Abstract
In the present work, macrorestriction analysis was applied to characterize 44 S. uberis field strains isolated from lactating cows suffering from mastitis in three dairy herds in Hesse State, Germany. Analysis of the obtained data by Pulse-Field Gel Electrophoresis (PFGE) showed that most of the isolates originating from different herds and cows were not related to each other. However, identical macrorestriction patterns were noted in 12 of 13 mastitic quarters in healing process, in three quarters even over the whole sampling period indicating persistent infection. In the present work, PFGE could detect variable levels of similarity ranging from 76 to 100%. The macrorestriction analyses revealed the presence of 10 S. uberis PFGE pattern with more than four bands difference. PFGE profiles with minor differences (only one to three bands) were considered to be subtypes. The use of sensitive genotyping methods like macrorestriction analyses by PFGE enables the differentiation among new and persistent infections. Nevertheless minor changes in macrorestriction profiles could occur which are clearly distinguishable from totally unrelated strains.
Keywords: Bovine mastitis, Milk, Pulsed-field gel electrophoresis, Streptococcus uberis
1. Introduction
Mastitis is one of the most costly problems in animal industry. The disease can be induced by more than 150 different bacterial species and subspecies. One of the most frequent environmental pathogens inducing bovine mastitis in dairy herds with an increasing tendency worldwide is the Streptococcus (S.) uberis [1]. Control programs concerning contagious mastitis pathogens were successfully performed in well-managed dairy herds [2]. In contrast S. uberis held responsible for a large proportion of both clinical and subclinical mastitis in lactating and non-lactating dairy cows [3], [4]. While it is considered mainly as an environmental pathogen isolated in large numbers from bedding material, faeces and water [5], a recent study indicated that a limited number of S. uberis clones are capable of contagious transmission among cows via milking machines [6], [7].
Ineffective treatment of acute udder infections caused by S. uberis may lead to chronic infections that can persist throughout and even for subsequent lactations [8], [9]. To improve infection control measures and to recognize persistent infections in dairy herds, the knowledge about the genetic diversity of S. uberis is an essential precondition. Field isolates of S. uberis were genotyped in previous studies using isolates from subclinical mastitis or environmental sources. For this purpose, various molecular tools were used such as Pulsed-Field Gel Electrophoresis (PFGE) or Multilocus-Sequence Typing (MLST) [2], [10], [11], [12]. In the present study, PFGE assay was applied in order to characterize S. uberis isolates from mild to moderate clinical mastitis at different time points post infection in German herds. The aim of the present work is to investigate genetic diversity among field S. uberis strains isolated from mastitic cows under treatment in order to differentiate between a persistent infection from re- or new infections. The study aimed also to explore the presence of dominant S. uberis clones in infected herds.
2. Materials and methods
2.1. Samples
In the present work, quarter milk samples were collected aseptically from 147 clinical mastitis cases at five times pre and post treatment (day 0, 7, 14, 28 and 56 after infection) to evaluate the bacteriological cure rate. The samples were obtained from three moderate size dairy herds (between 75 and 300 dairy cows) in Hesse State, Germany. The mean distance between each farm was about 60 km. Dairy cows in both herds (A and B) were kept in cubicle yards while those in herd (C) were kept in a deep litter house.
2.2. Bacteriological and biochemical identification of the field isolates
Colonies suspected to be S. uberis were subcultured for further identification and characterization where one colony per sample was randomly selected for this purpose. From three cows (C177, C408 and B357) two colonies were subcultured at the same point of investigation to detect a coinfection with different strains of S. uberis. The isolates were collected according to the procedure recommended by the International Dairy Federation as described by Werner et al. [13], [14]. In Brief, the collected isolates were re-cultured on Columbia esculin blood agar (Merck, Darmstadt, Germany) to examine their culturing ability and haemolytic characteristics, followed by biochemical characterization to species level using the API 20 STREP® system (BioMérieux, Nürtingen, Germany). The results were interpreted according to the manufacturers’ instructions. Additional S. uberis control strains were included in the study; the strains were obtained from the strain collection of the LHL, Germany.
2.3. Molecular identification and macrorestriction analysis of the isolates
In order to confirm the data obtained from the phenotypic identification, all cultured S. uberis isolates were genotypically characterized using tDNA-ILP-PCR as described by Zschöck et al. [15]. Confirmed isolates to be S. uberis field strains (n = 44, isolated from 13 lactating cows) were then subjected to fingerprinting based on macrorestriction analysis of the bacterial chromosomal DNA for genotyping of the field isolates according to Soedarmanto et al. [16].
The preparation of whole bacterial DNA was carried out in agarose gel plugs followed by the digestion of the bacterial genome with the restriction enzyme SmaI. This step was then followed by fragment separation by PFGE using the pulse time described by Baseggio et al. [17]. The interpretation of the restriction patterns was performed as described by Tenover et al. [18]. Briefly, bacterial isolates yielding the same PFGE pattern were considered identical. Isolates differing by one up to three bands were defined as closely related. Bacterial isolates containing more than four bands difference were considered unrelated. A dendrogram analysis of the restriction patterns was additionally performed using Bionumerics software 5.1 (Applied Maths, Belgium) with the DICE coefficient and the unweighted pair group method with arithmetic means (UPGMA) with a band position tolerance of 5%.
3. Results
In the present work 147 mastitic quarter milk samples were involved. Bacteriological examination of the milk samples could identify S. uberis as the causative agent in 42% (n = 38) of the samples on day (0) of the investigation. Repeated sampling during the course of the treatment showed that 25 (65.8%) of these cases cured and became bacteriologically negative. However, in the rest (13 out of the 38 infected quarters), S. uberis could be re-isolated from at least one additional sample post treatment. The 13 infected quarters belonged to 13 different cows (2 from herd A, 2 from herd B, and 9 from herd C). Follow up bacteriological examination of the 13 cows at the 7th day could detect S. uberis in 11 samples. The number of positive reactors decreased to 8 cows at the 14th day, and only 7 cows after 21 days. Meanwhile, the infection persisted in 5 cows till the 56th day of infection. The grown 44 S. uberis colonies (13 + 11 + 8 + 7 + 5 isolates) and the three colonies of the double investigations at one sampling-date were subjected to fingerprinting.
Monitoring of the persistence of S. uberis in dairy cattle under treatment showed that in one cow the S. uberis could be isolated in the days 0,7,14 and 56 but not in the days 28. In another two cows the pathogen could be detected in the 0 and 28 days but not in the 7, 14 or 56 day (Table 1).
Table 1.
Distribution and recovery of PFGE-patterns of Streptococcus uberis isolates (n = 47) from bovine clinical mastitis cases with regard to herd, cow, gland and infections’ days.
| PFGE patterns recovered |
|||||||
|---|---|---|---|---|---|---|---|
| Herd | Cow | Gland | Primarily isolated pattern | Post1 7 | Post 14 | Post 28 | Post 56 |
| A | 30 | LF | AI | AI | – | – | – |
| A | 42 | LF | AII | AII | AII | AII | – |
| B | 357 | LF | BI/BI | BII | BIIa | BIII | BIIIa |
| B | 344 | LR | BIV | BIV | x | – | x |
| C | 105 | RF | CI | x | – | – | – |
| C | 177 | RR | CII | CII | CII | CII | CII/CII |
| C | 251 | RR | CIII | – | – | CIV | – |
| C | 254 | RR | CIIIa | CIVa | CIVa | – | – |
| C | 264 | LF | CV | CV | CV | CV | CV |
| C | 293 | LR | CVI | – | – | CVI | – |
| C | 298 | LR | CVII | CVII | CVII | CVIII | CVIII |
| C | 321 | LF | CIX | CIX | x | – | – |
| C | 408 | LR | CX/CX | CXa | – | – | – |
Abbreviation: LF, left front; LR, left rear; RF, right front; RR, right rear; – = no growth; x = S. uberis isolate (not possible to evaluate macrorestriction profile; A, B, C = designation of herds; AI-CX = Designation of different PFGE patterns of herds A to C (e.g. BI = first pattern detected in herd B; BIa = subtype of B with a 5% difference; BI/BI = two identical pattern from samples taken at the same time).
Days after infection.
Further PFGE typing could be successfully performed from 43 out of the 47 isolates as four isolates were not possible to evaluate. A total of 16 S. uberis PFGE pattern with more than four bands difference could be distinguished. Five of the PFGE patterns showed minor differences (only one up to three bands) and were classified as subtypes.
For the description of the degree of the relationship among the isolates, different terms were used. The cluster (PFGE-type with a 100%match in their pattern as well as PFGE subtypes with a minimum 95% match rate were grouped into a cluster amount). The PFGE-type (a special pattern of a field-isolate, if the restriction pattern is indistinguishable it might be the same PFGE-type. A field isolate with a maximum difference of 5% (95% accordance of bands of a pattern) to a pattern of another field isolate was assumed to be the same PFGE-type. This is usually the case when the PFGE-pattern differs max. in two to three bands. It might be caused by a single genetic event. A field isolate might be closely related to the outbreak strain). The PFGE-Subtypes are those PFGE patterns with a relationship up to 80% between the field-strain pattern. The obtained results of PFGE-typing are summarized in (Table 1 and Fig. 1).
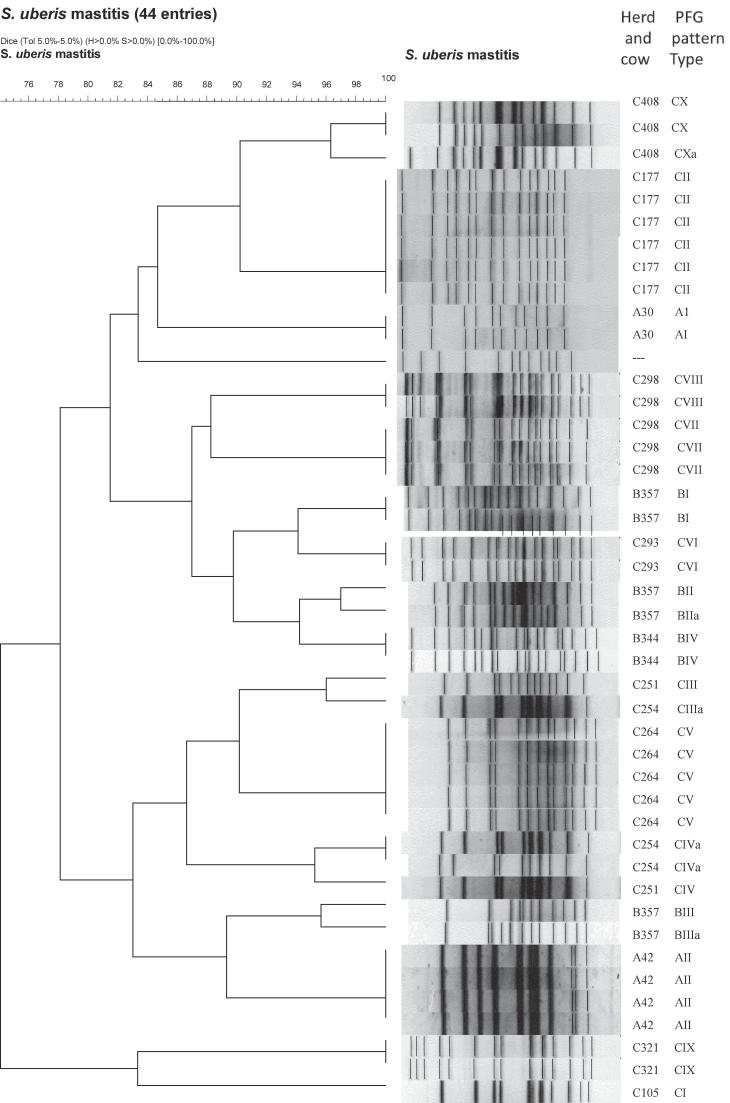
Fig. 1.
Pulsed field gel electrophoresis (PFGE) patterns of 44 Streptococcus uberis isolated from clinical milk samples of three dairy farms (designed as A, B, and C), digested with the SmaI restriction enzyme. The dendrogram was constructed with the BioNumerics software 5.1 (Applied Maths NV, Sint-Martens-Latem, Belgium) choosing the Dice coefficient setting with a band position tolerance of 5%. The horizontal scale on the left side (100 to 74) indicates the level of similarity in per cent among fingerprints. The 95% of similarities indicates the minimal levels for defining clusters and subtypes, respectively.
Evaluation of the dendrogram analysis showed levels of similarity between 74 and 100% (Table 2). The similarity between strains from different herds was in some cases higher than the in-herd similarity. PFGE-type BI (Herd B) had 94% relationship to PFGE-type CVI (Herd C). Other strains with a high genetic accordance from different herds were AI:CII (85%), AII:BIII (89%), BI:CIII (87%). The relationship (Dice coefficent) between the obtained PFGE-types (n = 16) of S. uberis isolates (n = 43) are illustrated in Table 2.
Table 2.
Relationship (Dice coefficent) between the identified PFGE-types (n = 16) of the investigated S. uberis isolates (n = 43).
| PFGE-pattern |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AI | AII | BI | BII | BIII | BIV | CI | CII | CIII | CIV | CV | CVI | CVII | CVIII | CIX | CX | ||
| PFGE-pattern | AI | 100* | 78 | 81,5 | 81,5 | 78 | 81,5 | 74 | 85 | 81,5 | 78 | 78 | 81,5 | 81,5 | 81,5 | 74 | 84,5 |
| AII | 78 | 100 | 78 | 78 | 89 | 78 | 74 | 78 | 83 | 83 | 83 | 78 | 78 | 78 | 74 | 78 | |
| BI | 81,5 | 78 | 100 | 90,5 | 78 | 90,5 | 74 | 81,5 | 87 | 78 | 78 | 94 | 87 | 87 | 74 | 81,5 | |
| BII | 81,5 | 78 | 90,5 | 100 | 78 | 94 | 74 | 81,5 | 78 | 78 | 78 | 90 | 87 | 87 | 74 | 81,5 | |
| BIII | 78 | 89 | 78 | 78 | 100 | 78 | 74 | 78 | 83 | 83 | 83 | 78 | 78 | 78 | 74 | 78 | |
| BIV | 81,5 | 78 | 90,5 | 94 | 78 | 100 | 74 | 87 | 78 | 78 | 78 | 90 | 87 | 87 | 74 | 81,5 | |
| CI | 74 | 74 | 74 | 74 | 74 | 74 | 100 | 74 | 74 | 74 | 74 | 74 | 74 | 74 | 83,5 | 74 | |
| CII | 85 | 78 | 81,5 | 81,5 | 78 | 87 | 74 | 100 | 78 | 78 | 78 | 81,5 | 81,5 | 81,5 | 74 | 90,5 | |
| CIII | 81,5 | 83 | 87 | 78 | 83 | 78 | 74 | 78 | 100 | 84 | 90,5 | 78 | 78 | 78 | 74 | 78 | |
| CIV | 78 | 83 | 78 | 78 | 83 | 78 | 74 | 78 | 86,5 | 100 | 86,5 | 78 | 78 | 78 | 74 | 78 | |
| CV | 78 | 83 | 78 | 78 | 83 | 78 | 74 | 78 | 90,5 | 86,5 | 100 | 78 | 78 | 78 | 74 | 78 | |
| CVI | 81,5 | 78 | 94 | 90 | 78 | 90 | 74 | 81,5 | 78 | 78 | 78 | 100 | 87 | 87 | 74 | 81,5 | |
| CVII | 81,5 | 78 | 87 | 87 | 78 | 87 | 74 | 81,5 | 78 | 78 | 78 | 87 | 100 | 88,5 | 74 | 81,5 | |
| CVIII | 81,5 | 78 | 87 | 87 | 78 | 87 | 74 | 81,5 | 78 | 78 | 78 | 87 | 88,5 | 100 | 74 | 81,5 | |
| CIX | 74 | 74 | 74 | 74 | 74 | 74 | 83,5 | 74 | 74 | 74 | 74 | 74 | 74 | 74 | 100 | 74 | |
| CX | 84,5 | 78 | 81,5 | 81,5 | 78 | 81,5 | 74 | 90,5 | 78 | 78 | 78 | 81,5 | 81,5 | 81,5 | 74 | 100 | |
Dice coefficient.
4. Discussion
In the present work, all involved field isolates (n = 47) were identified phenotypically as well as genotypically as S. uberis. For the typing of isolates at their strain level, PFGE was used due to its high discriminatory index in distinguishing field strains of S. uberis as recommended previously [1], [19] This was confirmed by the delivered data in the present work where a weak relationship among the detected genotypic profiles could be noticed. The data showed that S. uberis isolated from different herds belonged to different genotypic groups with various sets of PFGE patterns. These results are in agreement with the results of previously published reports [8], [12]. On the other hand strains with a high genetic similarity could be found in different herds (e.g.: PFGE-type BI and CVI: dice coefficient 94%, Table 2). This underlines that S. uberis is a causative agent for environmental mastitis predominantly.
Differences among genetic profiles of S. uberis field strains were not only seen among isolates originating from different herds. The present work, and in contrast to the data presented by Phuektes et al. [2], reports the presence of many genetic profiles of S. uberis within the same herd. The analysis of the obtained PFGE profiles showed great genetic variability among the isolates as reported previously by Reinoso et al. [19]. Some of the detected PFGE patterns were unique and sporadic; others from the farm C were closely related and even identical. In farm C, two closely related strains (with the PFGE pattern CIII/CIIIa) could be isolated from two different cows. However, the overall analysis of the pattern indicates the absence of predominant endemic/epidemic clones of S. uberis. This conclusion contradicts that of Phuektes et al. and Khan et al. [2], [10] who suggested the dominance of certain contagious clones of S. uberis in infected herds.
Indeed, the heterogeneity of PFGE patterns in one hand and the close relationship among other strains on the other hand (Fig. 1) confirm the epidemiological role of S. uberis in mastitis induction via cow-to-cow transmission in agreement with previous reports [1], [6]. However, this suggestion counteracts the widespread believe that S. uberis infections usually result from environmental infections [2], [8], [12], [17].
In two out of the 13 sampled quarters (cows 177, 264), the same S. uberis strain was isolated in all milk samples collected up to 8 weeks after infection suggesting the persistence of this S. uberis clone in the udder. The cows were treated with cloxacillin, but a treatment failure has to be stated in these cases. Conversely, in another mastitic udder quarter (cows no. 251, 254), an obvious pathogen-change could be stated. The S. uberis isolate showed another PFGE-profile on day 28 after infection as that recovered at the first sampling and followed by two samplings where no bacteria could be isolated indicating a new or super infection. However, McDougall et al. [8] considered the possibility that strains of various macrorestriction types of S. uberis were present in one milk sample on day of infection, but only one of them could be identified depending on the laboratory technique used. Another explanation was provided by Pryor et al. [20] who experimentally induced intramammary mixed infection with multiple strains of S. uberis with the result that in 70% of the udder quarters, a single strain seemed to be responsible for the development of infection because it predominated in strain typing using repetitive extragenic palindromic (REP)-PCR. A coinfection with different strains of S. uberis in samples of three cows (B357, C177, C408) was not observed.
The macrorestriction profile of that isolate changed at least four times within the sampling period. However, these differences were relatively minor as these isolates were closely related to each other. These findings lead to the assumption that genetic changes had also occurred in persisting isolates and that these S. uberis types were rather udder-associated than from environmental sources.
5. Conclusions
In summary, the findings of the present study confirmed mainly the epidemiological knowledge of S. uberis. Using sensitive methods for investigating genetic relationships between different bacterial isolates like marcrorestriction analysis, a persistent infection is clearly distinguishable from a new infection. Cow to cow infections with S. uberis could occur in dairy herds.
Competing interests
All authors declare no competing interests.
Acknowledgements
The endurance of the farmers in permitting repeated sampling of cows suffering from mastitis within their herds is gratefully acknowledged.
Footnotes
Peer review under responsibility of Faculty of Veterinary Medicine, Cairo University.
References
- 1.Loures R., de Pádua Pereira U, de Carvalho Castro G, Mian G., da Costa Custódio D., da Silva J. Genetic diversity and virulence genes in Streptococcus uberis strains isolated from bovine mastitis. Semina: Ciênc Agrár. 2017;38:2595–2606. [Google Scholar]
- 2.Phuektes P., Mansell P.D., Dyson R.S., Hooper N.D., Dick J.S., Browning G.F. Molecular epidemiology of Streptococcus uberis isolates from dairy cows with mastitis. J Clin Microbiol. 2001;39:1460–1466. doi: 10.1128/JCM.39.4.1460-1466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDougall S. Efficacy of two antibiotic treatments in curing clinical and subclinical mastitis in lactating dairy cows. N Z Vet J. 1998;46:226–232. doi: 10.1080/00480169.1998.36094. [DOI] [PubMed] [Google Scholar]
- 4.Patel D., Almeida R.A., Dunlap J.R., Oliver S.P. Bovine lactoferrin serves as a molecular bridge for internalization of Streptococcus uberis into bovine mammary epithelial cells. Vet Microbiol. 2009;137:297–301. doi: 10.1016/j.vetmic.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Benavides M.G., Williamson J.H., Pullinger G.D., Lacy-Hulbert S.J., Cursons R.T., Leigh J.A. Field observations on the variation of Streptococcus uberis populations in a pasture-based dairy farm. J Dairy Sci. 2007;90:5558–5566. doi: 10.3168/jds.2007-0194. [DOI] [PubMed] [Google Scholar]
- 6.Zadoks R.N., Gillespie B.E., Barkema H.W., Sampimon O.C., Oliver S.P., Schukken Y.H. Clinical, epidemiological and molecular characteristics of Streptococcus uberis infections in dairy herds. Epidemiol Infect. 2003;130:335–349. doi: 10.1017/s0950268802008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Sayed A., Awad W., Abdou N., Vázquez H. Molecular biological tools applied for identification of mastitis causing pathogens. Int J Vet Sci Med. 2017;5:89–97. doi: 10.1016/j.ijvsm.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDougall S., Parkinson T.J., Leyland M., Anniss F.M., Fenwick S.G. Duration of Infection and Strain Variation in Streptococcus uberis Isolated from Cows’ Milk. J Dairy Sci. 2004;87:2062–2072. doi: 10.3168/jds.S0022-0302(04)70024-7. [DOI] [PubMed] [Google Scholar]
- 9.Oliver S.P., Almeida R.A., Calvinho R.F. Virulence factors of Streptococcus uberis isolated from cows with mastitis. Zentralbl Veterinärmed B. 1998;45:461–471. doi: 10.1111/j.1439-0450.1998.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 10.Khan U., Hassan A.A., Abdulmawjood A., Lämmler C., Wolter W., Zschöck M. Identification and epidemiological characterization of Streptococcus uberis isolated from bovine mastitis using conventional and molecular methods. J Vet Sci. 2003;4:213–223. [PubMed] [Google Scholar]
- 11.Rato M.G., Bexiga R., Florindo C., Cavaco L.M., Vilela C.L. Antimicrobial resistance and molecular epidemiology of streptococci from bovine mastitis. Vet Microbiol. 2013;161:286–294. doi: 10.1016/j.vetmic.2012.07.043. [DOI] [PubMed] [Google Scholar]
- 12.Wang L., Chen W., Zhang L., Zhu Y. Genetic diversity of Streptococcus uberis isolates from dairy cows with subclinical mastitis in Southern Xinjiang Province, China. J Gen Appl Microbiol. 2013;59:287–293. doi: 10.2323/jgam.59.287. [DOI] [PubMed] [Google Scholar]
- 13.Werner C., Sobiraj A., Sundrum A. Efficacy of homeopathic and antibiotic treatment strategies in cases of mild and moderate clinical mastitis. J Dairy Res. 2010;77:460–467. doi: 10.1017/S0022029910000543. [DOI] [PubMed] [Google Scholar]
- 14.International Dairy Federation. Laboratory methods for use in mastitis work 1981. Document No 132 IDF, Brussels, Belgium.
- 15.Zschöck M., Manhold-Maurer S., Wescher A., Merl K., Khan I., Lämmler C. Evaluation of tRNA intergenic spacer length polymorphism analysis as a molecular method for species identification of streptococcal isolates from bovine mastitis. J Dairy Res. 2005;72:333–337. doi: 10.1017/S0022029905000956. [DOI] [PubMed] [Google Scholar]
- 16.Soedarmanto I., Pasaribu F.H., Wibawan I.W., Lämmler C. Identification and molecular characterization of serological group C streptococci isolated from diseased pigs and monkeys in Indonesia. J Clin Microbiol. 1996;34:2201–2204. doi: 10.1128/jcm.34.9.2201-2204.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baseggio N.P., Mansell P., Browning W.J., Browning G.F. Strain differentiation of isolates of streptococci from bovine mastitis by pulsed-field gel electrophoresis. Mol Cell Probes. 1997;11:349–354. doi: 10.1006/mcpr.1997.0126. [DOI] [PubMed] [Google Scholar]
- 18.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinoso E., Lasagno M., Odierno L. Genetic patterns of Streptococcus uberis isolated from bovine mastitis. Rev Arg de Microbiol. 2015;47:108–111. doi: 10.1016/j.ram.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Pryor S.M., Cursons R.T., Williamson J.H., Lacy-Hulbert S.J. Experimentally induced intramammary infection with multiple strains of Streptococcus uberis. J Dairy Sci. 2009;92:5467–5475. doi: 10.3168/jds.2009-2223. [DOI] [PubMed] [Google Scholar]