Abstract
Background
Gene fusions and fusion products have been proven to be ideal biomarkers and drug targets for cancer. Even though a comprehensive study of cervical cancer has been conducted as part of the Cancer Genome Atlas (TCGA) project, few recurrent gene fusions have been found, and none above 3% of frequency.
Methods
We believe that chimeric fusion RNAs generated by intergenic splicing represent a new repertoire of biomarkers and/or therapeutic targets. However, they would be missed when only genome sequences and fusions at DNA level are considered. We performed extensive data mining for chimeric RNAs using both our and TCGA cervical cancer RNA-Seq datasets. Multiple criteria were applied. We analyzed the landscape of chimeric RNAs at various levels, and from different angles.
Findings
The chimeric RNA landscape changed as different filters were applied. 15 highly frequent (>10%) chimeric RNAs were identified. LHX6-NDUFA8 was detected exclusively in cervical cancer tissues and Pap smears, but not in normal controls. Mechanistically, it is not due to interstitial deletion, but a product of cis-splicing between adjacent genes. Silencing of another recurrent chimera, SLC2A11-MIF, resulted in cell cycle arrest and reduced cellular proliferation. This effect is unique to the chimera, and not shared by the two parental genes.
Interpretation
Highly frequent chimeric RNAs are present in cervical cancers. They can be formed by intergenic splicing. Some have clear implications as potential biomarkers, or for shedding new light on the biology of the disease.
Fund
Stand Up To Cancer and the National Science Foundation of China.
Keywords: Chimeric RNA, Gene fusion, Bioinformatics, RNA-Seq, Cervical cancer
Research in context.
Evidence before this study
Few recurrent gene fusions have been reported in cervical cancer, and none of them were above 3% of frequency. We believe that chimeric fusion RNAs generated by intergenic splicing represent a new repertoire of biomarkers and/or therapeutic targets. However, they would be missed when only the genome sequences and gene fusions at DNA level are considered.
Added value of this study
Here we report 15 highly frequent (>10%) chimeric RNAs identified by analyzing RNA-Seq data. One of them is exclusively detected in cervical cancer tissues (>60%) and Pap smears (>40%), but not in healthy controls. Another one regulates cervical cancer cellular proliferation, a function not found in either of the parental genes.
Implications of all the available evidence
Taken together, we have shown that highly frequent chimeric RNAs are present in cervical cancers. Instead of chromosomal rearrangement at DNA level, they can be formed by intergenic splicing at RNA level. Some have clear implications as potential biomarkers, or for shedding new light on the biology of the disease.
Alt-text: Unlabelled Box
1. Introduction
Cervical cancer remains one of the leading causes of cancer-related deaths. It accounts for 10-15% of cancer-related deaths in women worldwide, with approximately 527,600 new cases and 265,700 deaths annually [[1], [2], [3]]. Although preventable, it is still the second most common cancer among women. Standard treatment is curative in more than 90% of women during the early stages, but for stage IIIb and above this rate drops to 50% or less [4]. Hence, early accurate detection, better management, and discovery of new therapeutic targets are needed in order to promote early diagnosis and improve prognosis in cervical cancer patients..
Chromosomal translocations and genes fusions are common in human cancers, especially in the subtypes of sarcomas and hematological cancers. The discovery of novel chromosomal translocations and gene fusions has been revolutionized by the rise of next-generation sequencing, advances in bioinformatics, and an increased capacity for large-scale computational biology. However, apart from gene fusions involving the ETS family of transcription factors in prostate cancers [5], highly recurrent gene fusions are hardly found in other common solid cancers. In the case of cervical cancer, a recent TCGA study revealed few recurrent gene fusions, including four cases having ZC3H7A–BCAR, three cases with CPSF6–C9orf3, two cases with ARL8B–ITPR1, and two with MYH9–TXN2 fusions, out of 178 samples [6]. On the other hand, chimeric fusion transcripts are being discovered in various cells and tissues, and at least some are shown to be the products of intergenic splicing instead of chromosomal rearrangement [[7], [8], [9], [10], [11]]. In this study, we analyzed a total of 212 cervical cancer RNA-Seq datasets, and some matched normal datasets, from which we characterized the landscape of chimeric RNAs at multiple levels, and validated 15 highly frequent chimeras. We then focused on two of them. LHX6-NDUFA8 was detected in about half of the cervical cancer tissue samples, as well as in the Pap smears of cervical cancer and cervical intraepithelial neoplasia (CIN) patients, making it a potential candidate biomarker. SLC2A11-MIF is critical for cancer cell growth, of which the effect is unique to the fusion, thus shedding new light on the biology of the cancer.
2. Materials and methods
2.1. Cell culture
Cervical cancer cell lines HeLa, SiHa, Ca Ski, and C33A were procured from ATCC. Cells were cultured in DMEM (Dulbecco's modified Eagle's medium) (Sigma) plus 10% FBS (Fetal Bovine Serum) (Invitrogen, Gaithersburg, MD). The cells were maintained in an incubator at 37°C, in a humidified 5% CO2 atmosphere. Cell culture development was assessed under an inverted phase microscope.
2.2. Clinical samples
Cervical cancer tissues, cervical intraepithelial neoplasia III (CIN III) and the normal cervical tissue samples were collected from Tongji hospital of Huazhong Science and Technology University, under a protocol approved by Huazhong Science and Technology University Institutional Review Boar. Written informed consents were obtained from the participants. Surgically resected specimens were snap frozen in liquid nitrogen and stored at -80°C.
2.3. Datasets
201 cervical cancer and some matched normal RNA-sequencing data were downloaded from the TCGA website. Our 11 RNA-sequencing dataset has been reported before [12].
2.4. RNA extraction, PCR, and qRT-PCR
Clinical samples were pulverized in liquid nitrogen. RNAs from both cell lines and clinical samples were extracted using TRIzol reagent (LifeTechnologies), following the manufacturer’s instruction. All of the RNA samples used in this study were treated with DNase I, followed by standard Reverse Transcription using AMV RT (NEB). PCR and qRT-PCR were performed as described [13,14]. Real-time PCR experiment was conducted using the ABI StepOne Plus system (Life Technologies) with Absolute Blue QPCR mix (Thermal Fisher, AB-4322). Primers are listed in Table S3. Following PCR and gel electrophoresis, all purified bands were submitted for Sanger sequencing.
2.5. Identification of chimeric fusion transcripts.
Chimeric RNA candidates were identified by the SOAPfuse algorithm(http://soap.genomics.org.cn/soapfuse.html ) as described before [15].
2.6. MTT assay and cell counting
Cells were plated on 96-well plates with 1,000 cells per well and transfected with various siRNAs after 24 hrs. Cell viability was measured by MTT (Sigma) at time points of day0 (after transfection), day1, day2, and day3 as described previously [16].
2.7. RNA interference
Cells were transfected with siRNA using RNAiMax (Invitrogen), following the manufacturer's instructions. All siRNAs were purchased from Invitrogen. Their sequences are as follows:
si-Con, CGUACGCGGAAUACUUCGA
SLC2A11 si: GGUAAUUAACUGACAGAAA
MIF1 si: GCGCAGAACCGCUCCUACA
S_M si1 : UGCACCGCGAUGUAACUAA
S_M si2: UUAGUUACAUCGCGGUGCA
2.8. Plasmid construction and transfection
The coding sequence of SLC2A11-MIF was amplified by PCR from Hela cDNA, and cloned into pQCXI-CMV (Clontech). The constructed vector or empty vector control was then transfected into Hela cells and Ca Ski cells using Lipofectamine 3000 (Invitrogen) in compliance with the manufacturer's guidelines.
2.9. Microarray
The Hela cells were maintained in DMEM high glucose medium with 10% FBS and 1% Pen/Strep at 37°C with 5% CO2. siRNAs against the fusion RNA SLC2A11-MIF, wild-type SLC2A11, MIF, or negative control siRNA were transfected into Hela cells. Cells were harvested 48hrs after siRNAs transfection. RNA was then extracted for microarray analyses in Macrogen (Korea) on the Illumina Human HT-22 v4 platform. QCs of all samples were analyzed using an Agilent Technologies 2100 Bioanalyzer, and given an RNA Integrity Number (RIN) value equal to or greater than 8. The differential expression levels were normalized to those in the siRNA negative control group.
2.10. Statistical analyses
The LHX6-NDUFA8 expression of clinical samples was calculated by chi-squared test or Fisher's exact test depending on the sample size and expected frequency, or Mann-Whitney U test. The other quantitative results were reported as the mean ± standard error of the mean (SEM). Statistical comparisons between groups were analyzed using the unpaired/two-tailed Student’s t-test, Mann-Whitney test, one-way ANOVA, or Kruskal-Wallis test. GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA) was used for statistical analyses. For all analyses, p < 0.05 was considered indicative of statistical significance. P values were labeled in figures as follows: *p<0.01, **p<0.001, ***P<0.0001.
2.11. Data access
Raw and processed microarray data are available at GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE114127.
3. Results
3.1. Discovery of chimeric RNAs in cervical cancer
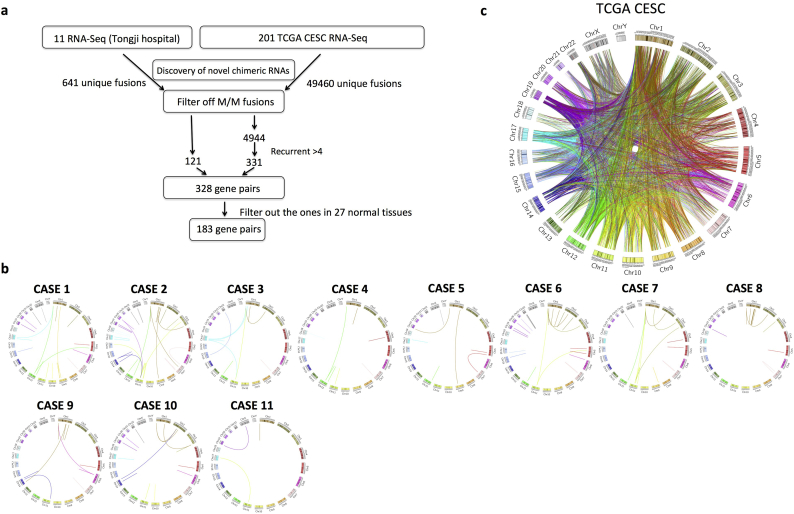
To identify recurrent chimeric RNAs in cervical cancer, we combined data from two sources: our RNA-sequencing of samples from 11 cervical cancer patients treated at Tongji Hospital [12], and the raw RNA-Seq data from TCGA cervical cancer study (CESC), which at the time of analysis contained 198 cervical cancer cases and three normal margins. We then used the bioinformatics software tool, SOAPfuse [15] to identify candidate chimeric transcripts. A total of 641 and 49,460 unique chimeric fusion transcripts were found in the two datasets respectively. We categorized the fusions according to the junction position relative to the exon of the parental genes: both sides being known exon/intron boundaries (E/E), both sides falling into the middle of exons (M/M), one side being exon/intron boundary and the other not (E/M or M/E). Based on our previous study, the fusion transcripts with at least one side of junction site being a known exon/intron boundary have much higher validation rates [17]. Therefore, in order to reduce the false discovery rate, we filtered out the M/M fusions. Furthermore, we aim to uncover frequent fusion RNAs, thus decided to focus on the ones that could be detected in at least five samples. After applying these filters, 425 unique fusions were uncovered, involving 328 gene pairs. We then examined this list of gene pairs against the list we previously generated from the analysis of around 300 RNA-Seq libraries covering 27 normal tissues [18]. 183 unique gene pairs were found only in the cervical cancer samples (Fig. 1A). Circos plots were used to depict the chimeric RNAs in 11 cases of our RNA-Seq data (Fig. 1B), and the ones in the TCGA CESC study (Fig. 1C).
Fig. 1.
Discovery of chimeric RNAs in cervical cancer.
(A) The pipeline for discovering cervical cancer chimeric RNAs. Two sources of RNA-Seq data were used: our own sequencing results and TCGA cervical cancer sequencing data. After filtering out “M/M” fusions, and setting the recurrent cutoff at five, the remaining 328 gene pairs were further narrowed down to 183 by their absence in 27 normal tissues. (B) Circos plot depicting chimeric RNAs from 11 of our own samples. Lines denote the chimeric RNAs connecting two parental genes. (C) Circos plot depicting all the chimeric fusion RNAs uncovered from the TCGA cervical cancer study.
3.2. The landscape of chimeric RNAs and parental genes in cervical cancer
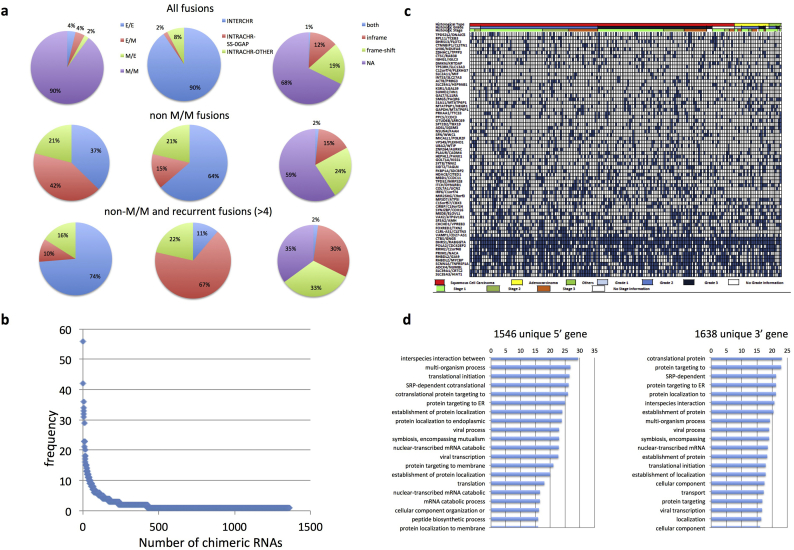
We then examined the landscape of these fusion RNAs from three angles, and at three different levels (Fig. 2A). First, as described above, based on the junction position relative to the exon of the parental genes, we categorized the chimeric RNAs into E/E, E/M or M/E, and M/M groups. Among all the fusions, the most prominent category was M/M fusions (90%), while E/E/ and E/M were about 4% each, and M/E only 2% (Fig. 2A). After we filtered out M/M, and less frequent fusions (<5), E/E fusions were significantly enriched (74%). Interestingly, M/E fusions became more abundant than E/M fusions in this population (16% vs. 10%).
Fig. 2.
The landscape of chimeric RNAs and their parental genes in cervical cancer.
(A) Distributions of chimeric RNAs from the TCGA data set. Chimeric RNAs are categorized based on their fusion junction position, fusion type, and fusion protein coding potential. When the criteria of “non-M/M”, and “recurrence” applied, more E/E, INTRACHR-SS-0GAP, and in-frame fusions were enriched. (B) The frequency of chimeric fusion RNAs detected in cervical cancer samples. (C) Integrative analysis of chimeric RNAs in TCGA cervical cancer cases. The most frequent chimeric RNAs are plotted here together with histological type, grade, and stage of the cervical cancer samples. (D) Gene ontology analyses of the 5′ and 3′ parental genes involved in non M/M fusion RNAs in cervical cancer. Plotted are statistical significance (-Log10(p-value)) of the top 20 terms.
We then characterized the fusions according to the chromosomal locations of their parental genes: parental genes located on different chromosomes (INTERCHR), neighboring genes transcribing the same strand (INTRACHR-SS-0GAP), and other fusions with parental genes on the same chromosome (INTRACHR-OTHER). For all of the fusions, INTERCHR is the most prominent group (90%). INTRACHR-SS-0GAP is the least common group (2%). However, as the M/M fusions were filtered out, the INTERCHR group shrunk (64%), and INTRACHR-SS-0GAP and INTRACHR-OTHER became more abundant (15% and 21% respectively). This trend became more obvious, when both “non M/M” and “recurrent fusion” filters were applied: INTRACHR-SS-0GAP became the largest group (67%), and INTERCHR became the smallest (11%).
Lastly, we categorized the fusions according to their reading frames: the known protein coding sequence of the 3’ gene uses a different reading frame than the 5’ gene (frame-shift); the known reading frame of the 3’ gene is the same as the 5’ gene (in-frame); no effect on the reading frame of the parental genes (NA) (this category includes fusion RNAs whose junction sequence fall into untranslated region or one or both parental genes is lncRNA). A very small population of fusions fell into the “both” category, which could be in-frame, or frame-shift depending on the alternative splicing isoforms of the parental genes. When all the fusions were examined, the number of NA is the largest (68%). Frame-shift fusions are more common than in-frame fusions (1.6 fold). After filtering out M/M fusions, the NA portion became smaller (59%), and the frame-shift fusions are still about 1.6 fold greater than the in-frame fusions. When both “non-M/M” and “recurrent” filters were used, the in-frame fusions were enriched, as the three groups became roughly the same size (35%, 33%, and 30%).
The majority of the chimeric RNAs were identified in one or two samples (Fig. 2B). We plotted the most frequent 68 chimeric RNAs against the histological type, grade, and stage of the cervical cancer samples (Fig. 2C). No obvious correlation was observed in regard to these clinical parameters for any of the chimeric RNAs. We searched gene ontology terms using Gorilla [19] for the parental genes involved in the non-MM fusion RNAs. Several terms related to viral processing, multi-organism interaction, and symbiosis were found to be enriched in the top 20 GO terms for both the 5’ gene and 3’ gene (Fig. 2D). For comparison, we analyzed RNA-Seq from 424 TCGA hepatocellular carcinoma (HCC) and 705 TCGA glioblastoma RNA-Seq datasets. More metabolic-related GO terms were enriched in HCC, together with viral processing, symbiosis, and interspecies interaction terms (Fig. S1). In contrast, no viral related terms were found in the glioblastoma RNA-Seq dataset (Fig. S2), consistent with the known involvement of viruses in both cervical cancer, and liver cancer, but not in gliomas.
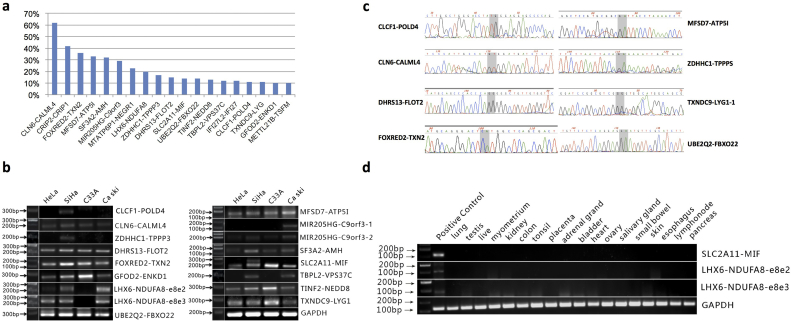
3.3. Validation of the highly frequent chimeric RNAs
We focused on the 19 recurrent chimeric RNAs that were detected in more than 10% of samples, but absent in the 27 normal tissues (Fig. 3A). Primers annealing to parental genes and flanking the fusion junction site were designed. 15 out of 19 chimeric RNAs were confirmed by RT-PCR and Sanger sequencing, with two forms for LHX6-NDUFA8 and MIR205HG-C9ORF3 (Fig. 3B and examples in Fig. 3C). We then examined the expression of the chimeric RNAs using a set of normal tissue RNA panels. Both forms of LHX6-NDUFA8 and SLC2A11-MIF were not found in any of the normal tissue samples (Fig. 3D). We focused on these fusion RNAs in the following study.
Fig. 3.
Validation of the highly frequent chimeric RNAs.
(A) The frequency of the19 recurrent chimeric RNAs (>10%) that are also absent in the 27 normal tissue dataset. (B) Gel image of RT-PCR product of the 15 candidate chimeric RNAs. The LHX6-NDUFA8 and MIR205HG-C9ORF3 fusions have two forms. (C) Examples of Sanger sequencing confirmation. Two base pairs at the fusion junction are highlighted in gray. (D) RT-PCR for fusion RNAs in normal tissue panels. Both forms of LHX6-NDUFA8 and SLC2A11-MIF were negative in these normal tissues. Positive RNA was extracted from Hela cells.
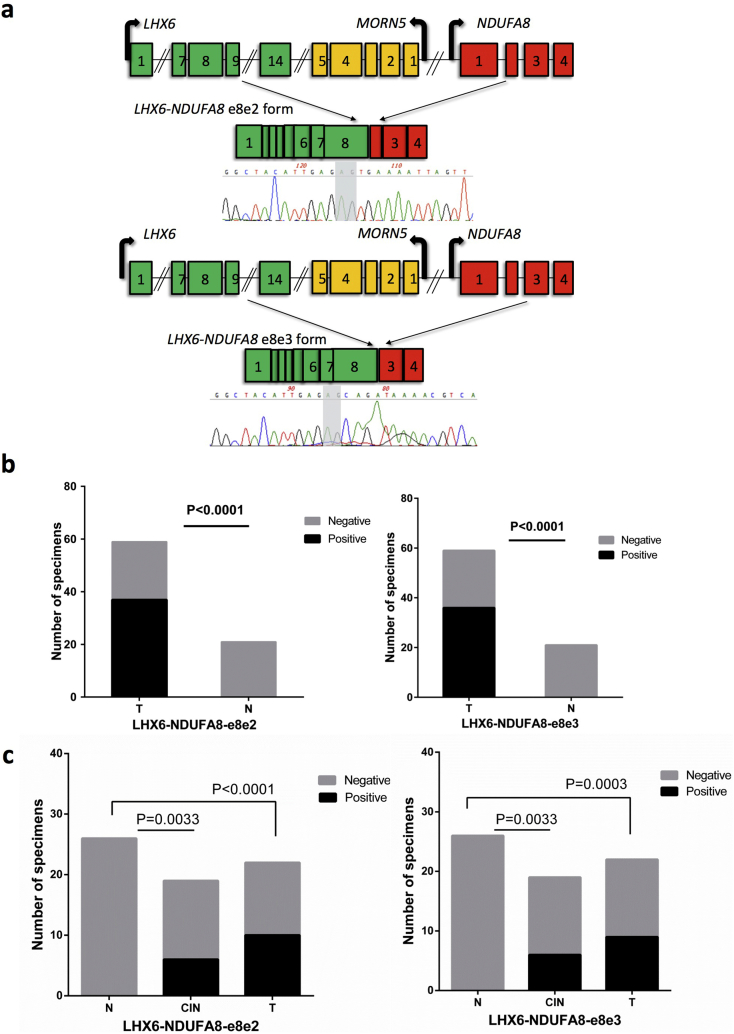
3.4. LHX6-NDUFA8 is positive in a high percentage of cervical cancer tissues and Pap smear samples
The LHX6-NDUFA8 fusion has two different isoforms: LHX6-NDUFA8-e8e2 and LHX6-NDUFA8-e8e3 (Fig. 4A). We first examined the frequency of both isoforms in cervical cancer tissues. LHX6-NDUFA8-e8e2 and LHX6-NDUFA8-e8e3 were detected in 37 (62.71%) and 36 of 59 cases (61.02%) respectively. In contrast, neither form was detected in any of the 21 non-cancer cervical tissues (Fig. 4B). Neither the detection nor the expression level of the e8e2 form correlates with lymph node metastasis, differentiation, FIGO stage, or pathology. However, the detection of the e8e3 form appears to be more enriched in squamous cell carcinomas than in adenocarcinomas (p=0.009, calculated by chi-squared test) (Table 1).
Fig. 4.
Both forms of LHX6-NDUFA8 are positive in a high percentage of cervical cancers.
(A) Structures of the two forms of the LHX6-NDUFA8 fusion and Sanger sequencing validation. Two base pairs at the fusion junction were highlighted in gray. (B) Both forms are specifically detected by RT-PCR in cervical cancer tissues (T), but not in control cervical tissues (N). (C) The RT-PCR detection rate of the two fusions in Pap smear samples. The fusions were detected in about half of the Pap smears from cervical cancer patients (T), about 1/3 of CIN III samples, but not from non-cancer patients (N), P value was calculated by chi-squared test.
Table 1.
The correlation between LHX6-NDUFA8 expression and clinical parameters for cervical cancer tissue samples
| LHX6-NDUFA8-e8e2 gene expression |
LHX6-NDUFA8-e8e3 gene expression |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable | No.ofpatients | Negative |
Positive |
Median of LHX6-NDUFA8-1 |
Negative |
Positive |
Median of LHX6-NDUFA8-2 |
||||||||||||
| No. | % | No. | % | aP value | relative expression | bP value | No. | % | No. | % | aP value | relative expression | bP value | ||||||
| Cervical cancer patients | |||||||||||||||||||
| Lympho node metastasis | |||||||||||||||||||
| Negative | 39 | 16 | 41.03 | 23 | 58.97 | 0.4071 | 0.2771 | 0.2417 | 17 | 43.58 | 22 | 56.41 | 0.3110 | 0.4113 | 0.5923 | ||||
| Positive | 20 | 6 | 30.00 | 14 | 70.00 | 0.4567 | 6 | 30.00 | 14 | 70.00 | 0.7476 | ||||||||
| Differentiation | |||||||||||||||||||
| Well or Moderately | 29 | 12 | 41.38 | 17 | 58.62 | 0.1337 | 0.3040 | 0.7436 | 12 | 41.38 | 17 | 58.62 | 0.1337 | 0.4862 | 0.6750 | ||||
| Poorly | 23 | 5 | 21.74 | 18 | 78.26 | 0.4063 | 5 | 21.74 | 18 | 78.26 | 0.4012 | ||||||||
| FIGO Stage | |||||||||||||||||||
| IA-IB | 29 | 10 | 34.48 | 19 | 65.52 | 0.6613 | 0.2744 | 0.1755 | 12 | 41.38 | 17 | 58.62 | 0.7106 | 0.4113 | 0.1214 | ||||
| IIA-III | 30 | 12 | 40.00 | 18 | 60.00 | 0.4749 | 11 | 36.67 | 19 | 63.33 | 0.7808 | ||||||||
| Pathology | |||||||||||||||||||
| Squamous cell carcionma | 48 | 15 | 31.25 | 33 | 68.75 | 0.0625 | 0.3419 | 0.0795 | 14 | 29.17 | 34 | 70.83 | 0.009 | ||||||
| Adenocarccionma | 9 | 6 | 66.67 | 3 | 33.33 | 0.0709 | 7 | 77.78 | 2 | 22.22 | |||||||||
| Normal cervical tissue verus cancer | |||||||||||||||||||
| Normal cervical tissue | 21 | 21 | 100.00 | 0 | 0.00 | <0.0001 | 21 | 100.00 | 0 | 0.00 | <0.0001 | ||||||||
| Cervical cancer | 59 | 22 | 37.29 | 37 | 62.71 | 23 | 39.98 | 36 | 60.02 | ||||||||||
P: calculated by chi-squared test or Fisher's exact test (values were underlined).
P: calculated by Mann-Whitney U test.
In the cervical cancer screening setting, cytology screening based on Pap smear is much more practical and commonly used than cervical biopsy. To determine whether the fusion RNAs have the potential for use in screening for cervical intraepithelial neoplasia (CIN) and cervical cancer via the Pap smear test platform, we examined the expression of the two forms of LHX6-NDUFA8 in Pap smear samples. Here, both forms had similar positive detection rate, with e8e2 form in 10 of 22 cases (45.45%), and the e8e3 form in 9 of 22 cases (40.91%). Both forms were detected in 6 out of 19 cases of CIN III, but neither form was detected in 26 Pap smear samples from non-cancer patients (Fig. 4C). The detection of neither form had any correlation with lymph node metastasis, differentiation, FIGO stage, or pathology of the cervical cancer case (Table S2 and S3), suggesting that the expression of these two chimeric RNAs may be an early event in the tumorigenesis of a subset of cervical cancers.
3.5. LHX6-NDUFA8 is a product of cis-splicing between adjacent genes (cis-SAGe)
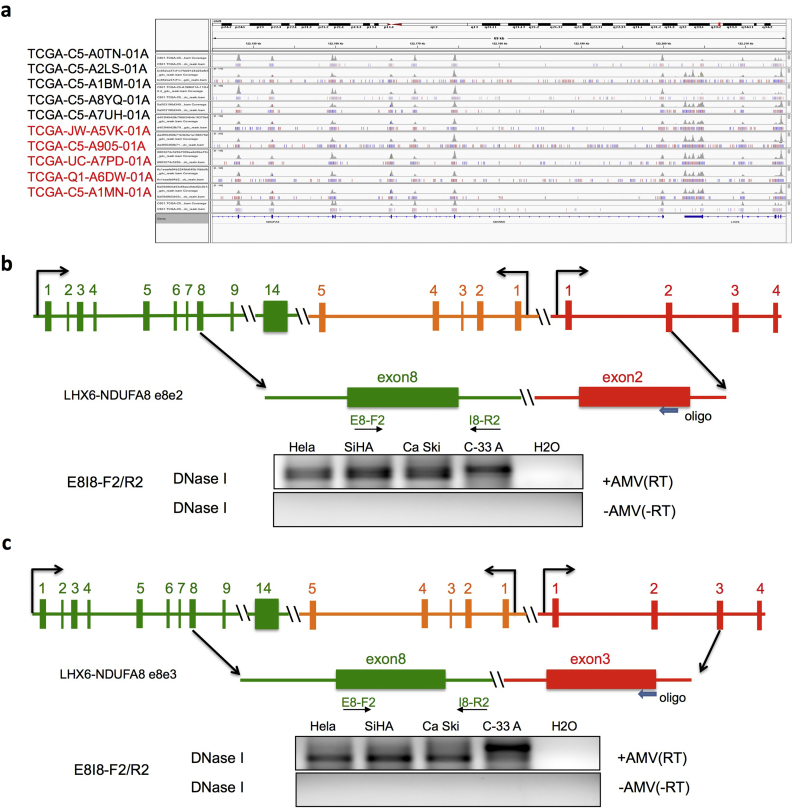
Both forms of LHX6-NDUFA8 were classified as INTRACHR-OTHER, as the two parental genes are located on the same chromosomal, and separated by a gene, MORN5. We first investigated whether LHX6-NDUFA8 chimeric RNAs are the product of interstitial deletion, which is the classic mechanism for gene fusions for this type of configuration in cancer. We examined whole-genome sequencing data from TCGA in the samples with or without the fusions. No copy number variation for the fragment covering the two parental genes and the middle gene MORN5 was found in either group, arguing against the mechanism of interstitial deletion (Fig. 5A).
Fig. 5.
LHX6-NDUFA8 fusions are the product of cis-splicing between adjacent genes.
(A) IGV view of the genomic region covering LHX6, NDUFA8 and the gene, MORN5 in between. Five fusion-positive (font red) and five fusion-negative (font black) cases were shown here. No deletion or copy number loss was observed in the region. (B) and (C) The two forms of the fusion involve the joining of the exon8 of LHX6 and the exon2 (B) or exon3 (C) of NDUFA8. Blocks represent exons. Lines represent introns or intergenic region. The arrowhead indicates the oligo used for reverse transcription. F and R primers anneal to exon8 and intron8 of LHX6 respectively. RNAs from four cervical cancer cell lines were first treated with DNaseI. They were then separated into two groups: with or without AMV RT enzyme. The correct product was only seen in the samples with AMV-RT enzyme.
Even though LHX6 and NDUFA8 are not immediate neighboring genes, the middle gene MORN5 transcribes on a different strand, making the fusion a possible candidate of cis-SAGe. To investigate, we designed an assay to detect the precursor read-through mRNA (Fig. 5C). In this experiment, a reverse primer annealing to exon2 or exon3 of NDUFA8 was used to perform reverse transcription. We then used a primer pair designed to amplify a fragment of cDNA covering exon8 and intron8 of LHX6. To eliminate DNA contamination, RNA was treated with DNaseI before the assay. To confirm that the signal was not due to remaining DNA contaminants, we included controls with no AMV-RT enzyme. We detected signals in four cervical cancer cell lines only in the presence of the AMV-RT enzyme (Fig. 5B and 5C). This confirms the presence of a precursor RNA transcribing from exon8 of LHX6 to exon2 or exon3 of NDUFA8. We, therefore, conclude that both forms of LHX6-NDUFA8 chimeric RNAs are products of cis-SAGe.
3.6. SLC2A11-MIF is crucial for cervical cancer cell proliferation
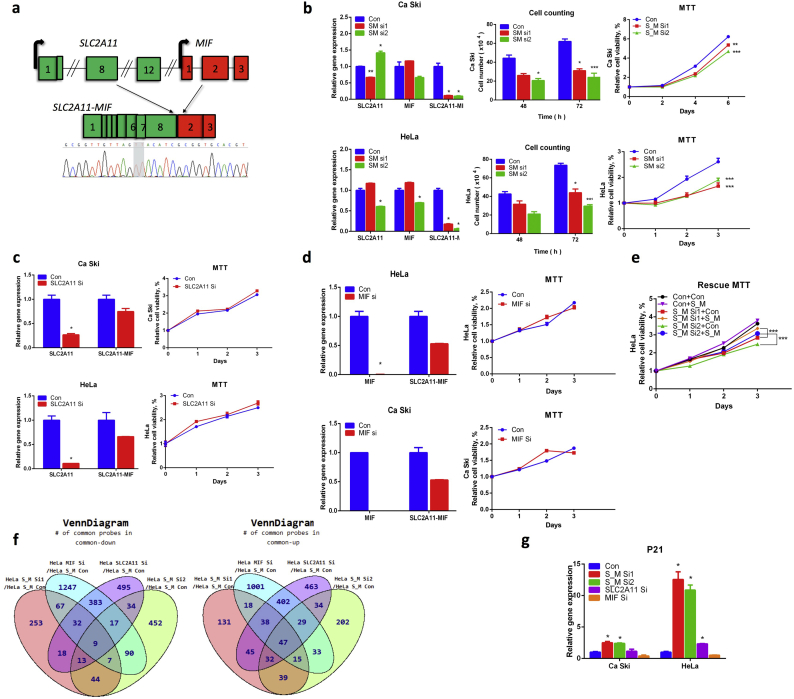
Similar to LHX6-NDUFA8, SLC2A11-MIF was not detected in the normal tissue panels (Fig. 3D). SLC2A11-MIF involves the joining of the 8th exon of SLC2A11 to the 2nd exon of MIF (Fig. 6A). We designed two siRNAs targeting the fusion junction site (S_M si1, and S_M si2). In two cervical cancer cell lines, Hela and Ca Ski, both effectively silenced the fusion transcripts, while having different effects on the parental genes (Fig. 6B). For instance, S_M si1 upregulated wild-type MIF expression, but S_M si2 downregulated it. When Ca Ski and Hela cells were transfected by the siRNAs, cellular proliferation was significantly reduced, evidenced by both cell counting and MTT assay (Fig. 6B). To confirm that the reduction in cell growth is due to the silencing of the fusion, we used siRNAs designed to silence the wild-type parental gene transcripts. Both dramatically silenced the parental transcripts. Notable, they also had some effect on the expression of the fusion transcripts, but to a much lesser extent than the S_M si1 and S_M si2. No obvious reduction in cellular growth was observed with these siRNAs (Fig. 6C and 6D). To further confirm that the reduction of the cellular growth is due to fusion RNA silencing, we performed rescue experiments. We infected Hela cells, which were transfected with siRNAs targeting the fusion, with virus expressing either the empty vector control construct, or a construct expressing SLC2A11-MIF. The fusion-expressing virus rescued the reduced cell viability in cells transfected with both siRNAs (Fig. 6E). HPV infection is responsible for the most majority of cervical cancers. Interestingly, SLC2A11-MIF was detected in C33A cells, which is HPV negative. When we silenced the fusion in C33A cells, obvious inhibition of cellular proliferation was also observed, similar to the HPV positive Hela and Ca Ski cells (Fig. S4).
Fig. 6.
SLC2A11-MIF is crucial for cervical cancer proliferation.
(A) Structure of the fusion. Blocks represent exons. Lines represent introns or intergenic region. Two base pairs at the fusion junction are highlighted in gray. (B) With siRNAs specific for the fusion (S_M Si1, and S_M Si2), proliferation in both Hela and Ca Ski cells was inhibited. qRT-PR measuring the efficiency of siRNA knocking down (left); cell counting assay (middle); MTT assay (right). P value was calculated by unpaired/two-tailed Student’s t-test. (C) An siRNA targeting the wild-type SLC2A11 (SLC2A11 Si) had no obvious inhibitory effect on cellular proliferation. (D) An siRNA targeting wild-type MIF1 (MIF Si) had no obvious inhibitory effect on cellular proliferation, either. P value was calculated by unpaired/two-tailed Student’s t-test. (E) The inhibitory effect caused by S_M Si1 and S_M Si2 can be rescued by introducing a fusion expression vector (S/M), but not an empty vector control (con). (F) Microarray analyses of Hela cells transfected with siRNAs targeting the fusion (S_M Si1, and S_M Si2), SLC2A11 (SLC2A11 Si), MIF (MIF Si), and siRNA control (S_M Con). Venn grams summarize the shared and unique targets that were up or downregulated by each transfection compared with control. (G) CDKN1A (p21) level was measured in both Ca Ski (left) and Hela (right) cells transfected with the five siRNAs. Fisher’s exact test was used. *p<0.01; **p<0.001; ***p<0.0001
To further investigate the functional mechanism of SLC2A11-MIF, we performed microarray analyses, comparing transcriptome profiles in the Hela cells transfected with siRNAs targeting the fusion and the siRNAs targeting the two wild-type parental genes, with that in the control siRNA transfected cells (Fig. 6F). We found a set of genes that are specifically up- or down-regulated in the two S_M siRNA groups, but not in the siRNAs targeting only the two wild-type parental genes. Top candidates were selected for validation via qRT-PCR. Examples are shown in Figures S5 and S6. However, the well-known targets for cervical cancer carcinogenesis, TP53 and RB levels were not consistently changed (Fig. S7). Among the candidates that were upregulated, CDKN1A (p21) was among the most dramatically changed, and its upregulation is specific to the fusion silencing. In both Hela and Ca Ski cells, silencing the fusion resulted in significant CDKN1A upregulation, whereas silencing the wild-type parental genes had no such effect (Fig. 6G).
4. Discussion
Gene fusions caused by chromosomal rearrangements are well-known cancer-causing genetic events, and are actively used in clinical cancer diagnosis. Some fusion products have also been shown to be effective targets of directed therapy [20,21]. Because of their potential as cancer-specific markers and therapeutic targets, gene fusions have been sought after ever since the report of the Philadelphia chromosome (translocation). However, in cervical cancer, few recurrent gene fusions have been reported, and their frequencies are rather low (<3%)(6). Even though gene fusions, and their fusion products, have been traditionally thought to be generated solely by chromosomal rearrangement, recent work on RNA trans-splicing [9,22,23] and intergenic cis-splicing [16,17,24] have defined a new paradigm for intergenic splicing processes, which can also generate fusion products. Such intergenically spliced chimeric RNAs represent a new repertoire of cancer biomarkers and/or therapeutic targets. In this study, we identified 15 highly frequent chimeric RNAs (>10%). Among them, LHX6-NDUFA8, is more frequently detected in both cervical cancer tissues and Pap smear samples than any other previously reported gene fusions. Instead of being a product of chromosomal rearrangement, this fusion RNA is a product of cis-SAGe.
We performed analyses on the landscape of cervical cancer chimeric RNAs on three levels, and from three angles. It seems that most of the chimeric RNAs are individualized, or only occurring in a small number of samples (<5). These less frequent chimeras also tend to be M/M fusions, and belong to the category of INTERCHR. Given the lower validation rate of these fusions [17], it is possible that a subset of them are false positives. However, since we do not have access to the original TCGA samples, we cannot formally test them.
Chimeric RNAs composed of exons from immediate neighboring genes transcribing from same strand (INTRA-SS-0GAP) are considered candidates of cis-SAGe fusion RNAs. In the case of LHX6-NDUFA8, it was originally grouped into the INTRACHR-OTHER category. However, even though the MORN5 gene sits in between LHX6 and NDUFA8, it is transcribed from the opposite strand, making the transcription reading through LHX6 and into NDUFA8 possible. Indeed, we demonstrated the presence of a primary transcript connecting the LHX6 and NDUFA8. These findings suggest that at least some cis-SAGe fusions may be wrongly clustered, and therefore missed. For instance, 39 out of the 425 recurrent fusion RNAs involve two genes transcribing from the same strand, and separated by only one gene. Manual examination on UCSC genome browser revealed that 37 might be true candidates for cis-SAGe fusions.
Both forms of LHX6-NDUFA8 can be frequently detected in cervical cancer and CIN patient Pap smear samples, supporting their potential as molecular biomarkers. No significant correlation of the detection of the fusion RNAs with clinical parameters was found, suggesting that LHX6-NDUFA8 expression may be an early event for at least some cervical cancer tumorigenesis. Both forms are predicted to be frame-shift chimeras, and siRNAs targeting the fusions had no obvious effect on cervical cancer cell growth (data not shown). In contrast, the SLC2A11-MIF fusion plays a significant role in cervical cancer cell growth. Silencing the fusion, but not the wild-type parental genes, showed significant cell cycle arrest, and reduction in cell growth. Consistently, CDKN1A was upregulated. These findings connect the SLC2A11-MIF fusion to the CDKN1A pathway. How the signaling axis works is one of the areas we are investigating.
Funding sources
This work was supported by Stand Up To Cancer SU2C-AACR-IRG0409 (HL), the National Science Foundation of China (Grants 81372806, 81630060)(PW and DM), and the National Key Research & Development Program of China (2016YFC0902901) (DM). The funding agencies had no roles in the experimental design, writing of the manuscript or the decision to submit. None of the authors were paid by any pharmaceutical company or other agency for the writing of the article. HL and DM had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Declarations of interests
None of the authors have any conflict to disclose.
Authors’ contributions
PW, SY, FQ, and LW performed experiments, conducted the analyses, and interpreted results. PW, SY, HL wrote and/or revised manuscript. SS and SK conducted bioinformatics analyses. DM and HL conceived and supervised the project and manuscript.
Acknowledgments
High-performance computing systems and services were provided by the Data Science Institute and the other Computation and Data Resource Exchange (CADRE) partner organizations at the University of Virginia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.059.
Contributor Information
Ding Ma, Email: dma@tjh.tjmu.edu.cn.
Hui Li, Email: hl9r@virginia.edu.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Cancer IAfRo A review of human carcinogen: biological agents. IARC. 2012:100B. [Google Scholar]
- 3.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Ramanathan P., Dhandapani H., Jayakumar H., Seetharaman A., Thangarajan R. Immunotherapy for cervical cancer: Can it do another lung cancer? Curr Probl Cancer. 2018;42(2):148–160. doi: 10.1016/j.currproblcancer.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Wang J., Mor G., Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321(5894):1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 8.Chase A., Ernst T., Fiebig A., Collins A., Grand F., Erben P. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95(1):20–26. doi: 10.3324/haematol.2009.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan H., Qin F., Movassagh M., Park H., Golden W., Xie Z. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3(12):1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 10.Wu C.S., Yu C.Y., Chuang C.Y., Hsiao M., Kao C.F., Kuo H.C. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24(1):25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zaphiropoulos P.G. Trans-splicing in Higher Eukaryotes: Implications for Cancer Development? Front Genet. 2012;2:92. doi: 10.3389/fgene.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z., Zhu D., Wang W., Li W., Jia W., Zeng X. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet. 2015;47(2):158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 13.Chwalenia K., Qin F., Singh S., Tangtrongstittikul P., Li H. Connections between Transcription Downstream of Genes and cis-SAGe Chimeric RNA. Genes (Basel). 2017;8(11) doi: 10.3390/genes8110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang R., Kumar S., Li H. Absence of Correlation between Chimeric RNA and Aging. Genes (Basel). 2017;8(12) doi: 10.3390/genes8120386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol.14(2):R12. [DOI] [PMC free article] [PubMed]
- 16.Zhang Y., Gong M., Yuan H., Park H.G., Frierson H.F., Li H. Chimeric Transcript Generated by cis-Splicing of Adjacent Genes Regulates Prostate Cancer Cell Proliferation. Cancer Discovery. 2012;2(7):598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 17.Qin F., Song Z., Babiceanu M., Song Y., Facemire L., Singh R. Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells. PLoS Genet. 2015;11(2) doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babiceanu M., Qin F., Xie Z., Jia Y., Lopez K., Janus N. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016;44(2):2859–2872. doi: 10.1093/nar/gkw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabbitts T.H. Chromosomal translocations in human cancer. Nature. 1994;372(6502):143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 21.Heim S., Mitelman F. Molecular screening for new fusion genes in cancer. Nat Genet. 2008;40(6):685–686. doi: 10.1038/ng0608-685. [DOI] [PubMed] [Google Scholar]
- 22.Li H., Wang J., Mor G., Sklar J. A Neoplastic Gene Fusion Mimics Trans-Splicing of RNAs in Normal Human Cells. Science. 2008;321(5894):1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 23.Finta C., Zaphiropoulos P.G. Intergenic mRNA molecules resulting from trans-splicing. J Biol Chem. 2002;277(8):5882–5890. doi: 10.1074/jbc.M109175200. [DOI] [PubMed] [Google Scholar]
- 24.Kumar-Sinha C., Kalyana-Sundaram S., Chinnaiyan A.M. SLC45A3-ELK4 Chimera in Prostate Cancer: Spotlight on cis-Splicing. Cancer Discovery. 2012;2(7):582–585. doi: 10.1158/2159-8290.CD-12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2