Abstract
Background
This study was designed to detect and analyze miR-146b-mediated circular RNA (circRNA) expression in hepatic stellate cells.
Material/Methods
The experiment was divided into a control group and a siRNA-miR-146b group. The interference efficiency of siRNA-miR-146b was confirmed by real-time quantitative reverse transcription PCR (qRT-PCR) and the cells were collected, and total RNA was collected for high flux sequencing. The miRNA-targeted carcass were predicted. Finally, the expression of 5 circRNAs was verified by qRT-PCR.
Results
miR-146b expression in the siRNA-miR-146b group was significantly lower than that in the control group. The quality of the original sequencing data and the processed data satisfied with the analysis, and the expression of circRNAs was modulated after the reduction of miR-146b. Among them, 18 circRNAs were upregulated, while 77 circRNAs were downregulated in the miR-146b group compared with the control group. The gene prediction showed that hsa_circ1887 was the largest contact point in miRNA and circRNA regulatory networks. qRT-PCR showed that rno-circRNA-469, rno-circRNA-1138, rno-circRNA-2168 and rno-circRAN-1907 were significantly reduced, while circRNA-1984 was significantly promoted in the siRNA-miR-146b group compared with the control group, which were consistent with the measurements by high-throughput sequencing technique.
Conclusions
miR-146b could regulate the expression of circRNAs in HSCs, which might take part in the formation and development of hepatic fibrosis.
MeSH Keywords: High-Throughput Nucleotide Sequencing, MicroRNAs, RNA Cap Analogs
Background
Liver fibrosis is a process of self-repair of the liver in response to chronic irritation injuries. However, liver fibrosis is also detrimental to liver, which may eventually lead to cirrhosis, liver failure, and even liver cancer [1,2]. How to delay or even reverse the development of liver fibrosis is particularly important. The pathogenesis of liver fibrosis is controlled by multiple cytokines and cell signaling pathways [3]. The increase of the extracellular matrix (ECM) caused by increased secretion or decreased degradation will cause a large amount of ECM deposition, which eventually contributes to liver fibrosis. Activated hepatic stellate cells (HSC) are majorly responsible for ECM production and there are several signaling pathways involved in HSC activation and formation of liver fibrosis [4,5].
MicroRNAs (miRNAs) are a type of highly conserved non-coding RNAs of 18–25 nucleotides in eukaryotes, which regulate mRNA expression through degrading transcript factors or inhibiting transcriptional activity [6–8]. miR-146b belongs to the miR-146 family and is located at chromosome 10 [9]. miR-146b-5p is the functional structure of miR-146b, which participants in the inflammatory regulation and plays an important role in the innate immunity [9]. The deletion of miR-146b likely contributes to chromosome fifth short arm partial deletion syndrome, which is closely related to the inflammatory response [10–12]. However, the mechanisms involved in the regulation of liver diseases by miR-146b are still not known.
Circular RNAs (circRNAs) are a new class of non-coding RNA without 5′ cap and 3′ tail [13,14]. With the development of RNA-Sequencing, especially single cell RNA-Sequencing technology, more functional circRNAs were distinguished in different species [15,16]. According to the source, circRNAs are divided into 3 categories: exonic circRNA, intronic circRNA, and exonic and intronic circRNA. circRNAs can regulate the expression of miRNA by RNA sponge [17,18]. With the further investigation of circRNAs, studies have implicated the important roles of circRNAs in liver diseases [19–21]. This study was designed to screen and obtain ideal markers for the diagnosis and treatment of liver fibrosis by reducing miR-146b expression.
Material and Methods
Cell culture and plasmid transfection
HSC-T6 cell line was purchased from Shanghai Cell Bank of Chinese Academy of Science (China) and cultured in Dulbecco’s Modified Eagle Medium (DMEM, high glucose) (Hyclone, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Shanghai, China) in 5% CO2 at 37°C. The cell confluence at 90% was used in plasmid transfection. The serum-free medium and Opti-MEM were reheated and balanced. The medium was replaced by a serum-free medium and the tube was sterilized to mix well with the plasmids (about 5 μg/well), each tube added 250 μL Opti-MEM and 10 μL Lipofectamine 2000. The mixture was added into the 6-well plate (500 μL/well) with the cells and incubated for 4 hours. Thereafter, the normal medium was changed back. 24 hours later, real-time quantitative reverse transcription PCR (qRT-PCR) was used to detect the interference efficiency. miR-146b vector was constructed in our lab according to the sequence. The cells with optimal interference efficiency were prepared for high-throughput sequencing experiment.
Illumina sequencing library
Total RNA was extracted following the instruction of TRIzol Reagent (Invitrogen). Nanodrop 2000 was used to detect the concentration and purity of the RNA. The integrity of RNA was detected by agarose gel electrophoresis, and RNA integrity number was measured by Agilent 2100. A database was established, and total RNA should satisfy with the following parameters: total level >5 μg, concentration ≥200 ng/μL, and OD260/280 between 1.8 and 2.0. Then Ribo-Zero Magnetic Kit (EpiCentre) was used to remove rRNA. RNase R (EpiCentre) was used to remove linear RNA. Paired-End sequencing library was constructed according to TruSeqTM Stranded Total RNA Library Prep Kit assay (Illumina). RNA sequence was measured by Hiseq4000 Platform.
Data analysis
SepPrep and Sickle software were used to evaluate the data quality, and the data were compared to the reference genome data by Bowtie. The circRNAs were predicted by KNIEF, and the expression of circRNA was calculated and classified as previously described [22,23].
Enrichment analysis of differentially expressed circRNAs
The differentially expressed genes underwent Gene Ontology (GO) function analysis (molecular function, cell component, biological processes) and were enriched and analyzed by Goatools software [24]. Thereafter, the data were analyzed with KOBAS for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and KEGG annotation.
Prediction of miRNA-targeted circRNAs
Miranda was used to predict the target genes of known miRNA.
qRT-PCR
qRT-PCR was used to verify the differential expression of circRNAs. According to the results of high-throughput sequencing, 5 differentially expressed circRNAs (rno-circRNA-1907, rno-circRNA-2168, rno-circRNA-1138, rno-circRNA-469 and rno-circRNA-1984) were screened, and the primers were designed with Primer 5 to verify the expression of the 5 circRNAs (Table 1). The expression of circRNAs was normalized to U6.
Table 1.
The primers for different circRNAs.
| circRNAs | Sequences of the primers | Length (bp) | Product length (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| rno-circRNA-1907 F | CTGTGTGCCCCCAGGTCTAC | 20 | 174 | 62.0 |
| rno-circRNA-1907 R | TATCATGGCCCGTCTCCAGC | 20 | ||
| rno-circRNA-2168 F | GCGCCTTGCTGGATACTGTG | 20 | 174 | 62.1 |
| rno-circRNA-2168 R | GCACAGCGAATGGATGGAGC | 20 | ||
| rno-circRNA-1138 F | CGTGGACGAGGAGCCAGTAG | 20 | 177 | 62.0 |
| rno-circRNA-1138 R | GGCCCAGCTCCTTTTTCTGC | 20 | ||
| rno-circRNA-469 F | TACCCTGCGTGTGGTCATCG | 20 | 137 | 62.1 |
| rno-circRNA-469 R | GTGTCGGTGACAGGAAGCCT | 20 | ||
| rno-circRNA-1984 F | AGGGGGTTGGTGTTGCTGAA | 20 | 145 | 62.0 |
| rno-circRNA-1984 R | GGTGAGGGAGGGAAAAGCCT | 20 | ||
| U6-F | CTCGCTTCGGCAGCACA | 17 | 94 | 60.0 |
| U6-R | AACGCTTCACGAATTTGCGT | 20 |
Results
Confirmation of the interference efficiency
qRT-PCR detected the expression of miR-146b after plasmid transfection. As shown in Figure 1, the expression of miR-146b decreased significantly in siRNA-miR-146b group compared with the control group.
Figure 1.
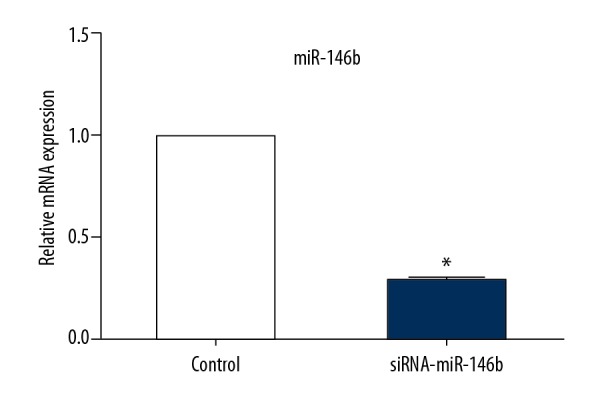
miR-146b siRNA reduces miR-146b expression. * P<0.05 compared with control.
Quality analysis of sequencing data
The number of sequences in the control group and siRNA-miR-146b group were 109 622 526 and 129 909 998, respectively. The bases with Phred value >20 accounted for about 98% of the total base group (Table 2), indicating that the original sequencing data were of good quality and could be used for the subsequent data analysis.
Table 2.
Quality analysis of the original data.
| Groups | Number of sequences | Bases (bp) | Error rate (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| Control | 109622526 | 16553001426 | 0.0115 | 98.30 | 95.72 |
| miR-146b | 129909998 | 19616409698 | 0.0115 | 98.24 | 95.62 |
Quality control analysis of sequencing data
The original data were processed to obtain clean data. After data quality control analysis, the processed data reached the quality requirement (Table 3) and the bases of Phred value greater than 20 accounted for more than 99% of the total base group. Therefore, the data could be used for subsequent analysis. The quality control data were compared to the reference sequence by the Read1 and Read2 of the original data (Table 4). The ratio of the comparison to the genome was over 64%, and the ratio of the alignment to the nonlinear shear loci was about 0.04%, and the ratio of the comparison to the ribosome was about 0.27%.
Table 3.
Quality control analysis.
| Groups | Number of sequences | Bases (bp) | Error rate (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| Control | 107124938 | 14224896706 | 0.0107 | 99.05 | 96.96 |
| miR-146b | 127157642 | 16863219046 | 0.0107 | 99.05 | 96.92 |
Table 4.
Comparison of the data.
| Samples | Genome mapping (%) | JUNC (%) | RIBO mapping (%) |
|---|---|---|---|
| miR-146b_Read1 | 64.38 | 0.05 | 0.29 |
| miR-146b_Read2 | 64.38 | 0.05 | 0.29 |
| Control_Read1 | 68.11 | 0.04 | 0.27 |
| Control_Read2 | 68.08 | 0.04 | 0.27 |
Identification and expression analysis of circRNAs
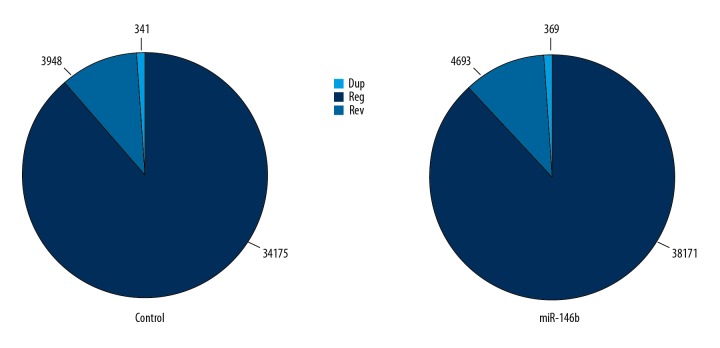
A total of 21 819 circRNAs were identified by data analysis, of which 12 466 circRNAs were identified in the miR-146b treatment group, and 9353 circRNAs were identified in the control group, suggesting that miR-146b might induce changes of circRNA expression in HSC-T6 cells. There were 3 major types of circRNAs identified (Figure 2). In the miR-146b treatment group, 38 171 reads were identified as reg (normal shear), 4693 reads belonged to rev (formed by 2 or more exons), and 369 reads belonged to dup (the circular shear formed by a single exon). In the control group, 34 175 reads belonged to reg, 3948 reads belonged to rev and 341 reads belonged to dup, and the proportion of 3 different types of circRNAs in the 2 groups was similar. The base information about the cicRNAs in the 2 groups were listed in Supplementary Data 1, available from authors on request.
Figure 2.
Types of circRNAs in control and siRNA-miR-146b groups.
Spliced reads per billion mapping (SPBRM) was used to express the number of reads in each transcript and the values indicated the expression of circRNAs. The gene expression was calculated with log10 (SPBRM+1) (Figure 3). The results showed that the gene expression was relatively uniform, and the sequencing data could be used for subsequent analysis, although the sequence data and gene expression in different samples were different.
Figure 3.
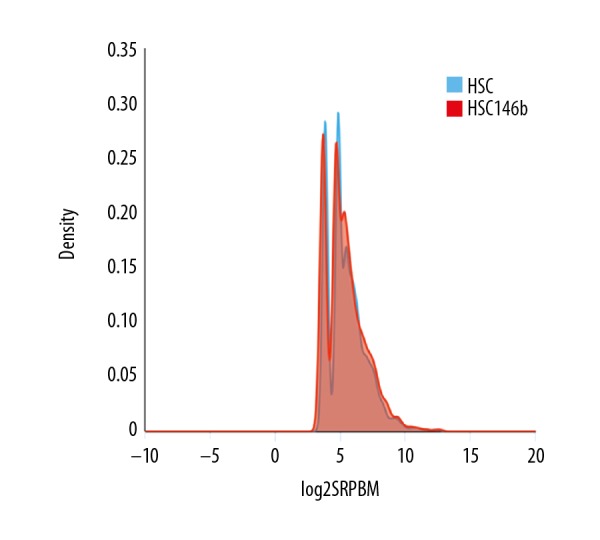
Expression of different types of circRNAs after inhibition of miR-146b.
Analysis of differential gene expression
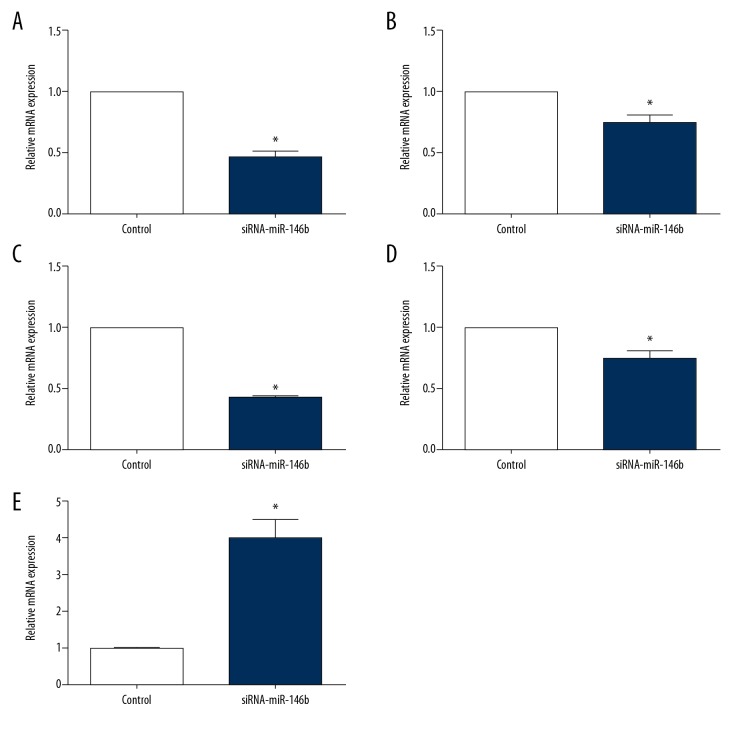
The multiplier of differentially expressed genes between samples more than 1, and the significant difference of P values in the samples less than 0.05 indicated differential expression. 95 differential genes were screened between the 2 groups (Figure 4), of which 18 circRNAs were up-regulated, 77 circRNAs were downregulated in the miR-146b group (Supplementary Data 2, available from authors on request). Especially, rno-circRNA-469, rno-circRNA-1138, rno-circRNA-2168, rno-circRNA-1907 and rno-circRNA-1984 were significantly altered.
Figure 4.
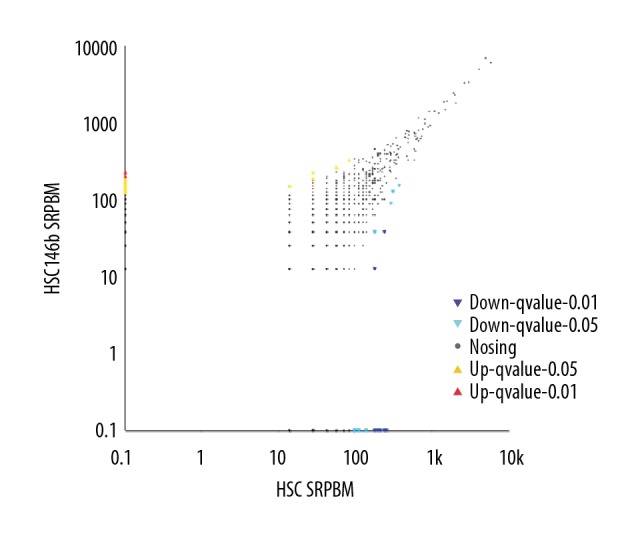
Differential gene analysis (HSC – Control, HSC146B for group miR-146b).
Prediction of miRNA-targeted circRNAs
Using Cytoscape to visualize the relationship between miRNA and circRNAs (Figure 5), the results showed that hsa_circ1887 was the largest contact point in miRNA and circRNA regulatory networks.
Figure 5.
miRNA and circRNA regulatory network. The rhombus represents the ring RNA, and the arrow represents the miRNAs associated with the circRNAs.
qRT-PCR confirmed the differential expression of circRNAs
Since high-throughput sequencing has a certain false positive, we screened 5 circRNAs using qRT-PCR according to the multiple relationships of differential genes. We found the expression of rno-circRNA-469, rno-circRNA-1138, rno-circRNA-2168 and rno-circRAN-1907 were significantly reduced in the siRNA-miR-146b group compared with the control group (Figure 6). While the expression of circRNA-1984 was significantly higher in the siRNA-miR-146b group compared with the control group. These data were consistent with the results of high-throughput sequencing.
Figure 6.
The expression of 5 circRNA by QRT-PCR. (A) rno-circRNA-469; (B) rno-circRNA-1138; (C) rno-circRNA-2168; (D) rno-circRNA-1907; (E) rno-circRNA-1984. * P<0.05 compared with control.
Discussion
HSCs are the main source of ECM production. The imbalance of synthesis and degradation of ECM will cause excessive deposition of hepatic fibrous, finally turning to cirrhosis. The mechanism of hepatic fibrosis is complex and not clear [25,26]. The central pathogenesis of liver fibrosis is HSC activation. Therefore, anti-fibrosis therapy is effective by inhibiting HSC activation. The methods mainly include the follows: drug intervention acts on HSC and inhibits its activation; removal of causes and inhibition of the secretion of cytokines in liver fibrosis (such as TGF-beta, PDGF, etc.), thus indirectly inhibiting the activation of HSC; promoting HSC apoptosis and reducing the numbers of active HSCs; inhibiting the synthesis and promoting the degradation of ECM [27].
As non-coding RNAs, circRNAs are considered to be a wide, diverse, rich, and stable RNA molecules [28]. The characteristics and functions of circRNA are emerging. After exposure to a variety of harmful substances, such as radiation and radiation proinflammatory cytokines, HSCs are activated, accompanied by excessive production of ECM, including alpha smooth muscle actin (α-SMA) and collagen, which eventually lead to liver fibrosis. In the current study, we identified a large number of differentially-expressed circRNAs between normal and siRNA-miR-146b-transfected HSCs from different genomic locations. It was found that in siRNA-miR-146b group, 18 circRNAs were upregulated, while 77 circRNAs were downregulated compared with control group. In particular, 5 circRNAs were significantly altered after miR-146b reduction. Therefore, we verified circRNA expression by qRT-PCR, and the results were consistent with microarray data, indicating the reliability of microarray analysis. Abnormal expression of circRNAs may be involved in the process of fibrosis. rno-circRNA-469, rno-circRNA-1138, rno-circRNA-2168 and rno-circRNA-1907 were positively related to the results of siRNA-miR-146b, indicating that these 4 circRNAs may play the same role as miR-146b for the modulation of HSC in the formation of liver fibrosis. By contrast, rno-circRNA-1984 was negatively related to siRNA-miR-146b, which indicates that this circRNA may play an opposite role in the regulation of HSC in the formation of liver fibrosis by miR-146b. For example, circRNA_000203 is upregulated in mouse cardiac fibroblasts and can enhance the expression of fibrosis related genes, including Col1a2, Col3a1, and α-SMA [29,30]. Nevertheless, the functions of circRNAs in liver fibrosis need further confirmation.
CircRNAs could be potential diagnostic biomarkers for malignant tumors (such as primary and metastatic ovarian cancer, acute myelocytic leukemia, non-small cell lung cancer and colorectal cancer) and non-malignant diseases (such as severe depression) [31,32]. Meanwhile, circRNAs competed with miRNA in cytoplasm, thus interfering with gene expression. Rong et al. analyzed the network between circRNAs and miRNAs [33]. One circRNA can interact with multiple miRNAs [18]. The regulatory role of miRNA in some pathological or physiological processes indicates that circRNAs may be the candidate regulatory factors in the process of liver fibrosis.
Conclusions
Our study demonstrates that miR-146b regulates circRNAs in HSCs. miR-146b and related circRNAs could provide a new theoretical basis and therapeutic targets for liver fibrosis and cirrhosis.
Footnotes
Source of support: This work was supported by grants from the National Natural Science Foundation of China (81760115), the Key Project of Jiangxi Provincial Department of Education (GJJ170004)
References
- 1.Nielsen SR, Quaranta V, Linford A, et al. Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat Cell Biol. 2016;18(5):549–60. doi: 10.1038/ncb3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebastiani G, Ghali P, Wong P, et al. Physicians’ practices for diagnosing liver fibrosis in chronic liver diseases: A nationwide, Canadian survey. Can J Gastroenterol Hepatol. 2014;28(1):23–30. doi: 10.1155/2014/675409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuppan D, Ashfaq-Khan M, Yang AT, Kim YO. Liver fibrosis: Direct antifibrotic agents and targeted therapies. Matrix Biol. 2018;68–69:435–51. doi: 10.1016/j.matbio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Zhou C, York SR, Chen JY, et al. Long noncoding RNAs expressed in human hepatic stellate cells form networks with extracellular matrix proteins. Genome Med. 2016;8(1):31. doi: 10.1186/s13073-016-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Zeng X, He Z, Zou Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM Trans Comput Biol Bioinform. 2017;14(4):905–15. doi: 10.1109/TCBB.2016.2550432. [DOI] [PubMed] [Google Scholar]
- 7.Marabita F, de Candia P, Torri A, et al. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17(2):204–12. doi: 10.1093/bib/bbv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10) doi: 10.3390/ijms17101712. pii: E1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou CK, Chen RF, Chou FF, et al. miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAF(V600E) mutation. Thyroid. 2010;20(5):489–94. doi: 10.1089/thy.2009.0027. [DOI] [PubMed] [Google Scholar]
- 10.Chou CK, Yang KD, Chou FF, et al. Prognostic implications of miR-146b expression and its functional role in papillary thyroid carcinoma. J Clin Endocrinol Metab. 2013;98(2):E196–205. doi: 10.1210/jc.2012-2666. [DOI] [PubMed] [Google Scholar]
- 11.Garcia AI, Buisson M, Bertrand P, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3(5):279–90. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu E, Zhao J, Ma J, et al. miR-146b-5p promotes invasion and metastasis contributing to chemoresistance in osteosarcoma by targeting zinc and ring finger 3. Oncol Rep. 2016;35(1):275–83. doi: 10.3892/or.2015.4393. [DOI] [PubMed] [Google Scholar]
- 13.Huang C, Shan G. What happens at or after transcription: Insights into circRNA biogenesis and function. Transcription. 2015;6(4):61–64. doi: 10.1080/21541264.2015.1071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Guo J, Chen Y, et al. Comprehensive CircRNA expression profile and selection of key CircRNAs during priming phase of rat liver regeneration. BMC Genomics. 2017;18(1):80. doi: 10.1186/s12864-016-3476-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Li L, Wang Q, et al. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–46. doi: 10.1159/000485953. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J, Yang Y, Jiang R. [Regulating mechanisms of circRNA and their relationship with cardiovascular diseases]. Zhonghua Xin Xue Guan Bing Za Zhi. 2016;44(4):364–66. doi: 10.3760/cma.j.issn.0253-3758.2016.04.021. [in Chinese] [DOI] [PubMed] [Google Scholar]
- 17.Jin X, Feng CY, Xiang Z, et al. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7(41):66455–67. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43(5):2525–34. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Zhou Y, Cheng Y, et al. Metformin-induced changes of the coding transcriptome and non-coding RNAs in the livers of non-alcoholic fatty liver disease mice. Cell Physiol Biochem. 2018;45(4):1487–505. doi: 10.1159/000487575. [DOI] [PubMed] [Google Scholar]
- 20.Fu L, Yao T, Chen Q, et al. Screening differential circular RNA expression profiles reveals hsa_circ_0004018 is associated with hepatocellular carcinoma. Oncotarget. 2017;8(35):58405–16. doi: 10.18632/oncotarget.16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Li P, Song Y, et al. Progress and prospects of circular RNAs in Hepatocellular carcinoma: Novel insights into their function. J Cell Physiol. 2018;233(6):4408–22. doi: 10.1002/jcp.26154. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Zheng YC, Kayani MUR, et al. Comprehensive analysis of circRNA expression profiles in humans by RAISE. Int J Oncol. 2017;51(6):1625–38. doi: 10.3892/ijo.2017.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen TB. Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol. 2018;6:20. doi: 10.3389/fcell.2018.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C, Zeng B, Huang J, et al. Genome-wide transcriptome and expression profile analysis of Phalaenopsis during explant browning. PLoS One. 2015;10(4):e0123356. doi: 10.1371/journal.pone.0123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angulo P, Keach JC, Batts KP, Lindor KD. Independent predictors of liver fibrosis in patients with nonalcoholic steatohepatitis. Hepatology. 1999;30(6):1356–62. doi: 10.1002/hep.510300604. [DOI] [PubMed] [Google Scholar]
- 26.Benhamou Y, Bochet M, Di Martino V, et al. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group. Hepatology. 1999;30(4):1054–58. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Jin Hong HU, Cai Z, et al. [Modulating the HSC-T6 activity by sodium ferulate in vitro]. Pharmaceutical Journal of Chinese Peoples Liberation Army. 2003;19(1):4–7. [in Chinese] [Google Scholar]
- 28.Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43(5):2525–34. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xuan L, Qu L, Zhou H, et al. Circular RNA: A novel biomarker for progressive laryngeal cancer. Am J Transl Res. 2016;8(2):932–39. [PMC free article] [PubMed] [Google Scholar]
- 30.Brucher BL, Li Y, Schnabel P, et al. Genomics, microRNA, epigenetics, and proteomics for future diagnosis, treatment and monitoring response in upper GI cancers. Clin Transl Med. 2016;5(1):13. doi: 10.1186/s40169-016-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, Bi ZY, Chen ZL, et al. Emerging landscape of circular RNAs in lung cancer. Cancer Lett. 2018;427:18–27. doi: 10.1016/j.canlet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Wang X, Lu W, et al. Comprehensive analysis of differentially expressed non-coding RNAs and mRNAs in gastric cancer cells under hypoxic conditions. Am J Transl Res. 2018;10(3):1022–35. [PMC free article] [PubMed] [Google Scholar]
- 33.Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–81. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]