prevalence of germline mutations. Reasons for the lower prevalence are multiple: Our methods removed mutations with MAFs of less than 40%, although most commercial laboratories would consider genomic DNA mutations of more than 25% MAF to be germline; only a subset of exons were sequenced for seven of the 16 genes; several well-described cancer susceptibility genes were not included in the Guardant360 panel; the cohort was highly enriched for patients with lung cancer, a type not commonly associated with hereditary cancer susceptibility; we did not evaluate germline copy number variation or intragenic rearrangements; and some mutations were excluded because of a high ctDNA fraction.
With regard to mutations excluded because of high ctDNA load, TP53, APC, RB1, and PTEN accounted for 93% of the total. These genes frequently are mutated in cancer27 and often are early somatic events.30,31 Therefore, somatic mutations in these genes are more likely to have higher MAFs, which leads to an inherent challenge in discerning germline from somatic mutations in the plasma of patients with advanced cancer. In contrast, no putative germline mutations in ATM, BRCA1, BRCA2, RET, MLH1, or SMAD4 were excluded as a result of high ctDNA load potentially because these genes are less likely to be early mutations (compared with TP53, APC, RB1, and PTEN).27 Therefore, putative germline findings in ATM, BRCA1, BRCA2, RET, MLH1, or SMAD4 from cfDNA analysis may represent more often a true germline mutation, especially if identified in cancers where somatic mutations in these genes are not common. Increasing cfDNA assay coverage of common SNPs in genes associated with hereditary cancer predisposition might improve discrimination of somatic versus germline mutations at high MAFs and improve incidental germline mutation detection rates.
Secondary analyses revealed many important associations and observations. Younger age at diagnosis has been associated with hereditary cancer predisposition germline mutations, and colorectal and breast cancer guidelines frequently recommend genetic risk assessment and consideration of genetic testing for individuals younger than 50 years of age with a cancer diagnosis.2,3 In the current study, rates of putative germline mutations were elevated in individuals younger than 50 years, consistent with patients who carry a germline cancer predisposition. In particular, putative germline mutations also were enriched in patients with ovarian cancer, consistent with the relatively high rates of germline mutations found in these patients,29 which supports recommendations for germline genetic counseling/testing for all individuals with epithelial ovarian cancer.3
We also demonstrate the value of gene copy number assessment in the evaluation of putative germline mutations in cfDNA. Because most cancer predisposition genes are autosomal tumor suppressors, two inactivating events generally are needed (one in each allele) to eliminate gene function and initiate tumorigenesis.32 Therefore, low allele fraction ctDNA abnormalities (mutations or copy number loss) in individuals with a putative germline mutation in the same gene may reveal the likely driver of the cancer.
With regard to therapy, restoration of germline-inactivated alleles (revertant mutations) identifiable with cfDNA NGS, which appear as a second alteration in genomic proximity to a germline or early somatic mutation, can either restore the open reading frame (in cases of frameshift mutations) or alter a nonsense mutation to a missense or synonymous alteration.33,34 Identification of germline mutations in such contexts is critical for proper therapy selection, particularly when considering poly (ADP-ribose) polymerase inhibitor therapy in the context of BRCA1 or BRCA2 mutations.8,23
Strengths of our study include large patient numbers and a wide spectrum of advanced cancers. The study was performed at a Clinical Laboratory Improvement Amendments–approved commercial laboratory35 with rigorous sample handling procedures and analytics. One limitation of the study design is that germline DNA samples (eg, from leukocytes, fibroblasts) for orthogonal validation of germline status were not available. However, all five patients with known ancillary clinical testing of leukocyte DNA had confirmed putative germline mutations, and findings from another six were supported by clinician report. The observed enrichment for known ancestral founder mutations also supports the accuracy of this study because such enrichment would not be expected for mutations of somatic or hematopoietic origin. Furthermore, most mutations were reported in ClinVar by major hereditary gene testing laboratories, and repeat sampling in 12 patients showed consistency of the putative germline mutation finding.
Another limitation is that the Guardant360 assay does not evaluate for BRCAness/homology–directed repair, microsatellite instability, or tumor mutational burden estimates, which may have been useful in identifying somatic correlations with germline variants. Furthermore, the de-identified methods used did not allow for provider recontact or collection of medical information beyond the test requisition form such that detailed pathology; health habits (eg, smoking history); family history; or race, ancestry, or ethnicity information were not available for study analyses.
The ability of ctDNA testing to identify incidental germline mutations is subject to the same challenges of reporting incidental germline findings as seen in patients who undergo tumor tissue sequencing.36 Because ctDNA laboratories likely will begin to include incidental germline findings, ordering providers must be knowledgeable about patient family genetic cancer risk assessment and genetic counseling resources (ie, local genetic counseling, telegenetics). Providers and patients also need to know that the majority of alterations in genes associated with cancer susceptibility identified through ctDNA analysis represent somatic events and that only a small fraction of identified alterations are germline. That said, before ordering ctDNA testing, providers will need to inform patients that ctDNA testing may identify incidental germline information. Furthermore, laboratories that provide incidental findings will need to distinguish somatic from probable germline findings clearly and include language about the need for confirmatory testing. They also may want to include information about genetic counseling services and/or education material in testing reports to ensure appropriate patient care. A reasonable paradigm for the return of results already exists in the context of universal screening for Lynch syndrome,37 wherein post-test referral and genetic counseling are acceptable practices. However, a need for some modifications to this paradigm exists given that the majority of patients who undergo ctDNA testing will have metastatic cancer.
Collectively, our findings and those previously published12-14 provide evidence that hereditary cancer predisposition germline mutations can be identified accurately from cfDNA. Despite this, incidental cfDNA germline evaluation should not replace validated hereditary cancer gene testing, but it may serve as an important supplement to increase the reach of genetic cancer risk assessment, particularly in populations with specific germline founder mutations or in cancers without clear hereditary predisposition genetic testing guidelines. Ideally, future studies will include prospective germline collection and companion analyses along with enhancements to the bioinformatics pipeline to allow for confident calls of germline findings. Internal validation in future studies will provide a more accurate estimate of the expected germline mutation prevalence by specific tumor type and enhance the understanding of limitations. Examples of improvements needed for accurate ascertainment of germline mutations include methods to identify germline mutations in cfDNA that account for gene copy number variation and high ctDNA load and germline mutations at low allele fractions in plasma. Furthermore, clinical translation of these findings will depend on establishing best practices to counsel patients about the possibility of incidental germline findings and to report potential germline results clearly to clinicians and/or patients in ways that enhance clinical care.
ACKNOWLEDGMENT
We thank Kara Maxwell, MD, PhD, for manuscript planning input.
Appendix
Glossary
American College of Medical Genetics and Genomics variant classification guidelines: standards and guidelines for the interpretation of DNA sequence variants on the basis of a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. The guidelines define five categories for the classification of germline variants (listed from most to least likely to be damaging to the protein): pathogenic, likely pathogenic, variant of uncertain significance, likely benign, and benign. Pathogenic and likely pathogenic variants often are referred to collectively as pathogenic variants or mutations.
Aneuploidy: the presence of an abnormal number of chromosomes in a cell.
Cell-free DNA: all nonencapsulated DNA in the blood stream.
Circulating tumor DNA: tumor-derived cell-free DNA.
Copy number variation: a variation in the number of copies of a DNA segment at a particular genomic position.
Genetic Cancer Risk Assessment: a clinical assessment of a patient’s personalized risk for cancer; it commonly includes genetic counseling and germline genetic testing of cancer predisposition genes and provides patient and family recommendations for cancer screening, risk-reducing strategies, and management as appropriate.
Mutant allele fraction: the relative frequency (commonly represented as a percentage) of a mutation at a particular genomic position (locus) determined by next-generation sequencing.
Mutation: a variation in the genetic sequence that usually involves a single nucleotide variation at a specific position in a gene and that is uncommon in the population and believed to disrupt the function of the protein product.
Next-generation sequencing: rapid, high-throughput genome sequencing.
Single nucleotide polymorphism: a variation in a single nucleotide at a specific position in the genome that, in contrast to a mutation, is common in the population.
Single nucleotide variant: a variation in a specific nucleotide at a specific position in the genome, which can represent a single nucleotide polymorphism or mutation.
The Cancer Genome Atlas: a database of DNA and RNA sequencing data and variants from a large collection of tumor samples from patients with a wide range of cancers.
Table A1.
Putative Germline Mutations From Individuals With One Sample
Table A2.
Putative Germline Mutations From Individuals With More Than One Sample
Footnotes
Supported by National Cancer Institute Grant No. P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was received from STOP CANCER (T.P.S., principal investigator), the American Cancer Society (J.N.W., principal investigator), and the Avon Foundation (J.N.W., principal investigator).
Presented at the 2017 American Society of Clinical Oncology Annual Meeting, Chicago, IL, June 2-6, 2017.
AUTHOR CONTRIBUTIONS
Conception and design: Thomas P. Slavin, Kimberly C. Banks, Darya Chudova, Geoffrey R. Oxnard, Justin I. Odegaard, Angel A. Rodriguez, Aditya Bardia, Richard B. Lanman, Razelle Kurzrock, Jeffrey N. Weitzel
Financial support: Thomas P. Slavin, Jeffrey N. Weitzel
Administrative support: Brian Leyland-Jones, Christine E. Lee, Jeffrey N. Weitzel
Provision of study materials or patients: Thomas P. Slavin, Massimo Cristofanilli, Angel A. Rodriguez, Mike F. Janicek, Guru Sonpavde, Richard B. Lanman
Collection and assembly of data: Thomas P. Slavin, Kimberly C. Banks, Darya Chudova, Rebecca J. Nagy, Kar Wing Kevin Tsang, Angel A. Rodriguez, Aditya Bardia, Brian Leyland-Jones, Mike F. Janicek, Michael Lilly, Guru Sonpavde, Richard B. Lanman, Jeffrey N. Weitzel
Data analysis and interpretation: Thomas P. Slavin, Kimberly C. Banks, Darya Chudova, Justin I. Odegaard, Susan L. Neuhausen, Stacy W. Gray, Massimo Cristofanilli, Angel A. Rodriguez, Aditya Bardia, Mike F. Janicek, Guru Sonpavde, Christine E. Lee, Richard B. Lanman, Funda Meric-Bernstam, Jeffrey N. Weitzel
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Identification of Incidental Germline Mutations in Patients With Advanced Solid Tumors Who Underwent Cell-Free Circulating Tumor DNA Sequencing
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Thomas P. Slavin
No relationship to disclose
Kimberly C. Banks
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Patents, Royalties, Other Intellectual Property: Guardant Health
Darya Chudova
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Patents, Royalties, Other Intellectual Property: Guardant Health
Geoffrey R. Oxnard
Honoraria: Chugai Pharma, Bio-Rad, Sysmex, Guardant Health
Consulting or Advisory Role: AstraZeneca, Inivata, Takeda Pharmaceuticals, Loxo Oncology, Ignyta, Dropworks, GRAIL
Patents, Royalties, Other Intellectual Property: Co-author of a patent pending titled “Non-invasive blood-based monitoring of genomic alterations in cancer” (Inst)
Justin I. Odegaard
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Rebecca J. Nagy
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Kar Wing Kevin Tsang
No relationship to disclose
Susan L. Neuhausen
No relationship to disclose
Stacy W. Gray
No relationship to disclose
Massimo Cristofanilli
Honoraria: Pfizer
Consulting or Advisory Role: Novartis, Merus
Angel A. Rodriguez
Employment: Austin Cancer Center
Aditya Bardia
Consulting or Advisory Role: Novartis, Genentech, Pfizer, Spectrum Pharmaceuticals, bioTheranostics
Brian Leyland-Jones
Stock and Other Ownership Interests: Catalyst Pharmaceuticals, ProGenix Health Solutions, Puma Biotechnology, Sucampo Pharmaceuticals, ARIAD Pharmaceuticals, Zogenix
Consulting or Advisory Role: GlaxoSmithKline, Amgen
Speakers’ Bureau: Genentech, Exelixis, Bayer AG
Research Funding: Takeda Pharmaceuticals, Tesaro
Expert Testimony: Amgen
Travel, Accommodations, Expenses: AKESOgen, National Foundation for Cancer Research
Mike F. Janicek
Stock and Other Ownership Interests: Intuitive Surgical
Honoraria: OncLive
Consulting or Advisory Role: Guardant Health, Ambry Genetics
Speakers’ Bureau: Clovis Oncology, AstraZeneca, Tesaro
Michael Lilly
Research Funding: Bavarian Nordic (Inst)
Guru Sonpavde
Honoraria: UpToDate
Consulting or Advisory Role: Bayer AG, Genentech, Sanofi, Merck, Novartis, Agensys, Eisai, AstraZeneca, Janssen Pharmaceuticals, Bristol-Myers Squibb, Exelixis, EMD Serono, Astellas Pharma
Speakers’ Bureau: Clinical Care Options, National Comprehensive Cancer Network, Physician Education Resource, OncLive, Research to Practice
Research Funding: Onyx Pharmaceuticals (Inst), Bayer AG (Inst), Boehringer Ingelheim (Inst), Celgene (Inst), Merck (Inst), Pfizer (Inst), Janssen Pharmaceuticals (Inst), Sanofi (Inst)
Other Relationship: Boehringer Ingelheim, AstraZeneca, Bristol-Myers Squibb
Christine E. Lee
Employment: Guardant Health
Stock and Other Ownership Interests: Guardant Health
Patents, Royalties, Other Intellectual Property: Work related to Guardant Health, minor role
Travel, Accommodations, Expenses: Guardant Health
Richard B. Lanman
Employment: Guardant Health, Veracyte
Leadership: Guardant Health, Biolase
Stock and Other Ownership Interests: Guardant Health, Biolase
Research Funding: Guardant Health
Funda Meric-Bernstam
Honoraria: Sumitomo Group, Dialectica
Consulting or Advisory Role: Genentech, Inflection Biosciences, Pieris Pharmaceuticals, Clearlight Diagnostics, DarwinHealth, Samsung Bioepis, Spectrum Pharmaceuticals
Research Funding: Novartis, AstraZeneca, Taiho Pharmaceutical, Genentech, Calithera Biosciences, Debiopharm Group, Bayer AG, Aileron Therapeutics, Puma Biotechnology, CytomX Therapeutics, Jounce Therapeutics, Zymeworks, Curis, Pfizer, eFFECTOR Therapeutics, AbbVie
Razelle Kurzrock
Leadership: CureMatch
Stock and Other Ownership Interests: CureMatch, IDbyDNA
Consulting or Advisory Role: Actuate Therapeutics, Loxo Oncology, XBiotech, NeoMed, Roche
Speakers’ Bureau: Roche
Research Funding: Guardant Health (Inst), Sequenom (Inst), Merck Serono (Inst), Genentech (Inst), Pfizer (Inst), Foundation Medicine (Inst), Incyte (Inst), Konica Minolta (Inst)
Jeffrey N. Weitzel
No relationship to disclose
REFERENCES
- 1.Lagos-Jaramillo VI, Press MF, Ricker CN, et al. : Pathological characteristics of BRCA-associated breast cancers in Hispanics. Breast Cancer Res Treat 130:281-289, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology V.1.2017: Genetic/familial high-risk assessment: Colorectal. https://www.nccn.org/professionals/physician_gls/default.aspx#genetics_colon.
- 3. National Comprehensive Cancer Network: NCCN clinical practice guidelines in oncology V.2.2017: Genetic/familial high-risk assessment: Breast and ovarian. https://www.nccn.org/professionals/physician_gls/default.aspx#genetics_colon. [DOI] [PubMed]
- 4. Childers CP, Childers KK, Maggard-Gibbons M, et al: National estimates of genetic testing in women with a history of breast or ovarian cancer. J Clin Oncol 35:3800-3806, 2017 [Erratum: J Clin Oncol 36:432, 2018] [DOI] [PMC free article] [PubMed]
- 5.Mandelker D, Zhang L, Kemel Y, et al. : Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA 318:825-835, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang KL, Mashl RJ, Wu Y, et al. : Pathogenic germline variants in 10,389 adult cancers. Cell 173:355-370, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schrader KA, Cheng DT, Joseph V, et al. : Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol 2:104-111, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George A, Kaye S, Banerjee S: Delivering widespread BRCA testing and PARP inhibition to patients with ovarian cancer. Nat Rev Clin Oncol 14:284-296, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Diaz LA, Jr, Bardelli A: Liquid biopsies: Genotyping circulating tumor DNA. J Clin Oncol 32:579-586, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brison N, Van Den Bogaert K, Dehaspe L, et al. : Accuracy and clinical value of maternal incidental findings during noninvasive prenatal testing for fetal aneuploidies. Genet Med 19:306-313, 2017 [DOI] [PubMed] [Google Scholar]
- 11.Odegaard JI, Vincent JJ, Mortimer S, et al. : Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res 24:3539-3549, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Alden RS, Odegaard JI, et al. : Discrimination of germline EGFR T790M mutations in plasma cell-free DNA allows study of prevalence across 31,414 cancer patients. Clin Cancer Res 23:7351-7359, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratajska M, Koczkowska M, Żuk M, et al. : Detection of BRCA1/2 mutations in circulating tumor DNA from patients with ovarian cancer. Oncotarget 8:101325-101332, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukuya T, Patel S, Shane-Carson K, et al. : Lung cancer patients with germline mutations detected by next-generation sequencing and/or liquid biopsy. J Thorac Oncol 13:e17-e19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanman RB, Mortimer SA, Zill OA, et al. : Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS One 10:e0140712, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zill OA, Mortimer S, Banks KC, et al: Somatic genomic landscape of over 15,000 patients with advanced-stage cancer from clinical next-generation sequencing analysis of circulating tumor DNA. J Clin Oncol 34, 2016 (suppl; abstr LBA11501)
- 17. doi: 10.1038/gim.2017.196. 10.1038/gim.2017.196 Weitzel JN, Chao EC, Nehoray B, et al: Somatic TP53 variants frequently confound germ-line testing results. Genet Med . [epub ahead of print on November 30, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. doi: 10.1158/1078-0432.CCR-18-0143. 10.1158/1078-0432.CCR-18-0143 Hu Y, Ulrich BC, Supplee J, et al: False positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res . [epub ahead of print on March 22, 2018] [DOI] [PubMed] [Google Scholar]
- 19.Pennisi E: Genomics. 1000 Genomes Project gives new map of genetic diversity. Science 330:574-575, 2010 [DOI] [PubMed] [Google Scholar]
- 20. National Heart, Lung, and Blood Institute: NHLBI Grand Opportunity Exome Sequencing Project (ESP), 2016. https://esp.gs.washington.edu/drupal.
- 21.Lek M, Karczewski KJ, Minikel EV, et al. : Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285-291, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richards S, Aziz N, Bale S, et al: Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405-424, 2015. [DOI] [PMC free article] [PubMed]
- 23.Benafif S, Hall M: An update on PARP inhibitors for the treatment of cancer. OncoTargets Ther 8:519-528, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartge P, Struewing JP, Wacholder S, et al. : The prevalence of common BRCA1 and BRCA2 mutations among Ashkenazi Jews. Am J Hum Genet 64:963-970, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Custódio G, Komechen H, Figueiredo FR, et al. : Molecular epidemiology of adrenocortical tumors in southern Brazil. Mol Cell Endocrinol 351:44-51, 2012 [DOI] [PubMed] [Google Scholar]
- 26. National Cancer Institute SEER Program: Common Cancer Sites, 2018. https://seer.cancer.gov/statfacts/html/common.html.
- 27.Meric-Bernstam F, Brusco L, Daniels M, et al. : Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol 27:795-800, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard CC, Mateo J, Walsh MF, et al. : Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375:443-453, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh T, Casadei S, Lee MK, et al. : Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc Natl Acad Sci U S A 108:18032-18037, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Méniel V, Megges M, Young MA, et al. : Apc and p53 interaction in DNA damage and genomic instability in hepatocytes. Oncogene 34:4118-4129, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolff RK, Hoffman MD, Wolff EC, et al. : Mutation analysis of adenomas and carcinomas of the colon: Early and late drivers. Genes Chromosomes Cancer 57:366-376, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knudson AG, Jr: Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci U S A 68:820-823, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norquist B, Wurz KA, Pennil CC, et al. : Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol 29:3008-3015, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. doi: 10.1200/PO.17.00176. 10.1200/PO.17.00176 Carneiro BA, Collier KA, Nagy RJ, et al: Acquired resistance to poly (ADP-ribose) polymerase inhibitor olaparib in BRCA2-associated prostate cancer resulting from biallelic BRCA2 reversion mutations restores both germline and somatic loss-of-function mutations. JCO Precision Oncol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz MK: Genetic testing and the clinical laboratory improvement amendments of 1988: Present and future. Clin Chem 45:739-745, 1999 [PubMed] [Google Scholar]
- 36.Raymond VM, Gray SW, Roychowdhury S, et al. : Germline findings in tumor-only sequencing: Points to consider for clinicians and laboratories. J Natl Cancer Inst 108:djv351, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vindigni SM, Kaz AM: Universal screening of colorectal cancers for Lynch syndrome: Challenges and opportunities. Dig Dis Sci 61:969-976, 2016 [DOI] [PubMed] [Google Scholar]