Abstract
Background
Previous literatures have investigated the change of miR-20a expression level in the progression of multiple cancers and its influence on patients' survival outcome, but results of now-available evidence are inconsistent.
Objective
To elucidate the prognostic value of circulating and tissue-based miR-20a for patients with various cancers.
Methods
A systematic search and review of eligible publications were carried out in three electronic databases including the Cochrane Library, PubMed, and Embase, and the methodological quality of included studies was assessed according to Newcastle-Ottawa Scale (NOS). Hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) for overall survival (OS), recurrence-free survival (RFS), disease-free survival (DFS), and progressive-free survival (PFS) of each study were pooled using a random effect model.
Results
In total, 24 studies involving 4186 samples of multiple cancers published in 20 articles were included in the statistical analysis. As for circulating miR-20a, five kinds of cancers containing gastric cancer, lymphoma, glioblastoma, prostate cancer, and non-small-cell lung cancer reported upregulated level in patients compared with normal healthy control, and overexpressed circulating miR-20a could confer an unfavorable factor for OS (HR = 1.71, 95% CIs: 1.43 -2.04, p < 0.01) and DFS (HR = 1.90, 95% CIs: 1.45-2.49, p < 0.01). As for tissue-based samples, 6 kinds of malignancies including colorectal cancer, salivary adenoid cystic carcinoma, gallbladder carcinoma, colon cancer, gastrointestinal cancer, and alveolar rhabdomyosarcoma revealed upregulated miR-20a expression level compared with paired nontumorous tissue, of which high expression of miR-20a was significantly associated with poor OS (HR = 2.74, 95% CIs: 1.38-5.42, p < 0.01) and DFS (HR = 2.68, 95% CIs: 1.32-5.45, p < 0.01); meanwhile, other 5 tumors containing breast cancer, cutaneous squamous cell carcinoma, hepatocellular carcinoma, oral squamous cell carcinoma, and epithelial ovarian cancer demonstrated downregulated miR-20a expression level compared with benign tissue, of which low miR-20a expression was significantly related to shorter OS (HR = 3.48, 95% CIs: 2.00-6.06, p < 0.01) and PFS/RFS (HR = 4.05, 95% CIs: 2.89-5.66, p < 0.01).
Conclusion
Change of circulating and tissue-based miR-20a expression possesses vital prognostic implication for human cancers. Augmented expression of circulating miR-20a predicts poor survival outcome for patients with cancers. Tissue-based miR-20a level may be upregulated or downregulated depending on cancer types; in the former condition, high expression of tissue miR-20a is a risk factor for unfavorable prognosis and in the latter condition low expression of tissue miR-20a is associated with shorter survival.
1. Introduction
MicroRNAs (miRNAs/miRs) are a set of single-stranded, nonprotein-coding RNAs approximately 19~24 nucleotides in length [1]. It is demonstrated that miRNAs are highly conservative in evolution and act as posttranslational inhibitors by binding to the complementary sequences in the 3′ untranslated regions (3′-UTR) of messenger RNAs (mRNAs) and therefore leading to translation regression or triggering decay factors of mRNAs [1, 2]. Due to the fact that majority of encoding sequences of miRNAs lie in cancer-related regions of genome [3], dysregulated miRNAs profile often plays a profound role in various tumor-associated biological processes such as proliferation, differentiation, migration, angiogenesis, stress response, metabolism, invasion, chemoresistance, and apoptosis [1, 3]. In these years, numerous miRNAs have emerged as candidates of molecular biomarkers for diagnosing human cancers and guiding treatment as well as predicting the metastasis, relapse, and prognosis [3–5].
MiR-20a is a typical and extensively investigated example of miRNAs originating from the miR-17~92 cluster, which is located at chromosomal locus 13q31.3 and able to encode five other mature miRNAs including miR-17, miR-18a, miR-19a/b, and miR-92a [6]. MiR-20a is identified in a wide range of clinical specimens (plasma, serum, tissue, feces, etc.) and the expression pattern of circulating and tissue-based miR-20a can characterize multiple human cancers [7, 8]. Zhang and his colleagues demonstrated that miR-20a level in cutaneous squamous cell carcinoma had a close relationship with tumor stage [9]; Sanfiorenzo et al. proposed that a six-miRNA plasma panel comprising miR-20a, miR-145, miR-24, miR-152, miR-25, and miR-199a was able to discriminate non-small-cell lung cancer (NSCLC) from chronic obstructive pulmonary disease (COPD) and indicate recurrence in resectable NSCLC [10]. Accumulating studies attempted to testify the clinical impact of the expression pattern of circulating and tissue-based miR-20a for human cancers, but their study designs were various, and results were inconsistent [11–18]. MiR-20a expression level is downregulated in several kinds of malignancy while upregulated in others; meanwhile, some studies exploited the expression level of miR-20a in serum or plasma, and some in tumorous and nontumorous tissues [7].
Although six mature miRNAs could be encoded by miR-17~92 cluster, the diverse sequence of each miRNA results in their specificity of target genes and separate physiobiological functions [17]. Moreover, studies appraising diagnostic significance of multiple miRNAs were often based on same group of population [13, 16–18]. Therefore, indiscriminately pooling data of all these miRNAs is inappropriate. On the other hands, if we make separative quantitative appraising of miRNAs related to miR-20a, these workloads are heavy, and the theme of this article will become ambiguous. Therefore, in this article, we only chose miR-20a, a widely investigated miRNA with controversy prognostic value in human cancers, as the target of interest.
Two previous meta-analyses with respect to the prognostic value of miR-17~92 cluster in various tumors were published in 2017[19, 20]. These analyses demonstrated that high expression of miR-17~92 cluster was significantly predictive of a poor prognosis in various cancers, with pooled risk ratios of 2.05 (95% confidence interval (CI) 1.58-2.65) and 1.71 (95% CI 1.39-2.11) for overall survival (OS), respectively. But these two meta-analyses only included 6 and 7 studies appraising prognostic value of miR-20a in cancer patients and evaluated all members of miR-17~92 cluster as a whole. Moreover, investigators did not distinguish different sources of tested samples (circulating or tissue-based) or perform subgroup analysis according to the trend of miR-20a change. Recently, an increasing amount of studies about circulating or tissue-based miR-20a have been published [9–11, 13–15, 17–20]. Therefore, we performed this updated systematic review and meta-analysis to authentically and comprehensively assess the value of miR-20a for monitoring therapeutic efficacy and prognosis of human cancers.
2. Materials and Methods
We followed the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement in the conduction of this study [21]; each process was performed by two investigators (Qingyu Zhang and Qiwei Wang) repeatedly and independently, and any disagreement was resolved by discussion or arbitrating by a third author (Wei Sun). Informed consent was not requisite because all data were extracted from published articles.
2.1. Literature Searching and Including
Three electronic databases including PubMed, Embase, and the Cochrane Library were searched for eligible literatures using following keywords: microRNA-20a OR miRNA-20a OR miR-20a OR microRNA-20a-5p OR miRNA-20a-5p OR miR-20a-5p. Then the reference lists of relevant articles were also checked by hands to retrieve other potentially qualified publications. Last search was updated on May 1, 2018, and no language limitations were imposed.
Eligible studies for this meta-analysis had to confirm following criteria: (1) population, patients with any kind of cancer; (2) the association between miR-20a expression level and prognosis was assessed; (3) the primary outcome was overall survival (OS) or relapse-free survival (RFS) or progression-free survival (PFS) or disease-free survival (DFS); (4) enough data were provided to obtain trustworthy hazard ratios (HRs) and corresponding 95% confidence intervals (CIs); and (5) nonoriginal articles (case reports, reviews, letters, meta-analyses, and editorials), meeting abstracts, and animal studies were excluded. If there were studies with duplicate cohorts, only the more comprehensive or recent one was enrolled.
2.2. Data Extraction
For included studies, collected information was as follows: first authors' surname, year of publication, original country, sample size, demographic information of participants (age, year, and so on), type and stage of cancer, source of sample, duration of follow-up, method of detecting miR-20a expression, level of miR-20a, survival outcome, and cut-off values. For each study, HRs and associated 95% CIs were extracted directly if they were provided explicitly in original articles or supplementary materials (available here); otherwise, they were calculated by using log-rank/Cox regression statistics by Tierney's methods [22]. Sample source was categorized into tissue, serum, and plasma, while sample sizes were divided into those of more than 100 and those less than 100. HRs calculated from multivariate regression analysis model were preferred to adjust the confounders and if not provided, those from univariate analysis were extracted. These data were filled into a predesigned excel file for further analysis and calculation.
2.3. Quality Assessment
Newcastle-Ottawa Scale (NOS) was used to appraise the methodological quality of included studies. This tool compromises nine items and one score was earned if information concerning this item was clearly reported in original studies. Studies of ≥ 7 were regarded as high-quality reporting.
2.4. Statistical Methods
HRs and corresponding 95% confidence intervals (CIs) of individual study were pooled to obtain the summary estimates by using a random effect model (DerSimonian and Laird method). The heterogeneity across studies was assessed using the Cochran Q and I2 index. I2 of < 25% represents small heterogeneity, 50% ≥ I2 ≥ 25% moderate heterogeneity, and I2 > 50% significant heterogeneity. One-way sensitivity analyses were conducted by removing publications individually to evaluate the stability of results. Meanwhile, subgroup analyses based on region of publication, cancer type, sample size, and calculation model of HR (multivariate or univariate) were performed. We utilized Egger's test to detect any possible publication bias (significant publication bias if a two-tailed p value < 0.05).
All analyses were performed with STATA, version 12.0 (StataCorp, College Station, TX, USA).
3. Results
3.1. Literature Selection and Study Characteristics
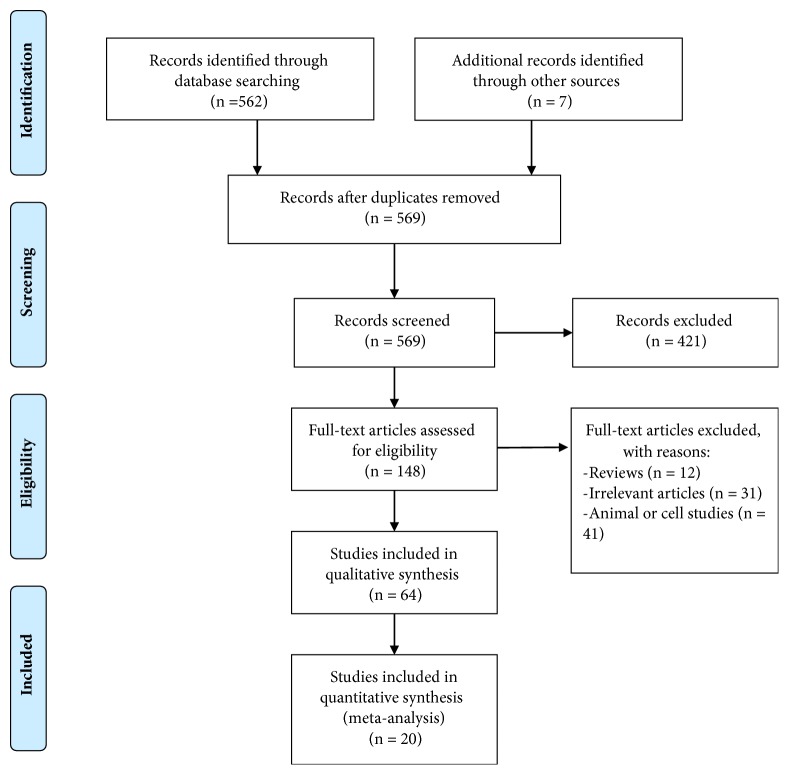
568 nonduplicated publications were retrieved by searching three databases and screening references of relevant articles, and eventually 20 [9–11, 13–15, 17, 18, 34] articles were included in statistical analysis. The literature search and selection processes were summarized in Figure 1 and the basic information of included studies was described in Tables 1 and 2. Among these studies, Chen and his colleagues validated prognostic impaction of miR-20a by analyzing colorectal cancer patients in Tumor Cancer Genome Atlas (TCGA) databases [26]; Zhang et al. randomly assigned patients to the training set, internal testing set, and independent validation set to testify the prognostic value of miR-20a in stage II~IV colon cancer [28]; meanwhile, Si et al. presented data about association between miR-20a and breast cancer patients' survival in three different cohorts (cohorts 1 and 2 from their own research center and cohort 3 from TCGA databases) [32]. These datasets from the same articles were collected simultaneously, and eventually this meta-analysis was established based on 24 studies.
Figure 1.
Flow diagram of literature research and selection process.
Table 1.
Main characteristic of included studies about circulating miR-20a.
| First author | Year | Country | Number of patients | Age, Median (range) | Cancer type | Stage range | Marker | Proposed target genes & pathways | Test method | Up- or down-regulated compared with healthy control | Number of high/low level of miR-20a | Cut-off value | Follow-up | Outcome | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xu et al [23] | 2018 | China | 196 | 59.5±10.31 | NSCLC | I-III | Plasma miR-20a | BID, TRAIL | RT-PCR | Up-regulated | NR | the median value | 56.7 months | OS, DFS | 7 |
| Yang et al [11] | 2017 | China | 55 | NR | gastric cancer | I-IV | Serum miR-20a | NFKBIB | microarray, qRT-PCR | Up-regulated | 27/28 | the median value | 34 months | OS | 7 |
| Khare et al [24] | 2017 | Israel | 25 | 18-85 | lymphoma | I-IV | Plasma miR-20a | PTEN, pi3k/AKT | qRT-PCR | Up-regulated | NR | the median value | 5 years | OS | 6 |
| Zhao et al [25] | 2017 | US | 106 | 58 | glioblastoma | NR | Plasma miR-20a-5p | TIMP-2 | qRT-PCR | Up-regulated | NR | the median value | 2 years | OS, DFS | 7 |
| Lin et al [9] | 2014 | Australia | 97 | 68(46-87) | prostate cancer | NR | Plasma/serum miR-20a | RUNX1, CSF1R | qRT-PCR | Up-regulated | NR | NR | 12 (3-62) months | OS | 7 |
| Sanfiorenzo et al [10] | 2013 | France | 62 | 65.1 | NSCLC | I-III | Plasma miR-20a | NR | microarray, qRT-PCR | Up-regulated | NR | NR | 18months~ | DFS | 8 |
| Wang et al [13] | 2012 | China | 65 | NR | gastric cancer | I-IV | Serum miR-20a | NR | qRT-PCR | Up-regulated | 34/31 | the median value | NR | OS | 6 |
NR, not reported; NSCLC, non-small-cell lung cancer; NR, not reported; qRT-PCR, quantitative real time-polymerase chain reaction; OS, overall survival; DFS, disease-free survival.
Table 2.
Main characteristic of included studies about tissue miR-20a.
| First author | Year | Country | Number of patients | Age, Median (range) | Cancer type | Stage range | Marker | Proposed target genes & pathways | Test method | Up- or down-regulated compared with normal tissue | Number of high/low level of miR-20a | Cut-off value | Follow-up | Outcome | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cheng et al [14] | 2016 | China | 544 | 65 | colorectal cancer | I~IV | miR-20a-5p | Smad4 | qRT- PCR | Up-regulated | 407/137 | NR | 6~12 years | OS, DFS | 8 |
| Chen et al [26] | 2015 | Asia/ Europe/ North America | 580 | 67.9±13.2 | colorectal cancer | I~IV | miR-20a-5p | BIM | qRT- PCR | Up-regulated | NR | NR | NR | OS | 7 |
| Mitani et al [17] | 2013 | US | 64 | NR | salivary adenoid cystic carcinoma | I~IV | micro-RNA-17-92 | NR | qRT- PCR | Up-regulated | 22/8 | 3 | NR | OS | 8 |
| Chang et al [27] | 2013 | China | 98 | NR | gallbladder carcinoma | NR | miR-17/20a | Smad7 | qRT-PCR | Up-regulated | 71/27 | NR | NR | OS | 7 |
| Zhang et al [28] | 2013 | China | 138(training set) | NR | colon cancer | II | miR-20a-5p | NR | qRT- PCR | Up-regulated | NR | NR | 66 months | DFS | 7 |
| 137(internal testing set) | NR | colon cancer | II | miR-20a-6p | qRT- PCR | Up-regulated | NR | NR | DFS | 7 | |||||
| 460 (independent validation set) | NR | colon cancer | II | miR-20a-7p | qRT- PCR | Up-regulated | NR | NR | DFS | 7 | |||||
| Valladares-Ayerbes et al [29] | 2011 | Spain | 38 | 62.5(45-76) | gastrointestinal cancer | I~IV | miR-17; miR-20a; miR-21 | E2F1, E2F2, E2F3, BIM, LRF, | qRT- PCR | Up-regulated | NR | NR | 153 weeks (2-388 weeks) | OS, PFS | 6 |
| Reichek et al [30] | 2011 | US | 123 | NR | alveolar rhabdomyosarcoma | NR | micro-RNA-17-92 | E2F | qRT- PCR | Up-regulated | NR | NR | NR | OS | 6 |
| Schetter et al [31] | 2008 | China | 84;113 | 64.6(32-87); 55.8(32-84) | colon adenocarcinoma | I~IV | miR-20 | NR | qRT- PCR | Up-regulated | NR | NR | NR | OS | 7 |
|
| |||||||||||||||
| Si et al [32] | 2017 | China | 66 (cohort 1) | 53.6 | breast cancer | I~IV | miR-20 | c-Myc | qRT- PCR | Down-regulated | 30/36 | NR | NR | OS | 7 |
| 40 (cohort 2) | 56.1 | breast cancer | I~IV | miR-21 | c-Myc | qRT- PCR | Down-regulated | 18/22 | NR | NR | OS | 7 | |||
| 716 (cohort 3 from TCGA database) | 59 | breast cancer | I~IV | miR-22 | c-Myc | qRT- PCR | Down-regulated | 334/382 | NR | NR | OS | 7 | |||
| Zhang et al [15] | 2015 | China | 152 | 53.9 | cutaneous squamous cell carcinoma | I-III | miR-20 | NR | qRT- PCR | Down-regulated | 54/98 | NR | NR | OS | 6 |
| Fan et al [33] | 2013 | China | 100 | 57.8/53.6 | hepatocellular carcinoma | I-III | miR-20 | Mcl-1 | qRT- PCR | Down-regulated | 50/50 | NR | NR | OS, RFS | 8 |
| Chang et al [27] | 2013 | China | 96 | NR | oral squamous cell carcinoma | I~IV | miR-20 | ITGb8 | qRT- PCR | Down-regulated | 34/33 | NR | NR | OS | 7 |
| Marchini et al [34] | 2011 | Italy | 144 | 9 | epithelial ovarian cancer | I | miR-20 | NR | qRT- PCR | Down-regulated | NR | NR | NR | OS, PFS | 7 |
NR, not reported; TCGA, Tumor Cancer Genome Atlas; NR, not reported; qRT-PCR, quantitative real time-polymerase chain reaction; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; RFS, relapse-free survival.
All 20 literatures were written in English. As for methodological quality of 24 included studies, four [10, 14, 17, 33] got 8 score and fifteen [9, 11, 18, 23, 25, 26, 28, 31, 32, 32, 34] studies had 7 score. Only five [13, 15, 24, 29, 30] studies received 6 score. Mean NOS score for these studies was 6.96 (range 6~8). The sample sizes ranged from 25 to 716 with a total of 4186 samples and 16 kinds of tumors were analyzed: gastric cancer, lymphoma, glioblastoma, prostate cancer, NSCLC, colorectal cancer, salivary adenoid cystic carcinoma, gallbladder carcinoma, colon cancer, gastrointestinal cancer, alveolar rhabdomyosarcoma, breast cancer, cutaneous squamous cell carcinoma, hepatocellular carcinoma, oral squamous cell carcinoma, and epithelial ovarian cancer. 11 types of tumors whose sample source was cancerous and noncancerous tissue were reported in 17 [14, 15, 17, 18, 26–30, 33, 34] studies (2 colorectal cancer, 1 salivary adenoid cystic carcinoma, 1 gallbladder carcinoma, 4 colon cancer, 1 gastrointestinal cancer, 1 alveolar rhabdomyosarcoma, 3 breast cancer, 1 cutaneous squamous cell carcinoma, 1 hepatocellular carcinoma, 1 oral squamous cell carcinoma, and 1 epithelial ovarian cancer); meanwhile, 7 [9–11, 13, 24, 25] studies testing circulating miR-20a expression involved a total of 5 kinds of tumors (2 gastric cancer, 1 lymphoma, 1 glioblastoma, 1 prostate cancer, and 1 NSCLC). All studies used real-time polymerase chain reaction (RT-PCR) to test the targeted miRNA. The cut-off value for high and low expression of miR-20a was various in included studies. As for the survival outcomes, 24 eligible studies could be divided into 30 datasets: 20 for OS, 7 for DFS, 2 for PFS, and 1 for RFS (see Table 2).
3.2. Circulating miR-20a and Cancer Prognosis
All 6 kinds of tumors involving circulating miR-20a shown upregulated miR-20a expression level in patients compared with the healthy control [9–11, 13, 25].
3.2.1. Circulating miR-20a and Overall Survival
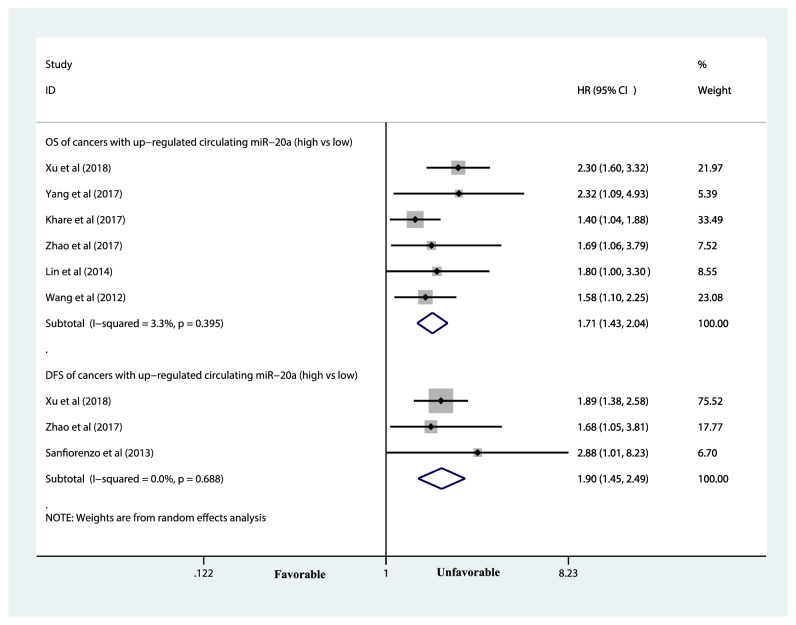
5 [9, 11, 13, 24, 25] studies provided HRs and corresponding 95% CIs for the association between circulating miR-20a expression and overall survival. Among those tumors, high expression of circulating miR-20a was significantly associated with unfavorable OS of cancer patients (HR = 1.71, 95% CIs: 1.43 -2.04, p < 0.01; p for heterogeneity = 0.395, I-square = 3.3%) (see Figure 2 and Table 3).
Figure 2.
Forest plot of the association between high expression of circulating miR-20a and survival in various cancers. The area of the square represented the weight of each study in the pooled results. The diamond indicated pooled HR and corresponding 95% CI. As depicted, the diamond in the right of the central vertical line represents an unfavorable prognosis in the former group in comparison with the latter group. CI, confidence interval; HR, hazard ratio.
Table 3.
Meta-analyses of tumors with upregulated circulating or tissue miR-20a.
| Circulating miR-20a | Tissue miR-20a | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of study | HR (95%CI) | p-value | I-square | p-value for heterogeneity | p-value of Egger' test | No. of study | HR (95% CI) | p-value | I-square | p-value for heterogeneity | p-value of Egger' test | |||
| OS | All | high vs low | 6[9, 11, 13, 23–25] | 1.71 (1.43 -2.04) | <0.01 | 3.3% | 0.395 | 0.369 | 7[14, 17, 26, 27, 29–31] | 2.74 (1.38-5.42) | <0.01 | 90.90% | <0.01 | 0.013 |
| Country | China (high vs low) | 3[11, 13, 23] | 1.94 (1.48-2.55) | <0.01 | 15% | 0.308 | / | 5[17, 26, 29–31] | 2.02 (1.14-3.58) | <0.01 | 77.60% | <0.01 | 0.001 | |
| Other (high vs low) | 3[9, 24, 25] | 1.50 (1.18-1.91) | <0.01 | 0.00% | 0.703 | / | 2[14, 27] | 4.63 (1.48-14.50) | <0.01 | 84.20% | 0.012 | / | ||
| Sample size | > 100 (high vs low) | 2[23, 25] | 2.13 (1.55-2.93) | <0.01 | 0.00% | 0.409 | / | 3[26, 27, 30] | 3.86 (1.28-11.63) | 0.016 | 84.10% | <0.01 | 0.903 | |
| < 100 (high vs low) | 4[9, 11, 13, 24] | 1.55 (1.27-1.91) | <0.01 | 0.00% | 0.613 | 0.031 | 4[14, 17, 29, 31] | 2.08 (1.04-4.19) | 0.039 | 82.80% | <0.01 | 0.002 | ||
| Cancer type | gastrointestinal cancer (high vs low) | 2[11, 13] | 1.69 (1.22-2.34) | <0.01 | 0.00% | 0.364 | / | 4[26, 27, 29, 31] | 2.51 (0.99-6.39) | 0.053 | 94.30% | <0.01 | 0.13 | |
| others (high vs low) | 4[9, 23–25] | 1.74 (1.34-2.26) | <0.01 | 30.9% | 0.227 | 0.702 | 3[14, 17, 30] | 2.95 (1.74-5.00) | <0.01 | 0.00% | <0.01 | 0.806 | ||
| Calculation model of HR | Multivariate (high vs low) | 5[9, 13, 23–25] | 1.69 (1.39-2.04) | <0.01 | 11.0% | 0.343 | 0.610 | 7[14, 17, 26, 27, 29–31] | 2.74 (1.38-5.42) | <0.01 | 90.90% | <0.01 | 0.013 | |
| Univariate (high vs low) | 1[11] | 2.32 (1.09-4.93) | / | / | / | / | / | / | / | / | / | / | ||
|
| ||||||||||||||
| DFS | All | high vs low | 2[10, 23, 25] | 1.90 (1.45-2.90) | <0.01 | 0.00% | 0.688 | / | 4[14, 28] | 2.68 (1.32-5.45) | <0.01 | 82.90% | <0.01 | 0.703 |
| Country | China (high vs low) | / | / | / | / | / | / | 4[14, 28] | 2.68 (1.32-5.45) | <0.01 | 82.90% | <0.01 | 0.703 | |
| Other (high vs low) | / | / | / | / | / | / | / | / | / | / | / | / | ||
| Sample size | > 100 (high vs low) | / | / | / | / | / | / | 4[14, 28] | 2.68 (1.32-5.45) | <0.01 | 82.90% | <0.01 | 0.703 | |
| < 100 (high vs low) | / | / | / | / | / | / | / | / | / | / | / | / | ||
| Cancer type | gastrointestinal cancer (high vs low) | / | / | / | / | / | / | 4[14, 28] | 2.68 (1.32-5.45) | <0.01 | 82.90% | <0.01 | 0.703 | |
| others (high vs low) | / | / | / | / | / | / | / | / | / | / | / | / | ||
| Calculation model of HR | Multivariate (high vs low) | / | / | / | / | / | / | 4[14, 28] | 2.68 (1.32-5.45) | <0.01 | 82.90% | <0.01 | 0.703 | |
| Univariate (high vs low) | / | / | / | / | / | / | / | / | / | / | / | / | ||
HR, hazard ratio; CI, confidence interval; OS, overall survival; DFS, disease free survival.
3.2.2. Circulating miR-20a and Disease-Free Survival
3 [10, 23, 25] studies provided HRs and corresponding 95% CIs for DFS, and high expression of miR-20a was associated with unfavorable pooled DFS (HR = 1.90, 95% CIs: 1.45-2.49, p < 0.01; p for heterogeneity = 0. 688, I-square = 0.0%), too (see Figure 2 and Table 3).
3.3. Tissue-Based miR-20a and Cancer Prognosis
6 kinds of tumors (colorectal cancer, salivary adenoid cystic carcinoma, gallbladder carcinoma, colon cancer, gastrointestinal cancer, and alveolar rhabdomyosarcoma) show upregulated miR-20a expression level in tumorous tissue compared with nontumorous tissue and other 5 cancers (breast cancer, cutaneous squamous cell carcinoma, hepatocellular carcinoma, oral squamous cell carcinoma, and epithelial ovarian cancer) demonstrated downregulated miR-20a expression level in tumorous tissue compared with paired nontumorous tissue.
3.3.1. Tissue miR-20a and Overall Survival
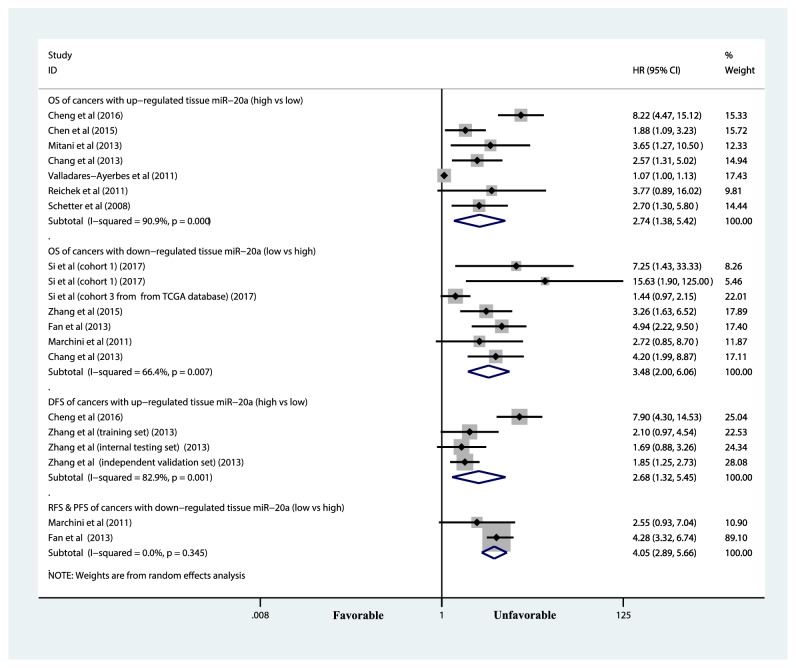
There were 14 [14, 15, 17, 18, 26, 27, 29, 30, 34] studies which reported HRs and corresponding 95% CI for the correlation between OS and expression of tissue-based miR-20a. Among those tumors with upregulated miR-20a level, highly expressed miR-20a was negatively associated with OS (HR = 2.74, 95% CIs: 1.38-5.42, p < 0. 01; p for heterogeneity < 0. 001, I-square = 90.9%) (see Figure 3 and Table 3). Meanwhile, among those tumors with downregulated miR-20a level, low expression of miR-20a notably indicated reduced OS (HR = 3.48, 95% CIs: 2.00-6.06, p < 0. 01; p for heterogeneity = 0. 007, I-square = 66.4%) (see Figure 3 and Table 4).
Figure 3.
Forest plot of the association between tissue-based miR-20a and survival in various cancers. The area of the square represented the weight of each study in the pooled results. The diamond indicated pooled HR and corresponding 95% CI. As depicted, the diamond in the right of the central vertical line represents an unfavorable prognosis in the former group in comparison with the latter group. CI, confidence interval; HR, hazard ratio.
Table 4.
Meta-analyses of tumors with downregulated tissue-based miR-20a.
| Summary estimate | Subgroup analysis | No. of study | HR (95% CI) | p-value | I-square | p-value for heterogeneity | p-value of Egger' test | |
|---|---|---|---|---|---|---|---|---|
| OS | All | low vs high | 7[15, 18, 32–34] | 3.48 (2.00-6.06) | <0.01 | 66.40% | <0.01 | 0.024 |
| Country | China (low vs high) | 6[15, 18, 32, 33] | 3.41 (1.80-6.47) | <0.01 | 68.40% | <0.01 | 0.04 | |
| Other (low vs high) | 1[34] | 4.2 (1.99-8.87) | / | / | / | / | ||
| Sample size | > 100 (low vs high) | 4[15, 32–34] | 2.70 (1.42-5.13) | <0.01 | 71.30% | 0.015 | 0.253 | |
| < 100 (low vs high) | 3[18, 32] | 5.21 (2.74-9.90) | <0.01 | 0.00% | 0.462 | 0.129 | ||
| Cancer type | gastrointestinal cancer (low vs high) | / | / | / | / | / | / | |
| others (low vs high) | 7[15, 18, 32–34] | 3.48 (2.00-6.06) | <0.01 | 66.40% | <0.01 | 0.024 | ||
| Calculation model of HR | Multivariate (low vs high) | 6[15, 32–34] | 3.41 (1.80-6.47) | <0.01 | 68.40% | <0.01 | 0.04 | |
| Univariate (low vs high) | 1[18] | 4.2 (1.99-8.87) | / | / | / | / | ||
|
| ||||||||
| PFS & RFS | All | low vs high | 2[33, 34] | 4.05 (2.89-5.66) | <0.01 | 0.00% | 0.345 | / |
| Country | China (low vs high) | 1[33] | 4.28 (3.32-6.74) | / | / | / | / | |
| Other (low vs high) | 1[34] | 2.55 (0.93-7.04) | / | / | / | / | ||
| Sample size | > 100 (low vs high) | 2[33, 34] | 4.05 (2.89-5.66) | <0.01 | 0.00% | 0.345 | / | |
| < 100 (low vs high) | / | / | / | / | / | / | ||
| Cancer type | gastrointestinal cancer (low vs high) | / | / | / | / | / | / | |
| others (low vs high) | 2[33, 34] | 4.05 (2.89-5.66) | <0.01 | 0.00% | 0.345 | / | ||
| Calculation model of HR | Multivariate (low vs high) | 2[33, 34] | 4.05 (2.89-5.66) | <0.01 | 0.00% | 0.345 | / | |
| Univariate (low vs high) | / | / | / | / | / | / | ||
HR, hazard ratio; CI, confidence interval; OS, overall survival; PFS, progression free survival; RFS, relapse free survival.
3.3.2. Tissue-Based miR-20a and Disease-Free Survival
4 [14, 28] studies investigated impact of tissue-based miR-20a on DFS of cancer (Zhang et al.'s article contains three independent studies). Among those tumors with upregulated miR-20a level, highly expressed miR-20a was associated with unfavorable OS (HR = 2.68, 95% CIs: 1.32-5.45, p < 0. 01; p for heterogeneity = 0. 001, I-square = 82.9%) (see Figure 3 and Table 3).
3.3.3. Tissue-Based miR-20a and Relapse-Free/Progression-Free Survival
2 [33, 34] studies provided HRs and corresponding 95% CI for the relationship between RFS/PFS of cancer patients and tissue-based miR-20a expression. Among those tumors with upregulated miR-20a level, low expression of miR-20a was significantly associated with unfavorable RFS/PFS (HR = 4.05, 95% CIs: 2.89-5.66, p < 0. 01; p for heterogeneity = 0. 345, I-square = 0.0%) (see Figure 3 and Table 4).
3.4. Subgroup Analysis and Sensitivity Analysis
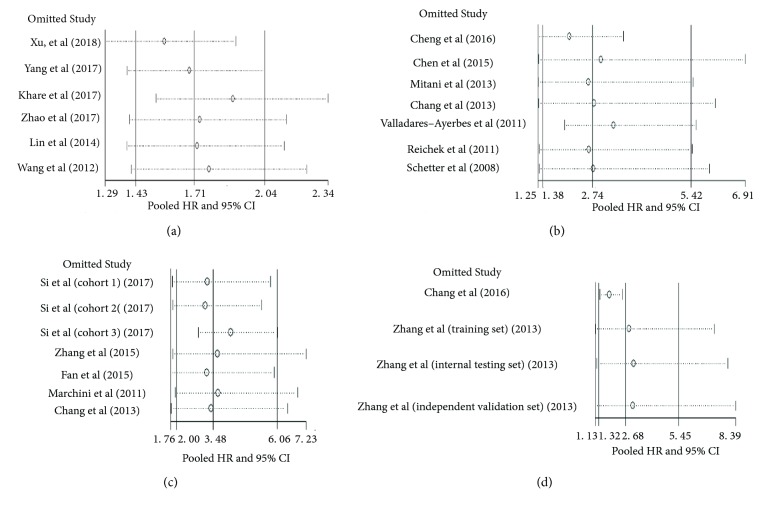
Above results were stable in the one-way sensitivity analysis when omitting studies one by one (see Figure 4). Meanwhile, subgroup analyses were conducted according to several factors such as region of publication (China or other countries), cancer type (gastrointestinal cancers or other cancers), sample size (< or > 100 participants), and calculation model of HR (multivariate or univariate), and the results demonstrated that these factors did not significantly change the relations between miR-20a and cancer prognosis (see Tables 3 and 4).
Figure 4.
One-way sensitivity analyses about the association between miR-20a expression and survival in various cancers. (a) high expression of circulating miR-20a and OS; (b) tissue-based miR-20a and OS in cancers with upregulated miR-20a expression (high versus low); (c) tissue-based miR-20a and OS in cancers with downregulated miR-20a expression (low versus high); (b) tissue-based miR-20a and DFS in cancers with upregulated miR-20a expression (high versus low). Sensitivity analyses demonstrated that results of these meta-analyses were stable when omitting one study in each turn.
4. Discussion
Identification of validated risk factors of various cancers could not only lead to a better understanding of molecular signaling pathways in the pathogenesis and progression of corresponding diseases but also offer new targets for clinical diagnosis and treatment [35]. Among alternative prognostic biomarkers such as plentiful proteins, RNAs, and DNAs, miRNAs seem to hold great promise and multiple profiling techniques (miRNA microarray, qRT-PCR, and next-generation sequencing) have been used for the measurement of circulating and tissue-based miRNAs [7]. MiRNA dysregulations in human cancer are mainly due to genomic changes including amplification, mutation, deletion, and disturbance of miRNA biogenesis enzymes, namely, Drosha, exportin 5, Dicer, and argonaute 2 (ARO2), and effects of these changes show cell/organ specificity [36–38].
Aberrant miR-20a level was observed in the progression of multiple human cancers but its efficacy as a prognostic biomarker remains inconsistent. In fact, a line of evidence has indicated that miR-20a could regulate a series of genes, where the effects may be either oncogenic or tumor-suppressive [39]. Moreover, the trend of miR-20a change in different kinds of cancer was variable. In addition, small sample sizes of pertinent studies lacking potent statistical power often lead to ambiguous conclusions. Two former meta-analyses suggested that high expression of miR-17~92 cluster is associated with unfavorable outcome of human cancers; however, authors did not specifically analyze miR-20a or made subgroup analysis based on tumor kinds or sample source, which undermine the credibility of these conclusions [20].
Circulating miRNAs were firstly identified in 2008 [40]. A vast majority of circulating miRNAs are derived from blood cells and endothelial cells and may act an important role in intercellular communication. During tumor progression, angiogenesis (formation of new vessels from preexisting vascular network) has been pathologically accelerated by multiple proangiogenic factors such as vascular growth factor (VEGF) and placenta growth factor (PIGF) [41]. Meanwhile, apoptotic and necrotic cells release miRNAs into the bloodstream and therefore some tissue-specific miRNAs are also detected in serum or plasma [42–44]. Circulating miRNAs are mainly capsulated in exosomes, cell-to-cell mediators of biological information, or binding to proteins, which makes these miRNAs highly stable in adverse physiological conditions [7, 45]. Moreover, compared with tissue miRNA, measuring plasma or serum miRNA is simple and injury-limited. However, it must be noted that perturbations of blood cells or hemolysis may greatly alter the level of circulating miRNAs [42] and therefore, blood-based phenomena should be taken into consideration when interpreting the results of circulating miRNAs.
Significant association between highly expressed circulating miR-20a and unfavorable OS/DFS of human cancers was identified in this study, which hinted that plasma/serum-based miR-20a acted as an oncogene. Previous studies have demonstrated overexpression of circulating miR-20a in a range of other malignant tumors such as colorectal cancer, cervical cancer, and hepatocellular carcinoma [46]. Circulating miR-20a could enhance tumor cell proliferation, migration, and invasion by targeting various pathways; for example, Du and his colleagues revealed that upregulated circulating miR-20a might suppress inhibitor β of nuclear factor- (NF-) κ B and therefore enhance activity of NF-κ B pathway and downstream molecules such as livin and survivin [47].
Unlike plasma/serum miR-20a, inconsistent results about the expression level and prognostic impaction of tissue-based miR-20a were reported in different kinds of tumors. In this investigation, a consistent tendency of tissue-based miR-20a expression change and survival was observed. Namely, for cancers with upregulated miR-20a level compared with paired nontumorous tissue, highly expressed miR-20a in tumor is associated with an unfavorable outcome, while for cancers with downregulated miR-20a level, high expression of miR-20a in tumorous tissue predicts a longer survival, which improves the conclusion of two former meta-analyses [19, 20]. MiR-20a could achieve diverse functions by targeting multiple mRNAs referred to as a targetome, and the cellular and genetic context may be decisive for the final effect of miRNAs [32, 39]. It is reported that upregulated miR-20a could promote colorectal cancer growth and progression by inhibiting multiple tumor-suppressive genes such as BIM and Smad4 [14, 48], while in breast cancer, miR-20a achieved inhibitory effect of cancer cell proliferation through directing targeting MAPK1/ERK2, a member of Ras/Raf/ERK pathway [32]. Impressively, existing evidences did not support aberrant circulating miR-20a in patients with breast cancer, cutaneous squamous cell carcinoma, hepatocellular carcinoma, oral squamous cell carcinoma, epithelial ovarian cancer, salivary adenoid cystic carcinoma, gallbladder carcinoma, and alveolar rhabdomyosarcoma compared with normal healthy controls [49]. Only in gastrointestinal tumors, upregulated miR-20a level in both serum/blood and tumorous tissue was confirmed and high expression of both circulating and tumorous miR-20a was associated with an unfavorable outcome.
These findings suggested miR-20a as a potential druggable target and provide an opportunity of using miR-20a mimics or miR-20a inhibitors (antimiR-20a) as innovative therapeutics for malignancies. With the advances of RNA biochemistry and delivery techniques, stable and effective miRNA-based agents have been put into clinical use [50, 51]. However, due to the complex interaction between receipt and donor cells and limited understanding of the underlying action mechanism and genomics of miR-20a in multiple cancer, clinical feasibility of the miR-20a as a therapeutic marker has a long way to go.
This investigation also revealed several directions of investigation for miR-20a. First, although multiple targeted genes and pathways of miR-20a have been identified in published studies, which one address key function in tumorigenesis and which factors resulted in the different change of miR-20a expression level in various cancers are undefined [9–11, 13–15, 17, 18, 34] (Tables 3 and 4). Second, some miRNAs (miR-17, miR-18a, miR-18b, miR-20b, miR-93, miR-106a, and miR-106b) are structurally homologous or expression/function-related to miR-20a [17, 52, 53]; polycistronic structures of these miRNAs may allow for reciprocal interactions. In the further, with the publication of more high-quality studies, meta-analyses with regard to other members of these miRNAs, especially miR-17 which was included in the same miRNA family with miR-20a [17, 53], may be conducted.
Some limitations of this meta-analysis merited consideration. First, inconsistent characteristics such as cut-off value and detection method may influence the outcomes. Second, many basic data were not provided in original studies. Third, although subgroup analyses were conducted, source of the heterogeneity was still not fully illustrated, indicating existence of negligible biased factors, i.e., surgical intervention, radiation, chemotherapy, mental state, and tumor characteristics. Fourth, only three electronic databases were searched. Although partial data also came from TCGA databases, some unpublished results may be missed. Last but not least, subgroup analysis, sensitivity analysis, and publication bias test were not available for some outcome measures due to the limited number of studies.
5. Conclusion
Elevated circulating miR-20a expression is significantly correlated with poor OS/DFS but the prognostic significance of tissue miR-20a has something to do with the trend of miR-20a change in different tumors. Highly expressed tissue-based miR-20a was associated with an unfavorable prognosis in cancers with upregulated miR-20a expression and favorable survival in malignancy with downregulated miR-20a expression. In a word, circulating and tissue-based miR-20a could serve as a reliable prognostic biomarker for patients with cancers. These results need additional validation for the limited number of studies.
Conflicts of Interest
All the authors declare that they have no any conflicts of interest.
Authors' Contributions
Qingyu Zhang and Qiwei Wang contributed to electronic searches, reference lists screening, study selection, data extraction, and assessment of risk of bias, analysis and interpretation of data, and drafting the manuscript. Wei S conceived the study, participated in study design, and revised the article. Fuqiang Gao and Lihua Liu contributed to study design and interpretation of data and drafting the manuscript. Liming Cheng and Zirong Li participated in study design and revised the article. All authors proofed and approved the submitted version of the article. Qingyu Zhang and Qiwei Wang have contributed equally to this work.
Supplementary Materials
The PRISMA Checklist for this systematic review and meta-analysis describing the page which every item located in.
References
- 1.Bhaskaran M., Mohan M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Veterinary Pathology. 2014;51(4):759–774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Calin G. A., Sevignani C., Dumitru C. D., et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Acadamy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moridikia A., Mirzaei H., Sahebkar A., Salimian J. MicroRNAs: Potential candidates for diagnosis and treatment of colorectal cancer. Journal of Cellular Physiology. 2018;233(2):901–913. doi: 10.1002/jcp.25801. [DOI] [PubMed] [Google Scholar]
- 5.Bahrami A., Aledavood A., Anvari K., et al. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. Journal of Cellular Physiology. 2018;233(2):774–786. doi: 10.1002/jcp.25813. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Z., Yin J., Fu W., et al. miRNA 17 Family Regulates Cisplatin-Resistant and Metastasis by Targeting TGFbetaR2 in NSCLC. PLoS ONE. 2014;9(4):p. e94639. doi: 10.1371/journal.pone.0094639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty C., Das S. Profiling cell-free and circulating miRNA: a clinical diagnostic tool for different cancers. Tumor Biology. 2016;37(5):5705–5714. doi: 10.1007/s13277-016-4907-3. [DOI] [PubMed] [Google Scholar]
- 8.Yau T. O., Wu C. W., Tang C., et al. microRNA-20a in human faeces as a non-invasive biomarker for colorectal cancer. Oncotarget . 2016;7(2) doi: 10.18632/oncotarget.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H. M., Castillo L., Mahon K. L. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. British Journal of Cancer. 2014;110(10):2462–2471. doi: 10.1038/bjc.2014.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanfiorenzo C., Ilie M. I., Belaid A., et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0054596.e54596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang R., Fu Y., Zeng Y., et al. Serum miR-20a is a promising biomarker for gastric cancer. Biomedical Reports. 2017;6(4):429–434. doi: 10.3892/br.2017.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng X., Xiang J., Wu M., et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046367.e46367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu W. Circulating miR-17-5p and miR-20a: Molecular markers for gastric cancer. Molecular Medicine Reports. 2012 doi: 10.3892/mmr.2012.828. [DOI] [PubMed] [Google Scholar]
- 14.Cheng D., Zhao S., Tang H., et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget . 2016;7(29):45199–45213. doi: 10.18632/oncotarget.9900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Zhang L., Xiang P., Han X., Wu L., Li X., Xiong Z. Decreased expression of microRNA-20a promotes tumor progression and predicts poor prognosis of cutaneous squamous cell carcinoma. International Journal of Clinical and Experimental Pathology. 2015;8(9):11446–11451. [PMC free article] [PubMed] [Google Scholar]
- 16.Battistella M., Romero M., Castro-Vega L.-J., et al. The high expression of the microRNA 17-92 cluster and its paralogs, and the downregulation of the target gene PTEN, is associated with primary cutaneous B-cell lymphoma progression. Journal of Investigative Dermatology. 2015;135(6):1659–1667. doi: 10.1038/jid.2015.27. [DOI] [PubMed] [Google Scholar]
- 17.Mitani Y., Roberts D. B., Fatani H., et al. MicroRNA Profiling of Salivary Adenoid Cystic Carcinoma: Association of miR-17-92 Upregulation with Poor Outcome. PLoS ONE. 2013;8(6):p. e66778. doi: 10.1371/journal.pone.0066778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C.-C., Yang Y.-J., Li Y.-J., et al. MicroRNA-17/20a functions to inhibit cell migration and can be used a prognostic marker in oral squamous cell carcinoma. Oral Oncology. 2013;49(9):923–931. doi: 10.1016/j.oraloncology.2013.03.430. [DOI] [PubMed] [Google Scholar]
- 19.Zhang K., Zhang L., Zhang M., et al. Prognostic value of high-expression of miR-17-92 cluster in various tumors: evidence from a meta-analysis. Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-08349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F., Zhang F., Li X., et al. Prognostic role of miR-17-92 family in human cancers: evaluation of multiple prognostic outcomes. Oncotarget . 2017;8(40) doi: 10.18632/oncotarget.19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberati A., Altman D. G., Tetzlaff J., et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. British Medical Journal. 2009;339 doi: 10.1136/bmj.b2700.b2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tierney J. F., Stewart L. A., Ghersi D., Burdett S., Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8, article 16 doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X., Zhu S., Tao Z., Ye S. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Medicine. 2018;7(1):21–31. doi: 10.1002/cam4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khare D., Goldschmidt N., Bardugo A., Gur-Wahnon D., Ben-Dov I. Z., Avni B. Plasma microRNA profiling: Exploring better biomarkers for lymphoma surveillance. PLoS ONE. 2017;12(11) doi: 10.1371/journal.pone.0187722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H., Shen J., Hodges T. R., Song R., Fuller G. N., Heimberger A. B. Serum microRNA profiling in patients with glioblastoma: A survival analysis. Molecular Cancer. 2017;16(1, article no. 59) doi: 10.1186/s12943-017-0628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Shi K., Wang Y., et al. Clinical value of integrated-signature miRNAs in colorectal cancer: miRNA expression profiling analysis and experimental validation. Oncotarget . 2015;6(35):37544–37556. doi: 10.18632/oncotarget.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y., Liu C., Yang J., et al. MiR-20a triggers metastasis of gallbladder carcinoma. Journal of Hepatology. 2013;59(3):518–527. doi: 10.1016/j.jhep.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J. X., Song W., Chen Z. H. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. The Lancet Oncology. Dec 2013;14(13):1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 29.Valladares-Ayerbes M., Blanco M., Haz M., et al. Prognostic impact of disseminated tumor cells and microRNA-17-92 cluster deregulation in gastrointestinal cancer. International Journal of Oncology. 2011;39(5):1253–1264. doi: 10.3892/ijo.2011.1112. [DOI] [PubMed] [Google Scholar]
- 30.Reichek J. L., Duan F., Smith L. M., et al. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: a report from the children's oncology group. Clinical Cancer Research. 2011;17(6):1463–1473. doi: 10.1158/1078-0432.CCR-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schetter A. J., Leung S. Y., Sohn J. J., et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. Journal of the American Medical Association. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Si W., Shen J., Du C., et al. A miR-20a/MAPK1/c-Myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death & Differentiation. 2017;25(2):406–420. doi: 10.1038/cdd.2017.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan M.-Q., Huang C.-B., Gu Y., Xiao Y., Sheng J.-X., Zhong L. Decrease expression of microRNA-20a promotes cancer cell proliferation and predicts poor survival of hepatocellular carcinoma. Journal of Experimental & Clinical Cancer Research. 2013;32(1, article no. 21) doi: 10.1186/1756-9966-32-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchini S., Cavalieri D., Fruscio R., et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. The Lancet Oncology. 2011;12(3):273–285. doi: 10.1016/S1470-2045(11)70012-2. [DOI] [PubMed] [Google Scholar]
- 35.Das V., Kalita J., Pal M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomedicine & Pharmacotherapy. 2017;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 36.Rupaimoole R., Calin G. A., Lopez-Berestein G., Sood A. K. MiRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discovery. 2016;6(3):235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S., Gregory R. I. MicroRNA biogenesis pathways in cancer. Nature Reviews Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha M., Kim V. N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 39.Rizzo M., Mariani L., Pitto L., Rainaldi G., Simili M. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. Journal of Cellular and Molecular Medicine. 2010;14(11):2633–2640. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chim S. S. C., Shing T. K. F., Hung E. C. W., et al. Detection and characterization of placental microRNAs in maternal plasma. Clinical Chemistry. 2008;54(3):482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 41.Li T., Kang G., Wang T., Huang H. Tumor angiogenesis and anti-angiogenic gene therapy for cancer. Oncology Letters. 2018;16(1):687–702. doi: 10.3892/ol.2018.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pritchard C. C., Kroh E., Wood B., et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prevention Research. 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams Z., Ben-Dov I. Z., Elias R., et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proceedings of the National Acadamy of Sciences of the United States of America. 2013;110(11):4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzenbach H., Hoon D. S. B., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Reviews Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 45.Vickers K. C., Remaley A. T. Lipid-based carriers of microRNAs and intercellular communication. Current Opinion in Lipidology. 2012;23(2):91–97. doi: 10.1097/MOL.0b013e328350a425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang G., Chen X., Cai Y., Wang X., Xing C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncology Reports. 2017;37(1):571–578. doi: 10.3892/or.2016.5278. [DOI] [PubMed] [Google Scholar]
- 47.du Y., Zhu M., Zhou X., et al. miR-20a enhances cisplatin resistance of human gastric cancer cell line by targeting NFKBIB. Tumor Biology. 2015 doi: 10.1007/s13277-015-3921-1. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchida A., Ohno S., Wu W., et al. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Science. 2011;102(12):2264–2271. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 49.Alunni-Fabbroni M., Majunke L., Trapp E. K., et al. Whole blood microRNAs as potential biomarkers in post-operative early breast cancer patients. BMC Cancer. 2018;18(1) doi: 10.1186/s12885-018-4020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Advanced Drug Delivery Reviews. 2015;81:75–93. doi: 10.1016/j.addr.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Fonsato V., Collino F., Herrera M. B., et al. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells. 2012;30(9):1985–1998. doi: 10.1002/stem.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olive V., Li Q., He L. Immunological Reviews. 2013;253(1):158–166. doi: 10.1111/imr.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong L., Lai M., Chen M., et al. The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Research. 2010;70(21):8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The PRISMA Checklist for this systematic review and meta-analysis describing the page which every item located in.