Abstract
Nonalcoholic Fatty Liver Disease (NAFLD) constitutes a wide spectrum of liver pathology with hepatic steatosis at the core of this pathogenesis. Variations of certain genetic components have demonstrated increased susceptibility for hepatic steatosis. Therefore, these inciting variants must be further characterized in order to ultimately provide effective, targeted therapies for NAFLD and will be the focus of this review. Several genetic variants revealed an association with NAFLD through Genome-wide Association Study, meta-analyses, and retrospective case–control studies. PNPLA3 rs738409 and TM6SF2 rs58542926 are the two genetic variants providing the strongest evidence for association with NAFLD. However, it remains to be determined if these genetic variants serve as the primary culprit which induces the pathogenesis of NAFLD. Prospective and intervention studies are urgently needed to firmly establish a cause-and-effect relationship between the presence of certain genetic variants and risk of NAFLD development and progression.
Abbreviations: ACC2, Acetyl-CoA Carboxylase 2; ACLY, ATP Citrate Lyase; BMI, Body Mass Index; CK-18, Cytokeratin 18; CT, Computed Tomography; FASN, Fatty Acid Synthase; GWAS, Genome-wide Association Study; 1H-MRS, Proton Magnetic Resonance Spectroscopy; HCC, Hepatocellular Carcinoma; LT, Liver Transplantation; miRNA, MicroRNA; NAFLD, Nonalcoholic Fatty Liver Disease; NASH, Nonalcoholic Steatohepatitis; SCD1, Stearoyl-CoA Desaturase 1; SNP, Single Nucleotide Polymorphism; US, Ultrasonography
Keywords: nonalcoholic fatty liver disease, genetic variants, genetic polymorphisms, single nucleotide polymorphisms, epigenetics
Nonalcoholic Fatty Liver Disease (NAFLD) encompasses a spectrum of liver disease that ranges from simple triglyceride (TG) accumulation, or steatosis, in the liver to necro-inflammation, fibrosis, cirrhosis, and hepatocellular carcinoma (HCC).1 Hepatic steatosis may be accompanied by other pathologic changes, such as lobular inflammation or hepatocellular ballooning, that define Non-alcoholic Steatohepatitis (NASH). Although the progression from isolated hepatic steatosis to NASH is unclear, cirrhosis and HCC remain common long-term complications found among patients with NASH.2, 3 In a longitudinal study of patients with NAFLD, fibrosis stage, but no other histologic features of steatohepatitis, were associated independently with long-term overall mortality, liver transplantation, and liver-related events.4
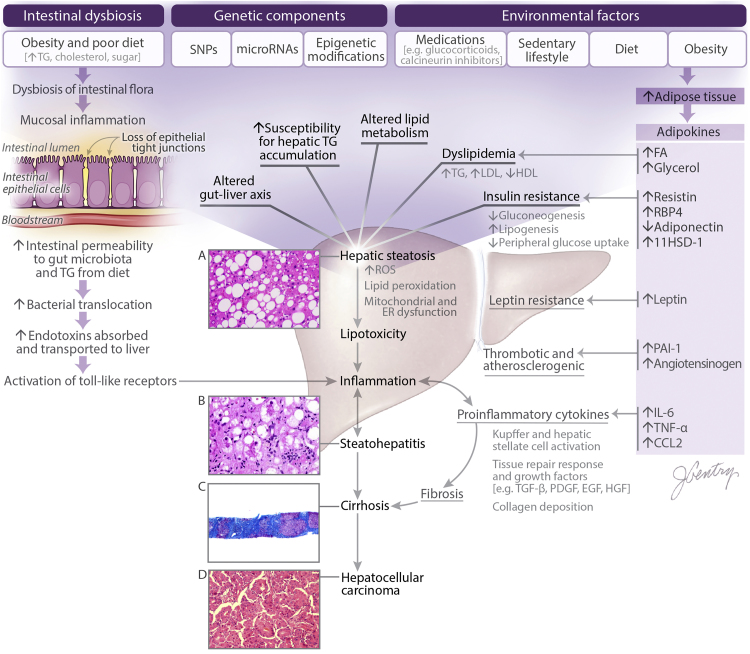
A complex interplay between cellular, genetic, and environmental factors is implicated in the pathogenesis of NAFLD (Figure 1). Hepatocytes play a central role in lipid metabolism, importing serum free fatty acids and manufacturing, storing and exporting lipids and lipoproteins. Accumulation of TG can occur in the liver as a result of abnormal fatty acid metabolism,5 with excessive delivery of free fatty acids to the liver compared to that which can be metabolized (obesity, rapid weight loss, TPN), an increased mitochondrial synthesis of fatty acids, or a failure of the synthesis or secretion of apolipoproteins or TGs.6 The primary metabolic abnormality catalyzing the transformation of hepatic steatosis to NASH is still unknown. Insulin resistance plays a key role, since it may influence several intracellular metabolic pathways.7 Higher levels of fasting serum insulin have been frequently noted in NASH patients.8 Conditions associated with peripheral insulin resistance, such as type 2 diabetes mellitus and obesity (in particular visceral adiposity), are frequently observed in patients with NAFLD. Insulin resistance has also been observed in patients with NASH who are not obese and those who have normal glucose tolerance.8 It is postulated that impaired capacity to adequately expand the peripheral adipose tissue compartments drives dyslipidemia and insulin resistance.9 Insulin resistance decreases peripheral glucose uptake and promotes a lipogenic state within hepatocytes.6, 10, 11, 12 After the introduction of excess hepatic TGs, increased lipid peroxidation and reactive oxidative species are generated which insidiously promotes dysfunction of the mitochondria and endoplasmic reticulum (ER).13, 14, 15, 16 This increased cellular stress defines the lipotoxicity which ultimately concludes with hepatocellular death, which further amplifies the inflammatory response within this milieu.17 Hepatic steatosis with ensuing lipotoxicity and inflammatory response set the stage for NASH. Several adipocytokines derived primarily from the adipose tissue depot such as leptin, TNF-alpha, IL-6, adiponectin, resistin has been implicated in the pathogenesis of NASH, however a detailed discussion of their mechanism is beyond the scope of this review, and has been recently reviewed by Polyzos et al.18 Adipokines serve as auxiliary catalysts at numerous points along the pathway of NAFLD pathogenesis.19 Finally, intestinal dysbiosis has been shown to exacerbate this entire process through diet-induced changes affecting the gut-liver axis.20, 21 It is imperative to note the underlying pathophysiology because it is the genetic variations within this mechanism that dictate functionality of proteins within these cellular processes. Thus, minor epigenetic changes or alterations within the genetic sequence can clearly predispose individuals to both the development and progression of NAFLD.
Figure 1.
Pathophysiology of NAFLD. Visualized here are the influences of environmental factors, genetic components, and the intestinal microbiome on the development of hepatic steatosis and the progression of NAFLD. (A) NAFLD is characterized as >5% triglyceride accumulation in hepatocytes. This can be isolated hepatic steatosis, or accompanied by minimal inflammation within the lobules. (B) Histology showing classic features of NASH such as steatosis, inflammation, and hepatocellular ballooning degeneration. (C) Trichrome stain revealing progression of NASH to cirrhosis. (D) Histology illustrating the progression of cirrhosis to HCC. 11HSD-1, 11β-Hydroxysteroid Dehydrogenase Type 1; CCL2, C-C Motif Chemokine Ligand 2; EGF, Epidermal Growth Factor; FA, Fatty Acid; IL-6, Interleukin 6; HCC, Hepatocellular Carcinoma; HDL, High Density Lipoprotein; HGF, Hepatocyte Growth Factor; LDL, Low Density Lipoprotein; PAI-1, Plasminogen Activator Inhibitor-1; PDGF, Platelet-Derived Growth Factor; RBP4, Retinol Binding Protein 4; ROS, Reactive Oxygen Species; SNP, Single Nucleotide Polymorphism; TG, Triglyceride; TGF-β, Transforming Growth Factor Beta; TNF-α, Tumor Necrosis Factor Alpha.
Although there are clearly numerous components at play, the individual genetic elements responsible for these cellular functions remain critical to the inner workings of this pathophysiology. Several genetic disorders that alter the lipoprotein levels such as, disorders of high density lipoprotein (familial hypoalphalipoproteinemia, Tangier disease, and LCAT deficiency); familial hypocholesterolemias (familial hypobetalipoproteinemia, abetalipoproteinemia, PCSK9 loss of function mutations, familial combined hypolipidemia, and chylomicron retention disease); β-sitosterolemia; cerebrotendinous xanthomatosis; and lysosomal acid lipase deficiency exists that potentially can predispose for development of NAFLD.22 Clear genetic association of NAFLD has been shown with LIPA gene mutation in lysosomal acid lipase deficiency, and a targeted therapies currently exists for this condition.23 The relevance of genetic factor in the context of NAFLD has been recently and elegantly outlined by twin studies.24 Particular variants or Single Nucleotide Polymorphisms (SNPs) across multiple genetic loci have been demonstrated to work synergistically with adipose tissue in order to augment the risk of NAFLD.25 SNPs and epigenetic modifications affecting the genes within this process have been demonstrated to increase the susceptibility for hepatic steatosis, a central crux of this pathology which this review will analyze in further detail.
Genetic Variants Implicated in NAFLD Pathogenesis
Genome Wide Association Studies
Genome-wide Association Studies (GWAS) have identified associations of several genetic variants with NAFLD26, 27, 28, 29, 30, 31, 32, 33, 34, 35 (Table 1). GWAS have become a novel, powerful tool in first assessing which genes are affiliated with a particular disease. The majority of these findings have incriminated genetic variants, such as SNPs, as potential mechanisms predisposing for NAFLD pathogenesis.
Table 1.
Genome-Wide Association (GWA) and Genome-Wide Exome Studies Identifying Genetic Variants Associated with NAFLD.
| Gene | SNP | Study population | Notable cohort characteristics | Diagnostic modality | NAFLD sample size |
|---|---|---|---|---|---|
| Romeo et al., 200826 | |||||
| PNPLA3 | rs738409 | American | %52 African American, 29% Caucasian, 17% Hispanic | 1H-MRS | 2111 total (controls vs NAFLD not reported) |
| Chalasani et al., 201027 | |||||
| COL13A1 | rs1227756 | American | 100% Caucasian females | Liver biopsy | 236 |
| FDFT1 | rs2645424 | ||||
| Speliotes et al., 201128 | |||||
| GCKR | rs780094 | American & Icelandic | 100% Caucasian Family studies included | CT and/or liver biopsy | 592 |
| LYPLAL1 | rs12137855 | ||||
| NCAN | rs2228603 | ||||
| PNPLA3 | rs738408 | ||||
| Kawaguchi et al., 201229 | |||||
| PNPLA3 | Several (but rs738409 not tested) | Japanese | – | Liver biopsy | 529 |
| Adams et al., 201330 | |||||
| LPPR4 | rs12743824 | Australian | Adolescents at 17 years of age | US | 126 |
| GC | rs222054 | ||||
| LCP1 | rs7324845 | ||||
| SLC38A8 | rs11864146 | ||||
| Kitamoto et al., 201331 | |||||
| PARVB | rs5764455, rs6006611 | Japanese | – | Liver biopsy | 392 |
| PNPLA3 | rs738409, rs2896019, rs3810622 | ||||
| SAMM50 | rs738491, rs2073082, rs3761472, rs2143571 | ||||
| Feitosa et al., 201332 | |||||
| ERLIN1 | rs2862954, rs1408579, rs10883451 | American | 100% Caucasian Family study | CT | 2767 |
| CHUK | rs11597086, rs11591741 | ||||
| CWF19L1 | rs17729876, rs17668255, rs17668357, rs12784396 | ||||
| PNPLA3 | rs738409 | ||||
| PPP1R3B | rs2126259 | ||||
| Kozlitina et al., 201433 | |||||
| TM6SF2 | rs58542926 | American | 48% African American, 32% Caucasian, 17% Hispanic | 1H-MRS | 2735 total [controls vs NAFLD not reported] |
| Shang et al., 201534 | |||||
| COL13A1 | rs1227756 | Chinese | Children aged 7–18 | US | 162 |
| EHBP1L1 | rs6591182 | ||||
| FDFT1 | rs2645424 | ||||
| GCKR | rs7800094 | ||||
| NCAN | rs2228603 | ||||
| PDGFA | rs343064 | ||||
| PNPLA3 | rs738409 | ||||
| DiStefano et al., 201535 | |||||
| PNPLA3 | rs4823173, rs2896019, rs2281135 | American | 100% Caucasian 81% female Subjects undergoing bariatric surgery | Histology from intraoperative liver biopsy | 1386 |
| SUGP1 | rs10401969 | ||||
CHUK, Conserved Helix-Loop Helix Ubiquitous Kinase; COL13A1, Collagen Type XIII, alpha1; CT, Computed Tomography; CWF19L1, CWF19-like Protein 1; EHBP1L1, EH Domain Binding Protein 1-like 1 Gene; ERLIN1, ER Lipid Raft Associated 1; FDFT1, Farnesyl Diphosphate Farnesyl Transferase 1; GC, Group-specific Component; GCKR, Glucokinase Regulator; 1H-MRS, Proton Magnetic Resonance Spectroscopy; LCP1, Lymphocyte Cytosolic Protein-1; LPPR4, Lipid Phosphate Phosphatase-Related Protein Type 4; LYPLAL1, Lysophospholipase-like 1; NCAN, Neurocan; PARVB, Parvin, Beta; PDGFA, Platelet-derived Growth Factor Alpha Polypeptide; PNPLA3, Patatin-like Phospholipase Domain Containing 3; PPP1R3B, Protein Phosphatase 1 Regulatory Subunit 3B; SAMM50, Sorting and Assembly Machinery Component 50; SLC38A8, Solute Carrier Family 38 Member 8; SUGP1, SURP and G Patch Domain Containing 1; TM6SF2, Transmembrane 6 Superfamily Member 2; US, Ultrasonography.
Romero et al. for the first time reported in a GWAS a relationship of NAFLD with a SNP identified in patatin-like phospholipase domain-containing 3 (PNPLA3) on chromosome 22, PNPLA3 rs738409, which has founded this variant as a prime candidate of genetic-associated NAFLD.26 The variant rs738409 (c.444 C>G, p.I148M), a non-synonymous cytosine to guanine mutation resulting in isoleucine to methionine conversion, correlates with increased hepatic lipid content and predisposes to fatty liver-associated liver disease, from simple steatosis to steatohepatitis, fibrosis and HCC.26, 36 Overexpression of the I148M variant in mouse liver promotes accumulation of triacylglycerol, increased synthesis of fatty acids and impaired hydrolysis of triacylglycerol.37 This genetic variant has been examined extensively in subsequent studies, and is analyzed in detail in this review. However, this study utilized an American cohort, of which African American subjects were predominant. This demographic composition may result in slightly skewed data because the prevalence of NAFLD is highest in Hispanics and Caucasians, while least common among African American individuals, respectively.38 The population with the highest prevalence was grossly underrepresented in this study, and therefore, may serve as a poor indicator of true genetic association. This GWAS analyzed cohort patients using Proton Magnetic Resonance Spectroscopy (1H-MRS) and fails to report the percentage of total patients with NAFLD as calculated via 1H-MRS, but only summarizes hepatic TG percentage within each ethnic group. Hepatic TG content was also stated as skewed, and a power transformation was applied to that trait prior to analysis. Nevertheless, this large-scale cohort was a landmark study that demonstrated a significant association between PNPLA3 rs738409 and NAFLD.
Chalasani et al. in another GWAS retrospectively analyzed data from the NAFLD database Study.27 The entire genome of 236 American subjects confirmed by histology via liver biopsy was analyzed. PNPLA3 was not verified in this cohort, but SNPs in the COL13A1 and FDFT1 genes were found to be associated with NAFLD. Despite providing convincing evidence, there was one major drawback of this study in that this cohort was purely composed of Caucasian females. A highly homogenous group such as this may not represent the most indicative genetic associations in the general population.
In the largest GWAS to date, Speliotes et al. not only was able to reproduce an association with PNPLA3 rs738409, but also established a new relationship between NAFLD and glucokinase regulatory protein GCKR rs780094 and NCAN rs2228603.28 This variant produces a GCKR with defective inhibitory function, leading to increased glucokinase activity and hepatic glucose uptake.39 The resultant unimpeded hepatic glycolysis reduces glucose levels, inducing malonyl-CoA synthesis, a substrate for lipogenesis that causes liver fat deposition and impairs mitochondrial β-oxidation. This cohort was comprised of populations in family-based studies from the Framingham Heart Study, Family Heart Study, Old Order Amish community, and Reykjavik Study from Iceland. All of the individuals in this cohort were Caucasian and from either America or Iceland. While the sheer number of individuals tested provides powerful data, this analysis will represent skewed information from within the same genetic pools, which could alter the validity of the GWAS as compared to the entire population. Also, the modality of NAFLD diagnosis was initially established with Computed Tomography (CT), and then if positive, followed up by liver biopsy. Utilizing this methodology, patients with lower levels of hepatic steatosis may have gone undetected in the initial screening process by CT, and thus, not confirmed on liver biopsy. Even though constraints certainly exist, this study was not only able to reproduce results on PNPLA3 rs738409, but also pioneered the association between NAFLD and GCKR rs780094 and NCAN rs2228603. Additionally, the multidimensional function of GCKR needs a mention here as a recent meta-analysis also suggests that the rs780094 polymorphism in GCKR is associated with elevated T2DM risk, which may indirectly influence the risk of developing NAFLD.40
Romero et al. in the first GWAS provided the framework for the remaining nine studies whose results are further detailed in Table 1. Three of the studies were strictly in Japanese and Chinese cohorts, the latter being primarily focused in a pediatric population.34 Of note, the individuals in the Chinese pediatric population were diagnosed with NAFLD based upon US. Another study was performed in Australia, which also used US for detection of hepatic steatosis.30 Patients with limited hepatic steatosis would potentially go undetected utilizing this modality. The remaining GWAS were performed in American cohorts, which were able to confirm the prevalence of SNPs from previous work as well as identify new potential genetic variants associated with NAFLD.
Candidate Gene Studies
After the explosion of information from numerous GWAS, many candidate gene studies have been performed in order to examine the influence of particular SNPs on NAFLD pathogenesis. The most prominent genetic variants41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55 exhibiting a predilection or progression of NAFLD are displayed in Table 2.
Table 2.
Meta-analyses and Prospective Evidence for the Association of Genetic Variants with NAFLD.
| Gene | SNP | Study type | Study population | Notable cohort characteristics | Sample size | Diagnostic modality |
|---|---|---|---|---|---|---|
| ADIPOQ | rs26672941 | Meta-analysis | Multiethnic (primarily Asian) | HB vs PB not reported | 9 retrospective case–control studies 1223 cases/1580 controls | Liver biopsy only in 3/9 studies |
| APOC3 | rs285411642 | Prospective case–control | Chinese | 67% male subjects | 300 cases/300 controls | US |
| rs2854116 and rs285411743 | Meta-analysisa | Multiethnic | 5 HB vs 2 PB controls; 1 study included pediatric subjects; 2 studies did not report gender % | 7 prospective case–control studies 1745 cases/1437 controls | Liver biopsy only in 2/7 studies 1H-MRS or US in others | |
| GCKR | rs78009444 | Meta-analysis | Multiethnic | 5 HB vs 2 PB controls; 1 of the Asian studies examines pediatric subjects | 5 retrospective case–control studies 2090 cases/3003 controls | Liver biopsy only in 3/5 studies |
| MTHFR | rs180113345 | Meta-analysis | Multiethnic | All 8 studies PB controls | 8 retrospective case–control studies 785 cases/1188 controls | Liver biopsy in 4/5 studies, not reported in Chinese and Ukrainian studies |
| rs1810113145 | Meta-analysis | Multiethnic | All 5 studies PB controls | 5 retrospective case–control studies 476 cases/679 controls | Liver biopsy in 3/4 studies, not reported in Ukrainian study | |
| MTTP | rs180059146 | Meta-analysis | Multiethnic | 6 HB vs 5 PB controls | 11 retrospective case–control studies 636 cases/918 controls | Not reported |
| PEMT | rs794647 | Meta-analysis | Multiethnic (primarily Asian) | 4 HB vs 2 PB controls; 1 study included pediatric subjects |
6 retrospective case–control studies 792 cases/2722 controls | Liver biopsy only in 2/6 studies |
| PNPLA3 | rs73840948 | Meta-analysis | Multiethnic | 11 HB vs 5 PB controls; 5 studies included pediatric subjects | 16 retrospective case–control studies 12,677 total | Liver biopsy only in 6/16 studies 1H-MRS, US, or CT in others |
| rs73840949 | Meta-analysis | Asian | 4 HB vs 8 PB controls | 12 retrospective case–control studies 4495 cases/7431 controls | Liver biopsy only in 5/12 studies | |
| rs73840950 | Meta-analysis | Multiethnic | 15 HB vs 8 PB controls | 23 retrospective case–control studies 5826 cases/10,796 controls | Liver biopsy only in10/23 studies | |
| PPARγ | rs1805192 and rs180128251 | Meta-analysisa | Caucasian and Asian | 4 HB vs 4 PB controls | 8 retrospective case–control studies 1697 cases/2427 controls | Liver biopsy in 5/9 studies |
| rs1805192 and rs180128252 | Meta-analysisb | Caucasian and Asian | HB vs PB not reported | 5 retrospective case–control studies 1225 cases/2016 controls | Not reported | |
| SREBF-2 | rs13329153 | Prospective cohort | Italian | 65% male subjects | 48 cases/127 controls | Liver biopsy |
| TM6SF2 | rs5854292654 | Meta-analysis | Multiethnic | 4 HB vs 3 PB vs 1 mixed controls; 1 of the 2 Italian studies in pediatric |
8 retrospective case–control studies 5537 total | Liver biopsy only in 3/8 studies |
| rs5854292655 | Prospective cohort | Multiethnic | Pediatric cohort | 957 total | Liver biopsy only in 11 patients |
This study demonstrated no association with NAFLD.
This study suggested association with NAFLD in the East Asian population, however not in the European population. ADIPOQ, Adiponectin; APOC3, Apolipoprotein C-III; CT, Computed Tomography; GCKR, Glucokinase Regulator; 1H-MRS, Proton Magnetic Resonance Spectroscopy; HB, Hospital-Based Controls; MTHFR, Methylenetetrahydrofolate REDUCTASE; MTTP, Microsomal Triglyceride Transfer Protein; PB, Population-Based Controls; PEMT, Phosphatidylethanolamine N-methyltransferase; PNPLA3, Patatin-like Phospholipase Domain Containing 3; PPARγ, Peroxisome Proliferator-Activated Receptor Gamma; SREBF-2, Sterol Regulatory Element-binding Factor 2; TM6SF2, Transmembrane 6 Superfamily Member 2; US, Ultrasonography.
PNPLA3 rs738409 remains the most highly proclaimed genetic variant incriminate in the pathogenesis of NAFLD. To date, three large-scale meta-analyses48, 49, 50 (Table 2) have been performed across a vast array of ethnicities and large sample populations, confirming the evidence for this relationship. Support from this data aligns with the intracellular actions of PNPLA3 as a lipase in both hepatocytes and hepatic stellate cells. Aberrant function of PNPLA3 rs738409 results in an absence of lipase activity, thus leading to intracellular TG or retinol accumulation in hepatocytes and hepatic stellate cells, respectively.56, 57, 58 However a recent study has argued against this mechanism as knockout mice fail to develop steatosis and it is more likely to be a gain-of-function mutation that modulates the composition of hepatic lipid, resulting in lipotoxicity.59
The genetic variant in TM6SF2 rs58542926 on chromosome 19 (19p13.11) has been reported to correlate with steatosis and increased risk of advanced fibrosis in NAFLD patients,33, 60 independently of other factors, including diabetes, obesity, or PNPLA3 genotype. TM6SF2, is a transmembrane protein localized in ER and ER–Golgi compartments and functions as a lipid transporter.61 The amino acid change E167K causes loss of function of TM6SF2 protein, and downregulation of TM6SF2 reduces lipoproteins and apolipoprotein B (APOB) levels, and increases hepatic deposition of TGs and the amount and size of lipid droplets. In contrast, the size and number of lipid droplets diminishes when TM6SF2 is overexpressed, indicating that TM6SF2 plays a role in regulating hepatic lipid efflux.33, 61 This genetic variant was first recognized after emerging as a leading candidate from the genome-wide exome study performed by Kozlitina et al.33 A myriad of studies have since validated its association with NAFLD, including one meta-analysis.54 Despite a large sample size only three out of the eight studies confirmed NAFLD on liver biopsy, and one study from Italy included pediatric subjects.55
Although the presence of several other genetic variants listed in Table 2 have proven a high propensity for NAFLD through retrospective evidence, only APOC3 rs2854116 and rs2854117 have been studied through meta-analysis of prospective data.43
PPARγ rs1805192 and 1801282 have been examined in two meta-analyses. In the first meta-analysis, these genetic variants were found to have no association with NAFLD.51 The second meta-analysis provides somewhat weaker methodology, in that the incidence of hospital-based and population-based controls, as well as the diagnostic modality for NAFLD, was not reported.52 Amidst these deficiencies, it was concluded that these genetic variants were associated with NAFLD in the East Asian population that was studied, but not the European population. Clearly, there is inconsistent data concerning the PPARγ rs1805192 and rs1801282 and further analysis is required regarding its role in NAFLD pathogenesis.
Meta-analysis provides the highest level of evidence to date, however there is a multitude of cross-sectional studies analyzing the role of specific genetic variants in their association to NAFLD. Selected retrospective studies62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85 from each genetic variant are detailed here in Table 3, which have been performed over a variety of populations and ethnicities. Certainly, there are many genetic variants embroiled in this disease process, and many of which, warrant further investigation. However, LYPLAL1 rs12137855 provides an interesting context to NAFLD pathogenesis. After being implicated in the landmark GWAS by Speliotes et al.,28 this genetic variant was shown to have no association with NAFLD in a Chinese study population through retrospective case–control analysis.84 Results of these studies potentially could be biased related to power of these studies and could have been confounded by the quality of the study design.
Table 3.
Retrospective Candidate Gene Studies Linking Genetic Variants to NAFLD.
| Gene | SNP | Study population | Notable cohort characteristics | Sample size | Diagnostic modality |
|---|---|---|---|---|---|
| ABCC2 | rs17222723, rs818771062 | Argentine | Poor selection of controls | 109 cases/58 controls | US |
| ACSL4 | rs788798163 | Finnish | Validated in 2 replication cohorts | 302 total with replication cohorts | 1H-MRS |
| ADIPOR2 | rs76787063 | Finnish | Validated in 2 replication cohorts | 302 total with replication cohorts | 1H-MRS |
| ADRB2 | rs104271464 | Japanese | – | 132 cases/119 controls | US |
| ADRB3 | rs499465 | Japanese | Exclusively NASH cases | 63 cases/100 controls | CT |
| AGTR1 | rs3772630, rs377262266 | Japanese | 80% males in NAFLD cases | 167 cases/435 controls | Liver biopsy |
| APOE | rs7412, rs42935867 | Turkish | – | 57 cases/245 controls | Liver biopsy |
| APPL1 | rs464052568 | Chinese | – | 280 cases/281 controls | Unreported |
| FADS1 | rs17455669 | American and Chinese | Analysis of explanted liver samples | 206 transplants | Histology of donor livers |
| GCLC/GCLM | rs1788390170 | Brazilian | – | 131 cases/141 controls | Liver biopsy |
| IL-1 | rs1694465 | Japanese | Exclusively NASH cases | 63 cases/100 controls | CT |
| IL-6 | rs180079571 | Italian | Poor selection of controls | 114 cases/79 controls | Liver biopsy in 59/114 cases |
| LEPR | rs670098672 | Egyptian | – | 90 cases/30 controls | Liver biopsy |
| rs1137100, rs113710173 | Malaysian, Chinese, Indian | – | 144 cases/198 controls | Liver biopsy | |
| LYPLAL1 | rs1213785584 | Chinese | – | 184 cases/114 controls | US |
| MBOAT7 | rs64173874 | American and European | – | 3854 total in Dallas Heart Study, 1149 total in Liver Biopsy Cohort | Liver biopsy and 1H-MRS |
| NCAN | rs222860375 | American | Bariatric cohort; 80% female subjects; Ethnicities not reported | 748 cases/344 controls | Liver biopsy |
| PARVB | rs5764455, rs600647376 | Chinese | 60% male subjects | 384 cases/384 controls | US |
| PPP1R3B | rs424062477 | American | Population-based | 4804 total | US |
| SAMM50 | rs738491, rs2143571, rs376147285 | Chinese | – | 340 cases/452 controls | US |
| SLC2A1 | rs4658, rs84185678 | Spanish | Poor selection of controls | 451 cases/304 controls | Liver biopsy |
| SOD2 | rs488079 | Chinese | – | 131 cases/90 controls | Liver biopsy |
| TCF7L2 | rs790314680 | Italian | ∼75% male subjects | 78 cases/156 controls | Liver biopsy in 34/78 cases |
| TLR4 | rs498679081 | Turkish | – | 119 cases/80 controls | Liver biopsy in 111/119 cases |
| TRAIL | rs1131568, rs113158082 | Chinese | – | 84 cases/80 controls | Unreported |
| UCP3 | rs1123597283 | Spanish | – | 39 cases | Liver biopsy |
| 11β-HSD1 | rs2235543, rs12565406, rs4844880111 | German | 100% Caucasian All subjects had family history of T2DM, BMI > 27, or impaired glucose tolerance testing |
327 total subjects | 1H-MRS |
| CD14 | C159T polymorphism112 | Indian | 64% male subjects | 200 NAFLD/50 controls | US |
| Omentin-1 | rs2274907113 | Iranian | Significantly lower BMI in control group | 94 NAFLD/188 controls | US |
| PTPRD | rs35929428114 | Japanese | Significantly lower age in control group | 36 NAFLD/27 controls | Liver biopsy |
| Resistin | rs1862513113 | Iranian | Significantly lower BMI in control group | 94 NAFLD/188 controls | US |
a These studies have demonstrated no association with NAFLD.
ABCC2, ATP-binding Cassette, Sub-family C, Member 2; ACSL4, Acyl-coA Synthase Long Chain Family Member 4; ADIPOR2, Adiponectin Receptor 2; ADRB2, Adrenergic Beta 2 Receptor; ADRB3, Adrenergic Beta 3 Receptor; AGTR1, Angiotensin II Type 1 Receptor; APOE, Apolipoprotein E; APPL1, Adaptor Protein, Phosphotyrosine Interacting with PH Domain and Leucine Zipper 1; CT, Computed Tomography; FADS1, Fatty Acid Desaturase 1; GCLC/GCLM, Glutamate-cysteine Ligase; 1H-MRS, Proton Magnetic Resonance Spectroscopy; IL-1, Interleukin-1; IL-6, Interleukin-6; LEPR, Leptin Receptor; LYPLAL1, Lysophospholipase-like 1; MBOAT7, Membrane Bound O-acyltransferase Domain-containing 7 Gene; NCAN, Neurocan; PARVB, Parvin Beta; PPP1R3B, Protein Phosphatase 1 Regulatory Subunit 3B; SAMM50, Sorting and Assembly Machinery Component 50; SLC2A1, Solute Carrier Family 2 Member 1; SOD2, Superoxide Dismutase 2, Mitochondrial; TCF7L2, Transcription Factor 7-like 2; TLR4, Toll-like Receptor 4; TRAIL, TNF-related Apoptosis-inducing Ligand; UCP3, Uncoupling Protein 3, Mitochondrial; 11β-HSD1, 11β-Hydroxysteroid Dehydrogenase Type 1; CD14, Coreceptor Cluster of Differentiation 14; PTPRD, Receptor-type Tyrosine-protein Phosphatase Delta; US, Ultrasonography.
Epigenetic Modifications Implicated in NAFLD Pathogenesis
Epigenetic changes consist in modifications at the transcriptional level affecting gene expression and phenotype. A number of epigenetic aberrations have been associated with NAFLD pathogenesis, causing alterations in lipid metabolism, Insulin Resistance (IR), dysfunction of ER and mitochondria, oxidative stress and inflammation. As opposed to targeting changes in the genetic sequence and analyzing SNPs associated with NAFLD, a growing interest in recent scientific studies have focused on mechanisms responsible for modified expression or translation of genes related to the disease process. Interestingly, it is unclear if the altered gene expression causes the disease or the disease itself drives the modifications of gene expression. Nevertheless, two major gene expression modifying mechanisms, MicroRNA (miRNA) and epigenetics are found to be associated with NAFLD.
Analysis of miRNA Mediated Modifications
miRNAs modulate gene expression via post-transcriptional mechanisms, regulating the main cellular processes, such as lipid metabolism, inflammation, apoptosis, cell growth and differentiation. Aberrant miRNA expression has been reported in a number of diseases including metabolic disorders,86, 87 whereas an increasing number of dysregulated miRNAs, implicated in fatty acid synthesis, uptake and storage of TGs or oxidation, have been recently identified in NAFLD.88 miRNAs have been the most comprehensively studied epigenetic mechanism in relation to NAFLD. There have been numerous miRNAs implicated in this pathology and demonstrated through mouse models, however only the associations found in humans have been listed in Table 4. In general, miRNAs listed here exhibit their action by modifying the expression or downstream effects of genes correlated with lipid metabolism, inflammation, and fibrogenesis. Presently, the most highly touted entity of this group has been miR-122, which constitutes over 70% of hepatic miRNA expression.89 One of its primary targets serves to decrease the expression of numerous genes critical for lipid metabolism, such as FASN, ACC2, SCD1, and ACLY.90 There have been three major studies examining the effects of this miRNA on NAFLD in human subjects. The first two demonstrated an association between miR-122 expression and NAFLD, however subsequent review exposed discordance between the levels miR-122 measured in hepatocytes and the serum.91, 92 This phenomenon remained relatively unchallenged until the landmark investigation by Pirola et al.,93 Serum levels of miR-122 were once again demonstrated to be elevated in individuals with isolated steatosis or NASH as compared to controls. However, the hepatic expression was significantly downregulated in NAFLD, particularly in more severe cases such as NASH. Immunohistochemical staining revealed that miR-122 expression was restricted to the periphery of lipid-laden hepatocytes, suggesting they were in the process of exporting out of the hepatocytes into the circulation. Not only was this finding independently significant, but the serum levels of miR-122 were also found to correlate with hepatocellular ballooning and fibrosis in addition to serving as a superior biomarker for NAFLD than aminotransferases and CK-18, which has gained traction as a novel biomarker for liver disease. These groundbreaking findings suggest the export of miR-122 from hepatocytes into the circulation detects the underlying lack of inhibition on lipogenic gene expression and NAFLD pathology. miRNAs, such as miR-122, still demand further investigation, yet provide novel targets for NAFLD therapy in the future.
Table 4.
Epigenetic and miRNA Targets Correlated with NAFLD.
| Gene | Study population | Notable cohort characteristics | Sample size | Diagnostic modality |
|---|---|---|---|---|
| miR-16, 34a, 12289 | American | 91% male Unreported ethnicities | 34 cases/19 controls | Liver biopsy |
| miR-12292 | American | 82% female 82% Caucasian | 25 NASH/25 controls | Liver biopsy |
| miR-122, 192, 37593 | Argentine | Ethnicities not reported | 16 NASH/16 isolated steatosis/16 controls | Liver biopsy |
| ACLY, GALNTL4, GRID1, IGF1, IGFBP2, IP6K3, PC, PLCG1, PRKCE94 | German and Swiss | Morbidly obese patients undergoing bariatric surgery | 45 cases/18 controls | Liver biopsy or histology from intraoperative sample |
| CASP1, FGFR2, MAT1A, MTHFD295 | American | ∼100% Caucasian | 56 cases/25 controls | Liver biopsy |
| Collagen 1A1, PDGFα, PPARα, PPARδ, TGFβ196 | British | 100% male | 17 cases (no controls, just comparing mild to severe NAFLD) | Liver biopsy |
| MT-ND697 | Argentine | Ethnicities not reported | 45 cases/18 controls | Liver biopsy |
| TET1, TET298 | Argentine | Ethnicities not reported | 67 cases/23 controls | Liver biopsy (only US in controls) |
| TFAM99 | Argentine | Ethnicities not reported | 63 cases/11 controls | Liver biopsy |
| lnc-JAM2-6101 | Argentine | None | 32 NAFL/32 NASH/32 controls | Liver biopsy |
| PPARγ100 | British | 2 subjects previously transplanted for “cirrhotic NAFLD” were included | 26 NAFLD/26 controls | Liver biopsy |
ACLY, ATP Citrate Lyase; CASP1, Caspase 1; Collagen 1A1, Collagen Type 1 Alpha 1; FGFR2, Fibroblast Growth Receptor 2; GALNTL4, UDP-N-acetyl-alpha-d-Galactosamine:Polypeptide N-Acetylgalactosaminyltransferase-like 4; GRID1, Glutamate Receptor, Ionotropic, Delta 1; HDAC3, Histone Deacetylase 3; HDAC8, Histone Deacetylase 8; IGF1, Insulin Like Growth Factor 1; IGFBP2, Insulin Like Growth Factor Binding Protein 2; IP6K3, Inositol Hexakisphosphate Kinase 3; MAT1A, Methionine Adenosyl Methyltransferase 1A, MT-ND6, Mitochondrially Encoded NADH Dehydrogenase 6; MTHFD2, Methylenetetrahydrofolate Dehydrogenase 2; PC, Pyruvate Carboxylase; PDGFα, Platelet Derived Growth Factor Alpha; PPARGC1α, PPARG Coactivator 1 Alpha; PLCG1, Phospholipase C Gamma 1; PPARα, Peroxisome Proliferator Activated Receptor Alpha; PPARδ, Peroxisome Proliferator Activated Receptor Delta, PRKCE, Protein Kinase C, Epsilon; SIRT1, Sirtuin 1; SIRT6, Sirtuin 6; TET1, Ten-Eleven Translocation Methylcytosine Dioxygenase 1; TET2, Ten-Eleven Translocation Methylcytosine Dioxygenase 2; TFAM, Transcription Factor A, Mitochondrial; TGFβ1, Transforming Growth Factor Beta 1; lncRNAs, Long Noncoding RNAs; PPARγ, Peroxisome Proliferator Activate Receptor Gamma; US, Ultrasonography.
Analysis of Epigenetic Modifications
While the seemingly small amount of data regarding miRNAs has yet provided new insight into this pathologic mechanism, epigenetic modifications have been characterized to an even lesser degree. Such epigenetic modifications are driven by DNA methylation. Defects within this epigenetic mechanism have not been studied as thoroughly as other genetic variants, but many have been correlated with NAFLD. Much like for the miRNAs, there has been a great deal of work in mouse models, but only those significant studies performed in human subjects89, 92, 93, 94, 95, 96, 97, 98, 99 have been outlined here in Table 4. The first study analyzed morbidly obese patients undergoing bariatric surgery through a retrospective case–control study.94 After histologic analysis of livers taken from both NAFLD and control patients, a correlation was found between the methylation of several genes with the existence of NAFLD. However, this data is very selective in that the subjects were morbidly obese and undergoing major surgical intervention. Furthermore, the control samples in this study were taken from subjects undergoing major oncologic surgery. This garners data in a very highly specific clinical scenario and may not accurately provide insight to the general population of patients with NAFLD. The next two major studies established a correlation between methylation of several target genes and the diagnosis of NAFLD based upon liver biopsy is presented in Table 4.95, 96 However, a great portion of this data vested its analysis in the epigenetic differences among various degrees of NAFLD severity, or increased fibrosis. Furthermore, it has recently been shown that DNA methylation as detected in the plasma can serve as an accurate biomarker for hepatic fibrosis in NAFLD.100 Certainly there is promise for NAFLD association with epigenetic mechanisms such as these, but many more studies with stronger methodology and a higher level of evidence must be completed in order to confirm this association.
Lastly, there has been novel exploration into the remaining genome and its influence on NAFLD development and severity. Analysis of Long Noncoding RNAs (lncRNA) have provided significant intrigue in regards to their transcriptional and epigenetic influence over this pathophysiology. Most recently, one specific lncRNA, lnc-JAM2-6, was associated with NAFLD and disease severity, potentially through interactions with oncogenes MAFK and JUND and the transcription factor CEBPB that is central to the inflammatory mechanism.101 This study represents a landmark trial in terms of genomic analysis among NAFLD subjects and may have pioneered methodology for future studies.
Emerging single-cell epigenomic methods are being developed with the exciting potential to transform our knowledge of gene regulation. High-throughput sequencing has revolutionized the field of epigenetics with methods for genome-wide mapping of DNA methylation, histone modifications, chromatin accessibility, and chromosome conformation. Initially, the input requirements for these methods meant that samples containing hundreds of thousands or millions of cells were required; but in the last couple of years this has changed with numerous epigenetic features now assayable at the single-cell level.102
Pharmacogenomics
Although genetic makeup of an individual does not fully explain the disease phenotype and natural history of NAFLD, the utility of genetic data on assessing the responsiveness to various therapeutic interventions in NAFLD is emerging. This personalized treatment, or pharmacogenomics, supplies a pragmatic treatment option in NAFLD given the heterogeneity of the disease phenotype, with contributions of host and environmental factors. Furthermore, the genotype of an individual has a potential to change the metabolism of dietary components and also the pharmacodynamics of the medications used for NAFLD patients. For example, de novo lipogenesis and hepatic fat content in patients with I148M variant, or GG polymorphism, of the PNPLA3 gene have been associated with intake of sugars and sweetened beverages103, 104 and hepatotoxicity due to imbalance of the ratio of omega-6 to omega-3 fatty acids.105 On the other hand, this variant was also associated with reduced hepatic steatosis and fat content with increased consumption of vegetables in diet106 and with weight loss.107 Although most studies have shown benefit of statins across the entire NAFLD spectrum, this benefit is blunted in patients with GG polymorphism of the PNPLA3 gene.108 Potential beneficial effects of statins in NAFLD and other chronic liver diseases are based on low quality RCTs, and needs further studies before can be routinely recommended.109 In another study, a marginal association was shown between V433M genotype of CYP4F2, a primary component of vitamin E metabolism, and improvement in NAFLD activity score without impact on vitamin E levels, suggesting another potential mechanism of this benefit in patients with this specific genotype.110 As the fields of genetic testing and therapies for NAFLD evolve, pharmacogenomics and personalized therapy is going to become increasingly more relevant in tailoring the diet, exercise regimen, and use of pharmacological treatment options for managing patients with NAFLD.
Conclusion
The exact mechanism of NAFLD pathogenesis still remains intricately obscure with interplay among environmental factors, individual genetic variants, or alterations in the intestinal microbiome to provide an environment which is susceptible for the development of NAFLD. Several genetic variants have been implicated in this disease process, however their specificity remains unknown. Certain genetic variants, such as PNPLA3 rs738409 and TM6SF2 rs58542926, have been repeatedly demonstrated to be closely linked in this disease process, however it is critical to note that causality cannot be established from these studies. While currently there is a great deal of unknown, identification of known genetic variants will help tailor treatment strategies for these high risk patients in the future. Rigorous prospective investigation of these genetic variants are needed in biopsy proven NAFLD patients in order to firmly establish which of those genetic components serve as the primary culprit of NAFLD pathogenesis, in addition to the progression of NASH to advanced fibrosis, cirrhosis and HCC.
Conflicts of Interest
SKS has received Grant/research support from Biotest, Conatus, Genfit, Gilead Sciences, Intercept, and Shire; served on the Advisory board or as consultant for Abbvie, Gilead Sciences, and Intercept; and on the speakers bureau for Intercept, and Alexion. The other authors declare no related conflicts of interest.
Acknowledgments
We would like to thank Jennifer N. Gentry, CMI for her brilliant work with the medical illustrations she was able to provide. We also wish to thank Dr. Pamela B. Sylvestre, MD of University of Tennessee Health Science Center for generously providing slides in order to visualize NAFLD histology. In addition, we would like to acknowledge Wilfried J.J. Karmaus, MD, MPH for his genetic and epidemiologic expertise in the editing of this paper.
References
- 1.Caldwell S., Argo C. The natural history of non-alcoholic fatty liver disease. Dig Dis. 2010;28:162–168. doi: 10.1159/000282081. [DOI] [PubMed] [Google Scholar]
- 2.Matteoni C.A., Younossi Z.M., Gramlich T. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–1419. doi: 10.1016/s0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 3.Adams L.A., Lymp J.F., St Sauver J. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Angulo P., Kleiner D.E., Dam-Larsen S. Liver fibrosis, but no other histologic features, associates with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alpers D.H., Sabesin S.M., White H.M. Fatty liver; biochemical and clinical aspects. In: Shiff L., Shiff E., editors. Disease of the Liver. Lippincott; Philadelphia: 1993. pp. 825–855. [Google Scholar]
- 6.Sanyal A.J., Campbell-Sargent C., Mirshahi F. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology. 2001;120:1183–1192. doi: 10.1053/gast.2001.23256. [DOI] [PubMed] [Google Scholar]
- 7.Marchesini G., Pagotto U., Bugianesi E. Low ghrelin concentrations in nonalcoholic fatty liver disease are related to insulin resistance. J Clin Endocrinol Metab. 2003;88:5674–5679. doi: 10.1210/jc.2003-031094. [DOI] [PubMed] [Google Scholar]
- 8.Chitturi S., Abeygunasekera S., Farrell G.C. NASH and insulin resistance: insulin hypersecretion and specific association with the insulin resistance syndrome. Hepatology. 2002;35:373–379. doi: 10.1053/jhep.2002.30692. [DOI] [PubMed] [Google Scholar]
- 9.Lotta L.A., Gulati P., Day F.R. Integrative genomic analysis implicates limited peripheral adipose storage capacity in the pathogenesis of human insulin resistance. Nat Genet. 2017;49:17–26. doi: 10.1038/ng.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gastaldelli A., Cusi K., Pettiti M. Relationship between hepatic/visceral fat and hepatic insulin resistance in nondiabetic and type 2 diabetic subjects. Gastroenterology. 2007;133:496–506. doi: 10.1053/j.gastro.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 11.Lomonaco R., Ortiz-Lopez C., Orsak B. Effect of adipose tissue insulin resistance on metabolic parameters and liver histology in obese patients with nonalcoholic fatty liver disease. Hepatology. 2012;55:1389–1397. doi: 10.1002/hep.25539. [DOI] [PubMed] [Google Scholar]
- 12.Pagano G., Pacini G., Musso G. Nonalcoholic steatohepatitis, insulin resistance, and metabolic syndrome: further evidence for an etiologic association. Hepatology. 2002;35:367–372. doi: 10.1053/jhep.2002.30690. [DOI] [PubMed] [Google Scholar]
- 13.Koek G.H., Liedorp P.R., Bast A. The role of oxidative stress in non-alcoholic steatohepatitis. Clin Chim Acta. 2011;412:1297–1305. doi: 10.1016/j.cca.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 14.Kujoth G.C., Hiona A., Pugh T.D. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 15.Lee J., Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem. 2014;289:1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozcan U., Cao Q., Yilmaz E. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs M., Sanyal A.J. Lipotoxicity in NASH. J Hepatol. 2012;56:291–293. doi: 10.1016/j.jhep.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Polyzos S.A., Kountouras J., Mantzoros C.S. Adipokines in nonalcoholic fatty liver disease. Metabolism. 2016;65:1062–1079. doi: 10.1016/j.metabol.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Flier J.S., Maratos-Flier E. Biology of obesity. In: Kasper D., Fauci A., Hauser S., editors. Harrison's Principles of Internal Medicine. 19the ed. McGraw-Hill Education; New York, NY: 2015. [Google Scholar]
- 20.Miele L., Valenza V., La Torre G. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 21.Vijay-Kumar M., Aitken J.D., Carvalho F.A. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shapiro M.D. Rare genetic disorders altering lipoproteins. In: De Groot L.J., Chrousos G., Dungan K., Feingold K.R., Grossman A., Hershman J.M., Koch C., Korbonits M., McLachlan R., New M., Purnell J., Rebar R., Singer F., Vinik A., editors. Endotext. MDText.com, Inc.; South Dartmouth, MA: 2000. [Google Scholar]
- 23.Erwin A.L. The role of sebelipase alfa in the treatment of lysosomal acid lipase deficiency. Therap Adv Gastroenterol. 2017;10:553–562. doi: 10.1177/1756283X17705775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomba R., Schork N., Chen C.H. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149:1784–1793. doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stender S., Kozlitina J., Nordestgaard B.G. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49:842–847. doi: 10.1038/ng.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo S., Kozlitina J., Xing C. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalasani N., Guo X., Loomba R. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology. 2010;139:1567–1576. doi: 10.1053/j.gastro.2010.07.057. 1576.e1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speliotes E.K., Yerges-Armstrong L.M., Wu J. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi T., Sumida Y., Umemura A. Genetic polymorphisms of the human PNPLA3 gene are strongly associated with severity of non-alcoholic fatty liver disease in Japanese. PLoS ONE. 2012;7:e38322. doi: 10.1371/journal.pone.0038322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams L.A., White S.W., Marsh J.A. Association between liver-specific gene polymorphisms and their expression levels with nonalcoholic fatty liver disease. Hepatology. 2013;57:590–600. doi: 10.1002/hep.26184. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto T., Kitamoto A., Yoneda M. Genome-wide scan revealed that polymorphisms in the PNPLA3, SAMM50, and PARVB genes are associated with development and progression of nonalcoholic fatty liver disease in Japan. Hum Genet. 2013;132:783–792. doi: 10.1007/s00439-013-1294-3. [DOI] [PubMed] [Google Scholar]
- 32.Feitosa M.F., Wojczynski M.K., North K.E. The ERLIN1-CHUK-CWF19L1 gene cluster influences liver fat deposition and hepatic inflammation in the NHLBI Family Heart Study. Atherosclerosis. 2013;228:175–180. doi: 10.1016/j.atherosclerosis.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozlitina J., Smagris E., Stender S. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang X.R., Song J.Y., Liu F.H. GWAS-identified common variants with nonalcoholic fatty liver disease in Chinese children. J Pediatr Gastroenterol Nutr. 2015;60:669–674. doi: 10.1097/MPG.0000000000000662. [DOI] [PubMed] [Google Scholar]
- 35.DiStefano J.K., Kingsley C., Craig Wood G. Genome-wide analysis of hepatic lipid content in extreme obesity. Acta Diabetol. 2015;52:373–382. doi: 10.1007/s00592-014-0654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dongiovanni P., Donati B., Fares R. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19:6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smagris E., BasuRay S., Li J. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61:108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalia H.S., Gaglio P.J. The prevalence and pathobiology of nonalcoholic fatty liver disease in patients of different races or ethnicities. Clin Liver Dis. 2016;20:215–224. doi: 10.1016/j.cld.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 39.Santoro N., Zhang C.K., Zhao H. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55:781–789. doi: 10.1002/hep.24806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Xu R., Peng X. Association of glucokinase regulatory protein polymorphism with type 2 diabetes and fasting plasma glucose: a meta-analysis. Mol Biol Rep. 2013;40:3935–3942. doi: 10.1007/s11033-012-2470-6. [DOI] [PubMed] [Google Scholar]
- 41.Wang J., Guo X.F., Yu S.J. Adiponectin polymorphisms and non-alcoholic fatty liver disease risk: a meta-analysis. J Gastroenterol Hepatol. 2014;29:1396–1405. doi: 10.1111/jgh.12562. [DOI] [PubMed] [Google Scholar]
- 42.Li M.R., Zhang S.H., Chao K. Apolipoprotein C3 (-455T>C) polymorphism confers susceptibility to nonalcoholic fatty liver disease in the Southern Han Chinese population. World J Gastroenterol. 2014;20:14010–14017. doi: 10.3748/wjg.v20.i38.14010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Chen L., Xin Y. Apolipoprotein c3 gene polymorphisms are not a risk factor for developing non-alcoholic Fatty liver disease: a meta-analysis. Hepat Mon. 2014;14:e23100. doi: 10.5812/hepatmon.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zain S.M., Mohamed Z., Mohamed R. Common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol. 2015;30:21–27. doi: 10.1111/jgh.12714. [DOI] [PubMed] [Google Scholar]
- 45.Sun M.Y., Zhang L., Shi S.L. Associations between methylenetetrahydrofolate reductase (MTHFR) polymorphisms and non-alcoholic fatty liver disease (NAFLD) risk: a meta-analysis. PLOS ONE. 2016;11:e0154337. doi: 10.1371/journal.pone.0154337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li L., Wang S.J., Shi K. Correlation between MTP -493G>T polymorphism and non-alcoholic fatty liver disease risk: a meta-analysis. Genet Mol Res. 2014;13:10150–10161. doi: 10.4238/2014.December.4.9. [DOI] [PubMed] [Google Scholar]
- 47.Tan H.L., Mohamed R., Mohamed Z. Phosphatidylethanolamine N-methyltransferase gene rs7946 polymorphism plays a role in risk of nonalcoholic fatty liver disease: evidence from meta-analysis. Pharmacogenet Genomics. 2016;26:88–95. doi: 10.1097/FPC.0000000000000193. [DOI] [PubMed] [Google Scholar]
- 48.Sookoian S., Pirola C.J. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 49.Zhang L., You W., Zhang H. PNPLA3 polymorphisms (rs738409) and non-alcoholic fatty liver disease risk and related phenotypes: a meta-analysis. J Gastroenterol Hepatol. 2015;30:821–829. doi: 10.1111/jgh.12889. [DOI] [PubMed] [Google Scholar]
- 50.Xu R., Tao A., Zhang S. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Sci Rep. 2015;5:9284. doi: 10.1038/srep09284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J., Guo X., Wu P. Association between the Pro12Ala polymorphism of PPAR-gamma gene and the non-alcoholic fatty liver disease: a meta-analysis. Gene. 2013;528:328–334. doi: 10.1016/j.gene.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 52.Lee Y.H., Bae S.C., Song G.G. Meta-analysis of associations between the peroxisome proliferator-activated receptor-gamma Pro12Ala polymorphism and susceptibility to nonalcoholic fatty liver disease, rheumatoid arthritis, and psoriatic arthritis. Genet Test Mol Biomarkers. 2014;18:341–348. doi: 10.1089/gtmb.2013.0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Musso G., Cassader M., Bo S. Sterol regulatory element-binding factor 2 (SREBF-2) predicts 7-year NAFLD incidence and severity of liver disease and lipoprotein and glucose dysmetabolism. Diabetes. 2013;62:1109–1120. doi: 10.2337/db12-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pirola C.J., Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: a meta-analysis. Hepatology. 2015;62:1742–1756. doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 55.Goffredo M., Caprio S., Feldstein A.E. Role of TM6SF2 rs58542926 in the pathogenesis of nonalcoholic pediatric fatty liver disease: a multiethnic study. Hepatology. 2016;63:117–125. doi: 10.1002/hep.28283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chamoun Z., Vacca F., Parton R.G. PNPLA3/adiponutrin functions in lipid droplet formation. Biol Cell. 2013;105:219–233. doi: 10.1111/boc.201200036. [DOI] [PubMed] [Google Scholar]
- 57.He S., McPhaul C., Li J.Z. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang Y., Cohen J.C., Hobbs H.H. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basantani M.K., Sitnick M.T., Cai L. Pnpla3/adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.L., Reeves H.L., Burt A.D. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahdessian H., Taxiarchis A., Popov S. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sookoian S., Castano G., Gianotti T.F. Polymorphisms of MRP2 (ABCC2) are associated with susceptibility to nonalcoholic fatty liver disease. J Nutr Biochem. 2009;20:765–770. doi: 10.1016/j.jnutbio.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Kotronen A., Yki-Jarvinen H., Aminoff A. Genetic variation in the ADIPOR2 gene is associated with liver fat content and its surrogate markers in three independent cohorts. Eur J Endocrinol. 2009;160:593–602. doi: 10.1530/EJE-08-0900. [DOI] [PubMed] [Google Scholar]
- 64.Iwamoto N., Ogawa Y., Kajihara S. Gln27Glu beta2-adrenergic receptor variant is associated with hypertriglyceridemia and the development of fatty liver. Clin Chim Acta. 2001;314:85–91. doi: 10.1016/s0009-8981(01)00633-7. [DOI] [PubMed] [Google Scholar]
- 65.Nozaki Y., Saibara T., Nemoto Y. Polymorphisms of interleukin-1 beta and beta 3-adrenergic receptor in Japanese patients with nonalcoholic steatohepatitis. Alcohol Clin Exp Res. 2004;28:106s–110s. doi: 10.1111/j.1530-0277.2004.tb03226.x. [DOI] [PubMed] [Google Scholar]
- 66.Yoneda M., Hotta K., Nozaki Y. Association between angiotensin II type 1 receptor polymorphisms and the occurrence of nonalcoholic fatty liver disease. Liver Int. 2009;29:1078–1085. doi: 10.1111/j.1478-3231.2009.01988.x. [DOI] [PubMed] [Google Scholar]
- 67.Sazci A., Akpinar G., Aygun C. Association of apolipoprotein E polymorphisms in patients with non-alcoholic steatohepatitis. Dig Dis Sci. 2008;53:3218–3224. doi: 10.1007/s10620-008-0271-5. [DOI] [PubMed] [Google Scholar]
- 68.Wang B., Wang B., Wang Y. Association of APPL1 gene polymorphism with non-alcoholic fatty liver disease susceptibility in a Chinese Han population. Clin Lab. 2015;61:1659–1666. doi: 10.7754/clin.lab.2015.150417. [DOI] [PubMed] [Google Scholar]
- 69.Wang L., Athinarayanan S., Jiang G. Fatty acid desaturase 1 gene polymorphisms control human hepatic lipid composition. Hepatology. 2015;61:119–128. doi: 10.1002/hep.27373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira C.P., Stefano J.T., Cavaleiro A.M. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2010;25:357–361. doi: 10.1111/j.1440-1746.2009.06001.x. [DOI] [PubMed] [Google Scholar]
- 71.Carulli L., Canedi I., Rondinella S. Genetic polymorphisms in non-alcoholic fatty liver disease: interleukin-6-174G/C polymorphism is associated with non-alcoholic steatohepatitis. Dig Liver Dis. 2009;41:823–828. doi: 10.1016/j.dld.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 72.Swellam M., Hamdy N. Association of nonalcoholic fatty liver disease with a single nucleotide polymorphism on the gene encoding leptin receptor. IUBMB Life. 2012;64:180–186. doi: 10.1002/iub.597. [DOI] [PubMed] [Google Scholar]
- 73.Zain S.M., Mohamed Z., Mahadeva S. Impact of leptin receptor gene variants on risk of non-alcoholic fatty liver disease and its interaction with adiponutrin gene. J Gastroenterol Hepatol. 2013;28:873–879. doi: 10.1111/jgh.12104. [DOI] [PubMed] [Google Scholar]
- 74.Mancina R.M., Dongiovanni P., Petta S. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150 doi: 10.1053/j.gastro.2016.01.032. 1219–1230.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorden A., Yang R., Yerges-Armstrong L.M. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75:34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu G., Wang K., Xue Y. Association of rs5764455 and rs6006473 polymorphisms in PARVB with liver damage of nonalcoholic fatty liver disease in Han Chinese population. Gene. 2016;575:270–275. doi: 10.1016/j.gene.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 77.Hernaez R., McLean J., Lazo M. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11 doi: 10.1016/j.cgh.2013.02.011. 1183-1190.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vazquez-Chantada M., Gonzalez-Lahera A., Martinez-Arranz I. Solute carrier family 2 member 1 is involved in the development of nonalcoholic fatty liver disease. Hepatology. 2013;57:505–514. doi: 10.1002/hep.26052. [DOI] [PubMed] [Google Scholar]
- 79.Huang Y.S., Chang C.H., Lin T.L. Genetic variations of superoxide dismutase 2 and cytochrome P450 2E1 in non-alcoholic steatohepatitis. Liver Int. 2014;34:931–936. doi: 10.1111/liv.12533. [DOI] [PubMed] [Google Scholar]
- 80.Musso G., Gambino R., Pacini G. Transcription factor 7-like 2 polymorphism modulates glucose and lipid homeostasis, adipokine profile, and hepatocyte apoptosis in NASH. Hepatology. 2009;49:426–435. doi: 10.1002/hep.22659. [DOI] [PubMed] [Google Scholar]
- 81.Kiziltas S., Ata P., Colak Y. TLR4 gene polymorphism in patients with nonalcoholic fatty liver disease in comparison to healthy controls. Metab Syndr Relat Disord. 2014;12:165–170. doi: 10.1089/met.2013.0120. [DOI] [PubMed] [Google Scholar]
- 82.Yan X., Xu L., Qi J. sTRAIL levels and TRAIL gene polymorphisms in Chinese patients with fatty liver disease. Immunogenetics. 2009;61:551–556. doi: 10.1007/s00251-009-0389-4. [DOI] [PubMed] [Google Scholar]
- 83.Aller R., De Luis D.A., Izaola O. Role of -55CT polymorphism of UCP3 gene on non alcoholic fatty liver disease and insulin resistance in patients with obesity. Nutr Hosp. 2010;25:572–576. [PubMed] [Google Scholar]
- 84.Yuan C., Lu L., An B. Association between LYPLAL1 rs12137855 polymorphism with ultrasound-defined non-alcoholic fatty liver disease in a Chinese Han population. Hepat Mon. 2015;15:e33155. doi: 10.5812/hepatmon.33155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen L., Lin Z., Jiang M. Genetic variants in the SAMM50 gene create susceptibility to nonalcoholic fatty liver disease in a Chinese Han population. Hepat Mon. 2015;15:e31076. doi: 10.5812/hepatmon.31076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rottiers V., Naar A.M. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jones A., Danielson K.M., Benton M.C. miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring) 2017 doi: 10.1002/oby.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferreira D.M., Simao A.L., Rodrigues C.M. Revisiting the metabolic syndrome and paving the way for microRNAs in non-alcoholic fatty liver disease. FEBS J. 2014;281:2503–2524. doi: 10.1111/febs.12806. [DOI] [PubMed] [Google Scholar]
- 89.Elmen J., Lindow M., Schutz S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 90.Esau C., Davis S., Murray S.F. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 91.Cermelli S., Ruggieri A., Marrero J.A. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheung O., Puri P., Eicken C. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pirola C.J., Fernandez Gianotti T., Castano G.O. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64:800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ahrens M., Ammerpohl O., von Schonfels W. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 95.Murphy S.K., Yang H., Moylan C.A. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:1076–1087. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeybel M., Hardy T., Robinson S.M. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenet. 2015;7:25. doi: 10.1186/s13148-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pirola C.J., Gianotti T.F., Burgueno A.L. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 98.Pirola C.J., Scian R., Gianotti T.F. Epigenetic modifications in the biology of nonalcoholic fatty liver disease: the role of DNA hydroxymethylation and TET proteins. Medicine (Baltimore) 2015;94:e1480. doi: 10.1097/MD.0000000000001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sookoian S., Rosselli M.S., Gemma C. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 100.Hardy T., Zeybel M., Day C.P. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66:1321–1328. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sookoian S., Rohr C., Salatino A. Genetic variation in long noncoding RNAs and the risk of nonalcoholic fatty liver disease. Oncotarget. 2017;8:22917–22926. doi: 10.18632/oncotarget.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Clark S.J., Lee H.J., Smallwood S.A. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davis J.N., Le K.A., Walker R.W. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. 2010;92:1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sevastianova K., Santos A., Kotronen A. Effect of short-term carbohydrate overfeeding and long-term weight loss on liver fat in overweight humans. Am J Clin Nutr. 2012;96:727–734. doi: 10.3945/ajcn.112.038695. [DOI] [PubMed] [Google Scholar]
- 105.Santoro N., Savoye M., Kim G. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS ONE. 2012;7:e37827. doi: 10.1371/journal.pone.0037827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nobili V., Liccardo D., Bedogni G. Influence of dietary pattern, physical activity, and I148M PNPLA3 on steatosis severity in at-risk adolescents. Genes Nutr. 2014;9:392. doi: 10.1007/s12263-014-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sevastianova K., Kotronen A., Gastaldelli A. Genetic variation in PNPLA3 (adiponutrin) confers sensitivity to weight loss-induced decrease in liver fat in humans. Am J Clin Nutr. 2011;94:104–111. doi: 10.3945/ajcn.111.012369. [DOI] [PubMed] [Google Scholar]
- 108.Dongiovanni P., Petta S., Mannisto V. Statin use and non-alcoholic steatohepatitis in at risk individuals. J Hepatol. 2015;63:705–712. doi: 10.1016/j.jhep.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 109.Kamal S., Khan M.A., Seth A. Beneficial effects of statins on the rates of hepatic fibrosis, hepatic decompensation, and mortality in chronic liver disease: a systematic review and meta-analysis. Am J Gastroenterol. 2017 doi: 10.1038/ajg.2017.170. [DOI] [PubMed] [Google Scholar]
- 110.Athinarayanan S., Wei R., Zhang M. Genetic polymorphism of cytochrome P450 4F2, vitamin E level and histological response in adults and children with nonalcoholic fatty liver disease who participated in PIVENS and TONIC clinical trials. PLOS ONE. 2014;9:e95366. doi: 10.1371/journal.pone.0095366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lutz S.Z., Peter A., Machicao F. Genetic variation in the 11beta-hydroxysteroid-dehydrogenase 1 gene determines NAFLD and visceral obesity. J Clin Endocrinol Metab. 2016;101:4743–4751. doi: 10.1210/jc.2016-2498. [DOI] [PubMed] [Google Scholar]
- 112.Kapil S., Duseja A., Sharma B.K. Genetic polymorphism in CD14 gene, a co-receptor of TLR4 associated with non-alcoholic fatty liver disease. World J Gastroenterol. 2016;22:9346–9355. doi: 10.3748/wjg.v22.i42.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kohan L., Safarpur M., Abdollahi H. Omentin-1 rs2274907 and resistin rs1862513 polymorphisms influence genetic susceptibility to nonalcoholic fatty liver disease. Mol Biol Res Commun. 2016;5:11–17. [PMC free article] [PubMed] [Google Scholar]
- 114.Nakajima S., Tanaka H., Sawada K. Polymorphism of receptor-type tyrosine-protein phosphatase delta gene in the development of non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2017 doi: 10.1111/jgh.13820. [DOI] [PubMed] [Google Scholar]