Abstract
Background:
Recent reviews have highlighted the potential use of blood-based methylation biomarkers as diagnostic and prognostic tools of current and future alcohol use and addiction. Due to the substantial overlap that often exists between methylation patterns across different tissues, including blood and brain, blood-based methylation may track methylation changes in brain, however, little work has explored the overlap in alcohol related methylation in these tissues.
Methods:
To study the effects of alcohol on the brain methylome and identify possible biomarkers of these changes in blood, we performed a methylome-wide association study in brain and blood from 40 male DBA/2J mice that received either an acute ethanol or saline intraperitoneal injection. To investigate all 22 million CpGs in the mouse genome, we enriched for the methylated genomic fraction using methyl-CpG binding domain (MBD) protein capture followed by next-generation sequencing (MBD-seq). We performed association tests in blood and brain separately followed by enrichment testing to determine if there was overlapping alcohol related methylation in the two tissues.
Results:
The top result for brain was a CpG located in an intron of Ttc39b (p=5.65×10−08) and for blood the top result was located in Espnl (p=5.11×10−08). Analyses implicated pathways involved in inflammation and neuronal differentiation, such as CXCR4, Il-7, and Wnt signaling. Enrichment tests indicated significant overlap among the top results in brain and blood. Pathway analyses of the overlapping genes converge on MAPKinase signaling (p=5.6×10−05) which plays a central role in acute and chronic responses to alcohol and glutamate receptor pathways, which can regulate neuroplastic changes underlying addictive behavior.
Conclusions:
Overall, we have shown some methylation changes in brain and blood after acute ethanol administration and that the changes in blood partly mirror the changes in brain suggesting the potential for DNA methylation in blood to be biomarkers of alcohol use.
Keywords: alcohol, epigenetics, glutamate, biomarker, methylation
INTRODUCTION
Growing evidence suggests that epigenetic modifications, which refer to changes in the DNA that do not change the underlying nucleotide sequence, may play a role in alcohol use and addiction (Robison and Nestler, 2011, Nieratschker et al., 2013). These modifications serve to provide phenotypic plasticity in response to the environment and include closely related processes of DNA methylation, histone modification and chromatin remodeling. In the case of DNA methylation, a methyl group is attached at the carbon 5 position of a cytosine, and is most often, although not exclusively, found in the sequence context of CG. Methylation could be relevant to the mechanisms underlying alcohol consumption and addiction. For example, repeated acute exposure to alcohol leads to chronic cycles of consumption and withdrawal that evoke long-lasting neuroplasticity thought to contribute to abusive consumption, dependence, sensitization, and increased risk for relapse even after years of abstinence (Nestler, 2001). These observations suggest lasting cellular and molecular modifications. DNA methylation, being the archetypal cellular information storage mechanism, has been heavily implicated as a chief mechanism for stable activity-dependent transcriptional alterations within the central nervous system (Heyward and Sweatt, 2015). Drugs of abuse are known to induce epigenetic changes that can persist over time and have lasting effects (Nestler, 2014), which may explain the observations above.
Recent reviews have highlighted the biomarker potential of blood-based DNA methylation as diagnostic and prognostic tools of current and future alcohol use and addiction (Andersen et al., 2015, Mahnke et al., 2017, Tulisiak et al., 2017) and noted several examples in the extant literature of blood-based methylation alcohol biomarkers (see, for example, (Beach et al., 2015, Heberlein et al., 2015)). These reviews also suggest that blood-based methylation may track methylation changes in brain (Tulisiak et al., 2017) due to the substantial overlap that often exists between methylation patterns across different tissues (Eckhardt et al., 2006). There is ample evidence supporting partly overlapping methylation profiles between human blood and post-mortem brain samples (Davies et al., 2012) as well as correlated drug-induced methylation changes in rodent blood and brain (Aberg et al., 2013). Studies in humans and rodents of other drugs of abuse show that overlap between brain and blood methylation does occur (Nestler, 2001, Nestler, 2014). However, little work has explored the overlap in alcohol related methylation in blood and brain. Barker and colleagues found that alcohol altered methylation in the promoter of Htr3a in a 5-day drinking in the dark mouse model in blood, hippocampus, dorsomedial striatum and dorsalmedial prefrontal cortex, but found no effect in six other brain regions (Barker et al., 2013). To our knowledge, the study by Barker and colleagues is the only study to have compared alcohol related methylation in blood and brain tissue, although that report only analyzed a single gene. Therefore, comprehensive investigations need to be conducted to identify alcohol related methylation sites in brain whose methylation profile is reflected in blood (Tulisiak et al., 2017).
Here, we interrogate all 22 million autosomal CpG sites of the mouse methylome for their response to acute ethanol exposure in both blood and brain to identify overlapping methylation in both tissues. For the brain, we chose the striatum because of the established connection between neurobiological and microcircuit changes to this region resulting from alcohol and/or drug addiction (Koob, 2014, Koob and Volkow, 2016). Simultaneous analysis of the blood and brain methylomes will determine the degree of methylation overlap between these tissues and establish a set of methylation sites in blood that can potentially be used as proxy for brain in alcohol methylation studies. To our knowledge, this is the first study to explore the effect of alcohol on DNA methylation on a methylome-wide scale in rodents and consider the overlap across tissues on such a scale.
MATERIALS & METHODS
Ethics Statement
All procedures were approved by Virginia Commonwealth University Institutional Animal Care and Use Committee under protocol number AM10332 and followed the NIH Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80–23, 1996).
Animals
Forty male DBA2/J (D2) mice from Jackson Laboratories (Bar Harbor, ME, United States) purchased at 10–13 weeks of age and group housed 4/cage. Mice were maintained in a temperature-controlled room (23°C±1) with 12 h light/dark cycles and free access to standard chow (Harlan Teklad #7912, Madison, WI, United States) and water. Cages and bedding (Harlan Sani-chips, #7090A, Harlan, Teklad, Madison, WI, United States) were changed weekly. All tests were carried out between 0900 and 1200 h. All mice were allowed to habituate to the animal facility for at least 1 week prior to testing.
Drugs
All injections were administered intraperitoneally (i.p.). Saline solutions were prepared by dissolving NaCl in sterile water to a concentration of 0.9% weight per volume. Ethanol solutions were prepared from 200-proof absolute anhydrous ethanol (Pharmco-Aaper brand, Brookfield, CT, United States). Ethanol was administered at 20% v/v in 0.9% saline.
Behavioral Testing and Material Collection
Animals received one of two possible i.p. injections: saline or 2.0 g/kg ethanol. They were then returned to their home cages and four hours following injection, they were killed by cervical dislocation followed by decapitation and collection of trunk blood. Brains were then extracted, and the striatum was excised as described previously (Kerns et al., 2005). The excised region was placed in an individual tube, flash-frozen in liquid nitrogen and stored at −80°C. Genomic DNA was extracted using the Gentra Puregene Blood/Tissue Kit (Qiagen, Valencia, CA) following the vendor’s instructions.
Methylation Assay
Our approach (Aberg et al., 2012, Chan et al., 2017) to assay the entire CpG methylome has been described previously. Briefly, genomic DNA was fragmented with ultrasonication (Covaris™ E220) to a median length of ~150 bp, we used MethylMiner (Invitrogen, Carlsbad, CA) which employs MBD protein-based enrichment of the methylated DNA fraction, followed by single-end sequencing (50 bp reads) on the SOLiD platform (Life Technologies). We eluted the captured methylation fractions with 0.5 NaCl to increase the relative number of fragments from CpG poor regions (Aberg et al., 2012), which otherwise would not be as well covered. Compared to other MBD kits, this protocol has a favorable noise to signal ratio and coverage of the methylome (Aberg et al., 2015).
Alignment and Quality Control
Reads were aligned to the D2 specific reference genome (Wang et al., 2010) using Cushaw3 (Liu et al., 2012). An overview of the quality control (QC) procedures can be found elsewhere (Aberg et al., 2012). We obtained an average of 32.4 million reads per sample (SD = 12.6 million), of which on average 77.9% (SD=7.7) aligned. After alignment, reads were further filtered on quality score and duplicate reads were removed.
In studies aimed at calling SNPs or estimating the absolute percentage of methylation at specific bases (e.g., WGBS), the total number of reads covering each base is a critical determinant of the precision of the methylation estimates. For example, if a base is sequenced at 10x coverage in WGBS, its methylation level can be estimated in only 10% increments since methylation status at a CpG is binary for any given DNA molecule and methylation is estimated as the number of reads/molecule (0…10), with methylated CpGs divided by the total number of reads. In enrichment-based sequencing, coverage itself is the measured variable, where precision is determined by total number of reads (or more precisely fragments) covering a CpG or what we call CpG score. The higher the level methylation is at a given locus (i.e., the percentage of cells in which that locus is methylated) the more likely it is to be captured by MBD protein pulldown. Thus, loci with higher methylation levels will obtain more reads than loci with low methylation levels. At the other extreme, loci that are non-methylated in all cells are not captured during MBD pulldown and are not represented in the sequencing library. The lack of coverage for these non-methylated loci is therefore expected and is not a consequence of poor sequencing performance. For this approach to distinguish between methylated loci versus non-methylated loci, it is critical that fragments with methylated sites are pulled down (e.g., if a methylated site is not pulled down, then we might erroneously conclude that it is not methylated). Thus, rather than the average number of reads covering CpGs, the critical QC parameter is the ratio of the average CpG score (van den Oord et al., 2013) to the average score of sites that are not CpGs (labeled as non-CpGs) and, therefore, cannot be methylated. Higher levels of this ratio indicate higher enrichment efficiency. Note that because not all CpGs are methylated this ratio is an underestimate of the enrichment efficiency. The opposite of this ratio, average non-CpG score / average CpG score, is a measure of the noise level with lower values indicating less noise.
The number of reads is important in the sense that if too few reads are sequenced there may not be enough to cover each CpG. Other authors have suggested that 30–60 million reads per human sample are sufficient for high quality methylation profiles (Bock et al., 2010, Chavez et al., 2010). Here we study mice. Given that the mouse genome is ~15% smaller than the human genome (Guenet, 2005), the equivalent need is 25.5–51 million reads per murine sample. In the present study we obtained an average of 32.4 million reads per sample, which is within the suggested range.
Akin to genome-wide association studies where SNPs are filtered based on minor allele frequency, the last step of the quality control procedure involved removing sites that were rarely methylated (CpG score <0.3). The main purpose for this filtering is because these sites lack biological variation and to avoid statistical problems caused by sparse data. This resulted in a total of 13,352,028 CpGs available for the blood and brain MWASs. Table S1 shows the distribution of CpGs in the MWAS by genomic feature.
In this study we observed an average CpG score (van den Oord et al., 2013) of 2.58 (SD=7.59), with an average CpG-to-nonCpG score ratio of 70.4 (SD=42.9) and average nonCpG-to-CpG score ratio (Shabalin et al., 2018b) of 0.024 (SD=0.020). Thus, the average CpG signal is strong, enrichment efficiency is high, and the background noise level is low, allowing for detection of differently methylated sites. Data are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.372c76c
Statistical Analyses
Methylation association testing
We regressed the methylation measurement of individual CpGs on treatment status (ethanol or saline) to determine the effect of ethanol on the methylome using RaMWAS (Shabalin et al., 2018a). Potential technical batch effects were controlled for by including them a covariate in the regression analysis. According to a stringent Bonferroni correction, p<5×10−8 were considered statistically significant. P<1×10−5 were considered suggestive. Separate MWASs were conducted for blood and brain, respectively.
Pathway Analyses
We used the Gene Set Knowledgebase (GSKB (Bares and Ge, 2015)) to perform pathway analyses based on Biocarta, MouseCyc, NetPath, PANTHER, and INOH. Genes located within ±20 kb from a CpG with p<1×10−5 were included in the pathway analysis. For each of 892 reference pathways present in GSKB, incorporating 4,463 known genes, a hypergeometric test was performed to study whether the overlap between the top MWAS genes and those present in each reference pathway was higher than expected by chance. Only pathways that contained at least two genes from our MWAS results were retained. A p-value threshold of 0.05 was used to determine significance.
Enrichment Testing for Genomic Features
To examine whether the top results from the blood and brain MWASs were enriched for specific genomic features we used the R package shiftR. To perform these tests, shiftR uses circular permutations (Cabrera et al., 2012) by shifting the mapping of the two data sets by a single random integer in each permutation. This approach generates the empirical test statistic distribution under the null hypothesis while preserving the correlational structure of the data. We used 1 million permutations for each test. Multiple thresholds (i.e., for our analyses we used the top 1%, 5% and 10%) were used to define “top” findings for the blood and brain MWAS. To account for this “multiple testing”, the same thresholds are used in the permutations where the test statistic distribution under the null hypothesis is generated from most significant combination of thresholds. For bivariate datasets, such as the genomic features, the presence versus absence of a feature defined the “top”. The genomic features investigated include overlap with genic, exonic, intronic, 3’UTR and 5’UTR regions as defined by RefSeq (Pruitt et al., 2014), gene promotors (within 8kb of a gene transcription start site), CpG Islands, CpG Shores (2kb flanking CpG Islands), high areas of conservation across eutherian mammals, and non-coding RNA (functional RNA molecule that is not translated into a protein).
Blood and Brain Overlap
We also used enrichment testing in shiftR to investigate whether top findings from the blood MWAS occur more frequently among the top findings of the brain MWAS (i.e., are enriched) than what is expected by chance. For this test, the two data sets were first mapped to each other based on chromosomal location where 20kb flanks were allowed in the blood MWAS to account for the fact that association signals may involve a region rather than a single base position. After this step, enrichment testing proceeded as described in the previous section.
After enrichment testing, we further investigated overlapping top results by performing pathway analyses as described in the previous section. For these pathway analyses, we took a conservative approach by including only overlapping CpGs that had the same direction of effect in both tissues and a p < 0.01 in both sets of results to avoid an extreme p-value in one set of findings dominating the results.
RESULTS
Methylome-wide association study
Two CpGs, one in each tissue, reached statistical significance (p< 5×10−8). The CpGs were located in the introns of Ttc39b and Espnl in brain and blood respectively. In addition, seven CpGs in brain and eight in blood were significant at the p<1×10−7 level (Table 1). For both brain and blood, these top findings tended to be located within gene boundaries and evolutionarily conserved regions. When suggestive sites were considered (p<1×10−7), we identified 40 and 84 CpGs in brain and blood respectively (See Tables S2 and S3 for an annotated list of all results with p<1×10−5 in brain and blood respectively). Pathway analyses were conducted on sites with p<1×10−5 located in genes, which corresponded to 39 and 77 genes being used as input for the pathway analyses in brain and blood respectively. In brain, pathway analyses related to responses to extracellular stimuli including CXCR4, Wnt, IL-7, and G-protein signaling (Table 2). Evidence for the importance of glutamate receptor pathways were found in both blood and brain.
Table 1.
Brain and blood individual CpG association results
| Chr | Coord | T-stat | P-value | Gene | Features |
|---|---|---|---|---|---|
| Brain | |||||
| 4 | 83260604 | −6.87 | 5.65E-08 | Ttc39b | Intron, Conserved |
| 13 | 99323071 | −6.79 | 7.08E-08 | Ptcd2 | Intron, Bidirectional promotor, Conserved |
| 4 | 83260612 | −6.66 | 1.04E-07 | Ttc39b | Intron, Conserved |
| 13 | 96999232 | 6.64 | 1.12E-07 | Conserved | |
| 13 | 44651605 | −6.40 | 2.28E-07 | Conserved | |
| 3 | 101902700 | −6.08 | 6.07E-07 | Slc22a15 | Intron, Conserved |
| 3 | 142688025 | 5.95 | 9.08E-07 | ||
| Blood | |||||
| 1 | 91329146 | −9.56 | 5.11E-08 | Espnl | Intron, Conserved |
| 10 | 88788436 | 8.11 | 4.63E-07 | Utp20 | Intron, Conserved |
| 3 | 111893685 | 7.88 | 6.74E-07 | Gm6602 | Intron, 3’UTR |
| 1 | 146200755 | 7.88 | 6.78E-07 | ||
| 4 | 141573986 | −7.85 | 7.11E-07 | Conserved | |
| 12 | 30711130 | 7.85 | 7.13E-07 | Conserved | |
| 1 | 190909965 | −7.81 | 7.62E-07 | Rps6kc1 | Intron, Conserved |
| 4 | 141573980 | −7.80 | 7.66E-07 | Conserved | |
Note: Chromosome (“Chr”), “Coord” is the coordinate of the CpG. “Gene” indicates which gene the CpG falls in the boundary of. “Feature” describes attributes overlapping the CpG. “Intron” designates overlap with RefSeq genes. “Conserved” indicates CpG lies with an evolutionarily conserved region; “Bidirectional promotor” indicates a CpG lies in a regulatory region that fall between pairs of genes, where the 5’ ends of the genes are positioned in close proximity to one another.
Table 2.
Pathway results with p-value < 1×10−05 for brain and blood individually
| Pathway | Significant genes in MWAS | Total | Gene No. | P-value |
|---|---|---|---|---|
| Brain | ||||
| CXCR4 signaling pathway | Ptk2b, Ptk2 | 14 | 2 | 0.003 |
| Wnt signaling pathway | Dixdc1, Fzd1, Fzd10, Prkce | 119 | 4 | 0.005 |
| IL-7 signaling | Mapk13, Prkce, Ptk2, Ptk2b | 180 | 4 | 0.020 |
| Heterotrimeric g-protein signaling pathway-gq alpha | Kcnj5, Prkce, Cacna1b | 109 | 3 | 0.025 |
| Alzheimer disease-presenilin pathway | Lrp3, Fzd1, Fzd10 | 113 | 3 | 0.028 |
| Ionotropic glutamate receptor pathway | Shank2, Cacna1b | 47 | 2 | 0.031 |
| Thyrotropin-releasing hormone receptor signaling pathway | Prkce, Cacna1b | 61 | 2 | 0.049 |
| Blood | ||||
| Metabotropic glutamate receptor group i pathway | Gnaq, Grik1 | 24 | 2 | 0.042 |
Note: MWAS, methylome-wide association study; Total is the total number of genes in the pathway; Gene No. is the number of genes in the pathway with p <1×10−05 in the MWAS.
Characteristics of CpGs associated with acute ethanol exposure
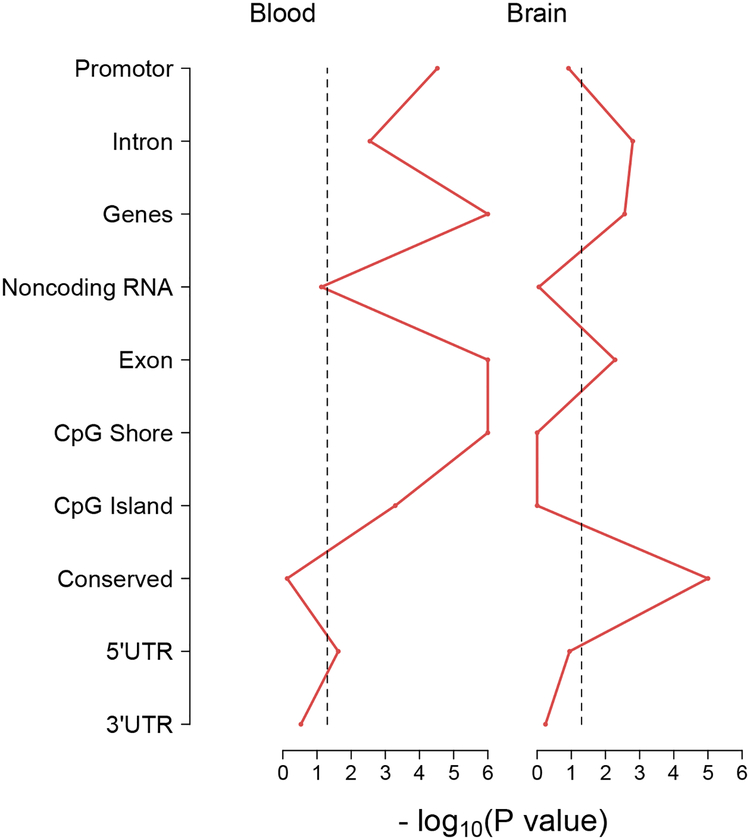
Given the observation that many of the top findings in both tissues were located in genes, we tested whether the top findings from the brain and blood MWASs were enriched for other genomic features. Results show (Figure 1) that top MWAS findings for brain tended to be located in introns and exons of genes and in evolutionarily conserved regions as indicated by the points for these features exceeding the significant threshold (vertical dashed line). For blood, the top MWAS findings were likely to be in introns and exons of genes, gene promotors, and CpG Islands and shores.
Figure 1.
Enrichment test results for blood and brain MWAS results against genomic features. The x-axis shows the -log10(P-value) for each enrichment test of blood and brain MWAS results against the corresponding genomic features on the y-axis. P-values falling to the right of the vertical dashed line (>1.3 -log10(P-value)) indicate significance at p-value < 0.05.
Overlap in CpGs between brain and blood results
The results of our enrichment testing indicated that there was a significant enrichment of top 10% blood MWAS findings among the top 10% brain MWAS findings (multiple testing corrected permutation p=0.021). This overlap corresponded to 370 sites with p < 0.01 in both MWASs and with the same direction of effect. The pathways of the 148 implicated genes overlapping between tissues (Table 3) were central to mitogen-activated protein kinase (MAPK) signaling and those upstream such as the Ras pathway, TNF/Stress related signaling, and growth factor signaling. Additional pathways implicate glutamatergic and adrenergic signaling.
Table 3.
Pathway results with p-value < 0.01 for overlapping brain and blood CpGs
| Pathway | Overlapping Genes | Total | Gene No. | P-value |
|---|---|---|---|---|
| MAPKinase signaling pathway | Map2k1, Map2k2, Map2k3, Map2k4, Map2k5, Map2k6, Map2k7 | 26 | 7 | 1.55×10−05 |
| Keratinocyte differentiation | Map2k1, Map2k3, Map2k4, Map2k6, Map2k7, Prkca, Ets1 | 37 | 7 | 1.78X10−04 |
| Ras pathway | Map2k1, Map2k2, Map2k3, Map2k4, Map2k7, Ets1, Rgl1, Sos2 | 66 | 8 | 0.001 |
| Egf receptor signaling pathway | Map2k1, Map2k2, Map2k4, Sos2, Ywhaz, | 28 | 5 | 0.002 |
| GPCR GroupI metabotropic glutamate receptor | Map2k1, Map2k2, Prkca, Plcb4, Prkce | 28 | 5 | 0.002 |
| Alpha adrenergic receptor signaling pathway | Prkca, Prkce, Plcb4, Plce1 | 20 | 4 | 0.004 |
| TNF/Stress related signaling | Map2k3, Map2k4, Map2k6, Cradd | 24 | 4 | 0.008 |
| Endothelin signaling pathway | Map2k1, Map2k2, Plcb4, Prkar1b, Prkca, Prkce, Prkg1 | 75 | 7 | 0.012 |
| Fgf signaling pathway | Map2k1, Map2k2, Map2k3, Map2k4, Prkca, Prkce, Sos2, Ywhaz, | 109 | 8 | 0.028 |
| Signaling pathway from G-protein families | Map2k1, Prkar1b, Prkca | 21 | 3 | 0.031 |
| NGF | Map2k1, Map2k2, Sos2 | 21 | 3 | 0.031 |
| PDGF signaling pathway | Map2k1, Map2k4, Prkca | 24 | 3 | 0.044 |
| Fas | Map2k4, Map2k7, Ywhaz | 24 | 3 | 0.044 |
Note: Total is the total number of genes in the pathway; Gene No. is the number of genes in the pathway with p <0.01 and the same direction of effect in both the brain and blood MWAS.
DISCUSSION
By screening the 22 million CpGs in the mouse methylome, we identified some methylation changes in brain and blood after acute ethanol administration. Several of the changes were located in genes or regions that have previously been associated with alcohol in humans. Espnl was found to be associated with initial alcohol sensitivity and acute withdrawal in a linkage study (Kuo et al., 2006). A variant in Utp20 was found to convey risk for age of onset of alcohol dependence (Kapoor et al., 2014). To further explore the potential overlap between our methylation results and human genetics results, we examined the fifteen genome-wide significant loci reported in recent human alcohol genome-wide association studies (Adkins et al., 2017, Clarke et al., 2017, Jorgenson et al., 2017, Mbarek et al., 2015, Schumann et al., 2016), 13 of which were located in genes with direct homologues in mice, and found that none of these genes were among the top tissue specific results (Table 1). Enrichment testing revealed that top findings from both the brain and blood MWASs tended to be located in intronic and exonic regions for both the brain and blood MWASs. The blood MWAS findings were also significantly enriched for CpG Islands. The enrichment findings are consistent with previous studies demonstrating that disease and environmental exposure related methylation is likely to occur outside of gene promotors and CpG Islands (Irizarry et al., 2009).
Pathway analyses based on the results of the individual MWASs showed multiple significant results in brain, implicating several related pathways involved in the acute response to ethanol. Many of these pathways have established roles in neuroadaptation following physical or chemical insults. For example, genes within the CXCR4 pathway are over-targeted by miRNAs in the pre-frontal cortex of human alcoholics (Lewohl et al., 2011). In adult rats, CXCR4 is expressed in striatal interneurons and may modulate cholinergic and dopaminergic signaling (Banisadr et al., 2002). The pro-inflammatory IL-7 pathway contributes to reactive gliosis and can induce neural progenitor cell differentiation (Moors et al., 2009). Thus, altered methylation of genes within the CXCR4 and IL-7 pathways may highlight an effect of acute ethanol on neuroinflammation and neuroplasticity.
Members of the Wnt signaling pathway (and the related presenilin pathway) were also heavily featured in our brain pathway results. The Wnt family of morphogens are key regulators of neural maturation and are essential for maintenance of dopaminergic neurons in the midbrain (Arenas, 2014). Acute ethanol exposure suppresses expression of WNT3A, WNT5A, and the Frizzled co-receptor LRP6 in human neural stem cells (Vangipuram and Lyman, 2012). Changes in DNA methylation among pathway members may facilitate ethanol-induced suppression in Wnt signaling.
The remaining significant pathways in brain revolved around members involved with glutamatergic, calcium and G-protein signaling. Pharmacological inhibition of the N-type voltage gated calcium channel Cav2.2 (Cacna1b) results in reduced ethanol intoxication, reinforcement, and reward in mice (Newton and Messing, 2009). Kcnj5, the Gβγ-activated inward rectifying potassium channel 4 (GIRK4) contains a binding site for and is activated by ethanol (Aryal et al., 2009), and is necessary for ethanol–induced conditioned place preference in mice (Tipps et al., 2016). Knockout of calcium-dependent Protein kinase C epsilon (Prkce), commonly featured in the enriched pathways, modulates ethanol consumption and sensitivity in mice (Choi et al., 2002). Within the brain, the NMDA and AMPA glutamate receptors are the highest affinity sites of ethanol-induced inhibition (Nagy, 2008). Furthermore, the gene promoters of the mGluRs have been shown to be more highly methylated in alcoholic patients (Meyers et al., 2015).
We also identified one significant pathway in blood implicating glutamatergic signaling. Of note, glutamate receptor ionotropic kainate 1 encoded by the pathway member Grik1 is also potently inhibited by ethanol (Valenzuela and Cardoso, 1999). Additionally, genetic polymorphisms in GRIK1 are contributors to alcohol dependence risk (Kranzler et al., 2009) and predictors of treatment outcomes (Kranzler and Edenberg, 2010). While it is unclear if the observed changes in methylation in glutamatergic pathways in blood are due to the inhibitory effect of ethanol, it remains plausible as many glutamate receptors are expressed in the periphery (Gill and Pulido, 2001). More importantly, from a biomarker perspective, affected biochemical pathways in the brain are also detected in blood following acute ethanol exposure.
Extending our pathway analyses beyond individual tissues, we further tested for enrichment between brain and blood, revealing a significant overlap among the top MWAS findings in the two tissues. Pathway analyses of the overlap between tissues indicated Mitogen-activated protein kinase (MAPK), growth factor, and G-protein signaling pathways. The predominant members of the enriched pathways are found at the MAP2K (MAPKK/MEK1/MEK2) level, upstream of p38/ERK1/ERK2. Treatment with MAP2K or p38 inhibitors leads to changes in ethanol consumption in mice trained to self-administer (Agoglia et al., 2015). This analysis also suggests G-protein coupled receptor signaling via α-adrenergic receptors may contribute to the acute effects of ethanol, evidenced by the number of Gαq and Ca2+ dependent members implicated (Plcb4, Plce1, Prkar1b, Prkg1, Prkca, Prkce). Extensively studied because of its involvement in the physical and motivational components of addiction (Walker et al., 2008), adrenergic receptors have been suggested as potential targets for alcohol dependence pharmacotherapies (Rasmussen et al., 2009). Indeed, preliminary trials with the α-adrenergic antagonist prazosin have shown promising results for treatment for alcohol dependence and craving (Simpson et al., 2018).
Our results showing overlapping methylation signals in blood and brain from the same animal suggests that alcohol related methylation in blood partly, but not fully, mirrors changes that occur in brain tissue. While our finding was from a model system, it is possible that some of these findings would extend to humans. This finding has important potential implications for the clinical application of blood biomarkers as it provides some evidence that methylation in blood might be able to be used as a proxy tissue to detect some processes that occur in brain as a result of ethanol, however, our findings require further validation and replication, preferably using a different method to measure methylation.
One form of validation would be to examine the overlapping genes for differential gene expression in brain tissue by comparing acute ethanol exposed to saline exposed mice. Using the results from a gene expression study where mice underwent a similar protocol to the one used in this study (Wolen et al., 2012), we annotated our overlapping gene list to indicate whether a gene was differentially expressed between acute ethanol and saline exposed mice (Table S4). In total, there were 23 genes from the overlapping gene list that were expressed in either the prefrontal cortex, nucleus accumbens or ventral mid-brain. However, the results reflect differential gene expression that occurred at the time of death of the mice and may not capture any differences that occurred before death. As such, there may be more differentially expressed genes than what is listed in Table S4. It should also be noted that not all methylation sites have an impact on expression of the closest gene. Many methylation sites are relatively far from the genes they influence and have other more complex regulatory effects than a negative correlation with the closest gene.
The MWASs performed here used DNA from bulk tissue (i.e., blood and brain). However, both of these tissues contain multiple cell types. While we did not consider the effect of acute ethanol administration in the different cell types, we recognize that knowing what cell type caused an association is key for further understanding the biological implications of findings and can be critical for designing proper (functional) follow-up studies. To further this aim, we have compiled expression levels for each of the overlapping genes from publicly available, cell-type specific gene expression mouse databases (Table S5). For blood, we used the “Mouse normal hematopoietic system” track from BloodSpot (Bagger et al., 2016) and for brain we utilized the database established by Zhang and colleagues (Zhang et al., 2014).
There are several natural extensions to the work presented here. First, our study focused on first contact with alcohol. However, methylation patterns may be different after repeated exposures or during other stages of addiction, such as the development of dependence after chronic administration, withdrawal when ethanol is discontinued and, after extended abstinence, the possibility of a persistent biological memory of past alcohol exposure that may prime mice to the effects of renewed alcohol exposure. Second, the mice in our studies were administered ethanol through intraperitoneal injections. Self-administration models of ethanol may yield different methylation patterns as methylation studies using forced administration models may pick up on the negative reinforcing effects of ethanol. Lastly, the landscape of the brain methylome is complex. For example, in the vast majority of mammalian somatic tissues methylation involves the addition of a methyl group to CG dinucleotides (Lister et al., 2009). However, the brain methylome contains other forms of methylation that rare or absent in other tissues. This includes methylation outside the CG context and hydroxymethylation. Future work should assess the impact of ethanol on these additional methylation forms.
In summary, our results show that there are some methylation changes occurring in both brain and blood after acute ethanol administration. By comparing brain and blood from the same animal, we have provided provisional evidence that alcohol related methylation in blood at least partly reflects methylation occurring in brain by demonstrating an enrichment of top brain findings among the top blood findings. Pathway analyses suggest that the overlapping genes may tag alcohol related processes. These findings provide preliminary evidence that blood-based methylation sites might be able to be used as proxy sites for brain when developing biomarkers for alcohol related outcomes but require further validation and replication.
Supplementary Material
Acknowledgements
This work was supported by NIH grants: K01 AA021266 (S.L.C.), R01 AA026057, R01 AA020634 and P50 AA022537.
REFERENCES
- Aberg KA, Mcclay JL, Nerella S, Xie LY, Clark SL, Hudson AD, Bukszar J, Adkins D, Consortium SS, Hultman CM, Sullivan PF, Magnusson PK & Van Den Oord EJ 2012. MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case--control samples. Epigenomics, 4, 605–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, Xie L, Chan RF, Zhao M, Pandey AK, Kumar G, Clark SL & Van Den Oord EJ 2015. Evaluation of Methyl-Binding Domain Based Enrichment Approaches Revisited. PLoS One, 10, e0132205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, Xie LY, Mcclay JL, Nerella S, Vunck S, Snider S, Beardsley PM & Van Den Oord EJ 2013. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics, 5, 367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins AE, Hack LM, Bigdeli TB, Williamson VS, Mcmichael GO, Mamdani M, Edwards AC, Aliev F, Chan RF, Bhandari P, Raabe RC, Alaimo JT, Blackwell GG, Moscati A, Poland RS, Rood B, Patterson DG, Walsh D, Collaborative Study of the Genetics of Alcoholism, C., Whitfield JB, Zhu G, Montgomery GW, Henders AK, Martin NG, Heath AC, Madden P. a. F., Frank J, Ridinger M, Wodarz N, Soyka M, Zill P, Ising M, Nothen MM, Kiefer F, Rietschel M, German Study of the Genetics of Addiction, C., Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer LA, Maher BS, Prescott CA, Dick DM, Bacanu SA, Mathies LD, Davies AG, Vladimirov VI, Grotewiel M, Bowers MS, Bettinger JC, Webb BT, Miles MF, Kendler KS & Riley BP 2017. Genomewide Association Study of Alcohol Dependence Identifies Risk Loci Altering Ethanol-Response Behaviors in Model Organisms. Alcohol Clin Exp Res, 41, 911–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoglia AE, Sharko AC, Psilos KE, Holstein SE, Reid GT & Hodge CW 2015. Alcohol alters the activation of ERK1/2, a functional regulator of binge alcohol drinking in adult C57BL/6J mice. Alcohol Clin Exp Res, 39, 463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen AM, Dogan MV, Beach SR & Philibert RA 2015. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes (Basel), 6, 991–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas E 2014. Wnt signaling in midbrain dopaminergic neuron development and regenerative medicine for Parkinson’s disease. Journal of Molecular Cell Biology, 6, 42–53. [DOI] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S & Slesinger PA 2009. A discrete alcohol pocket involved in GIRK channel activation. Nat Neurosci, 12, 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, Winther O, Rapin N & Porse BT 2016. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res, 44, D917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banisadr G, Fontanges P, Haour F, Kitabgi P, Rostène W & Mélik Parsadaniantz S 2002. Neuroanatomical distribution of CXCR4 in adult rat brain and its localization in cholinergic and dopaminergic neurons. European Journal of Neuroscience, 16, 1661–1671. [DOI] [PubMed] [Google Scholar]
- Bares V & Ge X 2015. gskb: Gene set data for pathway analysis in mouse. R package version 1.2.0 ed. [Google Scholar]
- Barker JM, Zhang Y, Wang F, Taylor JR & Zhang H 2013. Ethanol-induced Htr3a promoter methylation changes in mouse blood and brain. Alcohol Clin Exp Res, 37 Suppl 1, E101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SR, Dogan MV, Lei MK, Cutrona CE, Gerrard M, Gibbons FX, Simons RL, Brody GH & Philibert RA 2015. Methylomic Aging as a Window onto the Influence of Lifestyle: Tobacco and Alcohol Use Alter the Rate of Biological Aging. J Am Geriatr Soc, 63, 2519–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG & Meissner A 2010. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol, 28, 1106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera CP, Navarro P, Huffman JE, Wright AF, Hayward C, Campbell H, Wilson JF, Rudan I, Hastie ND, Vitart V & Haley CS 2012. Uncovering networks from genome-wide association studies via circular genomic permutation. G3 (Bethesda), 2, 1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RF, Shabalin AA, Xie LY, Adkins DE, Zhao M, Turecki G, Clark SL, Aberg KA & Van Den Oord E 2017. Enrichment methods provide a feasible approach to comprehensive and adequately powered investigations of the brain methylome. Nucleic Acids Res, 45, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, Herwig R & Adjaye J 2010. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res, 20, 1441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D-S, Wang D, Dadgar J, Chang WS & Messing RO 2002. Conditional Rescue of Protein Kinase C ε Regulates Ethanol Preference and Hypnotic Sensitivity in Adult Mice. The Journal of Neuroscience, 22, 9905–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ & Mcintosh AM 2017. Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry, 22, 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk LC & Mill J 2012. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol, 13, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K & Beck S 2006. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet, 38, 1378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS & Pulido OM 2001. Glutamate receptors in peripheral tissues: current knowledge, future research, and implications for toxicology. Toxicol Pathol, 29, 208–23. [DOI] [PubMed] [Google Scholar]
- Guenet JL 2005. The mouse genome. Genome Res, 15, 1729–40. [DOI] [PubMed] [Google Scholar]
- Heberlein A, Buscher P, Schuster R, Kleimann A, Lichtinghagen R, Rhein M, Kornhuber J, Bleich S, Frieling H & Hillemacher T 2015. Do changes in the BDNF promoter methylation indicate the risk of alcohol relapse? Eur Neuropsychopharmacol, 25, 1892–7. [DOI] [PubMed] [Google Scholar]
- Heyward FD & Sweatt JD 2015. DNA Methylation in Memory Formation: Emerging Insights. Neuroscientist, 21, 475–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S & Feinberg AP 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet, 41, 178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C & Choquet H 2017. Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry, 22, 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL, Budde J, Agrawal A, Almasy L, Bucholz K, Dick DM, Harari O, Xiaoling X, Hesselbrock V, Kramer J, Nurnberger JI Jr., Rice J, Schuckit M, Tischfield J, Porjesz B, Edenberg HJ, Bierut L, Foroud T & Goate A 2014. Genome-wide survival analysis of age at onset of alcohol dependence in extended high-risk COGA families. Drug Alcohol Depend, 142, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW & Miles MF 2005. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci, 25, 2255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF 2014. Neurocircuitry of alcohol addiction: synthesis from animal models. Handb Clin Neurol, 125, 33–54. [DOI] [PubMed] [Google Scholar]
- Koob GF & Volkow ND 2016. Neurobiology of addiction: a neurocircuitry analysis. Lancet Psychiatry, 3, 760–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR & Edenberg HJ 2010. Pharmacogenetics of alcohol and alcohol dependence treatment. Curr Pharm Des, 16, 2141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzler HR, Gelernter J, Anton RF, Arias AJ, Herman A, Zhao H, Burian L & Covault J 2009. Association of markers in the 3’ region of the GluR5 kainate receptor subunit gene to alcohol dependence. Alcohol Clin Exp Res, 33, 925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Webb BT, Sullivan PF, Vittum J, Patterson DG, Thiselton DL, Van Den Oord EJ, Walsh D, Kendler KS & Prescott CA 2006. Identification of susceptibility loci for alcohol-related traits in the Irish Affected Sib Pair Study of Alcohol Dependence. Alcohol Clin Exp Res, 30, 1807–16. [DOI] [PubMed] [Google Scholar]
- Lewohl JM, Nunez YO, Dodd PR, Tiwari GR, Harris RA & Mayfield RD 2011. Up-regulation of microRNAs in brain of human alcoholics. Alcohol Clin Exp Res, 35, 1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B & Ecker JR 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature, 462, 315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schmidt B & Maskell DL 2012. CUSHAW: a CUDA compatible short read aligner to large genomes based on the Burrows-Wheeler transform. Bioinformatics, 28, 1830–7. [DOI] [PubMed] [Google Scholar]
- Mahnke AH, Miranda RC & Homanics GE 2017. Epigenetic mediators and consequences of excessive alcohol consumption. Alcohol, 60, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, De Moor MH, Jansen R, Gelernter J, Sherva R, Willemsen G, Boomsma DI, Penninx BW & Vink JM 2015. The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet B Neuropsychiatr Genet, 168, 739–48. [DOI] [PubMed] [Google Scholar]
- Meyers JL, Salling MC, Almli LM, Ratanatharathorn A, Uddin M, Galea S, Wildman DE, Aiello AE, Bradley B, Ressler K & Koenen KC 2015. Frequency of alcohol consumption in humans; the role of metabotropic glutamate receptors and downstream signaling pathways. Transl Psychiatry, 5, e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moors M, Vudattu NK, Abel J, Kramer U, Rane L, Ulfig N, Ceccatelli S, Seyfert-Margolies V, Fritsche E & Maeurer MJ 2009. Interleukin-7 (IL-7) and IL-7 splice variants affect differentiation of human neural progenitor cells. Genes Immun, 11, 11–20. [DOI] [PubMed] [Google Scholar]
- Nagy J 2008. Alcohol related changes in regulation of NMDA receptor functions. Curr Neuropharmacol, 6, 39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ 2001. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci, 2, 119–28. [DOI] [PubMed] [Google Scholar]
- Nestler EJ 2014. Epigenetic mechanisms of drug addiction. Neuropharmacology, 76 Pt B, 259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton PM & Messing RO 2009. The N-type calcium channel is a novel target for treating alcohol use disorders. Channels, 3, 77–81. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Batra A & Fallgatter AJ 2013. Genetics and epigenetics of alcohol dependence. J Mol Psychiatry, 1, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, Mcgarvey KM, Murphy MR, O’leary NA, Pujar S, Rajput B, Rangwala SH, Riddick LD, Shkeda A, Sun H, Tamez P, Tully RE, Wallin C, Webb D, Weber J, Wu W, Dicuccio M, Kitts P, Maglott DR, Murphy TD & Ostell JM 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res, 42, D756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen DD, Alexander LL, Raskind MA & Froehlich JC 2009. The alpha1-adrenergic receptor antagonist, prazosin, reduces alcohol drinking in alcohol-preferring (P) rats. Alcohol Clin Exp Res, 33, 264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ & Nestler EJ 2011. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci, 12, 623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hallfors J, Harris SE, Hieber S, Hofer E, Hottenga JJ, Johansson A, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikainen LP, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud AC, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, De Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen AL, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin MR, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimaki T, Liu Y, Madden PA, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, et al. 2016. KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A, 113, 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA, Hattab MW, Clark SL, Chan RF, Kumar G, Aberg KA & Van Den Oord E 2018a. RaMWAS: Fast Methylome-Wide Association Study Pipeline for Enrichment Platforms. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA, Hattab MW, Clark SL, Chan RF, Kumar G, Aberg KA, Van Den Oord E & Birol I 2018b. RaMWAS: Fast Methylome-Wide Association Study Pipeline for Enrichment Platforms. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TL, Saxon AJ, Stappenbeck C, Malte CA, Lyons R, Tell D, Millard SP & Raskind M 2018. Double-Blind Randomized Clinical Trial of Prazosin for Alcohol Use Disorder. Am J Psychiatry, appiajp201817080913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipps ME, Raybuck JD, Kozell LB, Lattal KM & Buck KJ 2016. G Protein-Gated Inwardly Rectifying Potassium Channel Subunit 3 Knock-Out Mice Show Enhanced Ethanol Reward. Alcohol Clin Exp Res, 40, 857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulisiak CT, Harris RA & Ponomarev I 2017. DNA modifications in models of alcohol use disorders. Alcohol, 60, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela CF & Cardoso RA 1999. Acute effects of ethanol on kainate receptors with different subunit compositions. J Pharmacol Exp Ther, 288, 1199–206. [PubMed] [Google Scholar]
- Van Den Oord EJ, Bukszar J, Rudolf G, Nerella S, Mcclay JL, Xie LY & Aberg KA 2013. Estimation of CpG coverage in whole methylome next-generation sequencing studies. BMC Bioinformatics, 14, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangipuram SD & Lyman WD 2012. Ethanol affects differentiation-related pathways and suppresses Wnt signaling protein expression in human neural stem cells. Alcohol Clin Exp Res, 36, 788–97. [DOI] [PubMed] [Google Scholar]
- Walker BM, Rasmussen DD, Raskind MA & Koob GF 2008. alpha1-noradrenergic receptor antagonism blocks dependence-induced increases in responding for ethanol. Alcohol, 42, 91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Agarwala R, Capra J, Chen Z, Church D, Ciobanu D, Li Z, Lu L, Mozhui K, Mulligan M, Nelson S, Pollard K, Taylor W, Thomason D & Williams R 2010. High-throughput sequencing of the DBA/2J mouse genome. BMC Bioinformatics, 11, O7. [Google Scholar]
- Wolen AR, Phillips CA, Langston MA, Putman AH, Vorster PJ, Bruce NA, York TP, Williams RW & Miles MF 2012. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS One, 7, e33575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA & Wu JQ 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci, 34, 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.