Abstract
The tumor-stroma ratio (TSR) reflected on hematoxylin and eosin (H&E)-stained histological images is a potential prognostic factor for survival. Automatic image processing techniques that allow for high-throughput and precise discrimination of tumor epithelium and stroma are required to elevate the prognostic significance of the TSR. As a variant of deep learning techniques, transfer learning leverages nature-images features learned by deep convolutional neural networks (CNNs) to relieve the requirement of deep CNNs for immense sample size when handling biomedical classification problems. Herein we studied different transfer learning strategies for accurately distinguishing epithelial and stromal regions of H&E-stained histological images acquired from either breast or ovarian cancer tissue. We compared the performance of important deep CNNs as either a feature extractor or as an architecture for fine-tuning with target images. Moreover, we addressed the current contradictory issue about whether the higher-level features would generalize worse than lower-level ones because they are more specific to the source-image domain. Under our experimental setting, the transfer learning approach achieved an accuracy of 90.2 (vs. 91.1 for fine tuning) with GoogLeNet, suggesting the feasibility of using it in assisting pathology-based binary classification problems. Our result also show that the superiority of the lower-level or the higher-level features over the other ones was determined by the architecture of deep CNNs.
Keywords: Epithelium and stroma, TSR, CNNs, deep learning, transfer learning
1. Introduction
The tumor-stroma ratio (TSR) has gained profound interest over the last decade in the disease prognosis of patients with breast, colon, lung and cervical cancer. The examination of the TSR typically involves the use of hematoxylin and eosin (H&E)-stained slides prepared to preserve the underlying tissue architecture10. Histopathological studies of breast cancer progression have shown that stroma-rich tumors were associated with a relatively poor prognosis14,21. The similar trend has been described in studies on early cervical carcinoma where the disease-free and overall survival rates were observed to be significantly higher in the stroma-poor patients than in the stroma-rich patients16, and also on the prognosis of lung cancer where the TSR has been suggested as a potential prognostic factor for survival31. Reliable evaluation of TSR could therefore provide a more solid basis for the pursuit of personalized medicine.
Although most current histological analysis is based on the subjective and professional opinions of pathologists, there is a clear need for automated image processing techniques that are able to provide high-throughput and quantitative analysis of the pathology images for precise assessment of tumor epithelium and stroma. To reduce the manual labor in the feature-extraction processes, deep learning techniques are now being developed to automatically process images for handling detection and classification tasks4,15,6. The promising aspect of deep learning is that these levels of features were learned automatically rather than being designed by human engineers through a laborious process.
A major hurdle to apply deep learning to many biomedical fields is the availability of a sufficiently large annotated data set for training. However, transfer learning could perhaps be adopted to relieve the requirement for sample size, which leverages deep convolutional neural networks (CNNs) that have been previously trained on a huge natural scene data set. The hypothesis of this method is that the features learned by the deep CNNs to distinguish classes in the natural scene data set could also be applicable to medical data sets with mildly compromised performance. Currently, there are mainly three strategies concerning transfer learning in medical domains, namely, (i) inheriting features learned during training the deep CNNs with a large collection of natural images (NI), followed by training classifiers with the inherited features11,29,35, (ii) fine-tuning the pre-trained networks on a target medical data set with a limited sample size2,8,9,20,28 and (iii) directly training deep convolutional neural networks (CNNs) with random initialization on target medical data sets5,24.
In the study28, Shin et al. implemented these three strategies in an interstitial lung disease classification task using the deep CNN of AlexNet and GoogLeNet, demonstrating that for the GoogLeNet, there existed an evident rise in accuracy after fine-tuning with random initialization compared to using the network as feature extractors. However, when using AlexNet, differences among the three scenarios associated with the implementation of the three strategies were negligible. This observation is partially inconsistent with the finding reported in9, which reported that the performance of the pre-trained TorontoNet at three different levels (pool5, fc6 and fc7) was profoundly improved by the fine-tuning step. In this study, it also documented that the fully connected layers (fc6 and fc7) had superior performance to the layer of pool5, that is, the max-pooled output of the network’s fifth convolutional layer. At the same layers, the result in2, however, shows that the layer of pool5 steadily exhibited a higher discriminatory power than that of the other two layers. Another interesting finding was provided in20: Transferring learning from a non-medical domain to a medical domain (from ImageNet to melanoma) is more advantageous than doing it between two highly related domains such as from the retina to melanoma. Overall, although transfer learning has recently been considered a promising research direction or tool in artificial intelligence, more application-oriented investigation and optimization is needed due to the presence of many important and interesting findings that are somewhat inconsistent in certain aspects.
With this study, we aimed to investigate the efficacy of distinguishing tumor epithelium from stroma with the techniques of transfer learning in the following two aspects: (i) which would contain natural image (NI) features that show better discrimination ability in stratification of the two tissue types, the lower or higher level of CNN architecture? And how would it be related to the chosen CNN architecture? (ii) To what extent would the classification performance be enhanced by fine-tuning the models, with the initialization weights obtained from a fine-tuning procedure on a highly related dataset (e.g., the source and target dataset were both pathology image (PI) datasets, denoted as PI-to-PI) or on a completely different dataset (e.g., from a natural image dataset to a pathology image dataset, denoted as NI-to-PI)?
2. Materials and Methods
2.1. Datasets
We used two datasets in our study, namely, Dataset I and Dataset II. Dataset I came from the Stanford Tissue Microarray Database (TMAD)37, including H&E-stained histological images (whose annotations by class were provided in the reference3) from two independent groups of breast cancer patients: the NKI and VGH. Dataset II included H&E-stained histological images acquired from ovarian cancer tissue from the OUHSC. The institutional review board (#5668) of this study was approved by the OUHSC on July 31, 2016. The requirement for obtaining informed consent from all participants or their legal guardians had been waived by the institutional review board. All experiments involving patients were performed in accordance with guidelines and regulations of the OUHSC.
The main preprocessing steps for both two image sources (from TMAD and OUHSC) include image enhancement and creation of sub-images (or epithelium and stroma patches). Image contrast was enhanced by a standard auto-contrast algorithm implemented in MATLAB7. To prepare sub-images for training and test, we adopted Multiresolution Segmentation algorithm in Definiens Developer XD that produced superpixels of epithelium and stroma and exported them into the workspace as image patches. More details are provided in supplementary material. The images were partitioned into sub-images featuring a coherent appearance and referred to as superpixels (Figure 1a), which were fed into deep learning models to be classified as either epithelium or stroma. A total of 19748 superpixel images (9874 for each class) were generated from 158 H&E-stained histological images from two institutions in TMAD database, while 16444 (8222 for each class) superpixel images were produced from 154 H&E-stained histological images from OUHSC. For both datasets, the total number of images (of which the number of epithelium and stroma were balanced) were randomly split into training and test sets (Table 1) with a split ratio of 0.6:0.4 with train_test_split function from sklearn package in Python. To avoid overfitting during the model development, we performed prevalidation to evaluate the model performance on the held-out fold from the training set with cross_val_score function. The training set was used for both training classifier (denoted as TC, meaning only training the classifier with the features extracted using the pre-trained deep CNNs) and end-to-end (ETE) fine-tuning tasks (followed by training the classifier at a certain layer). For the training time, it required 2.8 and 6.0 hours when using the Alexnet model without (TC) and with fine-tuning (ETE) on Intel Core i5–6600 CPU @ 3.30 GHz RAM 8.00 GB 64 bit. Training time for GoogLeNet without and with fine-tuning with GPU (NVIDIA Tesla K40) were 0.6 and 7.5 hours.
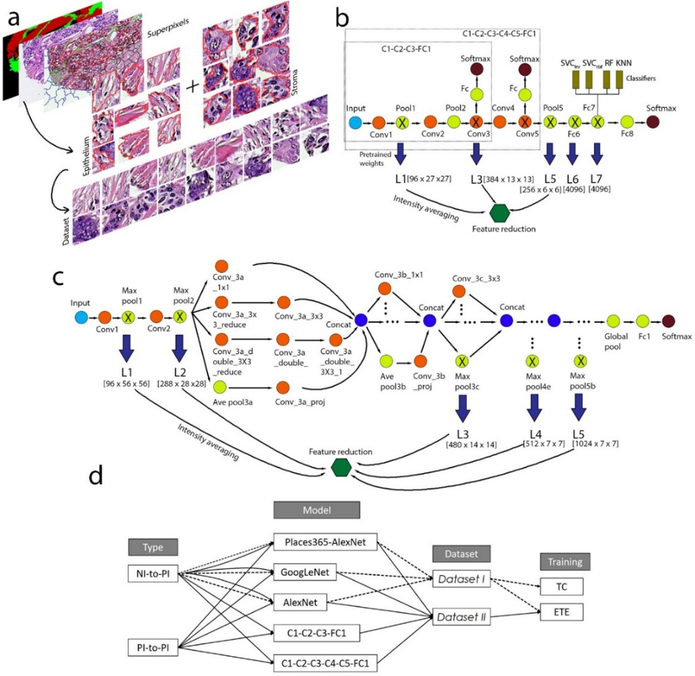
Figure 1.
Overview of experimental design. (a) Preparing image data via generation of the ROIs (of epithelium and stroma). (b) The architectures of the AlexNet (or Places365-AlexNet) and its two simplified networks: C1–C3 and C1–C5. (c) The architecture of the GoogLeNet model. (d) A summary of experimental procedures.
Table 1.
Sample sizes of the two data sets and the splitting of each data set into training and test sets
| Group | Images | Superpixels | Training | Test |
|---|---|---|---|---|
| Dataset I | ||||
| NKI | 107 | 19700 | 11800 | 7900 |
| VGH | 51 | |||
| Dataset II | ||||
| OUHSC | 154 | 16400 | 9800 | 6600 |
2.2. Transfer learning
We utilized natural-image (NI) features (compared to hand-engineered features) learned by the pre-trained deep CNNs from millions of source-task images to properly stratify our datasets into the epithelium and stroma classes. This procedure, known as transfer learning, can be beneficial for a small amount of task data. The deep CNNs used in our study were AlexNet13 (Figure 1b), Places365-AlexNet36 (Figure 1b), GoogLeNet12 (Figure 1c), and two modified AlexNet models (Figure 1b). These models were downloaded from Caffe Model Zoo (https://github.com/BVLC/caffe/wiki/Model-Zoo), and implemented in Python. Please refer to the supplementary material for a description of the implementation procedure.
These deep CNNs were employed in two distinct ways: (i) delivering the NI features without fine-tuning, with which the classifiers were trained, referred to as TC (i.e., training the classifiers only), and (ii) end-to-end fine-tuning of these architectures, followed by training the classifiers at certain layers, referred to as ETE.
The first way (or (i)) was to straightforwardly use the models to compute the output that contained an NI feature set at each layer. We impartially chose five sets of the NI features across all levels (including low-, mid- and high-level CNN features) of the architecture in view of the fact that no decisive conclusion has ever been reached regarding which layers should hold richer semantic information that is readily generalized. As for (ii), we performed end-to-end fine-tuning (1000 iterations; base learning rate: 0.001, gamma: 0.1, momentum: 0.9, weight decay: 5e-04) of the whole structure, which allows the entire architecture to be trained. After fine-tuning, we harnessed the feature set at the intermediate layer (AlexNet and Places365: L7; GoogLeNet: L5; C1–C3: L3; C1–C5: L5) and trained the classifiers with them. With fine-tuning of these networks, we used two disparate strategies of transfer learning (Figure 1d), namely, NI-to-PI and PI-to-PI. In the NI-to-PI, we directly fine-tuned the networks on Dataset I with initial weights optimized by the natural image classification task, which involved transfer learning from a natural image dataset to a pathology image dataset. As for PI-to-PI, we fine-tuned the networks on Dataset II with initial weights transferred from the NI-to-PI, where the source and target dataset were both pathology image datasets.
To eliminate variations in the performance of a classifier among multiple runs, we adopted four different classifiers (which were aimed at enhancing the discriminatory power of the feature set), namely, Support vector machines (SVMs) for classification with a kernel that is ‘linear’ (SVClnr), SVMs with an ‘rbf’ kernel (SVCrbf), random forest (RF) and k-nearest neighbors (KNN). They were implemented in Python using R package svm for SVClnr and SVCrbf, ensemble for RF classifier, neighbors for KNN. Please refer to the supplementary material for the setting. ROC curves were generated using roc_curve function of metrics package in R, and the other performance scores (ACC, TPR, PPV and F1) were calculated using confusion matrix obtained with confusion_matrix function of metrics package.
2.3. Statistical analysis
Data analysis was conducted using R package. Specifically, a pairwise t test (one tailed) with pooled SD was deployed to compare the TC and ETE results with the matched levels of CNN architecture based on the alternative hypothesis that the accuracy of the TC result was significantly less than that of the ETE result. We applied a pairwise t test (two tailed) to all five models to examine if there are significant differences in their mean values. The Welch Two Sample t-test was used to compare the discriminatory power between the higher- and lower-level features extracted from the pre-trained AlexNet, Places365-AlexNet and GoogLeNet models.
3. Results
3.1. Assessing transferability of NI features prior to fine-tuning
Table 2 shows the classification performance of the lower- and higher-level features extracted when using the different deep CNNs. Considering the AlexNet and Places365-AlexNet, the lower-level (L1–L3) features outperformed the higher-level features (L4, L5) in terms of almost all the statistical measures. This trend was observable in the results of the three classifiers (SVClnr, RF and KNN). With respect to the AlexNet, for example, training SVFlnr with the features from the L3 layer yielded an overall accuracy of 89.7% compared to 80.5% attained by using those of the L7 layer. Similarly, the discrepancy in the discrimination capability between these two layers was also reflected in the AUC value. For example, when using SVClnr, the L3 and L7 layer features of the AlexNet contributed to the AUC values of 0.96 and 0.89, respectively, of which the ROC curves are shown in Figure 3a. Moreover, the superiority of the lower-level features over the high-level features was manifested consistently in all five statistical parameters. This can be illustrated by the use of the RF classifier for the Place365s-AlexNet model; changing from the features of the L3 layer to those of the L7 layer led to a decrease in the ACC from 86.7% to 80.6%, in the TRP from 84.5% to 77.1% and in the PPV from 88.3% to 82.8%. Such a decreasing trend is also notable in the remaining two measures.
Table 2.
Testing result (Dataset I) of the four classifiers trained with the low- and high-level features extracted by the three pre-trained deep CNNs (the maximum was highlighted in each row).
| Score╲Model | AlexNet | Places365-AlexNet | GoogLeNet | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | L3 | L5 | L6 | L7 | L1 | L3 | L5 | L6 | L7 | L1 | L2 | L3 | L4 | L5 | ||
| ACC (%) | SVClnr | 88.3 | 89.7 | 88.5 | 81.3 | 80.5 | 87.9 | 89.8 | 86.7 | 80.4 | 82.4 | 87.6 | 89.0 | 90.0 | 90.4 | 90.2 |
| SVCrbf | 72.2 | 75.3 | 85.9 | 84.5 | 88.8 | 50.0 | 78.9 | 87.9 | 87.9 | 85.6 | 86.2 | 85.1 | 86.1 | 89.8 | 89.4 | |
| RF | 86.4 | 87.0 | 83.6 | 82.8 | 83.9 | 87.0 | 86.7 | 83.2 | 81.4 | 80.6 | 86.6 | 86.4 | 86.9 | 87.4 | 87.0 | |
| KNN | 87.0 | 87.1 | 86.8 | 85.8 | 85.8 | 86.4 | 87.0 | 84.6 | 83.0 | 82.0 | 86.7 | 86.3 | 88.0 | 89.8 | 89.1 | |
| AUC | SVClnr | 0.953 | 0.963 | 0.954 | 0.895 | 0.887 | 0.953 | 0.961 | 0.944 | 0.889 | 0.909 | 0.949 | 0.956 | 0.964 | 0.967 | 0.963 |
| SVCrbf | 0.868 | 0.902 | 0.930 | 0.920 | 0.952 | 0.500 | 0.915 | 0.945 | 0.948 | 0.935 | 0.940 | 0.931 | 0.939 | 0.961 | 0.959 | |
| RF | 0.933 | 0.933 | 0.910 | 0.901 | 0.909 | 0.935 | 0.932 | 0.904 | 0.892 | 0.889 | 0.935 | 0.931 | 0.936 | 0.939 | 0.938 | |
| KNN | 0.944 | 0.942 | 0.940 | 0.930 | 0.932 | 0.938 | 0.943 | 0.925 | 0.914 | 0.904 | 0.939 | 0.938 | 0.948 | 0.961 | 0.955 | |
| TPR (%) | SVClnr | 88.7 | 90.4 | 87.6 | 80.8 | 79.9 | 88.3 | 90.1 | 87.1 | 80.6 | 82.4 | 87.4 | 89.4 | 90.4 | 90.9 | 90.2 |
| SVCrbf | 51.5 | 57.1 | 90.7 | 90 | 88.6 | 44.4 | 64.4 | 88.6 | 88.6 | 86.5 | 86.5 | 85.1 | 86 | 90.2 | 89.4 | |
| RF | 85.6 | 84.6 | 81.1 | 79.4 | 81.1 | 85.6 | 84.5 | 80.4 | 78.6 | 77.1 | 85.6 | 84.2 | 85.3 | 86.4 | 85.2 | |
| KNN | 88.6 | 89.1 | 84.2 | 85.4 | 86.1 | 87.8 | 90.5 | 88 | 85.1 | 84 | 88 | 89.7 | 90.4 | 89.8 | 89.0 | |
| PPV (%) | SVClnr | 87.9 | 89.1 | 89.1 | 81.5 | 80.8 | 87.5 | 89.5 | 86.3 | 80.2 | 82.3 | 87.7 | 88.6 | 89.6 | 89.9 | 90.1 |
| SVCrbf | 87.7 | 89.6 | 82.7 | 81.0 | 88.9 | 49.9 | 90.6 | 87.3 | 87.3 | 84.9 | 85.9 | 85.0 | 86.1 | 89.4 | 89.3 | |
| RF | 86.9 | 88.8 | 85.3 | 85.1 | 85.8 | 88.0 | 88.3 | 85.1 | 83.2 | 82.8 | 87.3 | 88.0 | 88.0 | 88.1 | 88.3 | |
| KNN | 85.8 | 85.6 | 88.7 | 86.0 | 85.5 | 85.3 | 84.5 | 82.3 | 81.6 | 80.7 | 85.7 | 83.9 | 86.2 | 89.7 | 89.1 | |
| F1 (%) | SVClnr | 88.3 | 89.8 | 88.4 | 81.2 | 80.4 | 88.0 | 89.8 | 86.8 | 80.4 | 82.4 | 87.5 | 89.0 | 90.0 | 90.4 | 90.2 |
| SVCrbf | 64.9 | 69.8 | 86.5 | 85.3 | 88.8 | 66.6 | 75.4 | 88.0 | 88.0 | 85.7 | 86.3 | 85.0 | 86.1 | 89.8 | 89.4 | |
| RF | 86.3 | 86.6 | 83.2 | 82.2 | 83.5 | 86.8 | 86.4 | 82.7 | 80.9 | 79.9 | 86.5 | 86.1 | 86.6 | 87.3 | 86.8 | |
| KNN | 87.2 | 87.4 | 86.4 | 85.7 | 85.9 | 86.6 | 87.4 | 85.1 | 83.3 | 82.4 | 86.8 | 86.8 | 88.3 | 89.8 | 89.1 | |
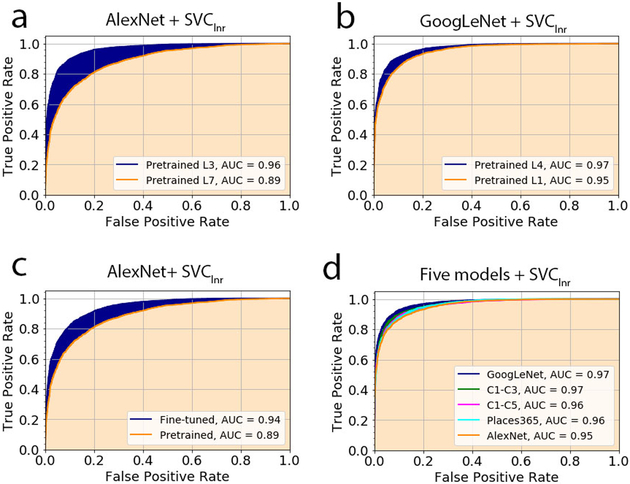
Figure 3.
The performance of epithelium and stroma classification for the typical scenarios. The curves, known as ROC curves, were generated based on the prediction that would yield ‘positive’ if the epithelium probability was larger than the threshold t (0≤t≤1). (a) The pre-trained AlexNet with the classifier of SVClnr at the layers of L3 and L7. (b) The pre-trained GoogLeNet with the classifier of SVClnr at the layers of L1 and L4. (c) The AlexNet prior to and after fine-tuning on Dataset I, with the classifier of SVClnr built at the L7 layer. (d) Comparison of all five models fine-tuned on Dataset II with the use of the classifier of SVClnr.
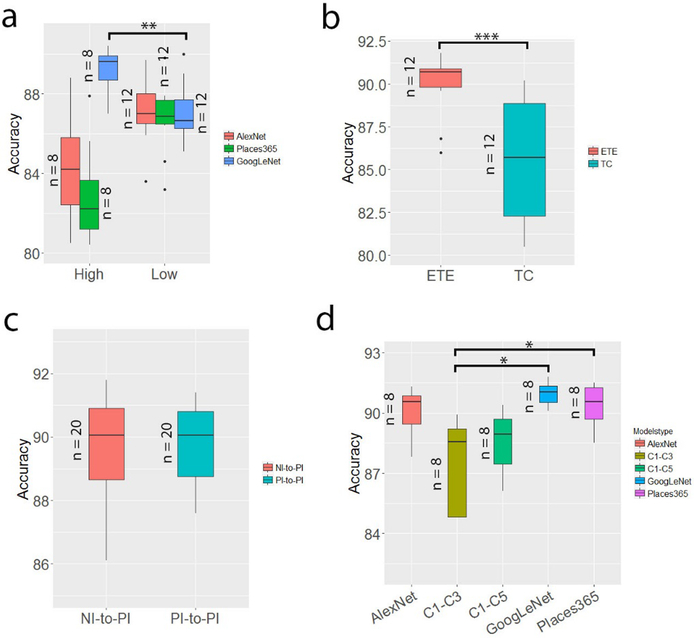
Conversely, in the GoogLeNet, the higher-level features consistently achieved superior performance to the lower-level features (P<0.01; Figure 2a) in almost all statistical measures and classifiers. The average accuracy associated with the higher-level (L4–L5) features was 89.14%, and the figure was 86.81% for the lower-level (L1–L3) features. Similarly, the AUC is 0.95 and 0.97 at the L1 and L4 layers for the classifier of SVClnr (Table 2 and Figure 3b), and the relevant TPR increases from 87.4% for the lower-level to 90.9% for the higher-level. Taken together, the superiority of the lower-level or the higher-level features over the others relied on the architecture itself. Even at a very high level of a deep CNN structure such as the GoogLeNet, the features extracted, which were reasonably specific to the target task, were generalizable to data of other fields.
Figure 2.
The statistics of transfer learning in classification of epithelium and stroma. (a) Comparison of the discriminatory capacity prior to fine-tuning between the higher-level features (from the L6 and L7 layers of AlexNet and of Places365-AlexNet and the L4 and L5 layers of GoogLeNet) and lower-level features (from the remaining three layers of the models (Table 2)). For each model, the sample sizes of the higher-level group and lower-level group for the t-test are 8 and 12, respectively. For example, the higher-level group contained two layers, each of which included four accuracy scores from the four classifiers. (b) Comparison of the performance of deep CNNs with fine-tuning (ETE) and without fine-tuning (TC). (c) Comparison of the performance of the two strategies of transfer learning. (d) Comparison of the performance of the fine-tuned five models. For each model, the results attained using the two strategies (NI-to-PI and PI-to-PI) and the four classifiers were included.
3.2. End-to-end fine-tuning of deep CNNs
As indicated in Table 3, implementing the NI-to-PI fine-tuning of the models (followed by training the classifiers at the intermediate layer) can significantly improve the classification performance of the models before fine-tuning. Specifically, it increased the accuracy by approximately 5.4% for the AlexNet, 8.3% for Places365-AlexNet and 1.7% for the GoogLeNet compared with the results of training only the classifiers given the pre-trained models. Taking the data from all three models together, the pairwise t test (Figure 2b) shows that the ETE group with a mean accuracy of 90.1% performed significantly better than the TC group with a mean accuracy of 85.44% (P<0.001). With the ETE, the minimum accuracy of 86.0% was achieved by the AlexNet and the maximum of 91.1% was achieved by the GoogLeNet (Table 3). The other four statistical parameters reflected the same trend. Specifically, when SVClnr was applied, the AUC of the AlexNet increased by 5% (or 0.94 vs 0.89), as demonstrated in Figure 3c. Moreover, our result suggested there was a negligible difference (Figure 2c) between using the NI-to-PI and PI-to-PI fine-tuning. Close observation of the data for the GoogLeNet and AlexNet shows that there is a change of only 0.18% for the GoogLeNet and −0.45% for AlexNet after switching from the NI-to-PI to the PI-to-PI strategy. A comparison of the classification performance of all five architectures (Table 4) indicates that the top two performers are the GoogLeNet and Places365-AlexNet (Figure 2d), with the mean accuracy of the former slightly higher than that of the latter. The performance of these two architectures was observed to be significantly better than the relatively shallow architecture, that is, the C1–C3. Their respective AUC curves are displayed in Figure 3d, where the GoogLeNet was associated with a very slightly higher AUC value. Considering both Table 3 and 4, the fine-tuning performance for Dataset I was highly consistent with that for Dataset II (Supplementary Figure S4).
Table 3.
Testing result (Dataset I) of the four classifiers trained with the features extracted before and after the NI-to-PI fine-tuning of the different deep CNNs.
| Score╲Model | AlexNet (L7) | Places365-AlexNet (L7) | GoogLeNet (L5) | ||||
|---|---|---|---|---|---|---|---|
| TC | ETE | TC | ETE | TC | ETE | ||
| ACC (%) | SVClnr | 80.5 | 86.0 | 82.4 | 86.8 | 90.2 | 91.1 |
| SVCrbf | 88.8 | 90.9 | 85.6 | 90.9 | 89.4 | 90.4 | |
| RF | 83.9 | 89.9 | 80.6 | 89.6 | 87.0 | 89.3 | |
| KNN | 85.8 | 90.6 | 82.0 | 90.8 | 89.1 | 90.8 | |
| AUC | SVClnr | 0.887 | 0.941 | 0.909 | 0.941 | 0.963 | 0.968 |
| SVCrbf | 0.952 | 0.973 | 0.935 | 0.966 | 0.959 | 0.965 | |
| RF | 0.909 | 0.956 | 0.889 | 0.952 | 0.938 | 0.951 | |
| KNN | 0.932 | 0.963 | 0.904 | 0.963 | 0.955 | 0.965 | |
| TPR (%) | SVClnr | 79.9 | 86.0 | 82.4 | 87.1 | 90.2 | 91.0 |
| SVCrbf | 88.6 | 91.7 | 86.5 | 90.7 | 89.4 | 90.7 | |
| RF | 81.1 | 89.2 | 77.1 | 88.3 | 85.2 | 88.3 | |
| KNN | 86.1 | 91.3 | 84 | 91.2 | 89 | 91.0 | |
| PPV (%) | SVClnr | 80.8 | 85.9 | 82.3 | 86.5 | 90.1 | 91.1 |
| SVCrbf | 88.9 | 90.2 | 84.9 | 91.0 | 89.3 | 90.1 | |
| RF | 85.8 | 90.4 | 82.8 | 90.6 | 88.3 | 90.0 | |
| KNN | 85.5 | 90.0 | 80.7 | 90.4 | 89.1 | 90.6 | |
| F1 (%) | SVClnr | 80.4 | 85.9 | 82.4 | 86.8 | 90.2 | 91.0 |
| SVCrbf | 88.8 | 90.9 | 85.7 | 90.9 | 89.4 | 90.4 | |
| RF | 83.5 | 89.8 | 79.9 | 89.4 | 86.8 | 89.1 | |
| KNN | 85.9 | 90.7 | 82.4 | 90.7 | 89.1 | 90.7 | |
Table 4.
Testing result (Dataset II) of the four classifiers trained with the features extracted after fine-tuning of the five different deep CNNs. NI-to-PI: transferring from the natural image dataset to the target pathology image dataset; and PI-to-PI: transferring from the non-target PI dataset to the target PI dataset. All three networks were fine-tuned on Dataset II. C1: first convolutional layer; FC1: fully connected layer. Each classifier was built on the C3 layer for the C1-C2-C3-FC1 network and on the C5 layer for the C1-C2-C3-C4-C5-FC1 network.
| Score╲Model | C1-C2-C3-FC1 | C1-C2-C3-C4-C5-FC1 | AlexNet | Places365-AlexNet | GoogLeNet | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NI-to-PI | PI-to-PI | NI-to-PI | PI-to-PI | NI-to-PI | PI-to-PI | NI-to-PI | PI-to-PI | NI-to-PI | PI-to-PI | ||
| ACC (%) | SVClnr | 89.9 | 89.8 | 89.3 | 89.5 | 88.7 | 87.8 | 88.5 | 88.8 | 91.8 | 91.3 |
| SVCrbf | 75.0 | 73.5 | 90.3 | 90.4 | 91.3 | 90.8 | 91.5 | 91.4 | 90.8 | 90.6 | |
| RF | 88.3 | 88.1 | 86.1 | 87.6 | 90.3 | 89.7 | 90.0 | 90.3 | 90.1 | 90.3 | |
| KNN | 89.0 | 88.8 | 87.0 | 88.6 | 90.8 | 91.0 | 91.2 | 90.8 | 91.5 | 91.3 | |
| AUC | SVClnr | 0.966 | 0.965 | 0.959 | 0.962 | 0.958 | 0.954 | 0.959 | 0.959 | 0.974 | 0.973 |
| SVCrbf | 0.919 | 0.922 | 0.964 | 0.965 | 0.973 | 0.972 | 0.973 | 0.973 | 0.970 | 0.970 | |
| RF | 0.947 | 0.951 | 0.937 | 0.945 | 0.961 | 0.958 | 0.959 | 0.959 | 0.961 | 0.961 | |
| KNN | 0.958 | 0.959 | 0.950 | 0.958 | 0.970 | 0.969 | 0.971 | 0.970 | 0.973 | 0.974 | |
| TPR (%) | SVClnr | 88.9 | 88.5 | 88.3 | 88.2 | 88.3 | 88 | 88.3 | 88.5 | 91.5 | 90.8 |
| SVCrbf | 53.7 | 49.5 | 89.3 | 88.4 | 90.2 | 88.6 | 90.7 | 90.7 | 90.6 | 89.5 | |
| RF | 85.8 | 84.5 | 83.4 | 85.1 | 88.3 | 88 | 88.0 | 88.7 | 87.9 | 88.6 | |
| KNN | 89.1 | 87.9 | 87.3 | 86.1 | 88 | 89.4 | 89.8 | 89.7 | 90.4 | 89.6 | |
| PPV (%) | SVClnr | 90.8 | 90.9 | 90.2 | 90.6 | 89.1 | 87.7 | 88.7 | 89.1 | 92.1 | 91.8 |
| SVCrbf | 94.0 | 95.7 | 91.2 | 92.1 | 92.3 | 92.7 | 92.2 | 92.0 | 91.0 | 91.6 | |
| RF | 90.4 | 91.1 | 88.3 | 89.7 | 92.0 | 91.2 | 91.7 | 91.7 | 92.0 | 91.8 | |
| KNN | 89.0 | 89.6 | 86.9 | 90.7 | 93.3 | 92.4 | 92.4 | 91.8 | 92.5 | 92.8 | |
| F1 (%) | SVClnr | 89.8 | 89.8 | 89.2 | 89.4 | 88.7 | 87.9 | 88.5 | 88.9 | 91.9 | 91.3 |
| SVCrbf | 68.3 | 65.3 | 90.3 | 90.2 | 91.3 | 90.6 | 91.5 | 91.4 | 90.9 | 90.6 | |
| RF | 88.0 | 87.7 | 85.8 | 87.3 | 90.1 | 89.5 | 89.9 | 90.2 | 90.0 | 90.2 | |
| KNN | 89.1 | 88.7 | 87.1 | 88.3 | 90.6 | 90.9 | 91.1 | 90.8 | 91.5 | 91.2 | |
3.3. Visualization of feature maps for gaining understanding of transfer learning
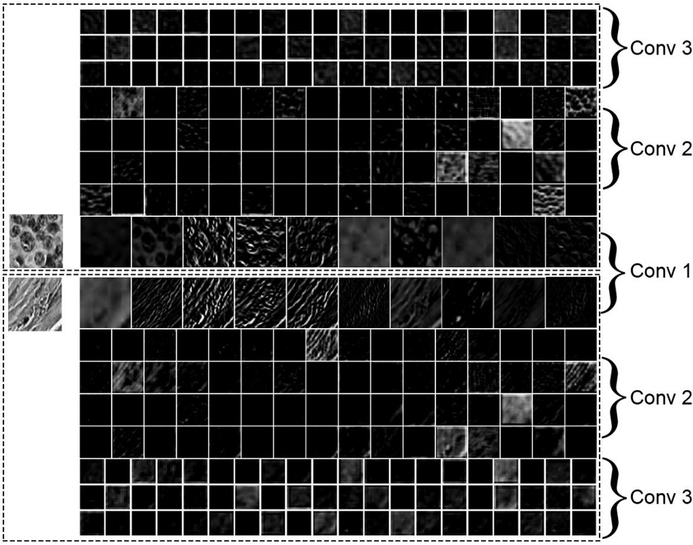
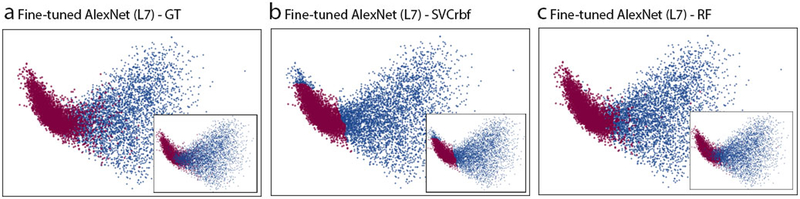
Deep CNN processes images to generate hierarchies of features, with higher-level features constructed by lower-level ones. Each convolutional layer acts as a filter, which takes the units in the previous layer as inputs to reveal morphological, textural, contextual descriptors that form higher-level feature maps. This computational operation involves a local weighted sum of the output from the previous layer, followed by setting a non-linear boundary with a ReLU. As illustrated in Figure 4, the first convolutional layer captured the contours of epithelial cells that appeared in the gray-scale example image of epithelium and oriented edges shown in the example image of stroma. Some of the feature patches in the first convolutional layer had a similar pattern as those produced by using the Sober operator or Prewitt operator (which are edge detectors) as weights when executing the convolution operation for the image (Supplementary Figure S5). As we go deeper into the CNN architecture, we can observe that the contextual information (that is, the distribution patterns of the brightness of the images) tend to be readily recorded. This figure was generated by displaying the computational results from the first to third convolutional layers (in a grid with a dimension of , n is the kernel size) given the gray-scale example images of epithelium and stroma. Figure 5 depicts the classification performance of the different classifiers trained with the features extracted after fine-tuning of the AlexNet on Dataset I. Each of the figures was produced using principle components analysis (PCA) to project all the features to a lower dimensional space. Panel a shows the ground truth of classification, in which the color indicates the true class, while panels b and c use the predicted result of the corresponding classifier to set the color for each data point. In Figure 5 we can also observe the closeness of the nearly 4000 features within the same group, and between the two different groups under the three scenarios. As we can see, the features extracted after fine-tuning became spatially distinguishable in two-dimensional space. The classifiers of SVCrbf and RF further stratified the features into the two classes with high accuracy.
Figure 4.
Visualization of feature maps. The image shows the output of the first three convolutional layers of the AlexNet model that was applied to the gray-scale example images of epithelium (upper) and stroma (lower). Each rectangular image displays one of the learned features at the layer, with the higher-level features composed of the lower-level ones. It is observable that the first convolutional layer captured the contours of epithelial cells that appeared in the epithelium image, and oriented edges are shown in the stroma image.
Figure 5.
Classification visualization at the L7 layer of the AlexNet after fine-tuning. The images were produced by taking the first two principal components in dimension reduction with the PCA that projects all the features to a lower dimensional space. (a) The ground truth (GT) of the representation of the features from the fine-tuned AlexNet (L7) on Dataset I. (b) and (c) display the classification result based on the features extracted from the fine-tuned model using the classifiers of SVCrbf and RF.
4. Discussion
In this study, we investigated the performance of transfer learning with the three different deep learning architectures in the classification of epithelial and stromal regions of histological images. Unlike similar studies, there are two unique characteristics in our investigation. First, we thoroughly analyzed the performance of the features extracted from both the lower and higher level of the three different deep learning networks. Second, we compared the efficacy of fine-tuning between the two strategies, i.e., from the NI dataset to the target PI dataset (or NI-to-PI) and from the non-target PI dataset to the target PI dataset (PI-to-PI).
With the use of the NI features prior to fine-tuning, a classification accuracy as high as 90.2% could be achieved using GoogLeNet at the layer of Max pool5b (L5). This figure is slightly better than that attained by using a set of 112 hand-engineered features and an epithelial/stromal classifier with L1 regularized logistic regression method that delivered an accuracy of 89%3. This result may suggest the potential use of the transfer learning as an alternative to manual extraction of a large quantity of imaging features for this binary classification problem. The comparison in discriminatory power between the higher-level features and lower-level ones revealed that the result was heavily dependent on the model used. For the AlexNet and Places365-AlexNet, the lower-level features outperformed the higher-level ones in nearly all statistical measures; however, it is not applicable to the situation using the GoogLeNet. The statistical analysis shows that there is a 3.3% and 5.8% improvement in accuracy if changing the higher-level (comprising L6 and L7) features to the lower-level ones (L1, L3 and L5) for AlexNet and Places365-AlexNet, respectively, and a 3.6% decline (P<0.01) for the GoogLeNet in the same scenario. It is therefore unsurprising to see that the densely connected layers (L6 and L7) exhibited superior performance to the layer of pool5 for the TorontoNet in9, whereas the situation in the opposite direction is documented in2, where the VGG16 network was used. Given that the higher-level features were more affected in the fine-tuning and thus more specific to the context of the source images than the lower-level ones, it may be concluded that the features that are highly specific or even exclusive to the source images are not necessarily less generalizable than those that are not.
By executing fine-tuning, accuracy was increased by 5.4% for the AlexNet, 8.3% for the Places365-AlexNet and 1.7% for the GoogLeNet. Our observation about the performance of fine-tuning mainly addresses three aspects. First, the pairwise t test that compares before and after fine-tuning shows that the improvement in quality of features by fine-tuning was very significant (P<0.001), resulting in an average accuracy of 90.1%. The observed effectiveness of fine-tuning was also documented for TorontoNet in9. However, in28, a similar result was only obtained for GoogLeNet, not AlexNet. The training parameters could be one factor that might account for the inconsistency between28 and our result. Particularly, a base learning rate of 0.01 was chosen in28, but we dropped that number to 0.001 with the same gamma value to reduce the divergence of the learning process (that is, large variations in the loss values). Second, our best result of fine-tuning, that is, an accuracy of 91.8% and an AUC value of 0.97 achieved by GoogLeNet, outperformed that of two similar studies27,32 that trained a self-constructed CNN structure. The highest accuracy reported by the former was 88.34%, whereas the highest AUC value documented by the latter was 0.92 for the original H&E stained histological images. Third, PI-to-PI fine-tuning did not show superiority over NI-to-PI fine-tuning. After switching from the NI-to-PI to the PI-to-PI strategy, there is a negligible increase in the overall accuracy for the GoogLeNet and a very slight fall-off for AlexNet. We suspect that the weights used in the NI-to-PI, which were optimized with millions of natural images, were probably already established at a global minima. The fine-tuning in the PI-to-PI case could not drive them away from this global minima.
Despite the encouraging results, there are several limitations. Our study was limited to the binary classification problem. A majority of the current studies using machine learning algorithms for specimen categorization or disease diagnosis focus on two-category problems, such as detection of Crohn’s disease and ulcerative colitis belonging to pediatric inflammatory bowel disease22, discrimination between patients with Parkinson’s disease and healthy subjects1, and binary decisions regarding whether the stroma is mature based on histological images26. However, it would be desirable to extend the approach to multiple-case classification of medical images, in particular in view of the great discriminatory power of transfer learning shown in this study – for instance, in situations such as staging fatty liver cases by distinguishing among steatosis, fibrosis and cirrhosis in ultrasound images23, classifying gastric biopsy specimens into carcinoma or positive or negative for a neoplastic lesion34, and three-class disease classification given Golub’s leukemia microarray data through gene feature selection with discriminant analysis33. Another important future direction for research will be to test transfer learning technologies on different modalities where the variety of machine learning algorithms have been considered, such as mammography for diagnosis of breast cancer based on microcalcification detection30, Computed Tomography18,25, and magnetic resonance imaging17,19. Overall, there is a wide range of potential medical applications for transfer learning methods. Given its performance in stratifying epithelium and stroma, which mark a certain degree of feature overlapping between the two, it seems that using it for multiple-class categorization of images from disparate modalities is viable.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the support from Oklahoma Center for the Advancement of Science & Technology (OCAST) grant HR15–016 and National Institutes of Health grant R01 CA197150. This research is also partially sponsored by one research award from Stephenson Cancer Center at the University of Oklahoma Health Sciences Center (OUHSC).
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Adeli E, Wu G, Saghafi B, An L, Shi F, and Shen D. Kernel-based Joint Feature Selection and Max-Margin Classification for Early Diagnosis of Parkinson’s Disease. Sci. Rep 7:41069, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antony J, McGuinness K, Connor NEO, and Moran K. Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks 1195–1200, 2016. at <http://arxiv.org/abs/1609.02469> [Google Scholar]
- 3.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, and Koller D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci. Transl. Med 3:108ra113–108ra113, 2011. [DOI] [PubMed] [Google Scholar]
- 4.Bengio Y, Courville A, and Vincent P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell 35:1798–1828, 2013. [DOI] [PubMed] [Google Scholar]
- 5.Carneiro G, Nascimento J, and Bradley AP. Unregistered multiview mammogram analysis with pre-trained deep learning models. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 9351:652–660, 2015. [Google Scholar]
- 6.Cireşan DC, Giusti A, Gambardella LM, and Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Med. Image Comput. Comput. Interv 16(Pt2):411–418, 2013. [DOI] [PubMed] [Google Scholar]
- 7.Roy Divakar. AUTO CONTRAST, 2009. at <http://www.mathworks.com/matlabcentral/fileexchange/10566-auto-contrast>
- 8.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, and Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girshick R, Donahue J, Darrell T, and Malik J. Region-Based Convolutional Networks for Accurate Object Detection and Segmentation. IEEE Trans. Pattern Anal. Mach. Intell 38:142–158, 2016. [DOI] [PubMed] [Google Scholar]
- 10.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, and Yener B. Histopathological Image Analysis: A Review. IEEE Rev. Biomed. Eng 2:147–171, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh BQ, Li H, and Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 3:34501, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ioffe S, and Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Arxiv 1–11, 2015.doi:10.1007/s13398-014-0173-7.2 [Google Scholar]
- 13.Krizhevsky A, Sutskever I, and Hinton GE. ImageNet Classification with Deep Convolutional Neural Networks. Adv. Neural Inf. Process. Syst 1–9, 2012.doi:http://dx.doi.org/10.1016/j.protcy.2014.09.007 [Google Scholar]
- 14.de Kruijf EM, van Nes JGH, van de Velde CJH, Putter H, Smit VTHBM, Liefers GJ, Kuppen PJK, Tollenaar RAEM, and Mesker WE. Tumor–stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients. Breast Cancer Res. Treat 125:687–696, 2011. [DOI] [PubMed] [Google Scholar]
- 15.LeCun Y, Bengio Y, and Hinton G. Deep learning. Nature 521:436–444, 2015. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, Liu J, Li J, Chen Y, Guan X, Wu X, Hao C, Sun Y, Wang Y, and Wang X. Tumor–stroma ratio is an independent predictor for survival in early cervical carcinoma. Gynecol. Oncol 132:81–86, 2014. [DOI] [PubMed] [Google Scholar]
- 17.Long X, Chen L, Jiang C, and Zhang L. Prediction and classification of Alzheimer disease based on quantification of MRI deformation. PLoS One 12:1–19, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu P, Barazzetti L, Chandran V, Gavaghan K, Weber S, Gerber N, and Reyes M. Highly accurate Facial Nerve Segmentation Refinement from CBCT/CT Imaging using a Super Resolution Classification Approach. IEEE Trans. Biomed. Eng 9294:1–1, 2017. [DOI] [PubMed] [Google Scholar]
- 19.Mehrtash A, Sedghi A, Ghafoorian M, Taghipour M, Tempany CM, Wells WM, Kapur T, Mousavi P, Abolmaesumi P, and Fedorov A. Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks, 2017.doi:10.1117/12.2277123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menegola A, Fornaciali M, Pires R, Avila S, and Valle E. Towards Automated Melanoma Screening: Exploring Transfer Learning Schemes 1–4, 2016at <http://arxiv.org/abs/1609.01228> [Google Scholar]
- 21.Moorman AM, Vink R, Heijmans HJ, van der Palen J, and Kouwenhoven EA. The prognostic value of tumour-stroma ratio in triple-negative breast cancer. Eur. J. Surg. Oncol 38:307–313, 2012. [DOI] [PubMed] [Google Scholar]
- 22.Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, and Ennis S. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci. Rep 7:2427, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owjimehr M, Danyali H, Helfroush MS, and Shakibafard A. Staging of Fatty Liver Diseases Based on Hierarchical Classification and Feature Fusion for Back-Scan–Converted Ultrasound Images. Ultrason. Imaging 39:79–95, 2017. [DOI] [PubMed] [Google Scholar]
- 24.Pan Y, Huang W, Lin Z, Zhu W, Zhou J, Wong J, and Ding Z. Brain Tumor Grading Based on Neural Network s and C onvolutional Neural Network s. Eng. Med. Biol. Soc. (EMBC), 2015 37th Annu. Int. Conf. IEEE 699–702, 2015.doi:10.1109/EMBC.2015.7318458 [DOI] [PubMed] [Google Scholar]
- 25.Pota M, Scalco E, Sanguineti G, Farneti A, Cattaneo GM, Rizzo G, and Esposito M. Early prediction of radiotherapy-induced parotid shrinkage and toxicity based on CT radiomics and fuzzy classification. Artif. Intell. Med, 2017.doi:10.1016/j.artmed.2017.03.004 [DOI] [PubMed] [Google Scholar]
- 26.Reis S, Gazinska P, Hipwell J, Mertzanidou T, Naidoo K, Williams N, Pinder S, and Hawkes DJ. Automated Classification of Breast Cancer Stroma Maturity from Histological Images. IEEE Trans. Biomed. Eng 1–1, 2017.doi:10.1109/TBME.2017.2665602 [DOI] [PubMed] [Google Scholar]
- 27.Sethi A, Sha L, Vahadane AR, Deaton RJ, Kumar N, Macias V, and Gann PH. Empirical comparison of color normalization methods for epithelial-stromal classification in H and E images. J. Pathol. Inform 7:17, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, and Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans. Med. Imaging 35:1285–1298, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran H, Phan H, Kumar A, Kim J, and Feng D. Transfer Learning of a Convolutional Neural Network for Hep-2 Cell Image Classification 2012:1208–1211, 2016. [Google Scholar]
- 30.Wang J, Yang X, Cai H, Tan W, Jin C, and Li L. Discrimination of Breast Cancer with Microcalcifications on Mammography by Deep Learning. Sci. Rep 6:27327, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xi K-X, Wen Y-S, Zhu C-M, Yu X-Y, Qin R-Q, Zhang X-W, Lin Y-B, Rong T-H, Wang W-D, Chen Y-Q, and Zhang L-J. Tumor-stroma ratio (TSR) in non-small cell lung cancer (NSCLC) patients after lung resection is a prognostic factor for survival. J. Thorac. Dis 9:4017–4026, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Luo X, Wang G, Gilmore H, and Madabhushi A. A Deep Convolutional Neural Network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 191:214–223, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang M, Li X, Li Z, Ou Z, Liu M, Liu S, Li X, and Yang S. Gene features selection for three-class disease classification via multiple orthogonal partial least square discriminant analysis and S-plot using microarray data. PLoS One 8:1–12, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida H, Shimazu T, Kiyuna T, Marugame A, Yamashita Y, Cosatto E, Taniguchi H, Sekine S, and Ochiai A. Automated histological classification of whole-slide images of gastric biopsy specimens. Gastric Cancer, 2017.doi:10.1007/s10120-017-0731-8 [DOI] [PubMed] [Google Scholar]
- 35.Zhang R, Zheng Y, Mak TWC, Yu R, Wong SH, Lau JYW, and Poon CCY. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features from Nonmedical Domain. IEEE J. Biomed. Heal. Informatics 21:41–47, 2017. [DOI] [PubMed] [Google Scholar]
- 36.Zhou B, Lapedriza A, Xiao J, Torralba A, and Oliva A. Learning Deep Features for Scene Recognition using Places Database. Adv. Neural Inf. Process. Syst. 27 487–495, 2014at <http://papers.nips.cc/paper/5349-learning-deep-features-for-scene-recognition-using-places-database.pdf> [Google Scholar]
- 37.Stanford Tissue Microarray Databaseat <https://tma.im/cgi-bin/home.pl>
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.