Abstract
Background
Gut microbiota alteration has been implicated in HIV infection and metabolic disorders. The relationship between gut microbiota and diabetes has rarely been studied in HIV-infected individuals, who have excess risk of metabolic disorders.
Methods
Our study during 2015–2016 enrolled predominantly African Americans and Hispanics in the Women's Interagency HIV Study. We studied 28 women with long-standing HIV infection under antiretroviral therapy and 20 HIV-uninfected, but at high risk of infection, women (16 HIV+ and 6 HIV- with diabetes). Fecal samples were analyzed by sequencing prokaryotic16S rRNA gene. Plasma metabolomics profiling was performed by liquid chromatography-tandem mass spectrometry.
Findings
No significant differences in bacterial α- or β-diversity were observed by diabetes or HIV serostatus (all P > .1). Relative abundances of four genera (Finegoldia, Anaerococcus, Sneathia, and Adlercreutzia) were lower in women with diabetes compared to those without diabetes (all P < .01). In women with diabetes, plasma levels of several metabolites in tryptophan catabolism (e,g., kynurenine/tryptophan ratio), branched-chain amino acid and proline metabolism pathways were higher, while glycerophospholipids were lower (all P < .05). Results were generally consistent between HIV-infected and HIV-uninfected women, and no significant modification effects by HIV serostatus were observed (all Pinteraction > 0.05). Anaerococcus, known to produce butyrate which is involved in anti-inflammation and glucose metabolism, showed an inverse correlation with kynurenine/tryptophan ratio (r = −0.38, P < .01).
Interpretation
Among women with or at high risk for HIV infection, diabetes is associated with gut microbiota and plasma metabolite alteration, including depletion of butyrate-producing bacterial population along with higher tryptophan catabolism.
Fund
NHLBI (K01HL129892, R01HL140976) and FMF.
Keywords: Gut microbiota, Metabolite, Diabetes, HIV
Research in context.
Evidence before this study
Gut microbiota, especially butyrate-producing bacteria which is involved in anti-inflammation, have been associated with diabetes in general population. Few studies have examined this relationship in HIV-infected individuals who have chronic immune activation and inflammation. Recent evidence from a study of European men (mostly men who have sex with men) showed that HIV-infected individuals with diabetes had reduced microbial composition along with increased tryptophan catabolism which reflects high levels of inflammation and immune activation.
Added value of this study
In the present study, we assessed the associations of gut microbiota and broad spectrum of metabolite profiles with diabetes in minority HIV-infected women and women at high risk for HIV infection. In women with diabetes, we observed reduced abundance of four bacterial genera (Finegoldia, Anaerococcus, Sneathia, and Adlercreutzia) and increased concentrations of plasma metabolites in tryptophan catabolism, branched-chain amino acid and proline metabolism. Butyrate producing Anaerococcus was inversely associated with tryptophan catabolism, providing clues about bacteria-mediated metabolic pathways.
Implications of all the available evidence
This study and previous studies consistently showed altered gut microbiota and related metabolite changes in HIV-infected individuals with diabetes. These data suggest a potential link between reduced butyrate-producing bacteria and increased tryptophan catabolism in the development of diabetes in the context of HIV infection, possibly through inflammation and immune activation.
Alt-text: Unlabelled Box
1. Introduction
Metabolic disorders such as insulin resistance and diabetes have been of significant concern in HIV-infected people with successful antiretroviral therapy (ART) [1]. Prior studies have shown that excess risk of chronic metabolic disorders in HIV-infected individuals might be due to complicated interactions of chronic HIV infection, long-term ART, factors unrelated to HIV infection, and the underlying inflammatory process. Understanding of this pathophysiology remains incomplete.
Emerging evidence from animal and human studies has suggested an important role of gut microbiota in the development of type 2 diabetes mellitus [[2], [3], [4], [5]]. Multiple observational studies with populations of diverse age, sex, and race/ethnicity suggested that Roseburia and Faecalibacterium prauznitzii were less abundant in individuals with diabetes, compared to those without diabetes [2,3]. It has been suggested that butyrate, a short-chain fatty acid produced by Roseburia and F. prauznitzii, may have a protective effect in the development of type 2 diabetes [[6], [7], [8]]. Moreover, previous studies have found that gut microbiota and some related metabolites (e.g., tryptophan-kynurenine metabolites) are altered in HIV-infected individuals [[9], [10], [11]], though concomitant influences remain to be unraveled such as HIV infection, low CD4+ T-cell counts, high viral load, ART treatment and sexual behavior [[12], [13], [14], [15]]. Several studies on gut microbiota reported lower bacterial diversity in HIV infected individuals with no ART, low CD4+ counts, or high viral loads [[16], [17], [18]]. The higher relative abundance of Prevotella in untreated HIV-infected has been observed in multiple studies [[13], [14], [15]], while a recent study suggested that the increase in Prevotella was associated with sexual preference, men who have sex with men (MSM), but was not driven by HIV infection [12].
Although gut microbiota and related metabolites have been implicated in diabetes and HIV infection, few studies have investigated the relationship between gut microbiota and diabetes in the context of HIV infection. One recent study reported that HIV-infected individuals with diabetes had reduced microbial diversity along with increased tryptophan catabolism, endothelial dysfunction and inflammation compared to HIV-infected only and healthy controls [19]. However, the majority of the HIV-infected participants included in the study were MSM [19] which might have different gut microbiota patterns [12]. Therefore, in this study, we aimed to examine the associations of gut microbiota and plasma metabolite profiles with diabetes among HIV-infected women and women with high risk of HIV infection from the Women's Interagency HIV Study (WIHS).
2. Methods
2.1. Study participants
The WIHS is an ongoing prospective cohort study established in 1994 to investigate the progression of HIV in women, with a comparison HIV-negative group with high-risk behaviors for HIV infection including injection drug use, sexually transmitted disease, unprotected sex with 3+ men or protected sex with >5 men, or having exchanged sex for drugs, money, or shelter [20]. Details on study design and methods have been described previously [20]. Every 6 months, WIHS participants undergo a core visit with a comprehensive physical examination, providing biological specimens, and completing interviewer-administered questionnaires. In this study, 50 women from the WIHS Bronx site provided fecal samples in 2015–2016 and we included 48 samples for analyses, excluding one sample of very low reads in 16S rRNA gene sequencing of the gut microbiome (497 total read counts) and one sample from the sole HIV-infected woman not receiving ART. All individuals provided informed consent, and the studies were approved by the Institutional Review Board of Albert Einstein College of Medicine.
2.2. Fecal sample collection
Fecal samples were collected using a home-based self-collection kit that was distributed to participants during their core WIHS visit. The kit had all of the materials needed, including a pre-addressed envelope that the participants used to return the specimen to our lab at Albert Einstein College of Medicine. Fecal samples were self-collected using a disposable paper in an inverted hat shape that goes over the toilet seat to catch the fecal samples. Fecal samples were deposited in a collection tube containing 4 ml of RNAlater (a nucleic acid stabilizer that preserves the microbiome composition for up to 14 days at room temperature [21]) (QIAGEN, Valencia, CA), which was placed into a bag with absorbent/desiccator material and shipped via mail to our lab within 2 days for immediate processing and storage at −80 °C. At the time of fecal sample collection, participants also completed a questionnaire regarding antibiotic use, gastrointestinal medications, commercial probiotics, steroids, drugs stimulating or suppressing immune system in the last 6 months.
3. DNA extraction, PCR amplification and high-throughput sequencing
Genomic DNA was extracted with the DNeasy PowerLyzer PowerSoil DNA Isolation Kit (QIAGEN, Valencia, CA), following the manufacturer's instructions. PCR amplification was performed on the 16S rRNA gene V4 hypervariable region using the primers 16SV4_515F (GTGYCAGCMGCCGCGGTA) and 16SV4_806R (GGACTACHVGGGTWTCTAAT), with a 12-bp unique Golay barcoding [22,23]. PCR reactions were performed with an initial denaturation of 95 °C for 5 min, followed by 15 cycles of 95 °C for 1 min, 55 °C for 1 min, and 68 °C for 1 min, followed by 15 cycles of 95 °C for 1 min, 60 °C for 1 min, and 68 °C for 1 min, a final extension for 10 min at 68 °C on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). The PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN) and quantified using a Qubit 2.0 Fluorometic High Sensitivity dsDNA Assay (Life Technologies). KAPA LTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA) was used on the purified PCR products according to the manufacturer's protocol and the size integrity of the amplicon with Illumina indices was validated with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) at the Genomics Core at Albert Einstein College of Medicine. High-throughput amplicon sequencing was conducted on a MiSeq (Illumina, San Diego, CA. RRID:SCR_016379) using 2 × 300 paired-end fragments at the Albert Einstein College of Medicine Sequencing Core.
4. Bioinformatics analysis
Illumina reads were preprocessed to remove bases that fell below PHRED quality score of 25 using prinseq [24]. Processed reads were then demultiplexed using sample specific barcode combinations using novobarcode [25]. Paired end reads were merged using pandaseq with default settings [26]. OTU clustering and quality filtering was performed using the Quantitative Insights Into Microbial Ecology (QIIME) software package, version 1.9 [27]. USEARCH [28] was used to remove sequencing noise and sequence chimeras. Sequences were demultiplexed and clustered into operational taxonomic units (OTUs) with a minimum cluster similarity of 97% using UCLUST [28]. Sequences were assigned using UCLUST with the Greengenes 13.8 microbial database [29]. The resulting biom table was rarefied to 7500 reads per sample and statistical analyses were performed after collapsing OTUs at the genus level. To characterize functional contents of microbiota, PICRUSt was implemented to predict the abundance of KEGG orthologs [30] and the butyrate pathway in KEGG was visualized to be integrated with differential abundance by diabetes status using R/pathview [31,32].
5. Targeted metabolomics analysis
Blood was drawn into CPT tube containing sodium citrate as anticoagulant at least 8 h of fasting at the core WIHS visit. The tube was gently inverted 8 times at room temperature and centrifuged at 1500g for 20 min within 6 h of blood draw. The plasma aliquot was stored at −80 °C. Targeted metabolomics profiling were performed on stored frozen plasma specimens that had been collected at the semiannual core study visit closest to the stool sample collection, using the AbsoluteIDQ p180 kit (BIOCRATES Life Sciences AG, Innsbruck, Austria). This targeted method simultaneously measures 188 metabolites with liquid chromatography and flow injection analysis-mass spectrometry, and the detailed assay procedures have been described previously [33,34]. The current analysis included 130 metabolites, passing the quality control criteria of coefficient of variation <30% and low rate of out of detection range (25%): 20 amino acids, 6 biogenic amines, 12 acylcarnitines, 78 glycerophospholipids, and 14 sphingolipids.
6. Assessment of HIV infection, diabetes, and other variables
Demographic, clinical, and laboratory variables were collected using standardized protocols at semi-annual core study visits [35]. HIV infection was ascertained by enzyme-linked immunosorbent assay and confirmed by Western blot. HIV-specific parameters included CD4+ T-cell counts, HIV-1 viral load, and detailed information on ART. Diabetes was defined based upon standardized data from semiannual visits: fasting plasma glucose ≥126 mg/dl, HbA1C ≥6.5%, or self-report of diabetes medication use.
7. Data statement
The datasets analyzed in the current study are not publicly available but are available upon request after approval from the WIHS.
7.1. Statistical analysis
Characteristics of the study sample by diabetes status and HIV serostatus were summarized as counts and percentages for categorical variables, and medians and interquartile ranges for continuous variables. The characteristics were compared by diabetes status or HIV serostatus using the Fisher's exact test for categorical variables and the Mann-Whitney U test for continuous variables. The α-diversity of microbiota was performed using three measures (observed richness at the OTU level, Chao 1 index and Shannon index in the genus level) and each was compared by diabetes status in a linear regression adjusting for age and HIV status. β-diversity measures (Bray-Curtis, unweighted UniFrac [36], and weighted UniFrac [37]) were estimated for dissimilarity of microbial community compositions between samples. Principal coordinates analysis (PCoA) for β-diversity was used to examine the clusters of samples by diabetes status and HIV serostatus. Permutational multivariate ANOVA (PerMANOVA) was performed to test the association of β-diversity with diabetes, adjusting for age and HIV serostatus. The differential abundance of each genus or upper hierarchy by diabetes was tested using a linear regression on arcsin transformed relative abundance, adjusting for age and HIV status or adjusting for age stratified by HIV status. False discovery rate corrected P-value was calculated using Benjamini and Hochberg's method [38]. The metabolite concentration by diabetes status was tested using the Mann-Whitney U test and the Spearman correlation between metabolites and relative abundance of genus was calculated. Predicted relative abundance of KEGG orthologs by PICRUSt was compared by diabetes status using t-test. All statistical tests were two-sided and were conducted using R (version 3.3.2) [39] with the phyloseq [40] and vegan [41] packages.
8. Results
8.1. Characteristics of study population
Table 1 shows the characteristics of study participants by diabetes status and HIV serostatus. There was no significant difference in race/ethnicity by diabetes status and HIV serostatus. Women with diabetes tended to be older (P = .06 [Mann-Whitney U test]) and have higher BMI (P = .06 [Mann-Whitney U test]) among HIV-uninfected women. For glycemic traits, women with diabetes tended to have higher glucose and hemoglobin A1c levels irrespective of HIV serostatus. Half of women with diabetes took antidiabetic medications, mostly metformin (8 out of 11). Antibiotic use in the past 6 months was more frequent among HIV-infected women compared to HIV-uninfected women (P = .06 [Fisher's exact test]). All the HIV-infected participants were currently receiving highly active ART, and HIV-infected participants with diabetes were taking ART longer than the HIV-infected women without diabetes (P = .006 [Fisher's exact test]). Most HIV-infected women (25/28) had suppressed viral load (< 40 copies/ml) and high CD4 counts (24 women with >500 counts). There were no significant differences in CD4 counts and viral load between women with and without diabetes (P > .13 [Mann-Whitney U test]).
Table 1.
Characteristics of study subjects with diabetes and without diabetes.
| HIV+ |
HIV- |
|||
|---|---|---|---|---|
| Diabetes | No Diabetes | Diabetes | No Diabetes | |
| Number of individuals | 16 | 12 | 6 | 14 |
| Age | 52 (51–55) | 51 (42–60) | 53 (52–57) | 49 (42–51) |
| Race | ||||
| Black | 11 (69) | 8 (67) | 2 (33) | 10 (71) |
| Hispanic | 3 (19) | 1 (8) | 2 (33) | 3 (21) |
| Other | 2 (13) | 3 (25) | 2 (33) | 1 (7) |
| BMI (kg/m2) | 32 (30–38) | 31 (27–37) | 33 (33–37) | 26 (22−32) |
| Antibiotic | 10 (71) | 6 (55) | 1 (25) | 4 (33) |
| Any antidiabetic medication | 7 (43.8) | – | 4 (66.7) | – |
| Glucose (mg/dl) | 105 (94–115) | 96 (87–105) | 109 (96–124) | 94 (88–99) |
| Hemoglobin A1C (%) | 6.2 (6.0–6.5) | 5.8 (5.7–6.0) | 6.3 (5.9–6.8) | 5.6 (5.5–5.8) |
| Hepatitis C infection | 5 (31) | 3 (25) | 2 (33) | 1 (7) |
| HIV-specific characteristics | ||||
| Current ART | 16 (100) | 12 (100) | – | – |
| Cumulative years of ART | 12.5 (8.0–15.9) | 4.8 (3.9–9.6) | – | – |
| CD4 counts | 810 (629–1124) | 678 (578–955) | – | – |
| Viral load (copies/ml) | <20 (20–30.2) | <20 (20−20) | – | – |
| Undetectable viral load (≤20 copies/ml) | 10 (62.5) | 11 (91.7) | – | – |
Data are presented as count (%) for categorical variables or median (IQR) for continuous variables.
8.2. Comparisons of microbial diversity and composition
A total of 1084 operational taxonomic units (OTUs) were found with a 97% similarity cutoff, after rarefication at 7500 counts. At the phylum level, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria were found in all participants, having median microbial abundances of 52%, 35%, 2.9%, and 0.8%, respectively (Fig. S1). At the genus level, 147 genera were identified with median relative abundance of 0.43% (IQR = 0.1–0.88%), and 91 genera showed at least 0.1% average relative abundance.
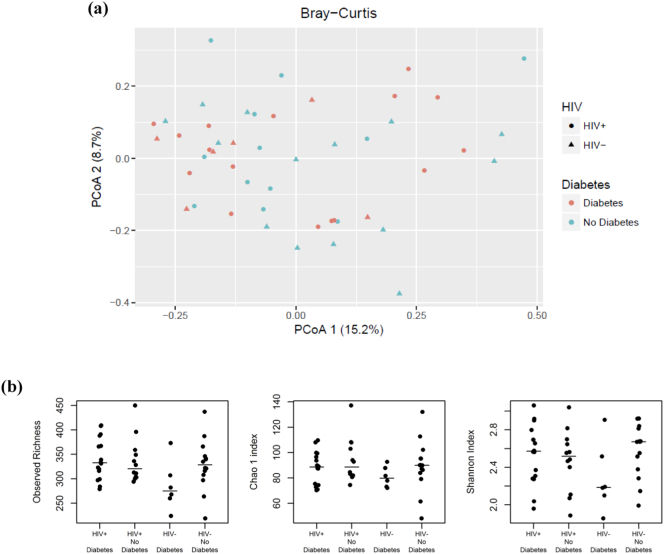
The microbiota profiles of our sample population were ordinated by principal coordinate analysis (PCoA) with the Bray-Curtis dissimilarity at genus level. The plot of the first and second principal coordinates in Fig. 1(a), explaining 15.2% and 8.7% of the total variance, showed no separation by diabetes status or HIV serostatus. In addition, PerMANOVA tests based on various sample-to-sample distances (Bray-Curtis, unweighted and weighted UniFrac) found no significant differences in microbiota profiles by age, HIV serostatus, diabetes status, or metformin use (all P > .10, R2 < 0.04, Table S1). Significant differences in Bray-Curtis dissimilarity and weighted UniFrac were observed by antibiotic use (P = .05, 0.01, Table S1, Fig. S2). The within-sample microbial diversity for each measure (observed OTU richness, Chao 1 index and Shannon index in genus level) was not significantly different by age, HIV, diabetes, antibiotic use, or metformin use (all P > .12 [linear regression], Fig. 1(b), Table S2).
Fig. 1.
Microbial composition and diversity by HIV serostatus and diabetes (a) Principal coordinates analysis of Bray-Curtis dissimilarity of microbiota by HIV serostatus and diabetes, (b) Alpha diversity of microbiota by HIV serostatus and diabetes. Observed richness is in the OTU level, and Chao 1 and Shannon index are in the genus level. A bar represents a median of the group.
8.3. Differentially abundant genera by diabetes
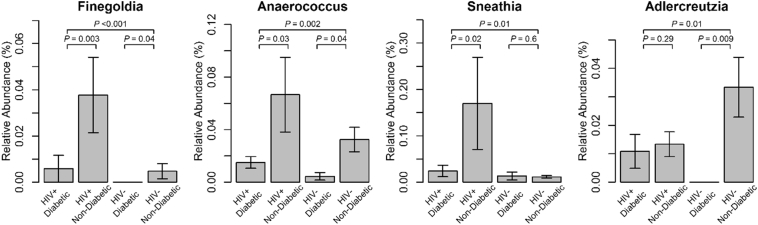
The differential abundance analysis identified four genera (Finegoldia, Anaerococcus, Sneathia, and Adlercreutzia) that were significantly associated with diabetes status (P < .05; FDR corrected P-value <.1 for Finegoldia and Anaerococcus. Table 2). These analyses adjusted for HIV status and age, examining 91 genera with at least 0.01% average relative abundance. These four differentially abundant genera were less abundant in women with diabetes compared to those without diabetes (Fig. 2). The four genera did not show statistically significant associations with HIV serostatus (Table S3). In the analysis stratified by HIV status (Table 2), Finegoldia and Anaerococcus showed significantly inverse association with diabetes in both HIV-infected and HIV-uninfected women, whereas there were suggestive but non-significant interactions between HIV infection and diabetes in relation to Sneathia and Adlercreutzia (PHIV×Diabetes = .09, 0.06, respectively). For those four genera that were differentially abundant by diabetes, we further examined the potential influences of antidiabetic medication use, antibiotic use and hepatitis C infection. No significant associations of four genera with these factors were observed (Table S4-S6), except for Sneathia which showed a nominal significant association with antibiotic use (Table S6). Sensitivity analyses indicated that the four genera exhibited consistent associations with diabetes after excluding the women with antibiotic use, metformin use, or hepatitis C infection, respectively, or adjusting for antibiotic use or hepatitis C infection (Table S7-S8).
Table 2.
Differentially abundant genera by diabetes status.
| Family | Genus | Alla |
HIV+ |
HIV- |
PHIV×Diabetesc | |||
|---|---|---|---|---|---|---|---|---|
| β (SE) | P | β (SE) | P | β (SE) | P | |||
| Tissierellaceae | Finegoldia | −0.010 (0.003) | <0.001b | −0.013 (0.004) | 0.003 | −0.005 (0.002) | 0.038 | 0.133 |
| Peptoniphilaceae | Anaerococcus | −0.012 (0.004) | 0.002b | −0.012 (0.005) | 0.026 | −0.012 (0.005) | 0.039 | 0.967 |
| Leptotrichiaceae | Sneathia | −0.015 (0.005) | 0.010 | −0.022 (0.008) | 0.015 | −0.002 (0.004) | 0.601 | 0.090 |
| Coriobacteriaceae | Adlercreutzia | −0.008 (0.003) | 0.010 | −0.004 (0.003) | 0.287 | −0.016 (0.005) | 0.009 | 0.057 |
Data are adjusted for age.
Additionally adjusted for HIV serostatus.
FDR corrected P-value <.1.
P for interaction by HIV serostatus on the association with diabetes status.
Fig. 2.
Relative abundance of four bacterial genera by diabetes status. A bar represents the mean relative abundances and an error bar represents 1 standard error of relative abundances. The topmost P-value is the significance level of differential abundances between the women with diabetes and without diabetes, tested by a linear regression adjusting for age and HIV serostatus, and P-values in the second line are the significance levels of differential abundances between the women with diabetes and without diabetes, tested by linear regressions adjusting for age among HIV+ (left) and among HIV- (right).
8.4. Associations of metabolites with diabetes and bacterial genera
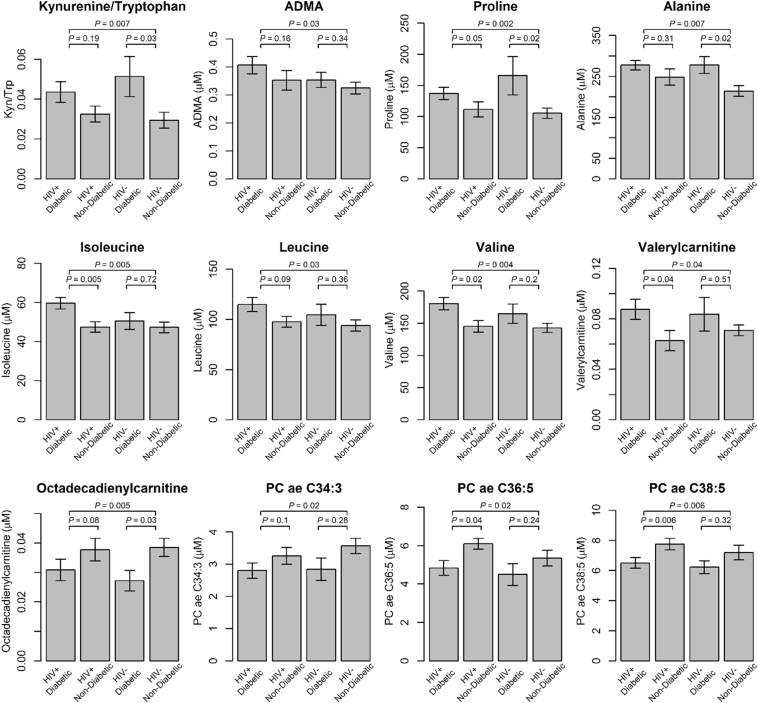
Plasma levels of 12 metabolites showed nominal significant associations with diabetes status (P < .05 [Mann-Whitney U test]; all FDR corrected P-value >.1. Fig. 3). In the women with diabetes compared to those without diabetes, we found higher concentrations of seven amino acids or biogenic amines (kynurenine/tryptophan ratio as a marker for kynurenine pathway activation, ADMA, proline, alanine, isoleucine, leucine, and valine) and elevated valerylcarnitine in acylcarnitines classes, while octadecadienylcarnitine in acylcarnitines and three glycerophospholipids (PC ae C34:3, PC ae C36:5 and PC ae C38:5) showed lower concentrations. To further investigate the engagement between metabolites and gut microbiome in relation to diabetes, we examined the correlations between the four diabetes-associated gut microbial genera and these 12 metabolites (Fig. S3). Anaerococcus was inversely correlated with kynurenine/tryptophan ratio and valine (r = −0.38, −0.3, P = .007, 0.04 [P-value for Spearman correlation]), and Adlercreutzia was inversely correlated with proline and alanine (r = −0.37, −0.32, P = .01, 0.03 [P-value for Spearman correlation]). On the other hand, all four diabetes-associated microbial genera showed some positive correlations with diabetes-associated glycerophospholipids (r = 0.12–0.36 for all three glycerophospholipids and r = 0.29–0.36 with P > .05 [P-value for Spearman correlation]). Across all metabolites, four diabetes-associated microbial genera tended to be positively correlated with glycerophospholipids (Fig. S4), though it should be noted that these glycerophospholipids are highly correlated with each other.
Fig. 3.
Metabolic concentration levels by diabetes status. A bar represents the mean concentrations (ratio for kynurenine/tryptophan ratio) and an error bar represents 1 standard error of concentrations (ratio for kynurenine/tryptophan ratio). The topmost P-value is the significance level of differential concentrations between the women with diabetes and without diabetes [Mann-Whitney U test], and P-values in the second line are the significance levels of differential concentrations between the women with diabetes and without diabetes [Mann-Whitney U test], among HIV+ (left) and among HIV- (right). All P for interaction of diabetes status and HIV serostatus >.18 [linear regression].
Of the pathways involved with the aforementioned metabolites, we then compared the microbial functional contents related to the pathways, predicted by PICRUSt, between women with diabetes and without diabetes (Table S9). Only Enoyl-CoA hydratase in butyrate pathway (K01692) and carbamoyl-phosphate synthase (K01955) in alanine, aspartate and glutamate metabolism showed nominally significant differences (P = .03, 0.006 [t-test]). In the butyrate pathway, a higher proportion of enzymes were decreased in diabetes, but most enzymes did not show statistically significant differences (Fig. S5).
9. Discussion
In this study of women with, or at high risk for HIV infection, we found no significant differences in the composition and the diversity of microbial communities between women with and without diabetes. These findings are generally consistent with most previous studies that examined the relationship between gut microbiota and diabetes in HIV-uninfected populations [2,42,43]. When examining HIV infection in relation to microbial diversity, our study did not find significant associations, while many previous studies found such differences by HIV infection to be statistically significant [12,13,16]. However, it should be noted that most previously reported alterations in microbial diversity were observed in newly HIV-infected, untreated patients, or MSM, compared to HIV-uninfected controls [12,16,19]. In our study, participants had long standing HIV-infection and ART use. Consistent with this, a prior study reported that individuals with chronic untreated HIV infection, but not those under long-term ART, had greater overall microbial diversity compared to HIV negative controls [14]. Another study comparing HIV-infected individuals who were receiving suppressive ART with HIV-uninfected individuals found no significant differences in alpha-diversity although clustering in beta-diversity was observed [44]. It still remains unclear how chronic HIV infection and long-term ART use influence overall gut microbial diversity.
At the genus level, we found that relative abundances of two genera, Finegoldia and Anaerococcus, were significantly lower in women with diabetes compared to those without diabetes, and the results were consistent for HIV-infected and HIV-uninfected individuals. Although there was little direct a priori evidence linking Finegoldia and Anaerococcus with diabetes, Anaerococcus is known to produce butyrate, a product of bacterial fermentation, which is involved in anti-inflammation, insulin sensitivity improvement, and diabetes amelioration [[45], [46], [47]]. The potential involvement of Anaerococcus in anti-inflammation is further supported by our results showing an inverse correlation between Anaerococcus and kynurenine/tryptophan ratio, an indicator of tryptophan catabolism. Tryptophan is catabolized into kynurenine through indoleamine 2,3-dioxygenase (IDO) which is an enzyme induced in many types of immune cells in response to inflammation and immune activation, while transcription of IDO can be down-regulated by sodium butyrate [10,[48], [49], [50], [51]]. In our study, we also found higher kynurenine/tryptophan ratio in diabetes, which is consistent with previous findings [19].
Indeed, two butyrate-producing gut microbes, Faecalibacterium and Roseburia, have been reported to be decreased in people with diabetes [2,3], and Roseburia was lower in metformin-untreated people with diabetes compared to those without diabetes [4]. In our study, we observed that Faecalibacterium was marginally reduced in women with diabetes (P = .07 [linear regression]), but Roseburia did not differ by diabetes status (P = .53 [linear regression]). In addition, PICRUSt analysis revealed a higher proportion of enzymes homologous to those for butyrate metabolism which were decreased in diabetes, although most of these orthologs did not show statistically significant differences, given the relatively small sample size of current analysis. In addition, a previous study reported beneficial effects on glucose metabolism by transplanting butyrate-producing bacteria Clostridium butyricum CGMCC0313.1 to diabetes mouse models [52]. These data suggest a beneficial role of butyrate-producing gut microbes in patients with diabetes. Further investigation is needed to examine the butyrate-producing microbes observed in this study (e.g., Anaerococcus) and their relationships with diabetes, especially in HIV-infected individuals.
In addition, another two genera, Sneathia and Adlercreutzia, showed suggestive inverse associations with diabetes in our study, with potential effect modifications by HIV infection. Interestingly, the HIV+ group without diabetes showed significantly higher relative abundance of Sneathia while the other three HIV/diabetes status groups showed lower and similar relative abundance of this genus. This may help illustrate why the inverse association between Sneathia and diabetes has been observed only in HIV+ groups. The genus Sneathia is one of the bacteria transmitted during vaginal delivery [53] and the disruption of natural transmission of the maternal microbiome to the newborn may be associated with type 1 diabetes, asthma, and obesity [[54], [55], [56], [57]]. However, no previous studies have examined the relationship between HIV infection and Sneathia. Our study suggested an inverse association between the Adlercreutzia genus and diabetes in HIV-uninfected individuals, while a previous study reported that the Adlercreutzia genus was decreased in fecal samples of HIV infected individuals with diabetes, compared to healthy controls and those with HIV infection only [19]. Although the potential effect modification by HIV infection needs further investigation, the suggestive association between Adlercreutzia and diabetes was supported by the inverse correlation between this genus and plasma proline observed in this study, elevated proline levels associated with diabetes in this and other studies [58,59] and potential involvement of this genus in the peroxisome proliferator-activated receptor gamma (PPAR-gamma) pathway [[60], [61], [62]]. However, our results should be interpreted with caution, because of the relatively small sample size. Future studies with larger sample sizes are warranted to clarify the associations between these genera and diabetes in the context of HIV infection.
One unique feature of this current study is the inclusion of women either with HIV-infection or at high risk for HIV-infection, who were of African American (65%) or Hispanic (29%) racial/ethnic backgrounds. Previous studies of the gut microbiota and diabetes focused primarily on Europeans or Chinese without HIV-infection [[2], [3], [4]]. The participants in a previous study on diabetes with HIV infection were Europeans, mostly men (88%) and MSM (75%), and had a relatively shorter time on stable ART (25.4 months (IQR = 13.2–37.7) for HIV infected with diabetes). Prior healthy control groups included convenience samples such as hospital staff, unlike our comparison group comprising a population based sample at high risk for HIV-infection [19]. In addition, dietary choices, access to health care, and overall health status of our study participants might be different from other studies due to demographic differences and exposure to HIV-infection [63]. These factors may influence microbiota compositions in relation to diabetes. HIV infection, including effective viral suppression by ART, might contribute to microbial dysbiosis, affecting the disruption of immune response and resulting in increased morbidity and mortality [6,9,13,64].
Our study has several limitations. The sample size is relatively small and multiple tests were performed in this study, and only two significant genera passed FDR correction. In the metabolomics analysis, although the associations between metabolites and diabetes did not pass FDR correction, many significant metabolites observed in our study (e.g. kynurenine/tryptophan ratio [19], branched-chain amino acids [65], proline [58,59]) have been previously reported to be associated with diabetes in HIV or non HIV populations. Thus, these results are unlikely to be false positive findings. In addition, our study did not collect information on dietary intake which has been closely related to the microbial communities [[66], [67], [68]]. Our plasma metabolomics does not cover the full spectrum of metabolites and especially short chain fatty acids were not measured (e.g., butyric acid). In addition, due to high sodium sulfate content in the samples collected with RNAlater, our fecal samples were not useful for metabolomics assay [69].
In summary, this study suggests decreased relative abundances of four gut microbial genera and related metabolite changes associated with diabetes in women with, or at high risk for HIV-infection. In particular, a known butyrate-producing bacterial genus had lower relative abundance, correlating with higher tryptophan catabolism, in diabetes. These findings suggest a potential interrelationship between gut microbiota, diabetes and HIV infection through inflammation and immune activation. Further studies with larger sample sizes and metagenomics data are needed to investigate specific microbes and their functional role in diabetes along with metabolic profiles in the context of HIV-infection.
Acknowledgements and funding
This study was supported by the National Heart, Lung, and Blood Institute (NHLBI) K01HL129892 and R01HL140976, and Feldstein Medical Foundation Research Grant to Q.Q. R.C·K was supported by NHLBI 5R01HL126543, R01 HL132794 and the National Institute on Mental Health (NIMH) 5R01MD011389-03, and J.R.K. was supported by NHLBI R01 HL132794 and K24 HL135413. Other funding sources include NHLBI R01HL083760, and R01HL095140, the National Institute of Allergy and Infectious Diseases (NIAID) U01 AI035004, and the Einstein Cancer Research Center (P30CA013330), the Einstein Liver Research Center (P30DK041296), the Einstein-Rockefeller-CUNY Center for AIDS Research funded by the NIAID (P30AI124414), and the Stable Isotope and Metabolomics Core Facility of the Einstein-Mount Sinai Diabetes Research Center (ES-DRC) of the Albert Einstein College of Medicine funded by National Cancer Institute (P60DK020541).
Data in this manuscript were collected by the Women's Interagency HIV Study (WIHS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). WIHS (Principal Investigators): UAB-MS WIHS (Mirjam-Colette Kempf and Deborah Konkle-Parker), U01-AI-103401; Atlanta WIHS (Ighovwerha Ofotokun and Gina Wingood), U01-AI-103408; Bronx WIHS (Kathryn Anastos and Anjali Sharma), U01-AI-035004; Brooklyn WIHS (Howard Minkoff and Deborah Gustafson), U01-AI-031834; Chicago WIHS (Mardge Cohen and Audrey French), U01-AI-034993; Metropolitan Washington WIHS (Seble Kassaye), U01-AI-034994; Miami WIHS (Margaret Fischl and Lisa Metsch), U01-AI-103397; UNC WIHS (Adaora Adimora), U01-AI-103390; Connie Wofsy Women's HIV Study, Northern California (Ruth Greenblatt, Bradley Aouizerat, and Phyllis Tien), U01-AI-034989; WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth Golub), U01-AI-042590; Southern California WIHS (Joel Milam), U01-HD-032632 (WIHS I – WIHS IV). The WIHS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute on Mental Health (NIMH). Targeted supplemental funding for specific projects is also provided by the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute on Alcohol Abuse and Alcoholism (NIAAA), the National Institute on Deafness and other Communication Disorders (NIDCD), and the NIH Office of Research on Women's Health. WIHS data collection is also supported by UL1-TR000004 (UCSF CTSA), UL1-TR000454 (Atlanta CTSA), and P30-AI-050410 (UNC CFAR).
Declaration of interest
Dr. Kizer reports stock ownership in Amgen, Johnson & Johnson, Gilead Sciences, and Pfizer. The remaining authors have no conflicts of interest to disclose.
Author contributions
Q.Q., R.C.K. and R.D.B. designed the study. J.-Y.M and Q.Q. performed literature search, analyzed and interpreted data, and drafted the manuscript. C.P.Z., Y.Q., J.R.K., I.J.K. K.A., R.C.K., R.D.B and Q.Q. contributed to data collection. C.P.Z., Z.W., M.U. and T.W. contributed to data analysis and interpretation. A.L.L contributed to literature search and data interpretation. All co-authors edited and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.10.037.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hernandez-Romieu A.C., Garg S., Rosenberg E.S., Thompson-Paul A.M., Skarbinski J. Is diabetes prevalence higher among HIV-infected individuals compared with the general population? Evidence from MMP and NHANES 2009-2010. Bmj Open Diab Res Ca. 2017;5:1. doi: 10.1136/bmjdrc-2016-000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin J.J., Li Y.R., Cai Z.M., Li S.H., Zhu J.F., Zhang F. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 3.Karlsson F.H., Tremaroli V., Nookaew I., Bergstrom G., Behre C.J., Fagerberg B. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 4.Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vrieze A., Van Nood E., Holleman F., Salojarvi J., Kootte R.S., Bartelsman J.F.W.M. Transfer of Intestinal Microbiota from Lean Donors increases Insulin Sensitivity in individuals with Metabolic Syndrome. Gastroenterology. 2012;143(4):913. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 6.Klatt N.R., Funderburg N.T., Brenchley J.M. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vos W.M., Genomics Nieuwdorp M. A gut prediction. Nature. 2013;498(7452):48–49. doi: 10.1038/nature12251. [DOI] [PubMed] [Google Scholar]
- 8.Gao Z., Yin J., Zhang J., Ward R.E., Martin R.J., Lefevre M. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams B., Landay A., Presti R.M. Microbiome alterations in HIV infection a review. Cell Microbiol. 2016;18(5):645–651. doi: 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- 10.Vujkovic-Cvijin I., Dunham R.M., Iwai S., Maher M.C., Albright R.G., Broadhurst M.J. Dysbiosis of the Gut Microbiota is Associated with HIV Disease Progression and Tryptophan Catabolism. Sci Transl Med. 2013;5(193) doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi Q., Hua S., Clish C.B., Scott J.M., Hanna D.B., Wang T. Plasma tryptophan-kynurenine metabolites are altered in HIV infection and associated with progression of carotid artery atherosclerosis. Clin Infect Dis. 2018;67(2):235–242. doi: 10.1093/cid/ciy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguera-Julian M., Rocafort M., Guillen Y., Rivera J., Casadella M., Nowak P. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mutlu E.A., Keshavarzian A., Losurdo J., Swanson G., Siewe B., Forsyth C. A Compositional look at the Human Gastrointestinal Microbiome and Immune Activation Parameters in HIV Infected Subjects. PLoS Pathog. 2014;10(2) doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozupone C.A., Li M., Campbell T.B., Flores S.C., Linderman D., Gebert M.J. Alterations in the Gut Microbiota Associated with HIV-1 Infection. Cell Host Microbe. 2013;14(3):329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dillon S.M., Lee E.J., Kotter C.V., Austin G.L., Dong Z., Hecht D.K. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7(4):983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McHardy I.H., Li X.X., Tong M., Ruegger P., Jacobs J., Borneman J. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1 doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowak P., Troseid M., Avershina E., Barqasho B., Neogi U., Holm K. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29(18):2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 18.Vesterbacka J., Rivera J., Noyan K., Parera M., Neogi U., Calle M. Vol. 7. 2017. Richer gut microbiota with distinct metabolic profile in HIV infected Elite Controllers. Sci Rep-Uk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoel H., Hove-Skovsgaard M., Hov J.R., Gaardbo J.C., Holm K., Kummen M. Impact of HIV and Type 2 diabetes on Gut Microbiota Diversity. Tryptophan Catabolism and Endothelial Dysfunction Sci Rep-Uk. 2018;8 doi: 10.1038/s41598-018-25168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacon M.C., von Wyl V., Alden C., Sharp G., Robison E., Hessol N. The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol. 2005;12(9):1013–1019. doi: 10.1128/CDLI.12.9.1013-1019.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song S.J., Amir A., Metcalf J.L., Amato K.R., Xu Z.Z., Humphrey G. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems. 2016;1(3) doi: 10.1128/mSystems.00021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Qian P.Y. Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. Plos One. 2009;4(10) doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27(6):863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hercus C. 2009. Novocraft short read alignment package. Website http://www novocraft com. [Google Scholar]
- 26.Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., Neufeld J.D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13(1):31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 29.Desantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langille M.G.I., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krumsiek J., Suhre K., Evans A.M., Mitchell M.W., Mohney R.P., Milburn M.V. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 2012;8(10) doi: 10.1371/journal.pgen.1003005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romisch-Margl W., Prehn C., Bogumil R., Rohring C., Suhre K., Adamski J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2012;8(1):133–142. [Google Scholar]
- 35.Hanna D.B., Post W.S., Deal J.A., Hodis H.N., Jacobson L.P., Mack W.J. HIV Infection is Associated with Progression of Subclinical Carotid Atherosclerosis. Clinical Infectious Diseases. 2015;61(4):640–650. doi: 10.1093/cid/civ325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microb. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
- 39.Core Team R. 2016. R: A Language and Environment for Statistical Computing. 3.3.2 ed. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 40.McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oksanen J., Kindt R., Legendre P., O'Hara B., Stevens M.H.H., Oksanen M.J. Vol. 10. 2007. The vegan package. Community ecology package; pp. 631–637. [Google Scholar]
- 42.Larsen N., Vogensen F.K., van den Berg F.W.J., Nielsen D.S., Andreasen A.S., Pedersen B.K. Gut Microbiota in Human adults with Type 2 Diabetes Differs from Non-Diabetic adults. Plos One. 2010;5(2) doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Shen D., Fang Z., Jie Z., Qiu X., Zhang C. Human gut microbiota changes reveal the progression of glucose intolerance. Plos One. 2013;8(8) doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dinh D.M., Volpe G.E., Duffalo C., Bhalchandra S., Tai A.K., Kane A.V. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ezaki T., Kawamura Y., Li N., Li Z.Y., Zhao L., Shu S. Proposal of the genera Anaerococcus gen. Nov., Peptoniphilus gen. Nov. and Gallicola gen. Nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51(Pt 4):1521–1528. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
- 46.Gao Z.G., Yin J., Zhang J., Ward R.E., Martin R.J., Lefevre M. Butyrate Improves Insulin Sensitivity and increases Energy Expenditure in mice. Diabetes. 2009;58(7):1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Correa-Oliveira R., Fachi J.L., Vieira A., Sato F.T., Vinolo M.A. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5(4) doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellor A.L., Munn D.H. Tryptophan catabolism and regulation of adaptive immunity. J Immunol. 2003;170(12):5809–5813. doi: 10.4049/jimmunol.170.12.5809. [DOI] [PubMed] [Google Scholar]
- 49.Platten M., Wick W., Van den Eynde B.J. Tryptophan Catabolism in Cancer: beyond IDO and Tryptophan Depletion. Cancer Res. 2012;72(21):5435–5440. doi: 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 50.Laurans L., Venteclef N., Haddad Y., Chajadine M., Alzaid F., Metghalchi S. Genetic deficiency of indoleamine 2,3-dioxygenase promotes gut microbiota-mediated metabolic health. Nat Med. 2018;24:1113–1120. doi: 10.1038/s41591-018-0060-4. [DOI] [PubMed] [Google Scholar]
- 51.Jiang G.M., He Y.W., Fang R., Zhang G., Zeng J., Yi Y.M. Sodium butyrate down-regulation of indoleamine 2, 3-dioxygenase at the transcriptional and post-transcriptional levels. Int J Biochem Cell B. 2010;42(11):1840–1846. doi: 10.1016/j.biocel.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 52.Jia L., Li D., Feng N., Shamoon M., Sun Z., Ding L. Anti-diabetic Effects of Clostridium butyricum CGMCC0313.1 through Promoting the growth of Gut Butyrate-producing Bacteria in Type 2 Diabetic mice. Sci Rep. 2017;7(1):7046. doi: 10.1038/s41598-017-07335-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller N.T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M.G. The infant microbiome development: mom matters. Trends Mol Med. 2015;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Algert C.S., McElduff A., Morris J.M., Roberts C.L. Perinatal risk factors for early onset of Type 1 diabetes in a 2000-2005 birth cohort. Diabetic Med. 2009;26(12):1193–1197. doi: 10.1111/j.1464-5491.2009.02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roduit C., Scholtens S., de Jongste J.C., Wijga A.H., Gerritsen J., Postma D.S. Asthma at 8 years of age in children born by caesarean section. Thorax. 2009;64(2):107–113. doi: 10.1136/thx.2008.100875. [DOI] [PubMed] [Google Scholar]
- 56.Huh S.Y., Rifas-Shiman S.L., Zera C.A., Edwards J.W.R., Oken E., Weiss S.T. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child. 2012;97(7):610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knip M., Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura H., Jinzu H., Nagao K., Noguchi Y., Shimba N., Miyano H. Plasma amino acid profiles are associated with insulin, C-peptide and adiponectin levels in type 2 diabetic patients. Nutr Diabetes. 2014;4:e133. doi: 10.1038/nutd.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y., Qiu L., Xiao Q., Wang Y., Meng X., Xu R. Obesity and diabetes related plasma amino acid alterations. Clin Biochem. 2013;46(15):1447–1452. doi: 10.1016/j.clinbiochem.2013.05.045. [DOI] [PubMed] [Google Scholar]
- 60.Maruo T., Sakamoto M., Ito C., Toda T., Benno Y. Adlercreutzia equolifaciens gen. Nov., sp nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Micr. 2008;58:1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- 61.Cho K.W., Lee O.H., Banz W.J., Moustaid-Moussa N., Shay N.F., Kim Y.C. Daidzein and the daidzein metabolite, equol, enhance adipocyte differentiation and PPAR gamma transcriptional activity. J Nutr Biochem. 2010;21(9):841–847. doi: 10.1016/j.jnutbio.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 62.Napolitano A., Miller S., Nicholls A.W., Baker D., Van Horn S., Thomas E. Novel Gut-based Pharmacology of Metformin in patients with Type 2 Diabetes Mellitus. Plos One. 2014;9(7) doi: 10.1371/journal.pone.0100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hunt P.W., Sinclair E., Rodriguez B., Shive C., Clagett B., Funderburg N. Gut Epithelial Barrier Dysfunction and Innate Immune Activation Predict Mortality in Treated HIV Infection. J Infect Dis. 2014;210(8):1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang T.J., Larson M.G., Vasan R.S., Cheng S., Rhee E.P., McCabe E. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Modi S.R., Collins J.J., Relman D.A. Antibiotics and the gut microbiota. J Clin Invest. 2014;124(10):4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jernberg C., Lofmark S., Edlund C., Jansson J.K. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol-Sgm. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 68.Singh R.K., Chang H.W., Yan D., Lee K.M., Ucmak D., Wong K. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z., Zolnik C.P., Qiu Y., Usyk M., Wang T., Strickler H.D. Comparison of Fecal Collection Methods for Microbiome and Metabolomics Studies. Front Cell Infect Microbiol. 2018;8:301. doi: 10.3389/fcimb.2018.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material