Abstract
Background
The pharmacological activation of thermogenesis in brown adipose tissue has long been considered promising strategies to treat obesity. However, identification of safe and effective agents remains a challenge. In this study, we addressed this challenge by developing a cellular system with a fluorescence readout, and applied in a high-throughput manner to screen for FDA-approved drugs that may activate endogenous UCP1 expression in adipocytes.
Methods
We have generated a Ucp1-2A-GFP reporter mouse, in which GFP intensity serves as a surrogate of the endogenous expression level of UCP1 protein; and immortalized brown adipocytes were derived from this mouse model and applied in drug screening. Candidate drugs were further tested in mouse models either fed with normal chow or high fat diet to induce obesity.
Findings
By using the cellular screening platform, we identified a group of FDA-approved drugs that can upregulate UCP1 expression in brown adipocyte, including previously known UCP1 activators and new candidate drugs. Further studies focusing on a previously unreported drug—sutent, revealed that sutent treatment could increase the energy expenditure and inhibit lipid synthesis in mouse adipose and liver tissues, resulting in improved metabolism and resistance to obesity.
Interpretation
This study offered an easy-to-use cellular screening system for UCP1 activators, and provided a candidate list of FDA-approved drugs that can potentially treat obesity. Further study of these candidates may shed new light on the drug discovery towards obesity.
Fund
National Key Research and Development Program and the Strategic Priority Research Program of the Chinese Academy of Sciences, etc. (250 words).
Keywords: Uncoupling protein 1, Brown adipose, FDA-approved drug library, Obesity, Sutent
Research in context.
Evidence before this study
The discovery of thermogenic brown adipose tissue in adult human highlights pharmacological activation of thermogenesis in brown adipose tissue as potential strategies to promote weight loss. Previously approaches have been adopted to screening for activators of UCP1, which applies as a surrogate of thermogenic activity. These approaches either use quantitative PCR examination of human UCP1 mRNA expression or use luciferase assay to determine the endogenous UCP1 protein level with an established Ucp1-2A-luciferase reporter cell line. However, the nature of PCR or luciferase assay makes the screening assays complicated and difficult to go high-throughput. Although numerous UCP1 activators have been identified so far, to our knowledge, a systematic search and analysis of FDA-approved drugs for potentially repurposing to treat obesity has not been performed.
Added value of this study
In this study, we developed a cellular system with a fluorescence readout, which can be applied in a high-throughput manner to identify chemicals that can activate endogenous UCP1 expression in adipocytes. Using this system, we screened a FDA-approved compound library and identified a group of FDA-approved drugs that can upregulate UCP1 expression in brown adipocytes. We further focused on the effect of sutent and revealed that sutent treatment could increase the energy expenditure and inhibit lipid synthesis in adipose and liver tissues, resulting in improved metabolism and resistance to obesity.
Implications of all the available evidence
Our study highlights an easy-to-use cellular screening system for UCP1 activators, and provides a candidate list of FDA-approved drugs that can potentially treat obesity.
Alt-text: Unlabelled Box
1. Introduction
Obesity is a major risk factor for the development of the metabolic syndrome, type 2 diabetes, and cardiovascular disease, posing a tremendous burden for patients and the public healthcare system. Current available drugs to treat obesity mostly center around the reduction of energy-intake, either through appetite-suppressing (e.g., phentermine), or inhibition in intestinal lipid absorption (e.g., orlistat) [1]. However, adverse side effects exist with these treatments; alternative strategies, for example, to target energy-expending pathways are therefore demanded, as potential complementary medications to treat obesity.
The discovery of thermogenic brown adipose tissue (BAT) in adult human [[2], [3], [4], [5], [6]], together with broadened understanding of the “browning” process of energy-storing white adipose tissue (WAT) to energy consuming beige adipose tissue [[7], [8], [9]], highlight pharmacological activation of BAT thermogenesis or induction of WAT browning as potential strategies to promote weight loss. A defining attribute of brown or beige adipocytes is their expression of uncoupling protein 1 (UCP1). UCP1 presents in the inner membrane of mitochondria. It disrupts the proton gradient generated in oxidative phosphorylation by increasing the permeability of the inner mitochondrial membrane. UCP1-mediated heat generation uncouples the respiratory chain, which allows for fast substrate oxidation with a low rate of ATP production [[10], [11], [12]]. Transgenic expression of UCP1 in adipose tissues, either in rodents or in pigs — the latter lack a functional UCP1 gene, decreases fat deposition and improves thermogenesis and metabolism [13,14].
Signaling pathways in regulating BAT activity and UCP1 expression have been intensely studied in recent years. BAT stimulation occurs predominantly through the action of norepinephrine, which is released by the sympathetic nervous system, in the adrenergic receptors, and specifically the adrenergic receptor beta 3 (AR-beta3) [15,16]. The formation of the 3′,5′ cyclic adenosine monophosphate (cAMP) and consequent stimulation of the production of protein kinase A (PKA) are later found to be crucial for AR-beta3 signaling in upregulating UCP1 expression and activating BAT [17]. cAMP/PKA then stimulates numerous pathways, among which, activation of p38 MAPK signaling pathway is essential for upregulating UCP1 expression as well as enhancing mitochondrial activity [[18], [19], [20], [21], [22]]. Several other pathways are also found to be important for BAT thermogenesis, including the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway [23,24], the vascular endothelial growth factor (VEGF) pathway [25] and the silent information regulator type 1 (SIRT1) pathway [26]. On the other hand, the browning process of the white adipose tissue presents a more sophisticated system since the first discovery of this phenomenon in 1984 [27]. However, similar to the role of adrenergic stimulation in activating BAT, the browning of WAT shares the most part of the norepinephrine-induced process and many key signaling pathways mentioned above [[7], [8], [9]]. Besides exploring of regulatory pathways in UCP1 regulation, searching for pharmacological approaches to increase UCP1 expression in adipose tissues offers a straightforward strategy to develop new treatments that may enhance whole-body thermogenic capacity and energy expenditure. To date, a number of compounds or cytokines, such as fibroblast growth factor 21 (FGF21) [28], artemisinin derivatives [29], berberine [30], adenosine [31], resveratrol [32,33], etc., have been demonstrated to be able to induce UCP1 expression in BAT or WAT. Several reviews have carefully summarized these reagents [34,35]. Among these known UCP1 activators, the discovery that some FDA-approved drugs, eg. artemisinin (approved for treating malaria), resveratrol (approved for cancer therapy), can stimulate UCP1 expression, enthuses about an idea to re-search FDA-approved drugs for potentially repurposing to treat obesity.
Success in identifying drugs relies on the development of screening assays. Different screening approaches have previously been adopted to identify chemicals to activate UCP1 expression. These approaches either use quantitative PCR examination of UCP1 mRNA expression in adipocytes [36]; or luciferase assay to determine the endogenous UCP1 protein level with an established Ucp1-2A-luciferase reporter cell line [37,38]. Both approaches successfully identified chemicals that can regulate UCP1 expression; however, the nature of PCR or luciferase assay makes the screening assays complicated and difficult to go high-throughput. To further facilitate the identification of pharmacological agents to modulate UCP1 expression in adipocytes, we have generated a Ucp1-2A-GFP reporter mouse, in which GFP intensity serves as a surrogate of the endogenous expression level of UCP1 protein. A chemical screen using immortalized brown adipocytes derived from this model and targeting the FDA-approved drug library revealed a group of drugs that may regulate UCP1 protein expression in brown adipocytes. Additional in vivo study focusing on one drug demonstrated significant weight loss and improved metabolism in mice treated with this drug, suggesting potential repurposing of this drug to treat obesity. Our study also highlights a useful cell model that enables high-throughput identification of pharmacological modulators of UCP1 expression in adipocytes.
2. Materials and methods
2.1. Animals
All animal procedures were performed according to guidelines by the Institutional Animal Care and Use Committee of the Shanghai Institutes for Biological Sciences. C57BL/6J mice were fed with normal chow diet (NCD) or high fat diet (HFD, 60% of energy from fat, Research Diets, Inc., D12492) for 16 weeks, and treated with sutent (20 mg/kg/day) or vehicle (10% PEG300, 0.5%Tween, H2O) as control by daily oral gavage. The body weight and food intake were monitored weekly. Body fat content was determined by nuclear magnetic resonance (Echo MRI).
2.2. Generation of Ucp1-2A-GFP reporter mice
The Ucp1-2A-GFP mouse was generated with a CRISPR-Cas9 approach. Single gRNA was screened targeting the stop codon locus of the endogenous Ucp1 gene (sequence: ATTGTACCACATAAGCAACT+TGG). Donor plasmid harboring a left arm of 2 kb before the stop codon of the Ucp1 gene, a right arm of 2 kb right after the stop codon of the Ucp1 gene flanking the 2A peptide (GSGATNFSLLKQAGDVEENPGP), and a GFP coding sequence were constructed. Primers used in donor plasmid construction and genotyping were listed in Supplemental Table 1.
2.3. Immortalization of primary Ucp1-2A-GFP adipocytes
The stromal vascular fraction from interscapular BAT of postnatal day 2 male mice was isolated [39]. Briefly, tissues were minced in isolation buffer (Supplemental Table 2). Digested tissues were filtered through a 70 μm cell strainer (Corning FALCON, 352350) and preadipoctyes were cultured in primary culture medium (Supplemental Table 2). Cells were then infected with a retrovirus expressing large T antigen (a kind gift from Dr. Dongning Pan) and selected in G418. Single colonies were then isolated and expanded for later study.
2.4. Adipocyte differentiation
Adipocyte differentiation was performed following a standard protocol. Two days after preadipocytes reaching confluence (day 0), differentiation was initiated by adding induction medium (Supplemental Table 2). After 3 days, the medium was replaced with maintenance medium (Supplemental Table 2). Medium was changed daily until day 8. For individual drug treatment, cells were treated by drugs either for the entire differentiation process or at day 8. Drugs used were as follows: sutent (0.5 μM, MCE, HY10255), isotretinoin (10 μM, TSBiochem, T1611), acitretin (10 μM, TSBiochem, T1330), tretinoin (10 μM, TSBiochem, T1051), bexarotene (10 μM, TSBiochem, T6410), tazarotene (10 μM, TSBiochem, T6696), adapalene (10 μM, TSBiochem, T1093) and isoproterenol (10 μM, Sigma, I6379).
2.5. Chemical screening
For high-throughput chemical screenings, the Ucp1-2A-GFP preadipocytes were differentiated in 96-well plates. At day 8 after differentiation, cells were treated with chemicals for 24 h and analyzed for GFP and DAPI signals using High Content Screening (HCS) (Cellomics ArrayScan VTI, Thermo Fisher Scientific). The ratio of GFP /DAPI signal of each well was used to evaluate the effect of each chemical. The chemical library was purchased from Selleck (L1300) and applied in a final concentration of 10 μM of each chemical in screening. Cells treated with DMSO and isoproterenol were used as controls.
2.6. Metabolic studies
Whole-body O2 consumption, CO2 production and physical activity were measured in metabolic cages (Columbus Instruments). Mice had access ad libitum to chow and water in respiration chambers. Rectal temperature was measured with a model BAT-12 thermometer (Physitemp Instruments).
2.7. Glucose and insulin tolerance tests
For the glucose tolerance test (GTT), mice were fasted for 14 h and injected with d-glucose (1.5 g/kg body weight, Sinopharm chemical reagent co., Ltd., 63,005,518) intraperitoneally. For the insulin tolerance test (ITT), mice were fasted for 4 h and injected with recombinant human insulin (0.75 U/kg body weight, Nono Nordisk) intraperitoneally. Blood glucose was monitored with a glucometer (Abbott) at various time points as indicated.
2.8. Measurements of blood and liver samples
Hepatic lipids were extracted using a chloroform methanol method. Briefly, liver extracts were homogenized in chloroform methanol mixture (2:1). Organic phase was transferred and air-dried. The residual liquid was resuspended in absolute ethanol plus 1% Triton X-100 for measurement. Total triglyceride (TG) and total cholesterol (TC) in serum or extracted hepatic samples were measured with TG and TC kit (Shanghai Shensuo UNF, 1030280, 1,040,280). TG and TC measured in liver extracts were further normalized to protein concentration.
2.9. RNA Isolation and real-time quantitative PCR
Total RNA was extracted from cells or tissues using TRIzol reagent (ThermoFisher, 15,596,018) and transcribed with the reverse transcription kit (Takara, RR047A). Quantitative real-time PCR was carried out on the 7900 System(ABI) using SYBR Green supermix (ABI, 4472908). Sequences of primers were listed in Supplemental Table 1.
2.10. Western analysis
Protein from cells or tissues was extracted by RIPA buffer (Millipore, 20,188) and subjected to regular western procedure. The primary antibodies used in the experiments were listed in Supplemental Table 2.
2.11. Histology
Mouse tissues were fixed and embedded in paraffin (for HE staining) or frozen (for oil red staining). Sections were stained with hematoxylin and eosin or oil red or UCP1 antibody (1:100, Abcam, ab10983) according to standard protocols.
2.12. RNA-Seq analysis
RNA-seq analysis was performed following standard procedures. Sequencing was performed in WuXi Apptec Co. Ltd. (Shanghai). For data analysis, differentially expressed genes were identified using FDR ≤ 0.05 as cutoff. GO term and pathway annotations were performed by homemade scripts of R and Python scripts using Fisher's exact test.
2.13. Statistics
The unpaired, two-tailed Student's t-test was used for experiments with two groups' comparison. One-way ANOVA test and post-hoc Bonferronic multiple-comparison test was used for experiments that contained more than two groups. All data are represented as means with SD.
2.14. Accessing numbers
Raw data were deposited in NCBI Gene Expression Omnibus(GEO) with the accession number: GSE118224.
3. Results
3.1. Generation of the Ucp1-2A-GFP reporter mouse and cell line
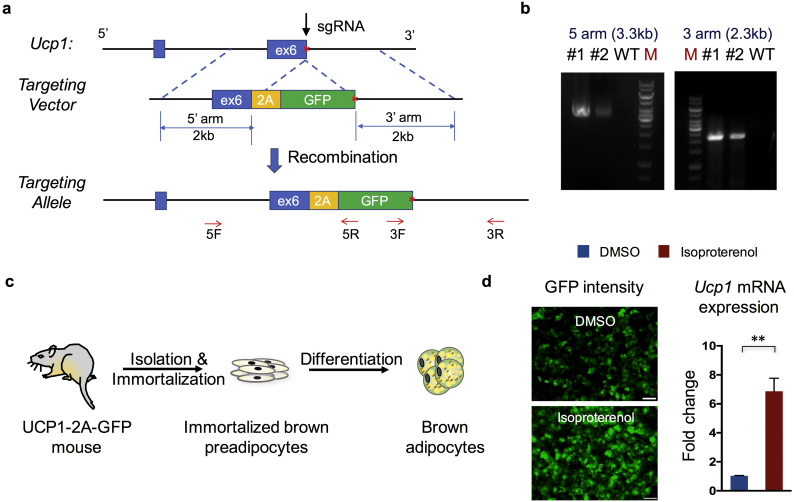
With the aim to determine the endogenous UCP1 expression level with a fluorescence signal, which is frequently used in high-throughput screening assays, we decided to knock in a 2A-GFP cassette immediately before the stop codon of the endogenous Ucp1 gene in mice (Fig. 1a). To generate this mouse model, we took a CRISPR/Cas9 approach [40]. We screened for an efficient single guide RNA targeting the site of the Ucp1 stop codon. A donor template harboring 2 kb homology arms on each side was constructed and injected simultaneously with Cas9 and sgRNA into mouse fertilized eggs. New-born mice were genotyped and sequenced to confirm the successful knock-in of the 2A-GFP cassette in the endogenous Ucp1 locus (Fig. 1b). To further obtain cells for in vitro screening, primary brown adipose tissue was collected from the day 2 postnatal Ucp1-2A-GFP mice. Brown pre-adipocytes were then isolated and immortalized through SV40 large T antigen overexpression (Fig. 1c). Several clones were established and validated for differentiation and GFP expression. Some of these clones displayed clear GFP expression under the normal culture condition and significantly increased GFP intensity in response to isoproterenol, which is a β adrenoreceptor agonist and stimulates UCP1 expression (Fig. 1d).
Fig. 1.
Generation of the Ucp1-2A-GFP reporter mice and adipocytes. (a) Schematic view of the targeting strategy to generate the Ucp1-2A-GFP mouse. The red star indicates the stop codon of the Ucp1 gene. Arrows indicate screening primers used for genotyping. (b) Representative PCR screening results in successfully targeted alleles. (c) Schematic view of the strategy to generate the Ucp1-2A-GFP immortalized adipocytes. (d) Representative images of GFP intensity in differentiated Ucp1-2A-GFP brown adipocytes treated with DMSO or isoproterenol (left). Scale bar = 200 μm. Gene expression analysis of Ucp1 mRNA expression in isoproterenol-treated Ucp1-2A-GFP brown adipocytes and control cells (right). (n = 3 for each treatment). Data are represented as means with SD. ** P < 0.01 (unpaired, two-tailed Student's t-test).
3.2. High-throughput screening with the FDA-approved drug library
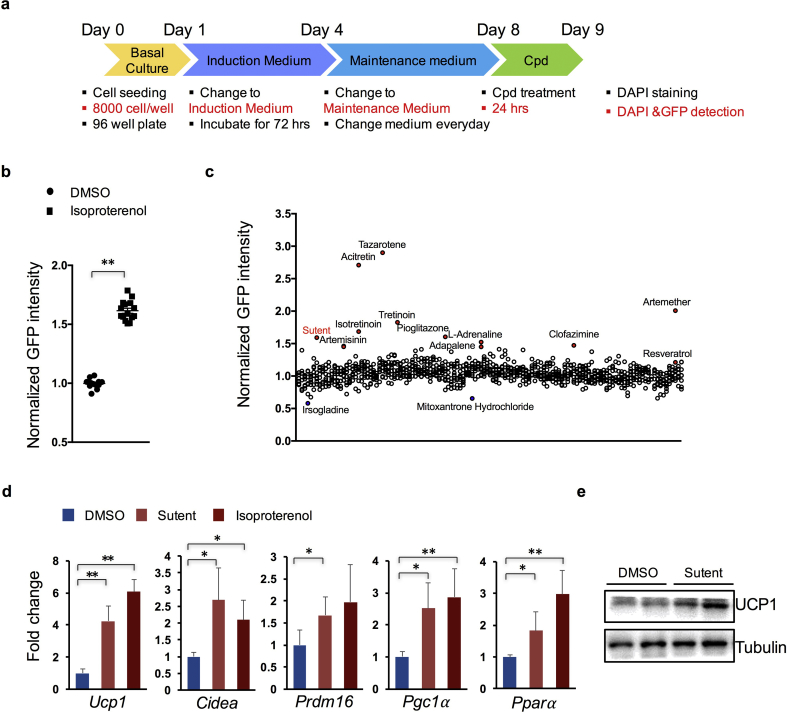
We next set up a high-throughput screening platform with the established Ucp1-2A-GFP brown adipocyte clones. To maximize the outcome, culture conditions applied in screening were first optimized. Two of the Ucp1-2A-GFP clones (#8 and #10) were picked up and first examined with isoproterenol treatment at different days (Supplementary Fig. 1a). Cells were also stained with DAPI to determine the cell number in each well, and the ratio of GFP/DAPI signal was used for evaluating the chemical effect. The relative GFP intensity at basal level displayed an increased trend, whereas fold changes upon isoproterenol treatment (isoproterenol/DMSO) did not increase significantly upon extended culture program. Between two different clones, clone #10 displayed in general higher GFP intensity at the basal level but less fold change upon stimulation, in comparison to clone #8 (Supplementary Fig. 1a). Therefore, #8 cells and cells differentiated for 8 days were used for subsequent compound screening. As the efficiency of adipocyte differentiation can also be significantly affected by cell density, different numbers of cells seeding at the beginning stage were further tested (Supplementary Fig. 1b). Results indicated that seeding with a density of 8000 cells/well in 96-well plates show best sensitivity to isoproterenol treatment, thus was used for later drug screening.
We next performed the screening assay using above optimized parameters (Fig. 2a). For each plate, DMSO and isoproterenol treatment were both included as internal controls. The FDA-approved library was selected as it contains ~1000 drugs that have already been approved in different medications in human, offering an appropriate library to identify drugs for repurposing. Consistent with previous tests, the ratio of isoproterenol treatment over control group was around 1.7 (Fig. 2b). Fold changes upon treatment over DMSO controls in each plate was then calculated for each compound. As resveratrol, a previously identified compound that can activate UCP1 expression [32,33], showed a modest ratio of 1.20, we decided that drugs with a calculated ratio over 1.20 were considered primary hits.
Fig. 2.
High-throughput screening with the FDA-approved drug library. (a) Schematic view of the screening strategy. Cpd, compoud. (b) Analysis of the relative GFP intensity change upon isoproterenol treatment in the screening. (n = 15 for each treatment). (c) Screening results displaying relative GFP intensity of individual compounds. Representative compounds that led to increased GFP intensity were highlighted in Red, and that led to decreased GFP intensity were highlighted in Blue. (n = 3 for each treatment). (d) mRNA expression analysis of genes in sutent, isoproterenol or DMSO -treated Ucp1-2A-GFP brown adipocytes. (n = 3 for each treatment). (e) Western blot analysis of UCP1 protein expression in sutent or DMSO -treated Ucp1-2A-GFP brown adipocytes. (n = 2 for each treatment). Data are represented as means with SD. * P < 0.05, ** P < 0.01 (one-way ANOVA test and post-hoc Bonferronic multiple comparison test).
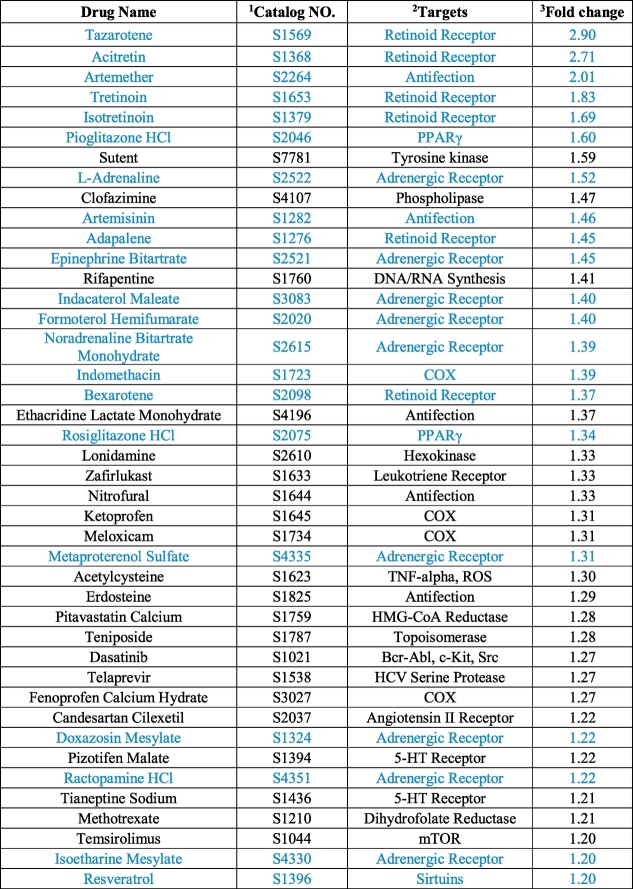
Using this criterion, we found 42 drugs that may activate UCP1 expression in brown adipocytes (Fig. 2c and Table 1). Of these drugs, drugs belong to retinoid receptors agonists family [[41], [42], [43], [44], [45], [46]], adrenergic receptors agonists family [47,48], and PPARγ agonists family [49,50] showed consistent effect in upregulating UCP1 expression (Table 1). Of note, consistent with a previous study, both artemether and artemisinin treatment led to a significant increase in UCP1 expression [29]. Besides, several inhibitors to cyclooxygenase (COX) also showed up in the primary hits, suggesting functional involvement of cyclooxygenase in UCP1 regulation. Interestingly, a very recent publication reported the function of indomethacin, one of COX inhibitors, in activating brown fat activity and UCP1 expression with both in vivo and in vitro studies [51]. Besides these known or reported activators, new candidates, for example, sutent (multi-target receptor tyrosine kinase inhibitor), clofazimine (phospholipase activator), acetylcysteine (reactive oxygen species, ROS inhibitor), suggested new potent drugs for UCP1 regulation in brown adipose, which warrant further investigation.
Table 1. Compounds with activating effect to UCP1 expression identified in the screening.
The blue indicates previously identifed compounds that showed effects of upregulating UCP1 in adipocytes. 1 Numbers categorized in Selleck.cn. 2 Established main targets. Other unlisted targets may also exist. 3 GFP/DAPI signals were first calculated for each well; Average fold changes of GFP/DAPI signal values upon treatment by each compound over in-plate DMSO controls were then calculated and listed in the table.
3.3. Sutent increases UCP1 expression in adipocytes
We next validated part of the screening results with individual tests of the retinoid receptors agonists family. We noticed significant increase in GFP intensity upon treatment with all drugs tested. Consistent with the screening results, treatment with tararotene and acitretin led to the strongest GFP intensity among all the agonists tested (Supplementary Fig. 2). We then decided to focus on one drug that showed the highest ratio of relative GFP intensity among the unreported drugs, which is named sutent, or sunitinib. Sutent belongs to receptor tyrosine kinase (RTK) inhibitor family, and is currently used to treat certain kidney, pancreatic, and gastrointestinal cancers in clinical [52]. We first validated the effect of sutent with in vitro cultured brown adipocytes. Results showed significantly increased expression levels of Ucp1 mRNA (Fig. 2d) and UCP1 protein (Fig. 2e) in brown adipocytes when cells were treated with sutent. Sutent treatment also stimulated expression of other thermogenic or brown differentiation genes, including Cidea, Pgc1a, Ppara, and Prdm16 (Fig. 2d).
3.4. Sutent promotes weight loss with increased thermogenesis and improved metabolism in vivo
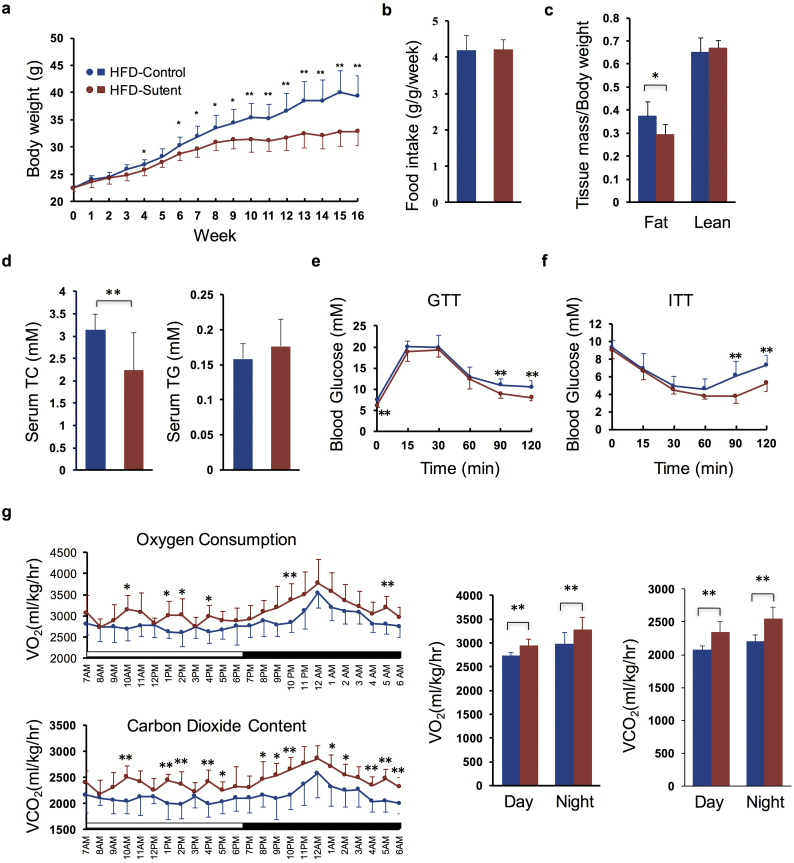
We next investigated the effect of sutent treatment in vivo. Mice were fed with normal chow diet (NCD) or high fat diet (HFD), and treated with sutent at a dose of 20 mg/kg per day through oral administration, which achieves drug exposures in mice comparable to 50 mg/day dose used in clinical patients [53,54]. Sutent treatment did not cause significant change in either body weight or body composition in mice under NCD (Supplementary Fig. 3a and c), but remarkably reduced the whole-body weight in mice under HFD (−16.34% after 16-week treatment) (Fig. 3a), which was mostly due to a reduction in fat mass (−35.82%) (Fig. 3c). Food intake monitoring did not show significant change after sutent treatment in either NCD or HFD groups (Fig. 3b and Supplementary Fig. 3b). Serum analysis indicated reduction in total cholesterol (−28.54%) but no significant change in total triglyceride level in mice treated with sutent under HFD (Fig. 3d). Consistent with body weight change, sutent treatment resulted in modest but significant improvement in both glucose tolerance test (GTT) and insulin sensitivity test (ITT), suggesting improved metabolism (Fig. 3e and f); whereas no significant change was observed in mice under NCD (Supplementary Fig. 3d and e). Mice were next housed in metabolic cages after 15 weeks' treatment to monitor energy expenditure. Sutent treatment did not affect physical activity (Supplementary Fig. 3g), but caused a significant increase in O2 consumption and CO2 content (Fig. 3g), indicating enhanced energy expenditure.
Fig. 3.
Sutent treatment enhances energy expenditure and improves metabolism in mice under HFD. (a) Body weights of mice treated with sutent or vehicle. (n = 8 for each treatment). (b) Food intake per mouse treated with sutent or vehicle measured over 16 week normalized by body weight. (n = 8 for each treatment). (c) Body lean and fat composition of mice determined by NMR after 14-week treatment with sutent or vehicle. (n = 8 for each treatment). (d) Serum total triglyceride (TG) and total cholesterol (TC) content of mice measured after 15-week treatment with sutent or vehicle. (n = 8 for each treatment). (e) Glucose tolerance test (GTT) performed in mice after 13-week treatment with sutent or vehicle. (n = 8 for each treatment). (f) Insulin tolerance test (ITT) performed in mice after 14-week treatment with sutent or vehicle. (n = 8 for each treatment). (g) Oxygen consumption and CO2 content measurement of mice in metabolic cages after 15-week treatment with sutent or vehicle. (n = 8 for each treatment). Results are means with SD. * P < 0.05, **P < 0.01 (unpaired, two-tailed Student's t-test).
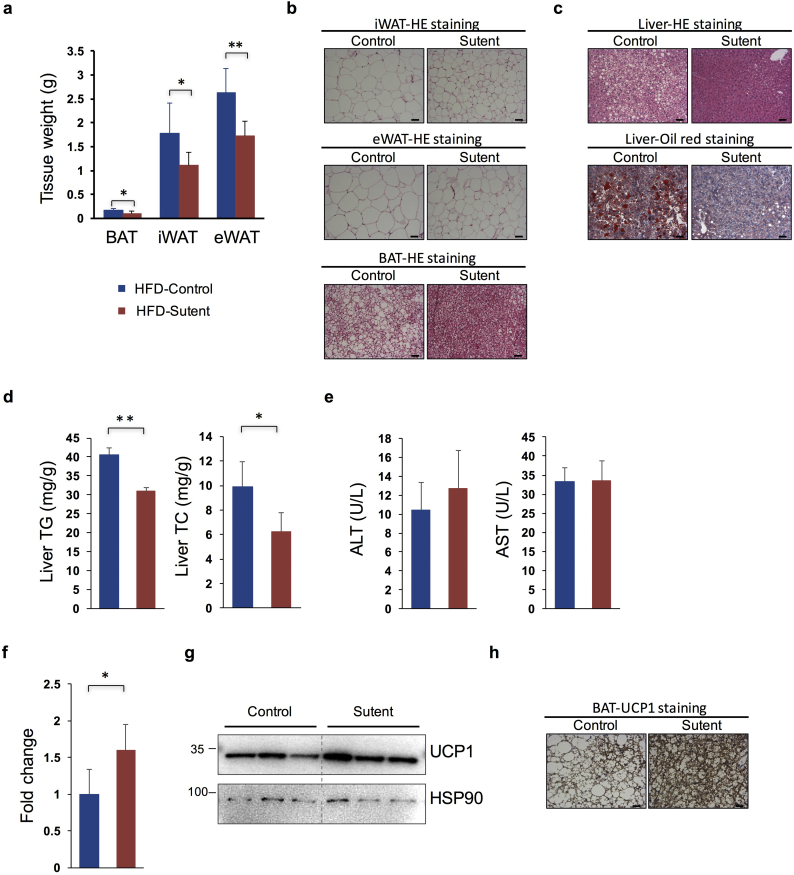
Mice were then dissected for further investigation. Total organ weights of BAT (−35.94%), eWAT (epididymal WAT, −34.47%) and iWAT (inguinal WAT, −37.81%) were reduced in sutent-treated mice under HFD (Fig. 4a), but not NCD (Supplementary Fig. 3f). Furthermore, histological analysis of adipose tissues displayed significantly reduced lipid content and adipocyte size in BAT, eWAT and iWAT (Fig. 4b). Meanwhile, sutent treatment remarkably ameliorated HFD induced lipid accumulation in mouse liver, as indicated both by HE staining and oil red analysis (Fig. 4c). This was also reflected in reduced liver triglyceride (−23.58%) and liver cholesterol content (−36.77%) using quantitative assays (Fig. 4d). We also analyzed serum AST and ALT levels to determine whether sutent treatment causes liver damage. Results showed no significant increase in sutent treated mice, which indicate that at least in the concentration applied in our experiment, sutent treatment cause no adverse effect to liver function (Fig. 4e). Besides, sutent treatment resulted in significant increase of UCP1 expression, both at the mRNA level and the protein level (Fig. 4f–h), indicating that the phenotypic differences observed in mice after sutent treatment are at least partly caused by enhanced UCP1 expression in brown adipose tissue.
Fig. 4.
Sutent treatment decreases lipid accumulation in adipose and liver tissues and increases UCP1 expression in brown adipose tissue. (a) Weights of different mouse adipose tissues after 16-week treatment with sutent or vehicle under HFD. (n = 8 for each treatment). (b) Representative HE staing images of eWAT, iWAT, and BAT. Scale bar = 100 μm. (c) Representative HE staing (upper) and oli-red staining (lower) images of the liver tissue. Scale bar = 100 μm. (d) Measurement of liver TG and TC content. (n = 8 for each treatment). (e) Measurement of serum AST and ALT levels. (n = 8 for each treatment). (f) The mRNA expression analysis of the Ucp1 gene in mouse brown adipose tissues. (n = 5 for each treatment). (g) Western blot analysis of UCP1 protein expression level in mouse brown adipose tissues. (h) Representative images of immunohistochemistry stainining of UCP1 in BAT. Scale bar = 50 μm. Results are means with SD. *P < 0.05, **P < 0.01 (unpaired, two-tailed Student's t-test).
3.5. Sutent activates STAT3 signaling pathway and thermogenesis genes
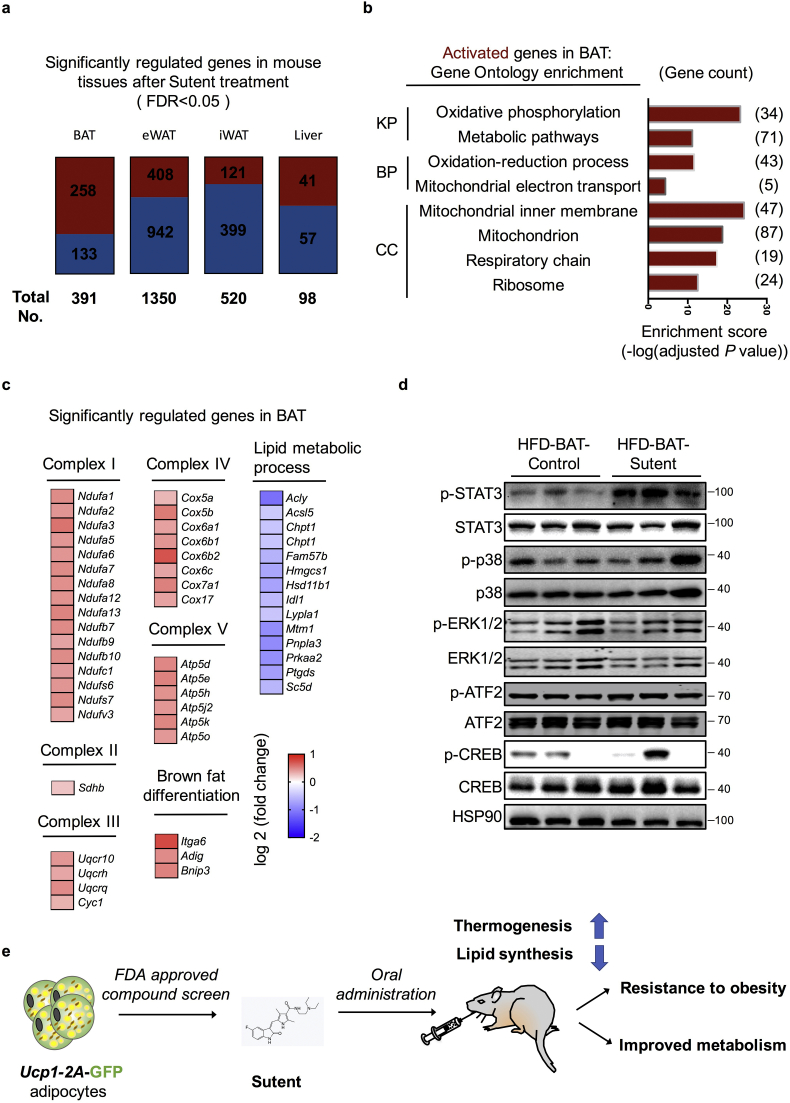
Sutent is supposed to be a multi-targeted RTK inhibitor. Among other RTK inhibitors that are also included in the FDA-approved library, only dasatinib popped out in our primary screening (Fig. 2c and Table 1), indicating the inhibition to receptor tyrosine kinases may not account for the primary effect of sutent treatment. To further understand the changes in mouse tissues after sutent treatment, we next did RNA-seq analysis of all four tissues (BAT, eWAT, iWAT and liver) in sutent or vehicle treated mice. Using a cutoff of FDR <0.05, there are 391, 1350, 520 and 98 genes that showed significant difference in expression in BAT, eWAT, iWAT and liver, respectively (Fig. 5a). A larger percentage (65.98%) of genes were being activated in BAT, in comparison to other tissues (30.22% in eWAT, 23.27% in iWAT, 41.84% in liver) (Fig. 5a). Analysis of the genes activated by sutent treatment in BAT revealed a strong enrichment for thermogenic pathways (Fig. 5b and c). A group of genes that are involved in lipid metabolic and lipid synthesis were down-regulated in BAT upon sutent treatment (Fig. 5c). This collectively indicates activated thermogenesis and reduced lipid synthesis in BAT after sutent treatment.
Fig. 5.
Sutent activates STAT3 signaling pathway and thermogenesis genes in brown adipose tissue. (a) The overview of genes upregulated and downregulated in different tissues after sutent treatment using a cutoff of FDR <0.05. Dark red, upregulated genes; dark blue, downregulated genes. (b) Gene Ontology and pathway analysis of upregulated genes in BAT after sutent treatment identified by RNA-seq. (KP, KEGG Pathway; BP, Biological Process; CC, Cellular Component). (c) Heatmap depicting up- or down- regulated genes identified in Gene Ontology analysis. (d) Western blot analysis of indicated proteins in mouse brown adipose tissues. (e) The schematic diagram of the strategy and discoveries in this study.
Analysis of gene expression changes in other tissues also revealed a significant reduction of gene expression levels involved in lipid biosynthesis in all eWAT, iWAT and liver tissues (Supplementary Fig. 4), indicating that lipid synthesis process was consistently affected in all adipose tissues and liver tissue after sutent treatment. Besides, we noticed significantly reduced expression of genes related to inflammatory response specifically in eWAT (Supplementary Fig. 4a), and a reduction of extracellular matrix gene expression specifically in iWAT (Supplementary Fig. 4b), suggesting improved adipose inflammation and tissue fibrosis. Collectively, gene expression analysis suggested that sutent treatment resulted in activated thermogenesis in BAT, reduced lipid synthesis, alleviated tissue inflammation and fibrosis in adipose tissues and liver tissue.
To get a better understanding of which pathway(s) may involve in the effect caused by sutent treatment, we next checked several signaling regulators that previously reported to involve in brown adipose function, including STAT3, p38, ATF2, ERK and CREB [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24],55]. (Fig. 5d). Only STAT3 phosphorylation signal displayed a significant and consistent increase in brown adipose from sutent-treated mice. Overexpression of constitutively active Stat3 in mouse brown adipose tissue was previously reported to improve BAT development and reduce body weights [23]. Although the underpinning mechanism of activated STAT3 signaling pathway by sutent treatment is so far unclear and in fact, surprising, we suspect this mediate at least part of the effect caused by sutent treatment.
4. Discussion
Obesity has become an epidemic in the modern society, proper and effective treatments are thus in urgent demand. Although we are obtaining significantly improved understanding of the function and development of brown or beige adipose, to identify safe and effective agents to activate brown or beige adipose remains a challenge. We addressed this challenge by developing a cellular system with a fluorescence readout, and screened a FDA-approved drug library for drugs that can be potentially repurposed to treat obesity. As a result, we identified a group of drugs that can upregulate UCP1 expression in brown adipocyte, demonstrating the feasibility of using this system to screen for UCP1 activators. Further studies focusing on a previously unreported drug—sutent, revealed that sutent treatment could increase the energy expenditure and inhibit lipid synthesis in mouse adipose and liver tissues with no obvious adverse effect, resulting in improved metabolism and resistance to obesity (Fig. 5e).
Transgenic or knock-in mouse models with a 2A-luciferase cassette under the control of Ucp1 genetic locus were previously generated, offering nice mouse models for directly evaluating UCP1 activity in vivo [37,38]. Immortalized adipocytes from these mouse models were also generated for compound screenings. Comparing to these models using luciferase signal as a readout, GFP expression allows a direct measure of UCP1 protein expression level through imaging, which can be easily adapted to high-throughput platforms. And this cellular system can also be potentially used for genome-wide genetic screenings to identify regulatory genes in UCP1 regulation using CRISPR libraries and a FACS sorting strategy [56]. Overall, Ucp1-2A-GFP knock-in mouse model offers a nice complementary model to the existing UCP1-studying models by allowing easier in vitro high-throughput chemical or genetic screenings with a fluorescence readout in adipose tissues. In contrast, as GFP signal does not penetrate well in vivo, to directly view the GFP signal in Ucp1-2A-GFP live animals requires high-end animal imaging system. To change GFP reporter to a different far-red fluorescence protein with better penetrance through skin (eg. Katushka) [57] may be of something interesting that can assist well in both in vivo and in vitro studies of UCP1.
In this study, we applied an FDA-approved drug library in UCP1 screening. Compounds in the library are structurally diverse, and currently being used in disease indications related to oncology, cardiology, anti-inflammatory, immunology, neuropsychiatry, etc. A few known UCP1 activators popped out in our primary hits, most among them are retinoid receptor agonists and adrenergic receptor agonists. It is interesting to notice that different agonists with modest structure changes can result in quite different UCP1 expression levels (Table 1 and Supplementary Fig. 2), which may offer useful information for further structure modifications if these drugs are repurposed to treat obesity. We also found a group of previously unreported drugs that showed effects in upregulating UCP1 expression in our screenings, providing an interesting set of candidates for further study.
We later focused on sutent for further in vivo investigation. We found that sutent treatment significantly decreased gain of whole-body weight due to HFD feeding and improved mouse metabolism. Consistent with the in vitro screening result, sutent treatment in vivo led to higher UCP1 expression in brown adipose tissue and more oxygen consumption, suggesting enhanced energy expenditure. However, cold exposure challenge experiment showed no significant change in rectal temperatures of mice with sutent treatment when compared to control mice (Supplementary Fig. 3h). We suspect that although sutent treatment led to higher UCP1 expression level (both in mRNA and protein levels) in brown adipocytes, substantially decreased BAT weight (−35.94%) may have neutralized this effect. Besides UCP1, sutent treatment also caused consistent up-regulation in genes in mitochondrial oxidative phosphorylation pathway in BAT, as well as down-regulation of genes in lipid synthesis pathway in eWAT, iWAT and liver (Fig. 5a–c and Supplementary Fig. 4). We therefore have good reason to believe that the improved metabolism after sutent treatment is only partly mediated by enhancing UCP1 expression in BAT; whereas decreased lipid synthesis in WAT and liver may have also contributed significantly to the overall phenotypic changes, and this may explain why the sutent effect is only marked in mice under HFD. Within a 16-week period in this study, sutent treatment did not display obvious adverse effect. However, side-effect observed in patients, such as nausea, diarrhea, and even cardiotoxicity [58], makes it impossible for immediate repurposing to treat obesity. The discovery of new drug delivery strategies, for example, local delivery through a microneedle patch [59], may help relieve some of the side-effect by directly targeting adipose tissue instead of systematic administration.
Among the limitations of our study is that the underlying mechanism that how sutent upregulates UCP1 in brown adipocytes is lacking. It is worth noting that the regular screening system, for example, our Ucp1-2A-GFP reporter system, at most times acts like a “black-box”, with only definitive input and output. While the advantage is clear that through high-through screening, we are able to identify novel “input” that leads to the favorable “output”, greater challenges may exist in exploring the “how the input causes output”, when compared to hypothesis-driven studies. Sutent belongs to a muti-target RTK inhibitor family. The nature that sutent has multiple potential targets makes it hard to identify the signaling pathway(s) and the direct target(s) that account for the effect of sutent in brown adipocytes. Within the pathways analyzed in our study (including p38, CREB, ATF2, ERK, STAT3), we only noticed significant increase in STAT3 phosphorylation upon sutent treatment. As overexpression of constitutively active Stat3 was previously reported to improve BAT function [23], we suspect that the effect of sutent may partly mediated by activated STAT3 signaling pathway. However, how sutent causes activation of STAT3 signaling is unclear. In fact, another RTK inhibitor, which was also identified in one screening for UCP1 activators, was reported to cause reduced phosphorylation of STAT3 in adipocytes [38], indicating rather different underlying mechanisms between axinitib and sutent, and also causing confusion on the regulation of UCP1 by STAT3 signaling pathway. Among other RTK inhibitors that are also included in our FDA-approved library, only dasatinib popped out in our primary screening (Fig. 2c and Table 1). It is interesting to note that a study focusing on screening for compounds to inhibit the phosphorylation of PPARγ at Ser273 (pS273), which is linked to obesity and insulin resistance [60,61], have identified sutent and dasatinib that both are able to inhibit pS273 [62]. However, this study did not report a direct up-regulation of UCP1 in brown adipocytes by inhibiting pS273. Whether the inhibitory effect to pS273 accounts for part of the sutent effect warrants further investigation. Overall, our study only has provided very limited mechanistic insight into the effect of sutent. Substantially more analysis will be needed to perform in order to understand how sutent contributes to the function of brown adipocytes.
The library used in our study comprises only part of the drugs being approved by FDA. Screening with different chemical libraries, for example, natural product library, clinical compound library, etc., also offer opportunities for the discovery of candidate drugs. Besides, we are enthused to apply or share this platform for high-throughput screenings with perhaps millions of chemicals if proper facilities are ready. In summary, we believe our study adds to the current effort in searching for agents that can activate UCP1 expression in adipose tissues. Specifically, we offered an easy-to-use cellular screening system for UCP1 activators and provided a list of FDA-approved drugs that can potentially treat obesity. Further detailed study of these drugs as well as the underlying mechanisms may shed new light on the drug discovery for obesity and new regulatory pathways in adipose function.
Acknowledgments
Acknowledgements
We thank Drs. Dongning Pan (Fudan University) and Qiong Wang (City of Hope) for helpful suggestions in the isolation and immortalization of primary brown adipocytes. We thank Hua Feng (Omics Core of Bio-Med Big Data Center, CAS-MPG Partner Institute for Computational Biology, SIBS, CAS) for assistance in data analysis. Dr. Qiurong Ding is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding sources
This work was supported by grants from the National Key R&D Program of China (2017YFA0102800, 2017YFA0103700), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16030402), the Key Research Program of the Chinese Academy of Sciences (ZDRW-ZS-2017-1, KFZD-SW-213), the National Natural Science Foundation of China (31670829, 81600650), China Postdoctoral Science Foundation (2016M601666), the CAS-TWAS program (M.F.), and the National Youth 1000 Talents Program (Q.D.). All these funders are not involved in study design, data collection, data analysis, interpretation or writing of this report.
Declarations of interest
No potential conflicts of interest relevant to this article were reported.
Author contributions
Y.L and Q.D. designed the research. Y.Q. and Y.S.performed all experiments. D.X. performed the drug screening, Y.Y., X.L., Y.W., Y.C., Z.F., S.L., M.F., Y.Z. and H.X. assisted with either experiments or data analysis. Q.Y. and Q.D. wrote the manuscript. Y.L and Q.D. supervised the project. All authors have read and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.019.
Contributor Information
Yuanzhi Lao, Email: laurence_ylao@163.com.
Qiurong Ding, Email: qrding@sibs.ac.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Kim G.W., Lin J.E., Blomain E.S., Waldman S.A. Antiobesity pharmacotherapy: New drugs and emerging targets. Clin. Pharmacol. Ther. 2014;95:53–66. doi: 10.1038/clpt.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 5.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 6.Sidossis L., Kajimura S. Brown and beige fat in humans: Thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 2015;125:478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kajimura S., Spiegelman B.M., Seale P. Brown and beige fat: Physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loft A., Forss I., Mandrup S. Genome-wide insights into the development and function of thermogenic adipocytes. Trends Endocrinol. Metab. 2017;28:104–120. doi: 10.1016/j.tem.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Nedergaard J., Cannon B. The browning of white adipose tissue: Some burning issues. Cell Metab. 2014;20:396–407. doi: 10.1016/j.cmet.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Nicholls D.G., Bernson V.S., Heaton G.M. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Experientia Suppl. 1978;32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- 11.Lowell B.B., Spiegelman B.M. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 12.Cannon B., Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 13.Kopecky J., Clarke G., Enerback S., Spiegelman B., Kozak L.P. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J. Clin. Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Q., Lin J., Huang J., Zhang H., Zhang R., Zhang X. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E9474–E9482. doi: 10.1073/pnas.1707853114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachman E.S., Dhillon H., Zhang C.Y., Cinti S., Bianco A.C., Kobilka B.K. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–845. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 16.Jimenez M., Leger B., Canola K., Lehr L., Arboit P., Seydoux J. Beta(1)/beta(2)/beta(3)-adrenoceptor knockout mice are obese and cold-sensitive but have normal lipolytic responses to fasting. FEBS Lett. 2002;530:37–40. doi: 10.1016/s0014-5793(02)03387-2. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann L.S., Etzrodt J., Willkomm L., Sanyal A., Scheja L., Fischer A.W. Stimulation of soluble guanylyl cyclase protects against obesity by recruiting brown adipose tissue. Nat. Commun. 2015;6:7235. doi: 10.1038/ncomms8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao W.H., Medvedev A.V., Daniel K.W., Collins S. Beta-adrenergic activation of p38 MAP kinase in Adipocytes – cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 2001;276:27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 19.Cao W.H., Daniel K.W., Robidoux J., Puigserver P., Medvedev A.V., Bai X. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robidoux J., Cao W.H., Quan H., Daniel K.W., Moukdar F., Bai X. Selective activation of mitogen-activated protein (MAP) kinase kinase 3 and p38 alpha MAP kinase is essential for cyclic AMP-dependent UCP1 expression in adipocytes. Mol. Cell. Biol. 2005;25:5466–5479. doi: 10.1128/MCB.25.13.5466-5479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Guo H., Deis J.A., Mashek M.G., Zhao M., Ariyakumar D. Lipocalin 2 regulates brown fat activation via a nonadrenergic activation mechanism. J. Biol. Chem. 2014;289:22063–22077. doi: 10.1074/jbc.M114.559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betz M.J., Enerback S. Human brown adipose tissue: What we have learned so far. Diabetes. 2015;64:2352–2360. doi: 10.2337/db15-0146. [DOI] [PubMed] [Google Scholar]
- 23.Derecka M., Gornicka A., Koralov S.B., Szczepanek K., Morgan M., Raje V. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab. 2012;16:814–824. doi: 10.1016/j.cmet.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi S.Y., Zhang W., Luk C.T., Sivasubramaniyam T., Brunt J.J., Schroer S.A. JAK2 promotes brown adipose tissue function and is required for diet- and cold-induced thermogenesis in mice. Diabetologia. 2016;59:187–196. doi: 10.1007/s00125-015-3786-2. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Martin R., Alexaki V.I., Qin N., Rubin De Celis M.F., Economopoulou M., Ziogas A. Adipocyte-specific HIF2alpha deficiency exacerbates obesity-induced brown adipose tissue dysfunction and metabolic dysregulayion. Mol. Cell. Biol. 2015;36:376–393. doi: 10.1128/MCB.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutant M., Joffraud M., Kulkarni S.S., Garcia-Casarrubios E., Garcia-Roves P.M., Ratajczak J. SIRT1 enhances glucose tolerance by potentiating brown adipose tissue function. Mol. Metab. 2015;4:118–131. doi: 10.1016/j.molmet.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young P., Arch J.R., Ashwell M. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett. 1981;167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- 28.Fisher F.M., Kleiner S., Douris N., Fox E.C., Mepani R.J., Verdeguer F. FGF21 regulates PGC-1alpha and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 2012;26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu P., Zhang F.C., Qian S.W., Li X., Cui Z.M., Dang Y.J. Artemisinin derivatives prevent obesity by inducing browning of WAT and enhancing BAT function. Cell Res. 2016;26:1169–1172. doi: 10.1038/cr.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Zhang H., Li B., Meng X., Wang J., Zhang Y. Berberine activates thermogenesis in white and brown adipose tissue. Nat. Commun. 2014;5:5493. doi: 10.1038/ncomms6493. [DOI] [PubMed] [Google Scholar]
- 31.Gnad T., Scheibler S., von Kugelgen I., Scheele C., Kilic A., Glode A. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature. 2014;516:395–399. doi: 10.1038/nature13816. [DOI] [PubMed] [Google Scholar]
- 32.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonet M.L., Oliver P., Palou A. Pharmacological and nutritional agents promoting browning of white adipose tissue. Biochim. Biophys. Acta. 2013;1831:969–985. doi: 10.1016/j.bbalip.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Wu J., Cohen P., Spiegelman B.M. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moisan A., Lee Y.K., Zhang J.D., Hudak C.S., Meyer C.A., Prummer M. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat. Cell Biol. 2015;17:57–67. doi: 10.1038/ncb3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galmozzi A., Sonne S.B., Altshuler-Keylin S., Hasegawa Y., Shinoda K., Luijten I.H.N. ThermoMouse: An in vivo model to identify modulators of UCP1 expression in brown adipose tissue. Cell Rep. 2014;9:1584–1593. doi: 10.1016/j.celrep.2014.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao L., Nie B., Nie T., Hui X., Gao X., Lin X. Visualization and quantification of browning using a Ucp1-2A-luciferase knock-in mouse model. Diabetes. 2017;66:407–417. doi: 10.2337/db16-0343. [DOI] [PubMed] [Google Scholar]
- 39.Fasshauer M., Klein J., Ueki K., Kriauciunas K.M., Benito M., White M.F. Essential role of insulin receptor substrate-2 in insulin stimulation of Glut4 translocation and glucose uptake in brown adipocytes. J. Biol. Chem. 2000;275:25494–25501. doi: 10.1074/jbc.M004046200. [DOI] [PubMed] [Google Scholar]
- 40.Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez R., de Andres J., Yubero P., Vinas O., Mampel T., Iglesias R. A novel regulatory pathway of brown fat thermogenesis. Retinoic acid is a transcriptional activator of the mitochondrial uncoupling protein gene. J. Biol. Chem. 1995;270:5666–5673. doi: 10.1074/jbc.270.10.5666. [DOI] [PubMed] [Google Scholar]
- 42.Larose M., Cassard-Doulcier A.M., Fleury C., Serra F., Champigny O., Bouillaud F. Essential cis-acting elements in rat uncoupling protein gene are in an enhancer containing a complex retinoic acid response domain. J. Biol. Chem. 1996;271:31533–31542. doi: 10.1074/jbc.271.49.31533. [DOI] [PubMed] [Google Scholar]
- 43.Rabelo R., Reyes C., Schifman A., Silva J.E. A complex retinoic acid response element in the uncoupling protein gene defines a novel role for retinoids in thermogenesis. Endocrinology. 1996;137:3488–3496. doi: 10.1210/endo.137.8.8754778. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez R., Checa M., Brun S., Vinas O., Mampel T., Iglesias R. Both retinoic-acid-receptor- and retinoid-X-receptor-dependent signalling pathways mediate the induction of the brown-adipose-tissue-uncoupling-protein-1 gene by retinoids. Biochem. J. 2000;345:91–97. [PMC free article] [PubMed] [Google Scholar]
- 45.Teruel T., Hernandez R., Benito M., Lorenzo M. Rosiglitazone and retinoic acid induce uncoupling protein-1 (UCP-1) in a p38 mitogen-activated protein kinase-dependent manner in fetal primary brown adipocytes. J. Biol. Chem. 2003;278:263–269. doi: 10.1074/jbc.M207200200. [DOI] [PubMed] [Google Scholar]
- 46.Nie B., Nie T., Hui X., Gu P., Mao L., Li K. Brown adipogenic reprogramming induced by a small molecule. Cell Rep. 2017;18:624–635. doi: 10.1016/j.celrep.2016.12.062. [DOI] [PubMed] [Google Scholar]
- 47.Collins S., Yehuda-Shnaidman E., Wang H. Positive and negative control of Ucp1 gene transcription and the role of beta-adrenergic signaling networks. Int. J. Obes. 2010;34:S28–S33. doi: 10.1038/ijo.2010.180. [DOI] [PubMed] [Google Scholar]
- 48.Cypess A.M., Weiner L.S., Roberts-Toler C., Elia E.F., Kessler S.H., Kahn P.A. Activation of human brown adipose tissue by a beta 3-adrenergic receptor agonist. Cell Metab. 2015;21:33–38. doi: 10.1016/j.cmet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nedergaard J., Petrovic N., Lindgren E.M., Jacobsson A., Cannon B. PPARgamma in the control of brown adipocyte differentiation. Biochim. Biophys. Acta. 2005;1740:293–304. doi: 10.1016/j.bbadis.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Ohno H., Shinoda K., Spiegelman B.M., Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao L., Kearns J., Scott S., Wu D.Y., Kodani S.D., Morisseau C. Indomethacin enhances brown fat activity. J. Pharmacol. Exp. Ther. 2018;365:467–475. doi: 10.1124/jpet.117.246256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motzer R.J., Hoosen S., Bello C.L., Christensen J.G. Sunitinib malate for the treatment of solid tumours: A review of current clinical data. Expert Opin. Investig. Drugs. 2006;15:553–561. doi: 10.1517/13543784.15.5.553. [DOI] [PubMed] [Google Scholar]
- 53.Haznedar J.O., Patyna S., Bello C.L., Peng G.W., Speed W., Yu X.M. Single- and multiple-dose disposition kinetics of sunitinib malate, a multitargeted receptor tyrosine kinase inhibitor: Comparative plasma kinetics in non-clinical species. Cancer Chemother. Pharmacol. 2009;64:691–706. doi: 10.1007/s00280-008-0917-1. [DOI] [PubMed] [Google Scholar]
- 54.Speed B., Bu H.Z., Pool W.F., Peng G.W., Wu E.Y., Patyna S. Pharmacokinetics, distribution, and metabolism of [C-14]sunitinib in rats, monkeys, and humans. Drug Metab. Dispos. 2012;40:539–555. doi: 10.1124/dmd.111.042853. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y., Li R., Meng Y., Li S.W., Donelan W., Zhao Y. Irisin stimulates browning of white adipocytes through mitogen-activated protein kinase p38 MAP kinase and ERK MAP kinase signaling. Diabetes. 2014;63:514–525. doi: 10.2337/db13-1106. [DOI] [PubMed] [Google Scholar]
- 56.Li S., Li M., Liu X., Yang Y., Wei Y., Chen Y. Genetic and chemical screenings identify HDAC3 as a key regulator in hepatic differentiation of human pluripotent stem cells. Stem Cell Rep. 2018;11:22–31. doi: 10.1016/j.stemcr.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shcherbo D., Merzlyak E.M., Chepurnykh T.V., Fradkov A.F., Ermakova G.V., Solovieva E.A. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 58.Chintalgattu V., Rees M.L., Culver J.C., Goel A., Jiffar T., Zhang J.H. Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci. Transl. Med. 2013;5 doi: 10.1126/scitranslmed.3005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y.Q., Liu Q.M., Yu J.C., Yu S.J., Wang J.Q., Qiang L. Locally induced adipose tissue browning by microneedle patch for obesity treatment. ACS Nano. 2017;11:9223–9230. doi: 10.1021/acsnano.7b04348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi J.H., Banks A.S., Estall J.L., Kajimura S., Bostrom P., Laznik D. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi J.H., Banks A.S., Kamenecka T.M., Busby S.A., Chalmers M.J., Kumar N. Antidiabetic actions of a non- agonist PPARg ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–481. doi: 10.1038/nature10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi S.S., Kim E.S., Jung J.E., Marciano D.P., Jo A., Koo J.Y. PPARγ antagonist Gleevec improves insulin sensitivity and promotes the browning of whiteadipose tissue. Diabetes. 2016;65:829–839. doi: 10.2337/db15-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material