Abstract
Background
Diagnostic screening of premalignant esophageal lesions is hampered by the absence of biomarkers indicative of metaplastic and/or malignant transformation. The aim of this exploratory study was to investigate the potential use of miRNAs as biomarkers capable of identifying patients with (pre)malignant lesions: Barrett’s esophagus (BE) metaplasia, high-grade dysplasia (HGD) or esophageal adenocarcinoma (EAC).
Methods
A total of 69 patients were included in the study. Six serum samples from each of four study groups, i.e., patients with normal squamous epithelium (SE), BE, HGD and EAC, were profiled using the Nanostring miRNA analysis platform. Differential miRNA expression patterns then were validated in 69 patient samples using qRT-PCR.
Results
miRNA expression profiling revealed seven miRNAs with a 2-fold change in expression level. Validation by qRT-PCR confirmed that serum miR-320e levels were significantly decreased in the BE group compared to the SE (P≤0.001, AUC 0.790) and HGD groups (P≤0.005, AUC 0.786). Serum miR-199a-3p levels were significantly decreased in the BE group compared to the SE group (P≤0.001), area under the curve (AUC) of 0.813.
Conclusions
The results of this study suggest that decreased serum miRNA levels of miR-199a-3p and miR-320e could help to identify patients with BE and HGD.
Keywords: Screening, biomarkers, cancer
Introduction
Esophageal adenocarcinoma (EAC) is a deadly disease with a rising incidence. While patients with early EAC lesions have a 5-year survival of 78%, this drops to around 47% in patients with locally advanced disease (1,2). Early detection significantly improves survival outcome (3,4), but the majority of EAC cases are diagnosed at an advanced stage. EAC is thought to develop through a metaplasia-dysplasia-carcinoma sequence. The metaplastic lesion is known as Barrett’s esophagus (BE). Patients with BE are advised to undergo regular endoscopic surveillance to detect early malignant transformation. Persistent gastro-esophageal reflux disease (GERD) is the main risk factor for BE development. Currently, BE is diagnosed by histologic examination of endoscopic biopsies, but the low absolute incidence of BE in GERD patients and the invasive nature of the endoscopy preclude its use as a screening tool in patients at risk for BE development (5,6). A screening strategy that at least partially mitigates these drawbacks could contribute to an early detection of BE, allowing surveillance and/or early treatment and so decrease morbidity and mortality.
Currently, no blood-based biomarkers are available for BE, high-grade dysplasia (HGD) or EAC. miRNAs are a class of small non-coding RNAs that act as negative regulators of gene expression by inhibition of mRNA translation. miRNAs are highly tissue-specific and various studies indicate that alterations in miRNA expression have key roles in carcinogenesis (7,8). Moreover, miRNAs are stable in the circulation and alterations in circulating miRNA levels have been detected in various solid malignancies (9). Circulating miRNAs might thus be attractive blood-based biomarkers of BE and EAC. Several studies showed significant differences in tissue miRNA expression patterns between normal squamous epithelium (SE), BE and EAC (10-13). However, it is currently not known whether these differences are also reflected in the circulating miRNA profile. The aim of this study was to investigate whether circulating miRNAs could be used to differentiate between patients with normal SE and patients with (pre)malignant changes in the esophageal epithelium (BE, HGD and/or EAC).
Methods
Patients and samples
Patients scheduled to undergo an esophagogastroduodenoscopy were prospectively recruited from 2012 up to and including 2014 into one of four study groups: the control group consisting of patients without endoscopic esophageal abnormalities (SE group), a group consisting of patients with BE without dysplasia, a group consisting of patients with columnar HGD or early stage (≤ T1a) EAC (HGD group) and a group consisting of patients with advanced EAC (stage ≥ T1b, EAC group). For each participant, the diagnosis was confirmed based on endoscopic appearance, pathological biopsy analysis and imaging [endoscopic ultrasound (EUS) and/or PET/CT for the HGD and EAC groups]. Each participant was asked about smoking history. General exclusion criteria were a history of previous malignancy in the past 5 years, or known chronic inflammatory disease. The study protocol was approved by the Ethics Committee of the University Medical Center Groningen. Written informed consent was obtained from all participants. The number of samples included in the study was based on the amount of available material.
Blood collection
Serum samples were collected from each participant. Using venipuncture, approximately 9 mL blood was collected from each participant in a BD vacutainer serum tube (BD, Breda, The Netherlands). The blood was left to clot for one hour at room temperature and serum was isolated by centrifugation and stored at −80 °C.
RNA extraction
Serum samples were thawed and filtered through a 0.2 µM filter (Corning Life Sciences, Amsterdam, The Netherlands) to eliminate any residual platelets or cell debris. Total RNA was isolated from 200 µL of serum using the miRNeasy micro kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocol, with the addition of Phase Lock Gel Heavy gel tubes (5 Prime, Hilden, Germany). RNA was eluted in 10 µL of RNase-free water and stored at −80 °C until further analysis.
Nanostring nCounter miRNA assay
A high-throughput serum miRNA expression profile was generated from a subset of study participants. We selected serum samples of six patients of each study group, resulting in a total of 24 samples. Patients from the different study groups were matched for sex, age and comorbidity, as much as possible given the limited number of patient samples. The number of samples included in the Nanostring assay was based on the number of assays on one Nanostring expression assay (N=12). To perform a comprehensive assay of circulating miRNAs in serum we used the nCounter® miRNA Expression Assay version 2 which contains probes for 800 mature human miRNAs (Nanostring Technologies, Seattle, USA). An equal volume of RNA eluate was used as input for each sample (5 µL). The Nanostring assay was performed by Nanostring Technologies.
Data analysis
Data analysis was performed using GeneSpring GX version 12.5 software (Agilent Technologies, Santa Clara, USA). Raw Nanostring data were normalized by percentile shift normalization to the 95th percentile. The upper range of expression for negative control oligo’s on the Nanostring platform was 30 counts. Therefore, a detection cutoff was defined as at least four out of six samples in at least one of the four study groups having a count number >30. This cutoff yielded expression levels above the background for 62 human miRNAs.
qRT-PCR
miRNA-specific cDNA synthesis was performed using the TaqMan miRNA transcription kit (Life Technologies, Bleiswijk, The Netherlands) in a multiplexed reaction as described previously (14) using specific miRNA reverse transcription primers as provided in the TaqMan microRNA assays: miR-16-5p (ID 000391), miR-144-5p (ID 002148), miR-150-5p (ID 000473), miR-199a-3p (ID 002304), miR-320e (ID 243005_mat), miR-451a (ID 001141) and RNU6B (ID 001093), all from Life Technologies. qRT-PCR was performed using the TaqMan Universal Master Mix II without UNG (Life Technologies) and PCR primers from the above mentioned TaqMan microRNA assays. All qPCR reactions were run in triplicates on the ABI7900HT thermo cycler (Life Technologies). Cycle threshold (Ct) values for all miRNAs were quantified using the Sequence Detection software (version 2.3, Life Technologies). Serum miRNA expression levels were normalized according to the geometric mean of the Ct values of the SE group.
Statistical analysis
Differences in patient characteristics were analyzed using ANOVA with Scheffé’s method. Differences in serum miRNA expression between the patient groups did not follow a normal distribution (evaluated using the D’Agostino-Pearson omnibus test) and were therefore examined using the Kruskal-Wallis test with Dunn’s multiple testing correction. For statistical analysis and receiver operating characteristic (ROC) curve construction of the qPCR data the Prism 5.0 statistical package was used (GraphPad Software, San Diego, USA). For all tests, a P value of <0.05 was considered significant.
Results
Patient groups
Patient characteristics are described in Table 1. The extended patient characteristics are described in Table S1. The only significant difference was observed in the occurrence of diabetes between the SE and HGD and between BE and HGD groups (P=0.036 and P=0.013 respectively).
Table 1. Patient characteristics for each study group.
| Study group | SE (N=19) | BE (N=27) | HGD (N=17) | EAC (N=6) | P value |
|---|---|---|---|---|---|
| Gender (male), n [%] | 8 [42] | 19 [70] | 13 [76] | 5 [83] | 0.067 |
| Mean age [range] | 54.3 [23.0–70.0] | 58.7 [25.0–75.0] | 65.1 [42.0–82.0] | 62.5 [46.0–73.0] | 0.086 |
| Smoking, n [%] | 4 [21] | 5 [19] | 5 [29] | 4 [67] | 0.790 |
| Cardiovascular disease, n [%] | 3 [16] | 6 [22] | 4 [24] | 2 [33] | 0.834 |
| Autoimmune disease, n [%] | 1 [5] | 1 [4] | 1 [6] | 1 [17] | 0.691 |
| Diabetes, n [%] | 1 [5] | 1 [4] | 6 [35] | 0 [0] | 0.004† |
†, after multiple testing correction SE study group vs. HGD study group: P=0.036, BE study group vs. HGD study group: P=0.013. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia; EAC, esophageal adenocarcinoma.
Table S1. Extended patient characteristics.
| Age (years) | Study group | Gender | Smoking | Cardiovascular disease | Autoimmune disease | Diabetes |
|---|---|---|---|---|---|---|
| 69* | SE | Female | Yes | No | No | No |
| 58* | SE | Male | No | No | No | No |
| 59* | SE | Male | No | No | No | No |
| 52* | SE | Male | Yes | Yes | No | No |
| 61* | SE | Male | No | No | No | No |
| 24* | SE | Male | No | No | No | No |
| 70 | SE | Male | No | Yes | No | Yes |
| 53 | SE | Female | Yes | No | No | No |
| 53 | SE | Female | No | No | No | No |
| 69 | SE | Male | No | No | No | No |
| 23 | SE | Female | No | No | No | No |
| 65 | SE | Female | No | No | No | No |
| 55 | SE | Female | No | No | No | No |
| 42 | SE | Female | No | No | No | No |
| 47 | SE | Male | No | No | No | No |
| 64 | SE | Female | No | Yes | No | No |
| 61 | SE | Female | No | No | No | No |
| 47 | SE | Female | Yes | No | No | No |
| 59 | SE | Female | No | No | Yes | No |
| 67* | BE | Male | No | No | No | No |
| 68* | BE | Male | No | No | No | No |
| 67* | BE | Male | No | Yes | No | No |
| 48* | BE | Male | Yes | No | No | No |
| 74* | BE | Male | No | Yes | No | No |
| 69* | BE | Female | Yes | Yes | No | No |
| 57 | BE | Female | No | No | No | Yes |
| 63 | BE | Female | Yes | No | No | No |
| 67 | BE | Female | No | No | No | No |
| 50 | BE | Female | No | No | No | No |
| 48 | BE | Male | Yes | No | No | No |
| 59 | BE | Male | No | No | No | No |
| 42 | BE | Female | No | No | No | No |
| 61 | BE | Male | No | No | No | No |
| 59 | BE | Male | No | No | No | No |
| 70 | BE | Male | No | No | No | No |
| 25 | BE | Male | No | No | No | No |
| 68 | BE | Female | No | No | No | No |
| 25 | BE | Male | No | No | No | No |
| 65 | BE | Male | No | No | No | No |
| 59 | BE | Female | No | No | No | No |
| 60 | BE | Male | No | Yes | No | No |
| 67 | BE | Male | No | Yes | No | No |
| 75 | BE | Male | No | Yes | No | No |
| 55 | BE | Male | No | No | No | No |
| 73 | BE | Male | No | No | Yes | No |
| 44 | BE | Male | Yes | No | No | No |
| 68* | HGD | Male | Yes | No | No | Yes |
| 53* | HGD | Female | Yes | No | No | No |
| 70* | HGD | Male | No | No | No | No |
| 68* | HGD | Male | No | No | No | No |
| 51* | HGD | Male | Yes | No | No | No |
| 69* | HGD | Male | No | No | No | No |
| 74 | HGD | Female | No | Yes | No | Yes |
| 62 | HGD | Female | No | No | No | Yes |
| 62 | HGD | Male | Yes | No | No | Yes |
| 67 | HGD | Male | No | Yes | No | Yes |
| 65 | HGD | Male | No | No | No | Yes |
| 71 | HGD | Male | No | No | No | No |
| 58 | HGD | Male | Yes | Yes | Yes | No |
| 42 | HGD | Female | No | No | No | No |
| 82 | HGD | Male | No | No | No | No |
| 69 | HGD | Male | No | Yes | No | No |
| 75 | HGD | Male | No | No | No | No |
| 61* | EAC | Male | No | No | No | No |
| 65* | EAC | Female | Yes | No | No | No |
| 73* | EAC | Male | No | No | Yes | No |
| 66* | EAC | Male | No | No | No | No |
| 64* | EAC | Male | Yes | Yes | No | No |
| 46* | EAC | Male | No | Yes | No | No |
*, samples used for the Nanostring array. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia.
Identifying differences in circulating miRNA levels
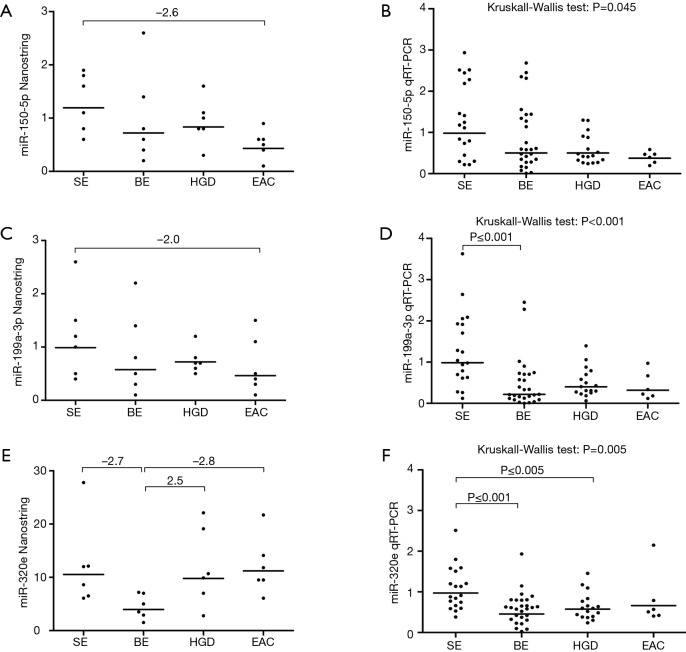
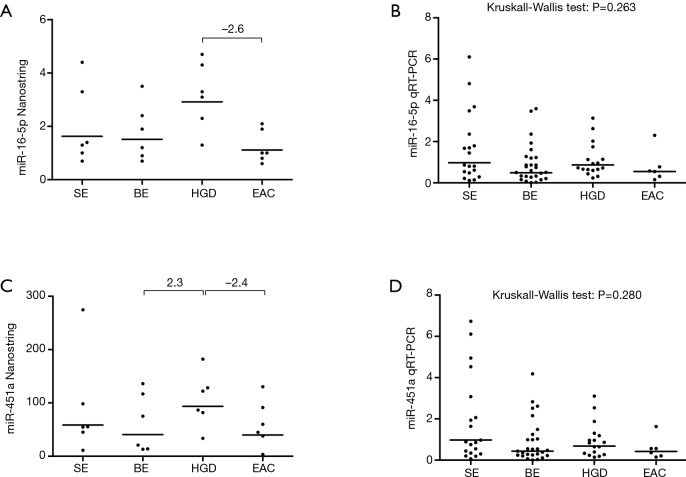
Using the Nanostring platform we identified 62 miRNAs with expression levels above the background (Figure S1). Due to the limited sample size we focused on miRNAs with a fold change >2 between the four study groups, which revealed seven differentially expressed miRNAs (Table 2). No Taqman assay was available for hsa-miR-4516. The remaining six miRNAs were used for validation by qRT-PCR in an extended patient group consisting of the 24 patients included in the Nanostring array and an additional 45 patients (13 SE, 21 BE and 11 HGD serum samples). Three miRNAs, i.e., miR-150-5p, miR-199a-3p and miR-320e, showed significant differences between the study groups based on the Kruskal-Wallis test (Figure 1). The expression of miR-199a-3p was significantly lower in the BE group compared to the SE group (P≤0.001) and the expression of miR-320e was significantly lower in the BE and HGD groups compared to the SE group (P≤0.001 and P≤0.005 respectively, Figure 1). The post-hoc test did not reveal significant differences in miR-150-5p levels between the study groups. No significant differences were observed for the miRNA levels between the four study groups for miR-16-5p and miR-451a (Figure S2). miR-144-3p was excluded from further analysis due to insufficient quality of the qRT-PCR assay data.
Figure S1.
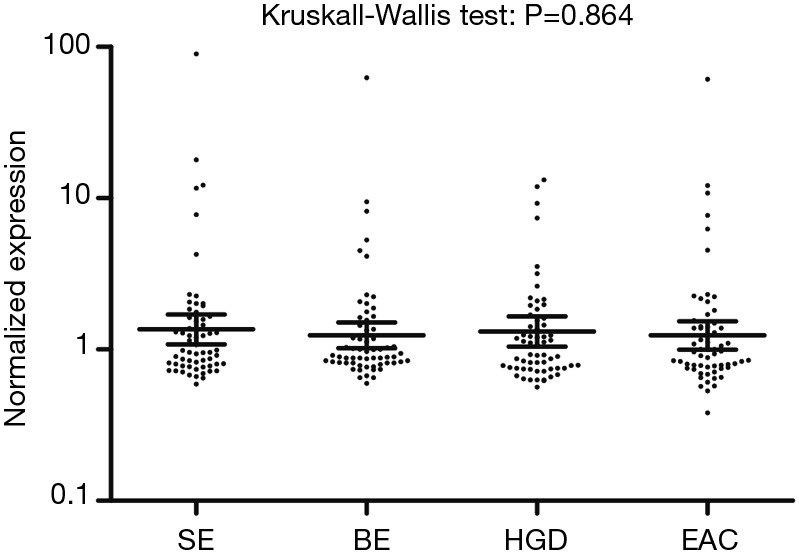
Normalized expression values of 62 serum miRNAs with expression above background in the Nanostring platform. Cutoff of expression was defined as 4 out of 6 of samples in at least one study group having >30 counts. Horizontal lines indicate the geometric mean and the error bars denote the 95% confidence interval. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia; EAC, esophageal adenocarcinoma.
Table 2. miRNAs with fold change >2 on the Nanostring assay.
| Study group | SE vs. BE | SE vs. HGD | SE vs. EAC | BE vs. HGD | BE vs. EAC | HGD vs. EAC |
|---|---|---|---|---|---|---|
| miR-16-5p | – | – | – | – | – | ↓ |
| miR-144-3p | – | – | ↓ | – | ↓ | ↓ |
| miR-150-5p | – | – | ↓ | – | – | – |
| miR-199a-3p† | – | – | ↓ | – | – | – |
| miR-320e | ↓ | – | – | ↑ | ↑ | – |
| miR-451a | – | – | – | ↑ | – | ↓ |
| miR-4516 | – | – | – | ↑ | – | – |
†, miR-199a-3p has the same sequence as miR-199b-3p. Arrows indicate fold change >2 based on the Nanostring assay. MiR-144-3p was excluded due to technical reasons, and no TaqMan assay was available for miR-4516. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia; EAC, esophageal adenocarcinoma.
Figure 1.
Serum expression of miR-150-5p, miR-199a-3p and miR-320e and validation in the expanded patient cohort. Results of the Nanostring analysis (A,C,E) in the original cohort of 24 serum samples (6 for each study group) and the qRT-PCR validation (B,D,F) in the expanded patient cohort including the same 24 serum samples and 45 additional samples. Shown are the three miRNAs that were significantly differentially expressed in the qRT-PCR validation experiments (Table 2), i.e., miR-150-5p (A,B), miR-199a-3p (C,D) and miR-320e (E,F). No significant differences in miR-150-5p expression were identified between the study groups in the post-hoc test (B). Compared to the SE group, serum levels of miR-199a-3p were significantly lower in the BE group (D) and serum levels of miR-320e were significantly lower in the BE and HGD groups (F). Horizontal bars denote geometric mean. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia; EAC, esophageal adenocarcinoma.
Figure S2.
Nanostring and full cohort graphs for miR-16-5p and miR-451a. Y-axis denotes normalized levels on the Nanostring platform (A,C) or RT-qPCR (B,D). Numbers indicate fold changes >2 observed on the Nanostring platform. Horizontal bars denote geometric mean. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia; EAC, esophageal adenocarcinoma.
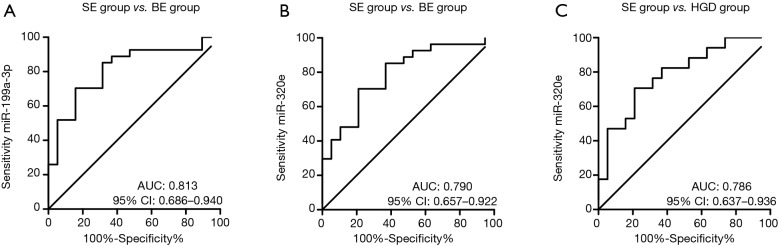
Potential use of the significantly decreased serum-derived miRNAs as disease biomarkers
Based on the significant differences in expression levels of miR-199a-3p and miR-320e between the SE and the BE/HGD groups, we investigated to what extend the expression of these miRNAs could be used to identify patients with (pre)malignant changes in the esophagus (Figure 2). Serum miR-199a-3p levels could discriminate between the SE and BE groups with an area under the curve (AUC) of 0.813 (optimal sensitivity of 85.2% and specificity of 68.4% at relative expression <0.766). Similarly, serum miR-320e levels could discriminate between the SE and BE groups with an AUC of 0.790 (optimal sensitivity of 85.2% and specificity of 63.2% at relative expression <0.844) and between the SE and HGD groups with an AUC of 0.786 (optimal sensitivity of 82.3% and specificity of 62.2% at relative expression <0.838).
Figure 2.
Receiver operating curves (ROC) for miR-199a-3p and miR-320e. (A) Serum miR-199a-3p levels could discriminate between the SE (N=19) and BE (N=27) groups with an area under the curve (AUC) of 0.813 (optimal sensitivity of 85.2% and specificity of 68.4% at relative expression <0.766); (B) serum miR-320e levels could discriminate between SE and BE with an AUC of 0.790 (optimal sensitivity of 85.2% and specificity of 63.2% at relative expression <0.844) and (C) between the SE and HGD (N=17) groups with an AUC of 0.786 (optimal sensitivity of 82.3% and specificity of 62.2% at relative expression <0.838). CI, confidence interval. SE, squamous epithelium; BE, Barrett’s metaplasia; HGD, high-grade dysplasia.
Discussion
The lack of easily accessible diagnostic biomarkers of BE and EAC is a significant contributor to the poor overall survival of EAC patients. miRNAs are attractive as potential biomarkers, but there is very limited data regarding circulating miRNAs that could differentiate between patients with normal SE and patients with (pre)malignant changes in the esophageal epithelium. Here, we studied the levels of serum miRNAs in patients at all stages of the metaplasia-dysplasia-carcinoma sequence using the Nanostring platform. We identified seven miRNAs with a fold change >2 between any of the different study groups. Using qRT-PCR in an extended group of patients we were able to validate decreased levels of circulating miR-199a-3p and miR-320e in patients with BE (both miRNAs) and HGD (miR-320e only) compared to the SE group.
miR-199a-3p is a known tumor suppressor miRNA. In vitro studies showed that miR-199a-3p reduces cell proliferation and migration through a direct repression of mTOR and c-MET (15-18) and miR-199a-3p tissue levels were decreased in endometrial, thyroid, prostate and hepatocellular carcinoma (15-19). In a previous study miR-199a-3p expression levels were significantly increased in EAC tissue samples compared to SE tissue samples, but not in BE or HGD tissue samples (20). miR-320e was previously found to be increased in EAC tissues compared to BE tissues (21) and high miR-320e tissue levels were associated with an adverse prognosis in colon cancer (22).
It is unclear why miR-199a-3p and miR-320e levels were decreased in the circulation while other studies report an increased expression in malignant tissue. However, similar findings have been observed previously for other miRNAs (22). Although a mechanistic explanation for increased tissue miRNA levels and a concurrent decrease in the circulating miRNA levels is missing, it seems unlikely that changes in the generally very small affected tissue areas (such as in BE and EAC) could lead to reduced circulating miRNA levels. Furthermore, the origin of serum miRNAs remains poorly understood. A study by Pritchard et al. (23) showed that the majority of previously identified cancer-associated circulating miRNAs, were highly expressed in hematopoietic cells and their levels correlated with blood count levels. This suggested the possibility that changes in blood cell composition and/or integrity (e.g., hemolysis) can affect circulating miRNA levels. Of note, these miRNAs did not include miR-199a-3p and miR-320e. It is also possible that changes in circulating miRNAs in cancer patients are not only influenced by the tumor itself, but also by systemic processes in the body, such as an inflammatory response or altered metabolism.
The lack of increased levels of any miRNAs during the metaplasia-dysplasia-carcinoma sequence of EAC is surprising, since increased levels of circulating miRNAs have been reported in other solid malignancies (7,9). A single previous study reported increased serum miRNA levels in BE (miR-194-5p and miR-451a) and EAC (miR-451a) (22). In our study, miR-194-5p was not differentially expressed, while miR-451a showed a fold change >2 between the BE and HGD and the HGD and EAC groups. However, no significant differences in miR-451a expression levels were observed in the extended cohort (Figure S2). The discrepancy between these findings could reflect differences in specificity of the assays or be due to the limited sample size in our study.
Of the 800 miRNA probes present in the Nanostring system, 62 showed expression levels above the cutoff. This relatively low number can be explained by the low yield of total RNA isolated from serum, the limited number of samples used in the discovery phase and/or the lack of an amplification step. We consider the lack of an amplification step an advantage because it eliminates the risk of selective amplification. However, the combination of a low RNA input in combination with the lack of an amplification step might have resulted in loss of potentially relevant miRNAs of which the expression levels did not reach the cutoff used in the Nanostring assay.
Finally, PCR validation results were not always consistent with the initial findings from the Nanostring assay; while the trend in expression was similar for miR-150-5p, miR-199a-3p and miR-320e, the fold changes were not statistically significant in the validation cohort. These inconsistencies can possibly be explained by heterogeneity of the patient samples and the limited number of samples used on the Nanostring assay.
Despite the above-mentioned limitations, we feel that the combination of serum samples from all stages of EAC carcinogenesis provides a thorough examination of the miR-199a-3p and miR-320e expression during the metaplasia-dysplasia sequence of EAC carcinogenesis and shown that decreased serum miRNA levels of miR-199a-3p and miR-320e could help to identify patients with BE and HGD, thereby increasing screening efficiency.
Acknowledgements
This work was supported by the van de Meer-Boerema foundation and the J.K. de Cock foundation.
Ethical Statement: The study had been approved by Medische Ethische Toetsingscommissie (METc) from the University Medical Center Groningen, Groningen, The Netherlands (No. 36064.042.11). All participants signed an informed consent form.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.van Hagen P, Hulshof MCCM, van Lanschot JJB, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 2012;366:2074-84. 10.1056/NEJMoa1112088 [DOI] [PubMed] [Google Scholar]
- 2.Qumseya BJ, Panossian AM, Rizk C, et al. Survival in esophageal high-grade dysplasia/adenocarcinoma post endoscopic resection. Dig Liver Dis 2013;45:1028-33. 10.1016/j.dld.2013.06.009 [DOI] [PubMed] [Google Scholar]
- 3.Fountoulakis A, Zafirellis KD, Dolan K, et al. Effect of surveillance of Barrett’s oesophagus on the clinical outcome of oesophageal cancer. Br J Surg 2004;91:997-1003. 10.1002/bjs.4591 [DOI] [PubMed] [Google Scholar]
- 4.Corley DA, Levin TR, Habel LA, et al. Surveillance and survival in Barrett’s adenocarcinomas: a population-based study. Gastroenterology 2002;122:633-40. 10.1053/gast.2002.31879 [DOI] [PubMed] [Google Scholar]
- 5.Shaheen N, Ransohoff DF. Gastroesophageal reflux, barrett esophagus, and esophageal cancer: scientific review. JAMA 2002;287:1972-81. 10.1001/jama.287.15.1972 [DOI] [PubMed] [Google Scholar]
- 6.Spechler SJ, Souza RF. Barrett’s esophagus. N Engl J Med 2014;371:836-45. 10.1056/NEJMra1314704 [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704-14. 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lujambio A, Lowe SW. The microcosmos of cancer. Nature 2012;482:347-55. 10.1038/nature10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzenbach H, Nishida N, Calin GA, et al. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol 2014;11:145-56. 10.1038/nrclinonc.2014.5 [DOI] [PubMed] [Google Scholar]
- 10.Wijnhoven BP, Hussey DJ, Watson DI, et al. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg 2010;97:853-61. 10.1002/bjs.7000 [DOI] [PubMed] [Google Scholar]
- 11.Revilla-Nuin B, Parrilla P, Lozano JJ, et al. Predictive value of MicroRNAs in the progression of barrett esophagus to adenocarcinoma in a long-term follow-up study. Ann Surg 2013;257:886-93. 10.1097/SLA.0b013e31826ddba6 [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Gu J, Wang KK, et al. MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res 2009;15:5744-52. 10.1158/1078-0432.CCR-09-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg 2008;135:255-60; discussion 260. 10.1016/j.jtcvs.2007.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kluiver J, Slezak-Prochazka I, van den Berg A. Studying microRNAs in lymphoma. Methods Mol Biol 2013;971:265-76. 10.1007/978-1-62703-269-8_15 [DOI] [PubMed] [Google Scholar]
- 15.Minna E, Romeo P, De Cecco L, et al. miR-199a-3p displays tumor suppressor functions in papillary thyroid carcinoma. Oncotarget 2014;5:2513-28. 10.18632/oncotarget.1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D, Huang H, He C, et al. MicroRNA-199a-3p regulates endometrial cancer cell proliferation by targeting mammalian target of rapamycin (mTOR). Int J Gynecol Cancer 2013;23:1191-7. 10.1097/IGC.0b013e31829ea779 [DOI] [PubMed] [Google Scholar]
- 17.Fornari F, Milazzo M, Chieco P, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res 2010;70:5184-93. 10.1158/0008-5472.CAN-10-0145 [DOI] [PubMed] [Google Scholar]
- 18.Qu KZ, Zhang K, Li H, et al. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 2011;45:355-60. 10.1097/MCG.0b013e3181f18ac2 [DOI] [PubMed] [Google Scholar]
- 19.Qu Y, Huang X, Li Z, et al. miR-199a-3p inhibits aurora kinase A and attenuates prostate cancer growth: new avenue for prostate cancer treatment. Am J Pathol 2014;184:1541-9. 10.1016/j.ajpath.2014.01.017 [DOI] [PubMed] [Google Scholar]
- 20.Streppel MM, Pai S, Campbell NR, et al. MicroRNA 223 is upregulated in the multistep progression of Barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res 2013;19:4067-78. 10.1158/1078-0432.CCR-13-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drahos J, Schwameis K, Orzolek LD, et al. MicroRNA Profiles of Barrett's Esophagus and Esophageal Adenocarcinoma: Differences in Glandular Non-native Epithelium. Cancer Epidemiol Biomarkers Prev 2016;25:429-37. 10.1158/1055-9965.EPI-15-0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bus P, Kestens C, Ten Kate FJ, et al. Profiling of circulating microRNAs in patients with Barrett's esophagus and esophageal adenocarcinoma. J Gastroenterol 2016;51:560-70. 10.1007/s00535-015-1133-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5:492-7. 10.1158/1940-6207.CAPR-11-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]