Abstract
NR4As are AML tumor suppressors that are frequently silenced in human AML. Despite their potential as novel targets for therapeutic intervention, mechanisms of NR4A silencing and strategies for their reactivation remain poorly defined. Here we show that NR4A silencing in AML occurs through blockade of transcriptional elongation rather than epigenetic promoter silencing. By intersection of NR4A-regulated gene signatures captured upon acute, exogenous expression of NR4As in human AML cells with in silico chemical genomics screening, we identify several FDA-approved drugs including dihydroergotamine (DHE) that reactivate NR4A expression and regulate NR4A-dependent gene signatures. We show that DHE induces NR4A expression via recruitment of the super elongation complex to enable elongation of NR4A promoter paused RNA polymerase II. Finally, DHE exhibits AML selective NR4A-dependent anti-leukemic activity in cytogenetically distinct human AML cells in vitro and delays AML progression in mice revealing its potential as a novel therapeutic agent in AML.
Introduction
AML (acute myeloid leukemia) is a heterogeneous disease associated with corruption of normal transcriptional and epigenetic control of myeloid cell differentiation and the emergence of transformed leukemic initiating cells (LICs) with aberrant self-renewal properties capable of sustaining leukemic expansion 1,2. Despite substantial improvements in genetic and molecular classification of AML, standard induction chemotherapy using anthracyclines and cytarabine remains a treatment mainstay but mainly targets bulk leukemic blasts rather than LICs 3. Although new therapies are urgently needed, genetic, phenotypic and functional heterogeneity among patient LICs presents a major challenge to their development 4,3, 5–7. Efficient therapeutic targeting of LICs to eradicate AML therefore requires a detailed understanding of the genetic, epigenetic and molecular pathway dependencies that distinguish AML LICs from normal hematopoietic stem and progenitor cells (HSPCs).
The NR4A subfamily of nuclear receptors consists of three structurally related transcription factors (NR4A1-3) that regulate cell context dependent cell fate decisions in response to extracellular signals including inflammatory, genotoxic, apoptotic and mitogenic stimuli 8, 9. NR4As are also diverse regulators of hematopoiesis including hematopoietic stem cell maintenance 10–12, T-lymphocyte development and function 13–17,18, and monocyte and macrophage maturation and inflammation 19–21.
NR4As also play key roles as tumor suppressors of both myeloid and lymphoid malignancies 11,22–24. In particular, NR4A1 and NR4A3 are functionally redundant tumor suppressors of AML and pre-AML malignancies. Codepletion of NR4A1 and NR4A3 in mice is sufficient to drive AML development while reduction of NR4A1/NR4A3 expression in NR4A1/NR4A3 hypoallelic mice leads to mixed myelodysplastic/myeloproliferative (MDS/MPN) disease 11, 22, 25. In human patients, NR4A1 and NR4A3 expression is reduced in MDS and silenced in AML bulk blasts and LIC enriched populations irrespective of patient cytogenetics 11, 26, 27. Further, forced expression of NR4A1 or NR4A3 in human AML cells inhibits their viability and reprograms a subset of gene signatures that distinguish primary human LICs from normal HSCs including suppression of a core oncogenic MYC driven gene signature25.
Given the widespread silencing of NR4As in AML patients and the sufficiency of their inactivation in causing AML, we hypothesized that NR4A silencing may be an obligate step in AML maintenance and that strategies directed toward NR4A reactivation may be of therapeutic benefit in treatment of AML. Here we address mechanisms of NR4A silencing in AML and we use an integrated systems approach combining NR4A target based genomics data with in silico chemical genomics screening to objectively identify small molecule activators (SMAs) of silenced NR4As. We show how this approach successfully identified the ergot alkaloid, dihydroergotamine (DHE), as a drug inducer of NR4As and NR4A-dependent gene signatures with anti-leukemic efficacy.
Results
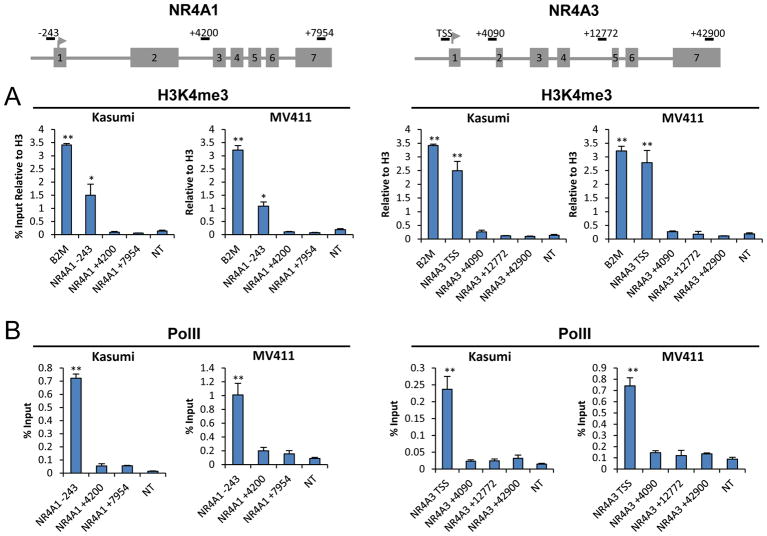
To disclose mechanisms of silencing of NR4As in AML cells, we examined the epigenetic status of NR4A1 and NR4A3 promoters. We found that both promoters are devoid of DNA methylation in cytogenetically distinct primary human AML HSPCs and AML cell lines similar to normal human HSPCs (Supplemental Figure S1). Next, we examined the trimethylation status of histone H3 lysine 4 (H3K4me3), a mark associated with active and poised promoters 28, 29. Using two cytogenetically distinct AML cell lines, Kasumi-1 and MV4-11, representing commonly occurring AML oncogenes (AML-ETO and mixed lineage leukemia (MLL) rearranged human AMLs), we found that both promoters contained high levels of H3K4me3 that were comparable to levels at the promoter of the highly expressed gene β2 microglobulin (B2M) (Figure 1A). Furthermore, query of publically available databases revealed that promoter enrichment of H3K4me3 at NR4As also extends to normal HSCs and primary HSPCs from AML patients of distinct cytogenetics (Supplemental Figure S2). Consistent with these findings, analysis of the status of RNA Pol II occupancy across NR4A1 and NR4A3 genomic loci revealed that both promoters contain relatively high levels of pre-associated Pol II that is largely dismissed from downstream intragenic regions (Figure 1B). These results indicate that NR4A promoters reside in an open chromatin context and actively recruit Pol II.
Figure 1. NR4A Promoters Reside in Open Chromatin.
(A) H3K4Me3 enrichment at NR4A1 and NR4A3 promoters compared to B2M. (B) Pol II occupancy at NR4A1 and NR4A3 loci. Gene loci numbers are relative to TSS. **p<0.01, *p<0.05 compared to non-transcribed control (NT).
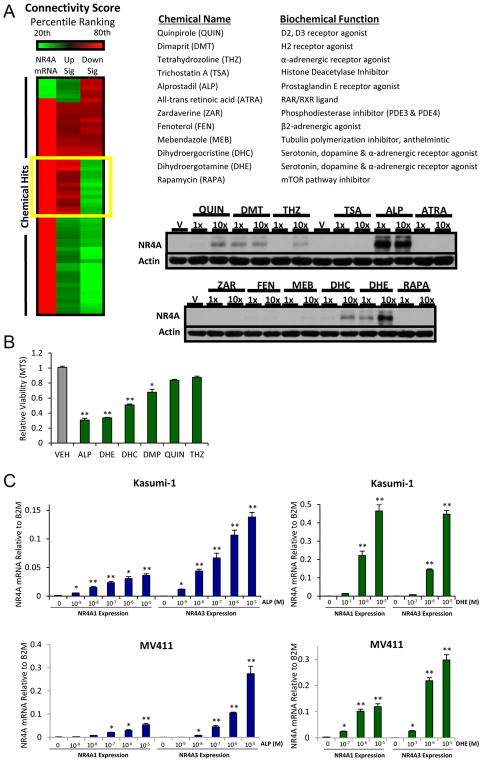
To objectively identify chemicals that can reactivate expression of silenced NR4As and restore their transcriptional activity, we used NR4A-regulated gene signatures in Kasumi-1 AML cells to conduct an in silico screen of the Connectivity Map (CMap) database 30. This generated 1229 connectivity scores in HL-60 AML cells. We examined 3 metrics of NR4A connectivity to identify chemicals for secondary in-vitro screening: NR4A-dependent upregulated genes connectivity, NR4A-dependent downregulated genes connectivity, and connectivity to upregulation of NR4A1 and NR4A3 mRNAs. Given the limitation of CMap HL60 data (e.g., no replicates), we used a simple non-statistical percentile ranking cutoff (80%) which yielded a list of 12 chemicals to pursue in empirical secondary screening (Figure 2A and Supplemental Figure S3). Visualization of NR4A connectivity with the 12 chemical signatures by both heatmap depiction (Supplemental Figure S3A) and CMap rank-ordered plotting (Supplemental Figure S3B) revealed significant connectivity with few conversely regulated genes.
Figure 2. Integration of an NR4A Activation Signature with CMAP Identifies Chemical Activators of NR4As with Anti-leukemic Activity.
(A) Heatmap showing top scoring chemicals with percentile ranking above 80% (highlighted in yellow) according to three independent connectivity scores: (1) NR4A mRNA induction, (2) NR4A UPregulated genes and (3) NR4A DOWNregulated genes connectivity. Chemical identities of top 12 chemicals. Western blot for NR4A protein in Kasumi-1 cells at 1x and 10x levels used in CMAP (10uM and 100uM, respectively). (B) Growth inhibitory activity of top chemicals (10uM) in Kasumi-1 cells (48h). (C) Alprostadil (ALP) and dihydroergotamine (DHE) are dose dependent activators of NR4A1 and NR4A3 mRNAs in Kasumi-1 and MV4-11 cells. **p<0.01, *p<0.05 compared to vehicle controls.
Next, we examined the ability of the 12 chemicals to induce NR4A protein expression and suppress cell viability in Kasumi-1 AML cells. We found that 6 of the 12 top chemicals induced measureable NR4A protein (Figure 2A) and also exhibited varying degrees of cell growth inhibition (Figure 2B). Since Alprostadil (ALP), a prostaglandin, and the ergot alkaloid, dihydroergotamine (DHE) performed best in these assays, we confirmed that both drugs are potent dose-dependent activators of NR4A1 and NR4A3 gene transcripts in Kasumi-1 and MV4-11 cells (Figure 2C). Thus, our integrative screen successfully identifies chemicals that induce NR4A expression and compromise the viability of human AML cells.
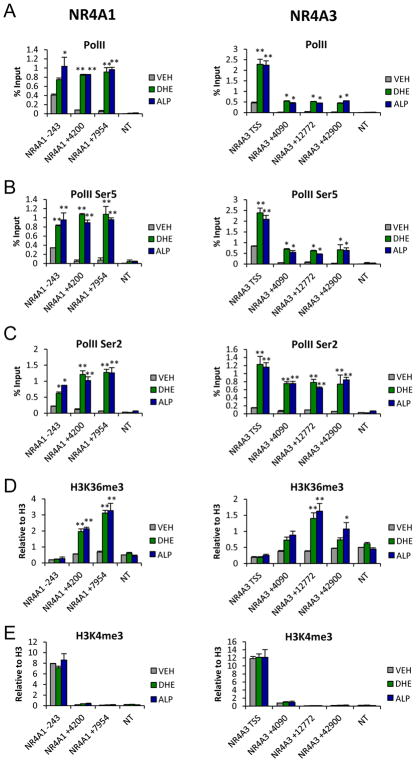
To understand how DHE and ALP facilitate NR4A activation, we monitored Pol II occupancy of the NR4A1 and NR4A3 loci in response to drug treatment using ChIP analysis. We found that both drugs elicited a moderate increase in bound Pol II at the NR4A1/3 promoter but stimulated significant increases in Pol II binding to intragenic regions of both genes (Figure 3A). This indicated that both drugs function as positive regulators of transcriptional elongation of promoter paused Pol II across the NR4A1 and NR4A3 genes in AML cells.
Figure 3. Chemical Inducers of NR4As Activate Transcriptional Elongation of the NR4A Loci.
MV4-11 cells were treated for 1hr with 10uM DHE or 1uM ALP followed by ChIP-qPCR analysis of (A) Pol II, (B) Pol II S5P, (C) Pol II S2P, (D) H3K36me3 and (E) H3K4me3 at NR4A1 and NR4A3 loci. Gene loci numbers are relative to TSS.**p<0.01, *p<0.05 compared to vehicle controls. NT = non-transcribed control.
Promoter clearance and elongation competence of Pol II require phosphorylation of Ser5 (S5-P) and Ser2 (S2-P) at its C-terminal domain (CTD). To examine the mechanisms by which DHE and ALP promote elongation competence of Pol II, we first investigated the phosphorylation status of Pol II CTD at the NR4A promoters and intragenic regions. We found low levels of S5-P and S2-P under basal conditions that were strongly induced upon treatment with ALP or DHE (Figure 3B,C). Pol II activation and processivity were also associated with acquisition of intragenic histone H3K36 trimethylation (H3K36Me3), a histone mark of transcription elongation, while high basal levels of promoter associated H3K4me3 activation mark were unchanged (Figure 3D,E).
Phosphorylation of Pol II is accomplished by several cyclin-dependent kinases (CDKs) that are components of a Pol II-associated preinitiation complex (PIC) composed of general transcription factors (GTFs) and Mediator complex, bridging transcription factors to Pol II and the super elongation complex (SEC) which is required for productive transcription elongation. CTD phosphorylation of Pol II at Ser2 is accomplished by the CDK9 catalytic subunit of positive transcription elongation factor b (P-TEFb), a component of the SEC complex critical for Pol II transcriptional elongation 31. CTD phosphorylation of Ser5 is deposited by CDK7, a kinase subunit of TFIIH and also by CDK8, a kinase component of the Mediator complex that also recruits P-TEFb to the promoter upon stimulus induction of IEG transcription elongation 32,33,34, 35. Consistent with increases in S5-P and S2-P, we also found significant increases in NR4A1 promoter bound transcriptional elongation factors including CDK9 and CDK8 (Supplemental Figure 4A,B) Further, both ALP and DHE dependent induction of NR4As was inhibited by the CDK9 antagonist, NVP-2 while constitutive expression of B2M was unaffected (Supplemental Fig 4C,D). Together these data indicate that DHE and ALP-dependent induction of NR4A expression occurs via regulation of transcription elongation and involves recruitment of CDK8 and P-TEFb leading to activation of elongation competent Pol II.
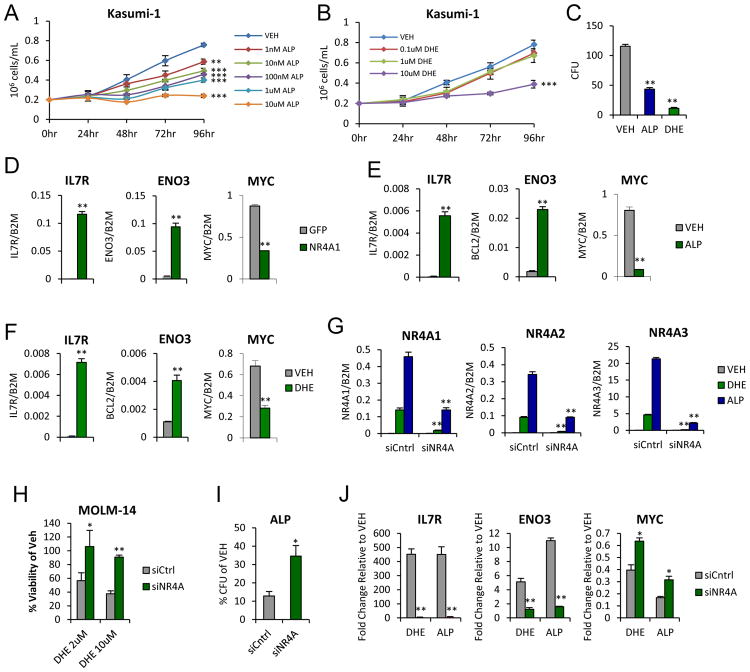
We previously demonstrated that rescue of NR4A expression in human AML cells (including Kasumi-1) inhibits their proliferation and leads to acute transcriptional regulation of a gene expression signature (GES) that includes suppression of a core oncogenic Myc signature that is common to AML LICs 25. We therefore asked whether chemical inducers of NR4As could mimic the cellular and molecular responses of AML cells observed after NR4A rescue in Kasumi-1 cells. We found that exposure to either ALP or DHE was sufficient to suppress cell viability in a dose-dependent manner (Figure 4A,B). Both drugs also strongly reduced AML colony-forming potential demonstrating potent long-term suppression of AML cell proliferation (Figure 4C).
Figure 4. Chemical Inducers of NR4As inhibit AML cell proliferation and restore NR4A-dependent transcriptional activity.
(A–B) Cell growth of Kasumi-1 cells treated with indicated doses of (A) ALP or (B) DHE. (C) AML colony forming units (CFUs) at 12 days after treatment with vehicle, 1uM ALP or 10uM DHE. (D) NR4A-dependent target gene regulation at 6 hours after NR4A1 rescue. (E–F) or treated with (E) 1uM ALP or (F) 10uM DHE. (G) siRNA-mediated suppression of drug-induced NR4A expression. (H) siRNA-mediated knockdown of NR4As restores MOLM-14 cell viability in liquid culture when treated for 48 hours with either 2 or 10uM DHE. (I) siRNA-mediated knockdown in Kasumi-1 partially overcomes ALP-dependent inhibition of AML colony forming ability. (J) Drug-dependent regulation of NR4A target genes in Kasumi-1 is compromised by siRNA-mediated NR4A knockdown. **p<0.01, *p<0.05 compared to vehicle (C–F) or siCntrl (G–J).
We next asked whether both drugs can regulate the expression of NR4A target genes. Using a subset of NR4A target genes regulated upon forced expression of NR4As in Kasumi-1 cells 36 (Figure 4D), we found that acute exposure to DHE or ALP (6h) mimicked NR4As in their ability to regulate the expression of these NR4A target genes including repression of c-MYC (Figure 4E,F). Finally, to determine whether the responses to DHE and ALP were dependent on reactivation of NR4As, we examined the consequences of siRNA-mediated depletion of all 3 NR4A family members (siNR4A1-3). Using a pool of scrambled siRNA controls or siNR4A1-3, we achieved selective siNR4A1-3 mediated knockdown of NR4A induction in response to DHE and ALP (Figure 4G, Supplemental Figure S5). Both DHE and ALP dependent AML cell growth inhibition were significantly overcome by NR4A knockdown (Figure 4H,I). Further, siNR4A1-3 mediated knockdown of NR4As strongly abrogated drug dependent regulation of NR4A-dependent target genes (Figure 4J) but had no off target effects on the basal level of expression of these genes in the absence of drug dependent induction of NR4As (Supplemental Figure S6).
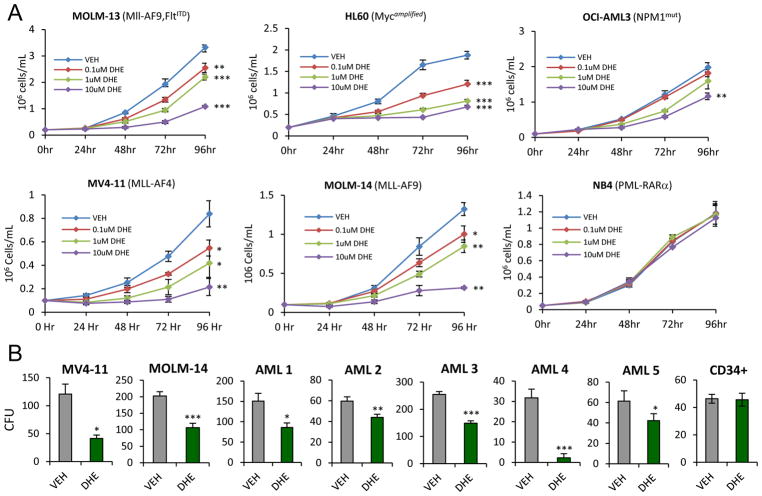
To address the AML subtype selectivity of NR4A activating drugs, we examined the growth suppressive effects of DHE on a range of established human AML cell lines representing diverse cytogenetic backgrounds. We focused on DHE since it is an FDA approved drug for treatment of migraine and has the potential for rapid repurposing for AML intervention. Dose responsive suppression of AML cell growth by DHE was observed to varying degrees in the majority of cell lines tested including HL60 (Myc amplified), OCI-AML3 (NPM1 mutated) and those containing mixed lineage leukemia (MLL) fusions, MLL-AF9 (MOLM-13, MOLM14) and MLL-AF4 (MV4-11) that represent more aggressive chemotherapy-resistant AML disease (Figure 5A). In contrast, DHE failed to suppress the growth of NB4 (PML-RARa) AML cells indicating that DHE-dependent cell growth suppression was cell selective (Figure 5A). This lack of growth suppression of NB4 cells by DHE correlated with a lack of DHE induction of NR4As and suppression of the NR4A target c-Myc in contrast to DHE responsive cell lines (Supplemental Fig S7). Using MV4-11 and MOML14 cells, we found that DHE also strongly suppressed their long-term colony forming potential (CFU) (Figure 5B). The growth suppressive effects of DHE also extended to patient derived primary AML samples of distinct cytogenetics (Supplemental Table S1) but DHE had no effects on the growth of normal CD34+ cells (Figure 5B and Supplemental Figure S8 and 9). Thus, the growth inhibitory effects of DHE are selective to AML cells.
Figure 5. DHE suppresses viability of cytogenetically distinct AML cell lines and primary AML samples but not normal CD34+ cells.
(A) HL-60, OCI-AML3, MV4-11, MOLM13, MOLM14 and NB4 cells counts at indicated doses of DHE. (B) CFUs 7 days after treatment of MV4-11 or MOLM14 cells and 10–14 days after treatment of primary AMLs or normal CD34+ cells with vehicle or 10uM DHE. *p<0.05, **p<0.01, ***p<0.001 compared to vehicle.
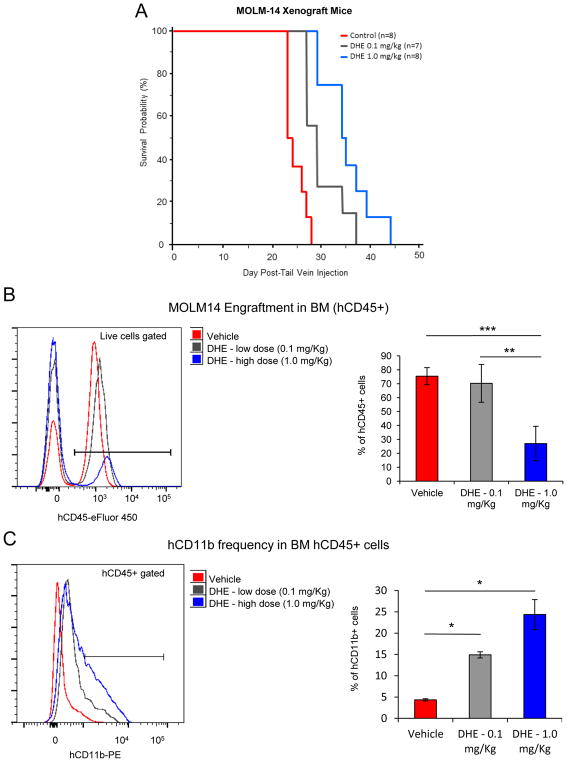
To address the anti-leukemic efficacy of DHE in vivo we used a xenograft mouse model of disseminated MLL-rearranged human AML. 2 ×106 MOLM14 cells were injected i.v. into the tail vein of NOD/SCID/gamma (NSG) immunocompromised mice, and mice were injected twice daily i.p. with vehicle (n=7) or DHE at two doses, 0.1mg/kg (n=7) or 1.0mg/kg (n=8) and monitored for overall NSG mouse survival. Dissemination of AML was confirmed at necropsy by bone marrow and peripheral blood smears (Supplemental Figure S10A–B). Treatment with DHE led to a dose-dependent increase in overall mouse survival that was significant at both doses of DHE (Figure 6A). Further, analysis of bone marrow (BM) engraftment of MOML14 derived human CD45+ blasts and the myeloid differentiation marker, human CD11b at necropsy revealed that DHE treatment leads to a decrease in BM tumor burden as well as increased myeloid differentiation of human AML blasts (Figure 6B and C).
Figure 6. DHE delays AML progression in mice.
(A) Kaplan-Meier survival plots of mice treated with vehicle (n=8), 0.1mg/kg DHE (n=7) or 1.0mg/kg DHE (n=8) twice daily (p=0.002, DHE 0.1mg/kg, P<0.001, DHE1.0mg/kg). (B) BM frequency of CD45+ cells at necropsy. (C) CD11b levels in CD45+ gated cells. Results expressed as mean ± SD, n=4 (Vehicle), n=3 (DHE 0.1 mg/Kg), n=4 (DHE 1.0 mg/Kg), * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001.
Finally, to address the in vivo effects of DHE on normal hematopoietic cell development, we examined the effects of daily exposure of normal mice to the high dose (1.0mg/kg) of DHE over a 30-day period. Chronic exposure of mice to DHE had no significant effects on frequencies of bone marrow HSPCs (Supplemental Figure S10A–D) or mature blood cells (Supplemental Table S2) relative to vehicle treated mice. Thus, the effects of DHE in vivo are selective to AML cells.
Discussion
We have revealed a novel approach for restoring expression of silenced NR4As in AML cells by integrating NR4A regulated GES with in-silico chemical genomics screening. This approach identified chemicals that restore NR4A expression, regulate NR4A-dependent gene signatures and exhibit anti-leukemic activity against cytogenetically distinct AML subtypes. In the absence of drug stimulation, silenced NR4A1 and NR4A3 promoters reside in open chromatin with recruited RNA Pol II that is stalled in an elongation incompetent state. Accordingly, chemical inducers of NR4As act not through promoter activation, but rather through recruitment of transcription elongation complex components to activate elongation competence of promoter stalled RNA Pol II. Together, our results disclose mechanisms of NR4A silencing and reactivation in AML, identify NR4A reactivating drugs with potential for drug repositioning for treatment of AML patients, and provide a general strategy for discovering chemical modulators of transcription factors.
Common mechanisms of tumor suppressor gene (TSG) silencing in cancers involve transcriptional repression through DNA hypermethylation and/or deposition of repressive histone marks primarily within the promoter regions of TSGs 37–39. The reversible nature of this repression has fueled development of drugs capable of inhibiting key factors involved in establishment of repressive chromatin, including DNA methyltransferases and histone deacetylases as a therapeutic strategy for epigenetic reactivation of silenced TSGs 40–42. However, profiling of genomic Pol II occupancy in mammalian cells has revealed that promoter proximal pausing of Pol II is a common regulatory step in transcriptional elongation of a large number of genes, including stimulus-responsive immediate early genes 29, 31, 35, 43. Further, comparative genome-wide analysis of the H3K4me3 epigenome in normal and tumor tissues has revealed that TSG expression in normal cells is often associated with a broad pattern of trimethylation of histone H3K4 across the promoter and TSS of these genes. In cancer cells, however, H3K4me3 at TSGs is frequently retained but more tightly restricted to the proximal promoter region, and this restricted H3K4me3 pattern is associated with TSG repression 44. These findings predict that additional mechanisms of TSG silencing may operate post recruitment of Pol II to active promoters at the elongation stage of transcription. Consistent with these findings, we observed that the promoter regions of silenced NR4As in AML cells are hypomethylated and contain active promoter marks (H3K4me3) with promoter proximally paused Pol II and therefore are unresponsive to epigenetic drugs targeted at reactivation of epigenetically silenced promoters.
Integration of NR4A GES data with chemical genomics screening provides a powerful objective approach to discover new drugs that reactivate NR4A TSGs in AML cells. This approach has several key advantages: First, it identifies chemicals inducers of NR4As that we can utilize as molecular tools to disclose mechanisms of NR4A silencing and reactivation in AML. Second, the NR4A-regulated GES is predictive of therapy, in that it corresponds to compromised AML cell viability. Third, by searching for chemicals that mimic the NR4A-dependent GES in contrast to simply inducing NR4A1 and NR4A3 mRNAs, we select for chemicals that recapitulate the tumor suppressive activities of NR4As in AML cells. Finally, since many of the top chemicals are FDA-approved drugs, the approach facilitates rapid reassignment of old drugs to new indications.
NR4A TSGs comprise subclass of nuclear receptors that are stimulus responsive immediate early genes (IEGs). Recent studies showed that IEG induction is primarily controlled at the level of transcription elongation 35, 43, 45, 46. Similarly, NR4A activating drugs, alprostadil and DHE rapidly induce NR4A expression via recruitment of P-TEFb to NR4A promoters, resulting in activation of elongation competence of Pol II via phosphorylation at Serine 2. Although NR4As respond to a variety of extracellular stimuli in a cell context-dependent manner under normal physiological conditions 8, NR4A induction in AML cells was drug selective. Out of over 1000 chemicals interrogated in CMap database only 6 chemicals, several of which were structurally related ergot alkaloids (supplemental Figure S9), scored positive for NR4A induction and GES connectivity.
In addition to inducing NR4A expression, the top scoring chemicals, DHE and alprostadil, regulated GES that displayed strong connectivity with NR4A regulated GES, including suppression of MYC which directs a core oncogenic signature in cytogenetically diverse human AML cells 47. Further, both drugs suppressed the growth of human AML cells in an NR4A dependent manner, indicating that GES connectivity is a reliable predictor of anti-leukemic response. We focused on DHE as it is FDA-approved for treatment of severe migraine and represents an ergot alkaloid drug class that demonstrated high connectivity with NR4A GES. DHE exhibited dose-dependent growth inhibitory activity against several cytogenetically distinct human AML cells lines and primary AML patient samples, including those representing MLL fusions which are highly dependent on MYC and represent aggressive chemotherapy resistant AMLs 48, 49. Importantly, the growth inhibitory activity of DHE was AML selective and DHE did not affect normal hematopoietic cell development. The lower effective dose of DHE in vivo (0.1mg/kg i.p. twice daily) is >200-fold lower than the lethal dose in mice (i.v. 44mg/kg, http://www.drugs.com/pro/d-h-e-45.html). Using the body surface area (BSA) recommended method of dose translation from mouse to humans 50, this represents a dose of approximately 0.01mg/kg daily in humans and is within the maximum dosage recommended for humans (0.012mg/kg per day). Thus DHE displays anti-leukemic efficacy in vivo at clinically relevant doses. DHE acts in the central nervous system (CNS) through binding to several serotonergic, dopaminergic and adrenergic G protein coupled receptors 51. Members of these receptor families are expressed in hematopoietic progenitors and regulate their mobilization 52–55. While the individual contributions of these receptors to the NR4A activating and anti-leukemic effects of DHE remain to be defined, analysis of the acute effects of DHE exposure on NR4A induction and MYC repression in primary patient samples together with preclinical screening in patient-derived xenografts should provide a powerful approach to stratify potential AML patient responders.
Materials and Methods
Human AML cell lines and Patient Samples
HL-60, Kasumi-1, MV4-11, MOLM-13, MOLM14, OCI-AML3, THP1 and NB4 cells were purchased from ATCC. Primary AML patient samples were collected after informed consent at Texas Children’s Hospital in accordance with the Declaration of Helsinki and institutional review board regulations. Normal cord blood CD34+ stem/progenitor cells were obtained from AllCells.
Connectivity Map (CMAP) Analysis
The NR4A1 minimal signature used to query CMAP was generated from our previously published NR4A1 regulated GES obtained in Kasumi-1 cells and defined as those probes whose q-value = 0.0 when subject to Rank Product Analysis (RPA) between NR4A1 and EGFP arrays 25. This represents the most conservative, and statistically rigorous NR4A signature (Supplemental Table S3). Probes comprising the NR4A minimal GES were mapped to CMAP’s Affymetrix U133A microarray platform 30. Querying CMAP resulted in 1229 connectivity scores with chemical signatures derived from HL60 AML cells. We used 3 independent metrics of connectivity: NR4A Up Gene connectivity, NR4A Down Gene connectivity and NR4A mRNA upregulation. Chemical signatures were classified as NR4A-connected signatures if they scored above the 80th percentile by all three metrics (Supplemental Table 3). DOWN connectivity scores are negative values. Thus, a low percentile ranking (<20th) was used in NR4A Down Gene signature analysis.
Cellular Growth Assays
Cell viability was assessed using CellTiter Cell Proliferation Assay (Promega). For colony-forming assays, 5,000 cells (MV4-11 and MOLM14), 500 cells (CD34+ human cord blood) or 10,000 – 1,000,000 cells (primary AML) were plated in triplicate in MethoCult H4100+10% FBS (MV4-11 and MOLM14) or H4434/H4534 (CD34+ cord blood and primary AMLs) (StemCell Technologies) and scored for CFUs after 7–14 days.
Chromatin Immunoprecipitation
ChIP-qPCR analysis was carried out as previously described 36. Details are in Supplemental Methods.
Xenograft Models of Human AML
MOLM14 cells (2×106) were injected into the tail vein of female (6–8 week) NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (The Jackson Lab, Bar Harbor, ME). Mice were treated twice daily with vehicle (n=8 mice), 0.1mg/kg DHE (n=7 mice) or 1.0mg/kg DHE (n=8 mice), sacrificed when moribund and bone marrow (BM) cellularity was determined by manual counts. Blood smears and bone marrow cytospins were stained with Wright–Giemsa Stain (Sigma, St Louis, MO, USA). Tumor burden in BM was measured by flow cytometry of human CD45+ cells and myeloid differentiation analyzed using human CD11b. Mouse experiments were approved by the BCM Animal Care and Use Committee.
Statistical Analysis
Mean values from experimental triplicates were analyzed for statistical significance using independent Student’s t-test (p-values of <0.05). For mouse survival experiments, statistical significance of mean latency of survival differences from Kaplan Meier plots was calculated using a Log-rank (Mantel-Cox) test using the survival R library.
Additional details are in Supplemental Methods
Supplementary Material
Statement of significance.
A chemical genomics strategy identifies dihydroergotamine as a novel activator of silenced NR4A tumor suppressors with repositioning potential for treatment of acute myeloid leukemia.
Acknowledgments
This work was supported by RO1 CA160747 from National Institutes of Health to OMC and by the Cell Sorting and Flow Cytometry shared resource of the Dan L. Duncan Comprehensive Cancer Center with funding from National Cancer Institute grant (P30CA125123). We acknowledge the joint participation by Adrienne Helis Melvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine. We also thank Dr. Terzah Horton and the Leukemia Research Interest Group at Texas Children’s Hospital for providing primary patient AML samples. Author contributions: SPB, RPD and SGC carried out experiments including Cmap chemical genomics integration, ChIP-experiments, bisulfite sequencing, and cell growth assays. PRF did flow cytometry analysis. LN did transplantation assays. PN, MSR and PRF were responsible for colony forming assays on primary AML samples. SPB and OMC conceived the strategy, designed experiments and wrote the paper.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994 Feb 17;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Jordan CT. The leukemic stem cell. Best Pract Res Clin Haematol. 2007 Mar;20(1):13–18. doi: 10.1016/j.beha.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kadia TM, Ravandi F, Cortes J, Kantarjian H. Toward Individualized Therapy in Acute Myeloid Leukemia: A Contemporary Review. JAMA Oncol. 2015 Sep;1(6):820–828. doi: 10.1001/jamaoncol.2015.0617. [DOI] [PubMed] [Google Scholar]
- 4.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May 30;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011 Jan 18;19(1):138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Wiseman DH, Greystoke BF, Somervaille TC. The variety of leukemic stem cells in myeloid malignancy. Oncogene. 2014 Jun 12;33(24):3091–3098. doi: 10.1038/onc.2013.269. [DOI] [PubMed] [Google Scholar]
- 7.Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O’Laughlin M, et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell. 2014 Mar 17;25(3):379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safe S, Jin UH, Morpurgo B, Abudayyeh A, Singh M, Tjalkens RB. Nuclear receptor 4A (NR4A) family - orphans no more. J Steroid Biochem Mol Biol. 2016 Mar;157:48–60. doi: 10.1016/j.jsbmb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamers AA, Hanna RN, Nowyhed H, Hedrick CC, de Vries CJ. NR4A nuclear receptors in immunity and atherosclerosis. Curr Opin Lipidol. 2013 Oct;24(5):381–385. doi: 10.1097/MOL.0b013e3283643eac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullican SE, Zhang S, Konopleva M, Ruvolo V, Andreeff M, Milbrandt J, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007 Jun;13(6):730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 12.Sirin O, Lukov GL, Mao R, Conneely OM, Goodell MA. The orphan nuclear receptor Nurr1 restricts the proliferation of haematopoietic stem cells. Nat Cell Biol. 2010 Dec;12(12):1213–1219. doi: 10.1038/ncb2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng LE, Chan FK, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997 Apr 15;16(8):1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994 Jan 20;367(6460):277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, Milbrandt J. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77) Science. 1995 Jul 28;269(5223):532–535. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 16.Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013 Mar;14(3):230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 17.Sekiya T, Kondo T, Shichita T, Morita R, Ichinose H, Yoshimura A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J Exp Med. 2015 Sep 21;212(10):1623–1640. doi: 10.1084/jem.20142088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowyhed HN, Huynh TR, Blatchley A, Wu R, Thomas GD, Hedrick CC. The nuclear receptor nr4a1 controls CD8 T cell development through transcriptional suppression of runx3. Sci Rep. 2015;5:9059. doi: 10.1038/srep09059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. 2011 Aug;12(8):778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacke R, Hilgendorf I, Garner H, Waterborg C, Park K, Nowyhed H, et al. The transcription factor NR4A1 is essential for the development of a novel macrophage subset in the thymus. Sci Rep. 2015;5:10055. doi: 10.1038/srep10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna RN, Shaked I, Hubbeling HG, Punt JA, Wu R, Herrley E, et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ Res. 2012 Feb 3;110(3):416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez-Herrick AM, Mullican SE, Sheehan AM, Conneely OM. Reduced NR4A gene dosage leads to mixed myelodysplastic/myeloproliferative neoplasms in mice. Blood. 2011 Mar 3;117(9):2681–2690. doi: 10.1182/blood-2010-02-267906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutsch AJ, Rinner B, Wenzl K, Pichler M, Troppan K, Steinbauer E, et al. NR4A1-mediated apoptosis suppresses lymphomagenesis and is associated with a favorable cancer-specific survival in patients with aggressive B-cell lymphomas. Blood. 2014 Apr 10;123(15):2367–2377. doi: 10.1182/blood-2013-08-518878. [DOI] [PubMed] [Google Scholar]
- 24.Wenzl K, Troppan K, Neumeister P, Deutsch AJ. The nuclear orphan receptor NR4A1 and NR4A3 as tumor suppressors in hematologic neoplasms. Curr Drug Targets. 2015;16(1):38–46. doi: 10.2174/1389450115666141120112818. [DOI] [PubMed] [Google Scholar]
- 25.Boudreaux SP, Ramirez-Herrick AM, Duren RP, Conneely OM. Genome-wide profiling reveals transcriptional repression of MYC as a core component of NR4A tumor suppression in acute myeloid leukemia. Oncogenesis. 2012;1:e19. doi: 10.1038/oncsis.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellagatti A, Cazzola M, Giagounidis AA, Malcovati L, Porta MG, Killick S, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006 Jul 1;108(1):337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 27.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007 Aug 2;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007 Jul 13;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 31.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015 Mar;16(3):167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999 Jan 28;18(4):1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- 33.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009 May 15;34(3):387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010 Feb;17(2):194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013 Jun 6;153(6):1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duren RP, Boudreaux SP, Conneely OM. Genome Wide Mapping of NR4A Binding Reveals Cooperativity with ETS Factors to Promote Epigenetic Activation of Distal Enhancers in Acute Myeloid Leukemia Cells. PLoS One. 2016;11(3):e0150450. doi: 10.1371/journal.pone.0150450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006 Feb;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 38.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007 Feb;39(2):232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 39.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008 Jun;40(6):741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 40.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007 Mar 15;13(6):1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 41.Wagner JM, Hackanson B, Lubbert M, Jung M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin Epigenetics. 2010 Dec;1(3–4):117–136. doi: 10.1007/s13148-010-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brien GL, Valerio DG, Armstrong SA. Exploiting the Epigenome to Control Cancer-Promoting Gene-Expression Programs. Cancer Cell. 2016 Apr 11;29(4):464–476. doi: 10.1016/j.ccell.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardini A, Baillat D, Cesaroni M, Hu D, Marinis JM, Wagner EJ, et al. Integrator regulates transcriptional initiation and pause release following activation. Mol Cell. 2014 Oct 2;56(1):128–139. doi: 10.1016/j.molcel.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet. 2015 Oct;47(10):1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009 Jul 10;138(1):129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brondfield S, Umesh S, Corella A, Zuber J, Rappaport AR, Gaillard C, et al. Direct and indirect targeting of MYC to treat acute myeloid leukemia. Cancer Chemother Pharmacol. 2015 Jul;76(1):35–46. doi: 10.1007/s00280-015-2766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011 Oct 27;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuber J, Radtke I, Pardee TS, Zhao Z, Rappaport AR, Luo W, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009 Apr 1;23(7):877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reagan-Shaw S, Nihal M, Ahmad N. Dose translation from animal to human studies revisited. FASEB J. 2008 Mar;22(3):659–661. doi: 10.1096/fj.07-9574LSF. [DOI] [PubMed] [Google Scholar]
- 51.Silberstein SD, McCrory DC. Ergotamine and dihydroergotamine: history, pharmacology, and efficacy. Headache. 2003 Feb;43(2):144–166. doi: 10.1046/j.1526-4610.2003.03034.x. [DOI] [PubMed] [Google Scholar]
- 52.Mendez-Ferrer S, Battista M, Frenette PS. Cooperation of beta(2)- and beta(3)-adrenergic receptors in hematopoietic progenitor cell mobilization. Ann N Y Acad Sci. 2010 Mar;1192:139–144. doi: 10.1111/j.1749-6632.2010.05390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lucas D, Bruns I, Battista M, Mendez-Ferrer S, Magnon C, Kunisaki Y, et al. Norepinephrine reuptake inhibition promotes mobilization in mice: potential impact to rescue low stem cell yields. Blood. 2012 Apr 26;119(17):3962–3965. doi: 10.1182/blood-2011-07-367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spiegel A, Shivtiel S, Kalinkovich A, Ludin A, Netzer N, Goichberg P, et al. Catecholaminergic neurotransmitters regulate migration and repopulation of immature human CD34+ cells through Wnt signaling. Nat Immunol. 2007 Oct;8(10):1123–1131. doi: 10.1038/ni1509. [DOI] [PubMed] [Google Scholar]
- 55.Yang M, Li K, Ng PC, Chuen CK, Lau TK, Cheng YS, et al. Promoting effects of serotonin on hematopoiesis: ex vivo expansion of cord blood CD34+ stem/progenitor cells, proliferation of bone marrow stromal cells, and antiapoptosis. Stem Cells. 2007 Jul;25(7):1800–1806. doi: 10.1634/stemcells.2007-0048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.