Abstract
Background:
Previous studies indicate that low initial sensitivity to alcohol may be a risk factor for later alcohol misuse. Evidence suggests that initial sensitivity is influenced by genetic factors, but few molecular genetic studies have been reported.
Methods:
We conducted a meta-analysis of two population-based genome-wide association studies of the Self-Rating of the Effects of Alcohol scale. Our final sample consisted of 7339 individuals (82.3% of European descent; 59.2% female) who reported having used alcohol at least 5 times. In addition, we estimated SNP-based heritability and conducted a series of secondary aggregate genetic analyses.
Results:
No individual locus reached genome-wide significance. Gene- and set-based analyses, both overall and using tissue-specific expression data, yielded largely null results, and genes previously implicated in alcohol problems and consumption were overall not associated with initial sensitivity. Only one gene-set, related to hormone signaling and including core clock genes, survived correction for multiple testing. A meta-analysis of SNP-based heritability resulted in a modest estimate of h2SNP=0.19 (SE=0.10), though this was driven by one sample (N=3683, h2SNP=0.36, SE=0.14, p=0.04). No significant genetic correlations with other relevant outcomes were observed.
Conclusions:
Findings yielded only modest support for a genetic component underlying initial alcohol sensitivity. Results suggest that its biological underpinnings may diverge somewhat from that of other alcohol outcomes, and may be related to core clock genes or other aspects of hormone signaling. Larger samples, ideally of prospectively assessed samples, are likely necessary to improve gene identification efforts and confirm the current findings.
Keywords: ALSPAC, genetics, GWAS, heritability, initial alcohol sensitivity, SRE
Introduction
Alcohol misuse is a common and costly human behavior, accounting for 3.3 million deaths worldwide in 2012 (World Health Organization, 2014) and over $220 billion in annual economic tolls in the US alone (Bouchery et al., 2011). Alcohol-related outcomes are influenced by genetic factors: the heritability of alcohol use disorder (AUD) was estimated at 0.50 in a meta-analysis of twin studies (Verhulst et al., 2015), and recent genome-wide association studies of alcohol consumption and symptoms have reported SNP-based heritabilities of 0.13 (Clarke et al., 2017) and 0.12 (Sanchez-Roige et al., 2017), respectively. Variation in genes involved in alcohol metabolism (e.g., alcohol and aldehyde dehydrogenases) is known to impact liability to problems (Edenberg, 2007) and consumption (Jorgenson et al., 2017); there is support for a role of genes outside of this metabolic pathway as well (Schumann et al., 2011; Schumann et al., 2016). However, much remains unclear about the mechanisms underlying genetic influences on alcohol outcomes, necessitating further study and consideration of precursors in addition to the outcomes themselves.
Sensitivity to alcohol’s effects, particularly during initiation of voluntary alcohol consumption, has been associated with later alcohol use and misuse (Schuckit, 1994; Schuckit et al., 2008a; Schuckit et al., 2008b). Under a model in which an individual consumes alcohol in part to experience its pleasant physiological effects (e.g., a “buzz”), it follows that those who are less sensitive to these effects will consume more than their peers (Trela et al., 2016). Higher consumption, in turn, is positively associated with alcohol problems (Barnett et al., 2014; Dick et al., 2011; Heath et al., 1999; Schuckit et al., 2007) in some but not all (Heath et al., 1999) studies, raising the possibility that those whose subjective response to alcohol is low have higher liability to later misuse.
The Self-Rating of the Effects of Alcohol (SRE) scale was developed by Schuckit and colleagues (Schuckit et al., 1997) to operationalize an individual’s alcohol sensitivity by quantifying the number of standard alcoholic drinks necessary to experience physiological effects of alcohol, such as dizziness and slurring; it does not capture all dimensions of intoxication. Higher scores reflect the need to consume a higher volume of alcohol to experience those effects – that is, lower sensitivity to alcohol per drink. Importantly, there is evidence that this sensitivity is heritable: Individuals with a family history of alcohol problems exhibit less pronounced alcohol sensitivity (Schuckit, 1980; Schuckit, 1984; Schuckit et al., 2003; Schuckit et al., 2000; Schuckit et al., 1996) across a variety of assessments, including the SRE (Schuckit et al., 2003). We are aware of only one twin study of alcohol response (Heath et al., 1999) in adulthood, which reported a heritability of 0.6. Furthermore, linkage studies have identified loci associated with SRE (Ehlers et al., 2010; Schuckit et al., 2001), and variants in GABRA2 were nominally associated with both subjective and objective measures of alcohol sensitivity in an Australian sample (Lind et al., 2008). A variety of gene sets are hypothesized to influence alcohol sensitivity (Schuckit, 2018), but in the absence of molecular genetic analyses, these remain speculative.
The current study aims to expand information available on genetic influences underlying initial sensitivity to alcohol. We conducted a meta-analysis of genome-wide association studies (GWAS) on SRE scores from two independent, population-based cohorts. While both samples are predominantly of European descent, one sample also included individuals of African (AFR) and American (AMR) descent. By elucidating the biological underpinnings of initial sensitivity to alcohol, we can improve existing models of mechanisms underlying risk of alcohol misuse, and potentially inform personalized intervention and prevention programming that might be used even before the first drink.
Materials and Methods
Samples
ALSPAC.
We used two population-based cohort samples: the Avon Longitudinal Study of Parents and Children (ALSPAC) and Spit for Science (S4S). ALSPAC recruited 14,541 pregnant women residing in Avon, UK, with expected dates of delivery April 1, 1991, to December 31, 1992; 14,541 is the initial number of pregnancies for which the mothers enrolled in the ALSPAC study and had either returned at least 1 questionnaire or attended a “Children in Focus” clinic by July 19, 1999. Of these initial pregnancies, there was a total of 14,062 live births and 13,988 children who were alive at 1 year of age. Subsequent phases of enrollment increased the sample size over time. The phases of enrollment are described in more detail elsewhere (Boyd et al., 2013; Fraser et al., 2013). For the current analyses, full or partial phenotypic data were available for 5,626 participants, in part reflecting the need for a subject to have had experience with alcohol in order to fill out the SRE. The study website contains details of all the data that is available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
S4S.
Spit for Science is an ongoing longitudinal study of college students enrolled in a large, urban university in the Mid-Atlantic (Dick et al., 2014). Briefly, incoming students age 18 or older were eligible to complete phenotypic assessments, which covered a wide range of topics but focused on alcohol use. Study data were collected and managed using REDCap electronic data capture tools (Harris et al., 2009) hosted at Virginia Commonwealth University. Follow-up assessments were completed in subsequent spring semesters. Individuals who did not participate in the first wave of data collection (including those who turned 18 after the end of the first wave of data collection) had the opportunity to join the study the following spring; those who participated during their first year were eligible to complete follow-up assessments each spring. Participants who completed the phenotypic assessments were eligible to provide a DNA sample. The current study includes three cohorts, which matriculated in Fall 2011 (N=2,714), 2012 (N=2,486), and 2013 (N=2,403), for a total N=7,603. Ethical approval was obtained from the local Institutional Review Board.
Phenotypes
Outcome.
The SRE consists of 4 items; for the current study, each item referred to the first five or so times a participant used alcohol. Participants were asked to report the number of standard drinks they consumed before they experienced signs of alcohol’s effect (from feeling any effect of the alcohol on to slurring words, feeling unsteady on their feet, to unwanted falling asleep). Responses were winsorized to account for outliers. Consistent with prior papers, responses were winsorized to limit extreme values and reduce the effect of possibly spurious outliers. SRE scores were calculated by summing drinks needed for effects across items and dividing by the number of the up to four effects experienced, as recommended by Schuckit and colleagues (Schuckit et al., 1997). The final score was used as a continuous outcome in subsequent GWAS.
Both ALSPAC and S4S participants were administered the SRE items across multiple assessments. For ALSPAC, we examined data from questionnaires/clinic visits at average ages 15.5, 16.5, and 17.5. S4S participants were also administered SRE items at average ages 18.5, 19.0, 19.9, and 21.0. For both samples, only participants who reported having initiated alcohol use were administered the SRE items; others were coded as “NA”. Where scores were available for a participant across multiple waves, the first score was used for GWAS, reasoning that this wave represented the assessment most temporally proximal to the initiation of alcohol use and therefore least susceptible to recall bias. The size of a standard drink, to which respondents are asked to refer when completing the SRE items, differs in the US and the UK (14g vs. 8g of ethanol, respectively). Therefore, raw SRE scores were standardized for GWAS analyses, ensuring that effect sizes observed across the ALSPAC and S4S samples were on the same scale.
Covariates.
Sex was included as a covariate (individuals whose self-reported gender was inconsistent with genetic sex were excluded from these analyses) in initial GWAS within both samples. For ALSPAC, wave at which the SRE items were first completed was included as a covariate; assessments are age-standardized but precise date of completion was unavailable. For S4S, age at which the SRE items were first completed was included as a covariate (mean [SD] age across all samples= 18.80 [0.79]). To account for population structure, 10 ancestry-informative principle components (PCs) were included in the ALSPAC GWAS, consistent with prior analyses using this sample (Edwards et al., 2017). S4S participants are of diverse ancestry, and were first assigned to empirically-based ancestry groups using principal components derived from 1000 Genomes (phase 3) reference populations, as described by Peterson and colleagues (Peterson et al., 2017). Subsequently, within-ancestry PCs were calculated to capture fine-grained stratification; PCs were retained as covariates in the GWAS using a stepwise regression approach.
Imputation and Quality Control Filters
Imputed genotypes for both samples were derived using the 1000 Genomes reference panels as previously reported (Edwards et al., 2017; Webb et al., 2017). Quality control procedures for genetic analyses of both the ALSPAC and S4S samples have been previously described (Fatemifar et al., 2013; Webb et al., 2017), and those within-sample approaches were applied for the current analyses (see Supplementary Information). Briefly, individual DNA samples and markers were excluded based on excess missingness (>5% for both samples), deviations from Hardy-Weinberg Equilibrium (p<5e-7 for ALSPAC, p<5e-6 for S4S), and minor allele frequency (<0.01 for ALSPAC, <0.005 for S4S). Cryptic relatedness was calculated using pi-hat and related individuals were excluded.
Genetic Analyses
GWAS and meta-analysis.
For ALSPAC, phenotypic (including outcome and covariate) and genetic data were available for N=3683 individuals, all of European ancestry. GWAS was run in Plink 1.9 (Chang et al., 2015). For S4S, we included only groups with N>=400 to reduce the likelihood of spurious results, yielding the following sample sizes: African (AFR)=892; American (AMR)=408; and European (EUR)=2356 (total S4S N=3656). The S4S GWAS were run in SNPTest version 2.5.2 (Marchini et al., 2007), separately by ancestry group, as described previously. Results across samples (total N=7339) were meta-analyzed using METAL (Willer et al., 2010), using inverse variance weighting by sample size. Markers with MAF<0.01 within sample/ancestry group and/or INFO<0.5 were excluded from further analysis.
Gene and gene-set analyses.
We applied two approaches to gene- and set-based analyses, FUMA (Watanabe et al., 2017) and JEPEGMIX (Lee et al., 2016). The former conducts gene-based tests across all markers followed by gene-set analysis. We submitted meta-analysis summary statistics to the FUMA pipeline, which requires selection of a reference panel in order to account for linkage disequilibrium (LD) among markers. We selected the EUR subsample of the 1000 Genomes reference panel as this group constituted >82% of the sample, and correcting for EUR LD is a more conservative approach than correcting for AFR LD (AFR being the next-largest component of the full sample). JEPEGMIX differs from FUMA in that its gene- and set-based analyses are tissue-specific: using GWAS summary statistics, it tests the joint effect of functional SNPs known to affect the expression of a gene, effectively predicting whether tissue-specific gene expression is associated with an outcome of interest (here, SRE score). The method can be extended to estimate the joint effects across gene sets.
Heritability and genetic correlations.
GCTA (Yang et al., 2011) was used to assess SNP-based heritability (h2SNP). Analyses were conducted within group (ALSPAC, AFR, AMR, and EUR), using unrelated individuals and markers with MAF≥0.01. To assess genetic correlations between SRE and other relevant traits assessed in independent samples, summary statistics were uploaded to LD Hub (Zheng et al., 2017). We tested whether SRE was genetically correlated with traits in selected categories: anthropometric, brain volume, cognitive, education, hormone, metabolites, personality, psychiatric, reproductive, and smoking behavior.
Polygenic risk scores.
Polygenic risk scores (PRS) were derived to test whether aggregate risk for SRE in the discovery set was associated with phenotype in the test set. We used weights from two discovery sets: i) the full meta-analysis; and ii) the ALSPAC-specific results, given evidence of heritability in ALSPAC but not S4S. The first test set involved participants (N=1,080; 61.1% male) with both genotype and phenotype data from the UCSF Family Alcoholism Study (Vieten et al., 2004). The sample was composed of small family pedigrees, which ranged in size from 3–20 individuals. The subsample used in the present study had an average age of 48.9 (SD=12.1) years. PLINK 1.9 (Chang et al., 2015) was used to derive PRS. We employed a linear mixed model with a kinship matrix fitted as a random effect to control for the relatedness among participants. Age, sex, and four ancestry PCs were included as covariates. The second dataset was from the Collaborative Study on the Genetics of Alcoholism (COGA). COGA is a multi-center study of families with alcohol dependence (AD). African American (AA) and European American (EA) subsamples of COGA were included in analysis. For each COGA subsample, all individuals (N=1,527 in AA; N=4,717 in EA) and COGA prospective samples (N=326 in AA: N=822 in EA), which were offspring of COGA families that were born after 1982 (Bucholz et al., 2017) to match the ALSPAC/S4S samples were tested separately. PRSice-2 (Euesden et al., 2015) was used to calculate PRS. Sex, the first four ancestry PCs, and genotyping array indicators were included as covariates. For analyses of all individuals, birth cohorts were also included as covariates. Linear mixed models were fit to adjust family clustering using SAS9.4 (SAS Institute INC., Cary, NC, USA). For both the UCSF and COGA samples, we tested for associations at 8 p-value thresholds: 0.001, 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5.
Results
Descriptive Statistics
Descriptive statistics of the sample, including mean SRE scores by group prior to transformation, are provided in Table 1. Scores were significantly higher in the S4S sample relative to ALSPAC (t=7.49, p<0.0001). Differences were also observed across S4S ancestry groups (F(2,3653)=10.12, p<0.0001). Men’s scores were higher than women’s (t=20.51, p<0.0001). SRE scores were moderately correlated with later alcohol consumption (r=0.20–0.25 in S4S; r=0.18–0.28 in ALSPAC; all p<0.0001), which was operationalized in S4S as grams of ethanol per month (derived from responses to alcohol use frequency and quantity (Salvatore et al., 2016)) and in ALSPAC as AUDIT-C scores (Bush et al., 1998).
Table 1.
Descriptive statistics by sample and sex.
| N | Mean (SD) Age | Mean (SD) SRE Score | |
|---|---|---|---|
| Sample | |||
| Combined | 7339 | 17.4 (1.58)1 | 5.30 (2.54) |
| ALSPAC | 3683 | 16.03 (0.74)1 | 5.08 (2.75) |
| S4S | 3656 | 18.78 (0.79) | 5.53 (2.29) |
| AFR | 892 | 18.82 (0.80) | 5.24 (2.41) |
| AMR | 408 | 18.76 (0.89) | 5.77 (2.43) |
| EUR | 2356 | 18.78 (0.77) | 5.59 (2.21) |
| Sex | |||
| Women | 4347 | 17.45 (1.54) | 4.81 (2.31) |
| Men | 2992 | 17.33 (1.62) | 6.02 (2.70) |
Because precise ages were not available for ALSPAC participants, expected average age for the wave of data collection was used for these values.
Meta-Analysis of GWAS Results
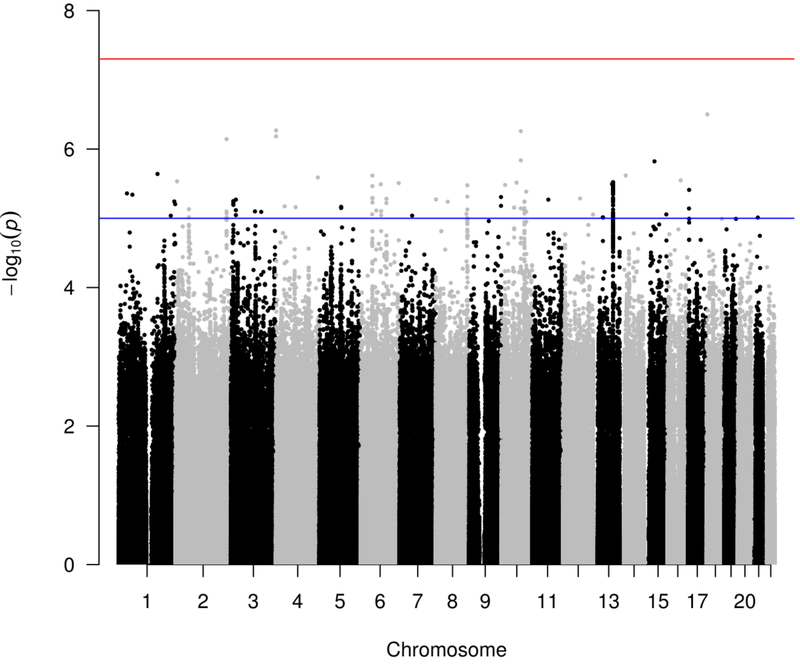
Results from the meta-analysis of SRE scores are displayed in Figures 1 and 2. In each group-level analysis and in the meta-analysis, there was no evidence of inflation due to population stratification (λ1000=0.99–1.00). A total of 15,642,250 markers were analyzed in the meta-analysis; 10,752,408 were assessed in at least 1000 individuals. No individual locus met genome-wide significance criteria (p<5×10−8). The top marker was rs146298733 (p=3.16×10−7), which maps to an intron in DLGAP1 on chromosome 18; this result may be spurious as surrounding markers do not have similar p-values. The minor allele was only of sufficient frequency to test in the ALSPAC and EUR groups (N=6039; MAF=0.02 in both samples).
Figure 1:
Manhattan plot of SRE scores for markers assessed in at least 1000 individuals. Red line indicates genome-wide significance threshold (p<5×10–8); blue line indicates suggestive significance (p<1×10–5).
Figure 2:
Quantile-quantile plot for each sample, as well as for meta-analysis results for markers assessed in at least 1000 individuals.
Meta-analysis summary statistics were uploaded to FUMA, which identified 35 lead SNPs based on p-value and linkage disequilibrium; these are presented in Table 2 alongside functional information derived from Combined Annotation Dependent Depletion (CADD (Kircher et al., 2014)) scores and RegulomeDB (Boyle et al., 2012). Overall, these markers are not predicted to be especially deleterious – only one marker has a CADD score >10 – nor are they predicted to have meaningful regulatory roles – no marker has a RegulomeDB score of 1a-1f.
Table 2.
Lead SNPs from Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA) and corresponding functional annotation.
| rsID | Chr | Position | P-value | Nearest Gene | Distance from Gene | Function | CADD Score | RDB |
|---|---|---|---|---|---|---|---|---|
| rs145005509 | 1 | 244472953 | 5.73E-06 | C1orf100 | 42984 | intergenic | 11.47 | 7 |
| rs10788734 | 1 | 248075398 | 6.26E-06 | OR2T8 | 8922 | intergenic | 1.717 | 7 |
| rs72806266 | 2 | 59501865 | 7.42E-06 | ENSG00000233891 | 0 | ncRNA_intronic | 0.442 | 6 |
| rs112834343 | 2 | 224599695 | 7.22E-07 | AP1S3 | 16708 | intergenic | 4.525 | 6 |
| rs17033567 | 3 | 10913738 | 5.65E-06 | SLC6A11 | 0 | intronic | 2.597 | 7 |
| rs2336522 | 3 | 22023520 | 5.40E-06 | ZNF385D-AS2 | 2200 | intergenic | 0.718 | 7 |
| rs112368179 | 3 | 133217908 | 8.09E-06 | ENSG00000214301 | 7559 | intergenic | 5.85 | 5 |
| rs75536499 | 4 | 536426 | 6.58E-07 | PIGG | 3108 | intergenic | 0.127 | 5 |
| rs115496994 | 4 | 86353705 | 6.92E-06 | ARHGAP24 | 42562 | intergenic | 7.664 | 7 |
| rs10020261 | 4 | 184171187 | 2.57E-06 | WWC2 | 0 | intronic | 8.42 | 4 |
| rs4869281 | 5 | 95651353 | 6.84E-06 | CTD-2337A12.1 | 0 | ncRNA_intronic | 3.662 | 3a |
| rs75886551 | 6 | 51028172 | 8.77E-06 | FTH1P5 | 147203 | intergenic | 0.435 | 4 |
| rs11465543 | 6 | 52108584 | 2.41E-06 | IL17F | 0 | intronic | 2.813 | NA |
| rs76563242 | 6 | 88277132 | 3.24E-06 | RARS2 | 0 | intronic | 0.719 | 7 |
| rs62421504 | 6 | 113654797 | 5.24E-06 | ENSG00000223811 | 23408 | intergenic | 0.663 | 7 |
| rs206972 | 6 | 167689552 | 3.11E-06 | UNC93A | 0 | intronic | 0.043 | 6 |
| rs73133463 | 7 | 55119501 | 9.18E-06 | EGFR | 0 | intronic | 3.336 | 5 |
| rs2100160 | 8 | 427140 | 5.35E-06 | ENSG00000272005 | 0 | ncRNA_exonic | 0.355 | NA |
| rs16905012 | 8 | 134905738 | 8.65E-06 | RP11–157E21.1 | 0 | ncRNA_intronic | 4.592 | 7 |
| rs11777857 | 8 | 138546064 | 3.35E-06 | ENSG00000254076 | 162903 | intergenic | 3.359 | 5 |
| rs28373932 | 9 | 139998042 | 4.94E-06 | MAN1B1 | 0 | intronic | 4.023 | 5 |
| rs76238752 | 10 | 16614401 | 3.30E-06 | RSU1 | 18209 | intergenic | 6.354 | 7 |
| rs10825405 | 10 | 56592865 | 7.00E-06 | PCDH15 | 0 | intronic | 1.107 | 5 |
| rs75752490 | 10 | 67293784 | 3.05E-06 | ENSG00000228065 | 36312 | intergenic | 5.932 | 6 |
| rs61866256 | 10 | 85682627 | 5.53E-07 | ENSG00000233258 | 9949 | intergenic | 4.181 | 7 |
| rs7076325 | 10 | 101868347 | 5.73E-06 | TPM4P1 | 5827 | intergenic | 0.899 | 6 |
| rs184338590 | 10 | 109779481 | 4.11E-06 | RP11–215N21.1 | 0 | ncRNA_intronic | 3.103 | 7 |
| rs75794081 | 11 | 71069255 | 5.39E-06 | AP002387.1 | 24392 | intergenic | 1.829 | 5 |
| rs41287003 | 13 | 41910631 | 9.71E-06 | NAA16 | 0 | intronic | 5.577 | 7 |
| rs9547398 | 13 | 86417640 | 3.01E-06 | SLITRK6 | 44017 | intergenic | 2.744 | 7 |
| rs1016246 | 14 | 26679400 | 2.42E-06 | CYB5AP5 | 1770 | intergenic | 2.935 | NA |
| rs116879015 | 15 | 45492952 | 1.51E-06 | SHF | 0 | exonic | 7.285 | 4 |
| rs146087183 | 16 | 57934080 | 2.83E-06 | CNGB1 | 0 | intronic | 1.436 | 4 |
| rs7214066 | 17 | 4678855 | 3.89E-06 | TM4SF5 | 0 | intronic | 2.927 | 7 |
| rs146298733 | 18 | 4114529 | 3.16E-07 | DLGAP1 | 0 | intronic | 2.39 | 7 |
Chr=chromosome; CADD=Combined Annotation Dependent Depletion; RBD=RegulomeDB
Gene and Gene-Set Analyses
Using FUMA, markers were mapped to 18,363 protein coding genes; none met genome-wide significance criteria (p<2.72×10−6). Complete results are available in Supplemental Table 1, while the top 10 genes are listed in Table 3. The FUMA pipeline also uses the complete distribution of SNP p-values to conduct a gene-set analysis (Nset=10,891) using MSigDB (Subramanian et al., 2005). Only one gene set had a corrected p-value of <0.05: regulation of intracellular steroid hormone receptor signaling pathway (corrected p=0.03). Complete results are available in Supplemental Table 2.
Table 3.
Top 10 gene-based results from FUMA.
| Symbol | Chr | Start BP | End BP | N SNPs | Z statistic | P-value |
|---|---|---|---|---|---|---|
| ZBTB44 | 11 | 130086572 | 130194581 | 397 | 4.2267 | 1.19e-05 |
| BHLHE40 | 3 | 5010801 | 5037008 | 88 | 4.2012 | 1.33e-05 |
| ISL1 | 5 | 50668921 | 50700564 | 77 | 3.8686 | 5.47e-05 |
| NDNF | 4 | 121946768 | 122004176 | 193 | 3.6830 | 1.15e-04 |
| ACTN4 | 19 | 39128289 | 39232223 | 370 | 3.5510 | 1.92e-04 |
| C1orf122 | 1 | 38262651 | 38285126 | 42 | 3.5426 | 1.98e-04 |
| CYSLTR2 | 13 | 49270951 | 49293498 | 83 | 3.5399 | 2.00e-04 |
| ATG4D | 19 | 10644571 | 10674094 | 113 | 3.5359 | 2.03e-04 |
| TMIGD1 | 17 | 28633351 | 28671077 | 90 | 3.4772 | 2.53e-04 |
| FAM159A | 1 | 53089016 | 53145355 | 181 | 3.4280 | 3.04e-04 |
Chr=chromosome; BP=base pair; N SNPs=number of single nucleotide polymorphisms
Using JEPEGMIX, we assessed whether tissue-specific expression for individual genes was predicted to be associated with SRE scores. No gene met the corrected significance threshold (Supplementary Table 3). We next tested whether expression levels of genes in canonical gene-sets was jointly predicted to be associated with SRE scores. No gene-set met the corrected significance threshold (Supplementary Table 4).
Heritability, Genetic Correlations, and Polygenic Risk Scores
Heritability estimates for each S4S ancestry group were not significantly different from 0 (h2SNP<0.001; SE=0.16–0.59; p=0.13–0.50). However, for ALSPAC, heritability was moderate (h2SNP=0.36, SE=0.14, p=0.04). Although the meta-analytic h2SNP was different from 0 (h2SNP=0.19, SE=0.10), this was clearly driven by the ALSPAC group. Because h2SNP exceeded 0 only in the ALSPAC sample, only ALSPAC-specific summary statistics (i.e., not the meta-analytic results) were uploaded to LD Hub. There were no significant genetic correlations between SRE and any of the traits assessed in LD Hub, though we note that the mean χ2 (1.005) was flagged by the program as potentially too low. Complete results are available in Supplemental Table 5. We also conducted bivariate GCTA between SRE and AUDIT-C and total scores at ages 16, 18, and 21; these analyses were limited to ALSPAC given null h2SNP estimates in S4S. Genetic correlations were not significant, but were largely positive (rGSNP=0.55–1.00) with one exception (SRE and age 21 AUDIT-C, rGSNP= −0.07, n.s.). Finally, we tested whether markers implicated at 8 p-value thresholds in the meta-analysis were associated with SRE scores in two independent samples. We derived the PRS using meta-analysis SNP weights and using ALSPAC-specific weights, due to the detection of significant h2snp in ALSPAC but not S4S groups. We observed several nominally significant associations (0.01 < p < 0.05) but no systematic effects.
Discussion
Initial sensitivity to the effects of alcohol has been associated, in the ALSPAC sample (Schuckit et al., 2008a; Schuckit et al., 2008b) among others, with later alcohol misuse and problems, such that individuals less sensitive to alcohol when they begin drinking are at higher risk of later misuse. Evidence from twin and family studies, alongside preliminary findings from gene identification efforts, has suggested that the association may be due in part to genetic influences on sensitivity. In the current study, we meta-analyzed GWAS of initial sensitivity to alcohol, using the first 5 times drinking Self-Rating of the Effects of Alcohol (SRE) scale in two large, population-based samples. Analyses yielded few genome-wide significant findings, and PRS were not consistently associated with SRE in two independent samples. Our limited positive results came from aggregate tests, which indicated moderate heritability (overall h2SNP=0.19, SE=0.10) and support for a role of genes involved in hormone signaling. Initial alcohol sensitivity may be more prominently environmentally influenced than previously thought. However, studies of other behavioral outcomes with heritable components have yielded null results until much larger sample sizes were amassed (e.g., (Wray et al., 2018)). Follow-up is warranted, preferably using samples assessed during adolescence, contemporaneous with initial alcohol use. Furthermore, assessment of multiple ancestry groups is critical for clarifying the extent to which phenotypic differences are influenced by genetic factors.
Although no marker met genome-wide significance criteria, this is not entirely unexpected given evidence that substantially larger sample sizes may be necessary to reliably identify loci of small effect in complex traits (Bacanu and Kendler, 2017; Sullivan et al., 2017). Suggestive loci localize to several genes of interest. For example, SLC6A11 is a GABA transporter preferentially expressed in brain (Fagerberg et al., 2014); variation in this gene has been associated with intellectual and behavioral aberrations (Dikow et al., 2014) and resistance to epilepsy pharmacotherapy (Kim et al., 2011). Given the role of the GABAergic system in alcohol response and sensitization (Camarini and Pautassi, 2016; Koob, 2013), the biological plausibility of SLC6A11 is compelling. While other GABAergic genes involved in alcohol-relevant processes had suggestive p-values (e.g., GABARAP, p=0.003; GABRB3, p=0.001), these did not survive a multiple testing correction. Genes implicated in recent large GWAS of alcohol-related outcomes (Clarke et al., 2017; Jorgenson et al., 2017; Sanchez-Roige et al., 2017; Schumann et al., 2016) were also not supported. Indeed, no locus implicated by lead SNPs or gene-based analyses has been previously associated with alcohol use/misuse; these novel candidates require further investigation to determine the mechanisms by which they may influence alcohol sensitivity. However, the dearth of loci meeting strict correction thresholds prevents extensive interpretation of the current findings.
As indicated by CADD scores, lead SNPs are overall not predicted to be deleterious. Only rs145005509 has a CADD score >10; this SNP is intronic to a predicted locus and upstream of an open reading frame, thus its functional significance is unclear. Importantly, we evaluated only common alleles, which are relatively infrequently deleterious. Perhaps more interestingly, lead SNPs are not predicted to have clear regulatory functions, in contrast with findings for schizophrenia (Roussos et al., 2014), major depression (Wray and Sullivan, 2017), and nicotine dependence (Zanger and Schwab, 2013). RegulomeDB annotations were only obtained for lead SNPs through the FUMA pipeline, leaving open the possibility that less strongly implicated markers have regulatory functions.
One gene set survived the multiple test correction threshold (regulation of intracellular steroid hormone reception signaling pathway), and scrutiny of the genes in that category (obtained from MSigDB) revealed a potentially interesting avenue for further inquiry: they include core clock genes involved in circadian rhythms and/or photoperiodism, which were among the top 10 most strongly implicated gene sets (Supplemental Table 2). Genes included in all three sets are CLOCK, CRY1, CRY2, and PER1; other clock genes are common to two of the three sets. CLOCK and PER1 have been associated with alcohol use disorders (Partonen, 2015), and there is evidence that clock genes may have a regulatory role in reward circuitry (Parekh et al., 2015). Furthermore, mice with various perturbations in clock genes exhibit aberrant alcohol-related phenotypes (Dong et al., 2011; Gamsby et al., 2013; Perreau-Lenz et al., 2009; Spanagel et al., 2005; Wang et al., 2012).
The heritability of SRE scores in ALSPAC was moderate (h2SNP=0.36) and differed significantly from 0, suggesting that aggregate genetic factors contribute substantially to initial alcohol sensitivity. However, the heritability estimates were effectively 0 for each S4S ancestry group. This pronounced difference may be due in part to assessment. ALSPAC participants were periodically assessed in the time frame during which they were likely to begin experimenting with alcohol: while 62% responded to SRE items in wave 1 (age ~15.5), the remainder had not used alcohol 5 or more times until a later assessment. In contrast, 79% of S4S participants’ reports were from wave 1 (age ~18.5) and it is likely that many were reporting on alcohol exposure several years in the past. This raises the possibility that the scores are quite sensitive to recall bias. Thus, it is unclear whether the null heritability estimates of SRE across S4S ancestry groups is due to a true absence of genetic influences on SRE in S4S, potential error introduced by retrospective reports, or other factors. We are further unable to determine whether ancestry-based differences in heritability exist.
Additional tests of aggregate genetic influences did not yield significant findings. The absence of association between PRS derived from meta-analysis and ALSPAC-specific SNP weights and SRE scores in independent samples could be due to assessment, i.e., recall bias within the older individuals in the samples. However, we cannot rule out the possibility that the signals from our GWAS were of insufficient precision for outcome prediction, or that non-genetic factors are simply more influential than genetic factors on initial alcohol sensitivity.
The analyses presented here suggest that genetic factors have a modest impact on initial sensitivity to alcohol, but are largely inconclusive with respect to underlying biology. This underscores the need for prospective assessments of large, diverse samples to clarify the biological mechanisms underlying alcohol sensitivity and how they may differ across ancestries. Prior evidence that low initial sensitivity is associated with later alcohol misuse (Barnett et al., 2014; Heath et al., 1999; Schuckit, 1994) suggests that SRE scores could be a useful risk indicator. Further elucidation of the biological processes impacting initial sensitivity is necessary, and could be accomplished in part by characterizing loci implicated in the current study in model systems to determine whether, and how, genetic manipulations effect ethanol sensitivity. Another avenue for potential research is the identification of specific environmental factors that account for the balance of phenotypic variance in SRE scores; examples may include diet, pubertal status, or psychological stressors with physiological consequences. While we understand a great deal about alcohol metabolism via alcohol dehydrogenases and other pathways (Lieber, 2005; Marshall and Chambers, 2005), the subjective experience of drinkers is likely influenced by a wider range of genetic factors, the identification of which is critical to developing a comprehensive model of risk.
Limitations
Despite this being the largest genetic study of initial alcohol sensitivity to date, it is possible that the retrospective SRE assessment in approximately half of the total sample compromised our statistical power to detect influential loci. Individuals in both samples for whom multiple waves of data were available generally reported increasing SRE scores in later assessments despite the reporting period being constant (i.e., the first five or so times they drank alcohol); this raises the possibility that current drinking experiences influence reporting of past sensitivity. This may have contributed to the discrepancy in h2SNP estimates across the ALSPAC and S4S samples. The moderate estimate in ALSPAC encourages us that genetic factors are, indeed, influential, though power analyses indicated <10% power to detect a h2SNP of 0.30 in the smallest S4S subgroup, AMR. The EUR samples were more adequately powered: In ALSPAC, we estimated 64% power to detect h2SNP of 0.20 and 99% power to detect h2SNP of 0.36 (the actual estimate), and 61% power to detect h2SNP of 0.30 the S4S EUR ancestry group. Despite the lack of genome-wide significant variants, the current report represents an important initial effort to improve our understanding of the biological underpinnings of alcohol sensitivity.
Genetic studies are frequently limited to samples of European ancestry, precluding opportunities to assess differential genetic effects across ancestry groups. Although this study included a diverse sample, the non-European groups were of modest sample size and we lacked sufficient power to directly test such effects. However, efforts to recruit diverse samples are increasing, and meta-analytic approaches will enable the current samples to be incorporated into larger analyses in the future. Results from such approaches raise issues regarding the incorporation of linkage disequilibrium in various secondary analyses; here, we elected to use European linkage disequilibrium for FUMA, as this is likely a conservative approach and is appropriate for the majority (>82%) of the sample, but other methods may be preferable. JEPEGMIX was designed to be robust to the inclusion of cosmopolitan samples.
Finally, genetic analyses have consistently benefitted from larger sample sizes. Here, we report initial progress toward the identification of genetic factors influencing alcohol sensitivity, but these efforts must be bolstered by combining data across samples to increase statistical power. Given its ease of use and evidence of validity, the SRE represents a potentially powerful tool to employ to that end. However, the SRE does not capture all dimensions of alcohol sensitivity, and the subjective nature of the measure introduces uncertainty into analyses that are sensitive to measurement error, as is the case for most complex behavioral traits. Studies of more objective measures, such as body sway or motor coordination, would complement studies employing the SRE.
In conclusion, we report evidence of modest genetic influences on initial sensitivity to alcohol. Suggestive loci have not been previously implicated in alcohol outcomes, suggesting that the biology of sensitivity is not entirely parallel to that of alcohol consumption or problems. However, gene set analysis supports a role for core clock genes in initial sensitivity. Assessment of sensitivity is likely superior when conducted temporally proximal to initial alcohol experimentation; ideally, future studies will involve diverse samples such as that included here. Further investigation of loci identified in the current study is warranted to determine their impact and optimally arrange them in a comprehensive model of risk for alcohol misuse.
Supplementary Material
Table 4.
Top 10 gene set-based results from FUMA.
| Full Set Name | N Genes | Beta | Beta SD | SE | P-value |
|---|---|---|---|---|---|
| GO bp:go regulation of intracellular steroid hormone receptor signaling pathway | 57 | 0.4570 | 0.0254 | 0.1000 | 2.51e-06 |
| Curated gene sets:dasu il6 signaling up | 58 | 0.4140 | 0.0232 | 0.1030 | 2.89e-05 |
| GO bp:go regulation of metal ion transport | 315 | 0.1770 | 0.0229 | 0.0444 | 3.41e-05 |
| Curated gene sets:kegg circadian rhythm mammal | 13 | 0.7670 | 0.0204 | 0.1930 | 3.47e-05 |
| GO bp:go regulation of cell proliferation involved in heart morphogenesis | 15 | 0.9090 | 0.0260 | 0.2300 | 4.00e-05 |
| GO bp:go cell cell recognition | 59 | 0.3640 | 0.0206 | 0.0943 | 5.73e-05 |
| GO bp:go protein alpha 1 2 demannosylation | 13 | 0.7530 | 0.0200 | 0.2040 | 1.13e-04 |
| GO bp:go protein demannosylation | 13 | 0.7530 | 0.0200 | 0.2040 | 1.13e-04 |
| GO bp:go positive regulation of hydrolase activity | 872 | 0.0995 | 0.0212 | 0.0271 | 1.22e-04 |
| GO bp:go photoperiodism | 23 | 0.5170 | 0.0183 | 0.1420 | 1.40e-04 |
GO=gene ontology; bp=biological process; SD=standard deviation; SE=standard error
Acknowledgements
We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We would like to thank the Virginia Commonwealth University students for making the S4S study a success, as well as the many VCU faculty, students, and staff who contributed to the design and implementation of the project.
Funding/Support: The UK Medical Research Council and Wellcome (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and ACE will serve as guarantor for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website. This research was specifically funded by NIH grants AA021399, AA018333, and AA022537. GWAS data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. The Spit for Science project is supported by P20AA017828, R37AA011408, K02AA018755, and P50AA022537 from the National Institute on Alcohol Abuse and Alcoholism; UL1TR000058, from the National Center for Advancing Translational Studies; and UL1RR031990 from the National Center for Research Resources and National Institutes of Health Roadmap for Medical Research. The UCSF study was supported by R01DA030976 and F31AA025516, in conjunction with funding from the State of California and the Ernest Gallo Clinic and Research Center for medical research on alcohol and substance abuse through the University of California at San Francisco. The authors have no conflicts of interest to report.
Funding: NIH grants AA021399, AA018333, AA022537, P20AA017828, R37AA011408, K02AA018755, P50AA022537, UL1TR000058, UL1RR031990, DA030976, AA025516. The UK Medical Research Council (MR/L022206/1) and Wellcome (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC.
This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
References
- Bacanu S- A & Kendler KS (2017) Method to estimate the approximate samples size that yield a certain number of significant GWAS signals in polygenic traits. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Barnett NP, Clerkin EM, Wood M, Monti PM, O’leary Tevyaw T, Corriveau D, Fingeret A & Kahler CW (2014) Description and predictors of positive and negative alcohol-related consequences in the first year of college. J Stud Alcohol Drugs 75:103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ & Brewer RD (2011) Economic costs of excessive alcohol consumption in the U.S., 2006. Am J Prev Med 41:516–24. [DOI] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S & Davey Smith G (2013) Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol 42:111–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM & Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Mccutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, Kuperman S, Nurnberger JI Jr., Salvatore JE, MA Schuckit, Bierut LJ, Foroud TM, Chan G, Hesselbrock M, Meyers JL, Edenberg HJ & Porjesz B (2017) Comparison of Parent, Peer, Psychiatric, and Cannabis Use Influences Across Stages of Offspring Alcohol Involvement: Evidence from the COGA Prospective Study. Alcohol Clin Exp Res 41:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Kivlahan DR, Mcdonell MB, Fihn SD & Bradley KA (1998) The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med 158:1789–95. [DOI] [PubMed] [Google Scholar]
- Camarini R & Pautassi RM (2016) Behavioral sensitization to ethanol: Neural basis and factors that influence its acquisition and expression. Brain Res Bull 125:53–78. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM & Lee JJ (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ & Mcintosh AM (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry 22:1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Meyers JL, Rose RJ, Kaprio J & Kendler KS (2011) Measures of Current Alcohol Consumption and Problems: Two Independent Twin Studies Suggest a Complex Genetic Architecture. Alcohol Clin Exp Res 35:2152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Nasim A, Edwards AC, Salvatore JE, Cho SB, Adkins A, Meyers J, Yan J, Cooke M, Clifford J, Goyal N, Halberstadt L, Ailstock K, Neale Z, Opalesky J, Hancock L, Donovan KK, Sun C, Riley B & Kendler KS (2014) Spit for Science: launching a longitudinal study of genetic and environmental influences on substance use and emotional health at a large US university. Front Genet 5:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikow N, Maas B, Karch S, Granzow M, Janssen JW, Jauch A, Hinderhofer K, Sutter C, Schubert-Bast S, Anderlid BM, Dallapiccola B, Van Der Aa N & Moog U (2014) 3p25.3 microdeletion of GABA transporters SLC6A1 and SLC6A11 results in intellectual disability, epilepsy and stereotypic behavior. Am J Med Genet A 164A:3061–8. [DOI] [PubMed] [Google Scholar]
- Dong L, Bilbao A, Laucht M, Henriksson R, Yakovleva T, Ridinger M, Desrivieres S, Clarke TK, Lourdusamy A, Smolka MN, Cichon S, Blomeyer D, Treutlein J, Perreau-Lenz S, Witt S, Leonardi-Essmann F, Wodarz N, Zill P, Soyka M, Albrecht U, Rietschel M, Lathrop M, Bakalkin G, Spanagel R & Schumann G (2011) Effects of the circadian rhythm gene period 1 (per1) on psychosocial stress-induced alcohol drinking. Am J Psychiatry 168:1090–8. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ (2007) The genetics of alcohol metabolism. Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research & Health 30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Heron J, Vladimirov V, Wolen AR, Adkins DE, Aliev F, Hickman M & Kendler KS (2017) The Rate of Change in Alcohol Misuse Across Adolescence is Heritable. Alcohol Clin Exp Res 41:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR, Schuckit MA & Wilhelmsen KC (2010) Genome-wide scan for self-rating of the effects of alcohol in American Indians. Psychiatr Genet 20:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euesden J, Lewis CM & O’reilly PF (2015) PRSice: Polygenic Risk Score software. Bioinformatics 31:1466–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, Von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F & Uhlen M (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics 13:397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemifar G, Hoggart CJ, Paternoster L, Kemp JP, Prokopenko I, Horikoshi M, Wright VJ, Tobias JH, Richmond S, Zhurov AI, Toma AM, Pouta A, Taanila A, Sipila K, Lahdesmaki R, Pillas D, Geller F, Feenstra B, Melbye M, Nohr EA, Ring SM, St Pourcain B, Timpson NJ, Davey Smith G, Jarvelin MR & Evans DM (2013) Genome-wide association study of primary tooth eruption identifies pleiotropic loci associated with height and craniofacial distances. Hum Mol Genet 22:3807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, Ring S, Nelson SM & Lawlor DA (2013) Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol 42:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamsby JJ, Templeton EL, Bonvini LA, Wang W, Loros JJ, Dunlap JC, Green AI & Gulick D (2013) The circadian Per1 and Per2 genes influence alcohol intake, reinforcement, and blood alcohol levels. Behav Brain Res 249:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N & Conde JG (2009) Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB & Martin NG (1999) Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med 29:1069–81. [DOI] [PubMed] [Google Scholar]
- Jorgenson E, Thai KK, Hoffmann TJ, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Mertens J, Weisner C & Choquet H (2017) Genetic contributors to variation in alcohol consumption vary by race/ethnicity in a large multi-ethnic genome-wide association study. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Kim MK, Cho YW, Kim YS, Kim WJ, Lee MG, Kim SE, Nam TS, Cho KH, Kim YO & Lee MC (2011) Association of a synonymous GAT3 polymorphism with antiepileptic drug pharmacoresistance. J Hum Genet 56:640–6. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM & Shendure J (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF (2013) Theoretical frameworks and mechanistic aspects of alcohol addiction: alcohol addiction as a reward deficit disorder. Curr Top Behav Neurosci 13:3–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Williamson VS, Bigdeli TB, Riley BP, Webb BT, Fanous AH, Kendler KS, Vladimirov VI & Bacanu SA (2016) JEPEGMIX: gene-level joint analysis of functional SNPs in cosmopolitan cohorts. Bioinformatics 32:295–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber CS 2005. Alcohol Metabolism: General Aspects In: PREEDY VR & WATSON RR (eds.) Comprehensive Handbook of Alcohol Related Pathology. San Diego, CA: Elsevier Academic Press. [Google Scholar]
- Lind PA, Macgregor S, Montgomery GW, Heath AC, Martin NG & Whitfield JB (2008) Effects of GABRA2 variation on physiological, psychomotor and subjective responses in the alcohol challenge twin study. Twin Res Hum Genet 11:174–82. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, Mcvean G & Donnelly P (2007) A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 39:906–13. [DOI] [PubMed] [Google Scholar]
- Marshall SJ & Chambers GK 2005. Genetic Aspects of Alcohol Metabolism: An Overview In: PREEDY VR & WATSON RR (eds.) Comprehensive Handbook of Alcohol Related Pathology. San Diego, CA: Elsevier Academic Press. [Google Scholar]
- Parekh PK, Ozburn AR & Mcclung CA (2015) Circadian clock genes: effects on dopamine, reward and addiction. Alcohol 49:341–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T (2015) Clock genes in human alcohol abuse and comorbid conditions. Alcohol 49:359–65. [DOI] [PubMed] [Google Scholar]
- Perreau-Lenz S, Zghoul T, De Fonseca FR, Spanagel R & Bilbao A (2009) Circadian regulation of central ethanol sensitivity by the mPer2 gene. Addict Biol 14:253–9. [DOI] [PubMed] [Google Scholar]
- Peterson RE, Edwards AC, Bacanu SA, Dick DM, Kendler KS & Webb BT (2017) The utility of empirically assigning ancestry groups in cross-population genetic studies of addiction. Am J Addict 26:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Mitchell AC, Voloudakis G, Fullard JF, Pothula VM, Tsang J, Stahl EA, Georgakopoulos A, Ruderfer DM, Charney A, Okada Y, Siminovitch KA, Worthington J, Padyukov L, Klareskog L, Gregersen PK, Plenge RM, Raychaudhuri S, Fromer M, Purcell SM, Brennand KJ, Robakis NK, Schadt EE, Akbarian S & Sklar P (2014) A role for noncoding variation in schizophrenia. Cell Rep 9:1417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore JE, Thomas NS, Cho SB, Adkins A, Kendler KS & Dick DM (2016) The role of romantic relationship status in pathways of risk for emerging adult alcohol use. Psychol Addict Behav 30:335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Fontanillas P, Elson SL, Andme Research T, Gray JC, De Wit H, Davis LK, Mackillop J & Palmer AA (2017) Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA (1980) Self-rating of alcohol intoxication by young men with and without family histories of alcoholism. J Stud Alcohol 41:242–9. [DOI] [PubMed] [Google Scholar]
- Schuckit MA (1984) Subjective responses to alcohol in sons of alcoholics and control subjects. Arch Gen Psychiatry 41:879–84. [DOI] [PubMed] [Google Scholar]
- Schuckit MA (1994) Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry 151:184–9. [DOI] [PubMed] [Google Scholar]
- Schuckit MA (2018) A Critical Review of Methods and Results in the Search for Genetic Contributors to Alcohol Sensitivity. Alcohol Clin Exp Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A & Foroud T (2001) A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res 25:323–9. [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko GP & Isacescu V (2003) Level of response to alcohol measured on the self-rating of the effects of alcohol questionnaire in a group of 40-year-old women. Am J Drug Alcohol Abuse 29:191–201. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Danko GP, Pierson J, Hesselbrock V, Bucholz KK, Kramer J, Kuperman S, Dietiker C, Brandon R & Chan G (2007) The ability of the Self-Rating of the Effects of Alcohol (SRE) Scale to predict alcohol-related outcomes five years later. J Stud Alcohol Drugs 68:371–8. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Kalmijn J, Tsuang J, Hesselbrock V & Bucholz K (2000) Response to alcohol in daughters of alcoholics: a pilot study and a comparison with sons of alcoholics. Alcohol Alcohol 35:242–8. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL & Tipp JE (1997) The Self-Rating of the Effects of alcohol (SRE) form as a retrospective measure of the risk for alcoholism. Addiction 92:979–88. [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim R, Heron J, Horwood J, Davis JM, Hibbeln JR & Team AS (2008a) The performance of elements of a ‘level of response to alcohol’-based model of drinking behaviors in 13-year-olds. Addiction 103:1786–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL, Trim RS, Heron J, Horwood J, Davis J, Hibbeln J & Team AS (2008b) The self-rating of the effects of alcohol questionnaire as a predictor of alcohol-related outcomes in 12-year-old subjects. Alcohol Alcohol 43:641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Tsuang JW, Anthenelli RM, Tipp JE & Nurnberger JI Jr. (1996) Alcohol challenges in young men from alcoholic pedigrees and control families: a report from the COGA project. J Stud Alcohol 57:368–77. [DOI] [PubMed] [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, De Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, De Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, Mccarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, Van Der Harst P, Van Der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, et al. (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A 108:7119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Segura Lepe M, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hallfors J, Harris SE, Hieber S, Hofer E, Hottenga JJ, Johansson A, Joshi PK, Kaartinen N, Laitinen J, Lemaitre R, Loukola A, Luan J, Lyytikainen LP, Mangino M, Manichaikul A, Mbarek H, Milaneschi Y, Moayyeri A, Mukamal K, Nelson C, Nettleton J, Partinen E, Rawal R, Robino A, Rose L, Sala C, Satoh T, Schmidt R, Schraut K, Scott R, Smith AV, Starr JM, Teumer A, Trompet S, Uitterlinden AG, Venturini C, Vergnaud AC, Verweij N, Vitart V, Vuckovic D, Wedenoja J, Yengo L, Yu B, Zhang W, Zhao JH, Boomsma DI, Chambers J, Chasman DI, Daniela T, De Geus E, Deary I, Eriksson JG, Esko T, Eulenburg V, Franco OH, Froguel P, Gieger C, Grabe HJ, Gudnason V, Gyllensten U, Harris TB, Hartikainen AL, Heath AC, Hocking L, Hofman A, Huth C, Jarvelin MR, Jukema JW, Kaprio J, Kooner JS, Kutalik Z, Lahti J, Langenberg C, Lehtimaki T, Liu Y, Madden PA, Martin N, Morrison A, Penninx B, Pirastu N, Psaty B, Raitakari O, et al. (2016) KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R, Pendyala G, Abarca C, Zghoul T, Sanchis-Segura C, Magnone MC, Lascorz J, Depner M, Holzberg D, Soyka M, Schreiber S, Matsuda F, Lathrop M, Schumann G & Albrecht U (2005) The clock gene Per2 influences the glutamatergic system and modulates alcohol consumption. Nat Med 11:35–42. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES & Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, Mathews CA, Nievergelt CM, Smoller JW, O’donovan MC & Psychiatric Genomics C (2017) Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry appiajp201717030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trela CJ, Piasecki TM, Bartholow BD, Heath AC & Sher KJ (2016) The natural expression of individual differences in self-reported level of response to alcohol during ecologically assessed drinking episodes. Psychopharmacology (Berl) 233:2185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B, Neale MC & Kendler KS (2015) The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 45:1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten C, Seaton KL, Feiler HS & Wilhelmsen KC (2004) The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcohol Clin Exp Res 28:1509–16. [DOI] [PubMed] [Google Scholar]
- Wang X, Mozhui K, Li Z, Mulligan MK, Ingels JF, Zhou X, Hori RT, Chen H, Cook MN, Williams RW & Lu L (2012) A promoter polymorphism in the Per3 gene is associated with alcohol and stress response. Transl Psychiatry 2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, Van Bochoven A & Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BT, Edwards AC, Wolen AR, Salvatore JE, Aliev F, Riley BP, Sun C, Williamson VS, Kitchens JN, Pedersen K, Adkins A, Cooke ME, Savage JE, Neale Z, Cho SB, Dick DM & Kendler KS (2017) Molecular Genetic Influences on Normative and Problematic Alcohol Use in a Population-Based Sample of College Students. Front Genet 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y & Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2014. Global status report on alcohol and health 2014. Luxembourg: World Health Organization. [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu SA, Baekvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschon HN, Bybjerg-Grauholm J, Cai N, Castelao E, Christensen JH, Clarke TK, Coleman JIR, Colodro-Conde L, Couvy-Duchesne B, Craddock N, Crawford GE, Crowley CA, Dashti HS, Davies G, Deary IJ, Degenhardt F, Derks EM, Direk N, Dolan CV, Dunn EC, Eley TC, Eriksson N, Escott-Price V, Kiadeh FHF, Finucane HK, Forstner AJ, Frank J, Gaspar HA, Gill M, Giusti-Rodriguez P, Goes FS, Gordon SD, Grove J, Hall LS, Hannon E, Hansen CS, Hansen TF, Herms S, Hickie IB, Hoffmann P, Homuth G, Horn C, Hottenga JJ, Hougaard DM, Hu M, Hyde CL, Ising M, Jansen R, Jin F, Jorgenson E, Knowles JA, Kohane IS, Kraft J, Kretzschmar WW, Krogh J, Kutalik Z, Lane JM, Li Y, Li Y, Lind PA, Liu X, Lu L, Macintyre DJ, Mackinnon DF, Maier RM, Maier W, Marchini J, Mbarek H, Mcgrath P, Mcguffin P, Medland SE, Mehta D, Middeldorp CM, Mihailov E, Milaneschi Y, Milani L, Mill J, Mondimore FM, Montgomery GW, Mostafavi S, Mullins N, Nauck M, Ng B, et al. (2018) Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR & Sullivan PF (2017) Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME & Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM & Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–41. [DOI] [PubMed] [Google Scholar]
- Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, Hemani G, Tansey K, Laurin C, Early G, Lifecourse Epidemiology Eczema C, Pourcain BS, Warrington NM, Finucane HK, Price AL, Bulik-Sullivan BK, Anttila V, Paternoster L, Gaunt TR, Evans DM & Neale BM (2017) LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33:272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.