Abstract
Introduction
We developed and validated a clinically applicable decision tree for the use of cerebrospinal fluid biomarkers in the diagnosis of Alzheimer's disease (AD).
Methods
Subjects with probable AD (n = 1004) and controls (n = 442) were included. A decision tree was modeled using Classification And Regression Tree analysis in a training cohort (AD n = 221; controls n = 221) and validated in an independent cohort (AD n = 783; controls n = 221). Diagnostic performance was compared to previously defined cutoffs (amyloid β 1-42 < 813 pg/ml; tau>375 pg/ml).
Results
Two cerebrospinal fluid AD biomarker profiles were revealed: the “classical” AD biomarker profile (amyloid β 1-42: 647-803 pg/ml; tau >374 pg/ml) and an “atypical” AD biomarker profile with strongly decreased amyloid β 1-42 (<647 pg/ml) and normal tau concentrations (<374 pg/ml). Compared to previous cutoffs, the decision tree performed better on diagnostic accuracy (86% [84-88] vs 80% [78-83]).
Discussion
Two cerebrospinal fluid AD biomarker profiles were identified and incorporated in a readily applicable decision tree, which improved diagnostic accuracy.
Keywords: Alzheimer's disease, CSF, Amyloid β 1-42, Tau, Clinical implementation, Decision tree, CART, Cutoff, Cerebrospinal fluid
Highlights
-
•
Classification And Regression Tree analysis revealed two Alzheimer's disease (AD) biomarker profiles based on cerebrospinal fluid amyloid beta 1-42 and tau.
-
•
A “classical AD-like” profile and an atypical AD profile were identified.
-
•
Combining both AD biomarker profiles led to a higher diagnostic accuracy.
-
•
Both biomarker profiles are incorporated in a readily applicable decision tree.
1. Introduction
Cerebrospinal fluid (CSF) biomarkers amyloid β 1-42 (Aβ42), total tau (tau), and phosphorylated tau (Ptau) can aid in the diagnosis of Alzheimer's disease (AD) [1]. According to the recent National Institute on Aging and Alzheimer's Association (NIA-AA) criteria and International Working Group (IWG)-2 criteria, an abnormal CSF Aβ42 in combination with an abnormal CSF tau or Ptau is considered to be an “AD-like” pathological profile [2], [3]. In daily practice, however, not all AD patients have a clear abnormal pattern of all three biomarkers. As previously shown, 8% of clinically diagnosed AD patients can have abnormal CSF Aβ42 alone without tau pathology [4]. Similarly, another study reported five AD subgroups with different CSF biomarker profiles with regard to Aβ42, tau, and ubiquitin [5]. The diversity of possible CSF biomarker profiles that can accompany AD dementia can be confusing for clinicians, contributing to the challenge of implementing CSF markers in daily clinical practice. Another complication is the requirement of current criteria for AD dementia to have both amyloid and tau pathologies present, with equal weight ascribed to each biomarker [2], [3].
The aim of the present study was to determine whether a statistical decision tree approach may aid in interpreting combinations of CSF biomarkers. An advantage of this approach is that it can identify and visually represent multiple CSF biomarker combinations in a decision tree, which can be readily used by the clinician, thereby facilitating decision-making in clinical practice. We further assessed the diagnostic and predictive performance of the decision tree.
2. Method
2.1. Subjects
We selected 1809 subjects with a diagnosis of AD (n = 1004), mild cognitive impairment (MCI) (n = 363), or subjective cognitive decline (SCD; n = 442) from the Amsterdam Dementia Cohort, who visited our outpatient clinic for diagnosis in the period from October 2000 until July 2015, and of whom CSF biomarker values were available. All subjects underwent standardized dementia screening at the baseline, including physical and neurological examinations, cognitive screening, an electroencephalogram, magnetic resonance imaging, and laboratory tests. Cognitive screening included at least a Mini-Mental State Examination, and often a comprehensive neuropsychological test battery (previously described elsewhere) [6], [7]. Diagnoses were made by consensus according to internationally established criteria in a multidisciplinary team without knowledge of CSF results [8]. All probable AD dementia patients met the core clinical NIA-AA criteria [9]. MCI was determined according to the criteria by Petersen et al. [10] and NIA-AA core clinical criteria [11]. All subjects with MCI received follow-up consultations with repeated medical and neuropsychological testing. The average follow-up period was (mean ± SD) 2.5 ± 1.6 years during which 143 MCI subjects progressed to AD-type dementia and 220 MCI subjects did not. MCI subjects who had progressed to a non-AD type dementia, that is, vascular dementia, Lewy body disease, possible AD, “other dementias” (e.g. due to Parkinson's disease) or were given a postponed diagnosis, were labeled as not having AD dementia at the follow-up. All controls consisted of SCD subjects who were labeled as such when results of all clinical examinations and test results were normal, that is, when the criteria for MCI or AD were not fulfilled, and there was no psychiatric diagnosis. Level of education was classified according to the Verhage system, ranging from 1 to 7 points (low to high education level) [12].
2.2. Standard protocol approvals, registrations, and patient consents
All subjects gave written informed consent for the use of clinical data for research purposes, and the use of clinical data was approved by the local ethical review board [8].
2.3. CSF biochemical analysis
CSF was obtained by lumbar puncture, using a 25-gauge needle, and collected in 10 mL polypropylene tubes (Sarstedt, Nümbrecht, Germany), which is in line with international consensus protocols [13]. Within two hours, CSF samples were centrifuged at 1800g for 10 minutes at 4° C. The CSF supernatant was transferred to new polypropylene tubes and stored at −20° C until further analysis (within two months). Baseline Aβ42, tau, and Ptau-181 were measured with commercially available ELISAs (INNOTEST β-AMYLOID(1-42), INNOTEST hTAU-Ag, and INNOTEST Phospho tau(181P) Fujirebio, Ghent, Belgium) on a routine basis as described before with intra-assay and interassay variations for all analysis of <3.2% and 10.9%, respectively [14]. The team performing the CSF analysis was not aware of the clinical diagnosis. Previous analysis has shown that there is a drift in Aβ42 results over the analysis years [15], [16]. We therefore applied a correction to the values to control for this drift, as previously described [17]. No such drift was observed for the tau and Ptau values.
2.4. Apolipoprotein E genotyping
DNA was isolated from 10 ml vacutainer tubes containing EDTA using the QIAamp DNA blood isolation kit from Qiagen (Venlo, The Netherlands). For genotype determination, the LightCycler ApoE mutation Detection Kit (Roche Diagnostics, GmbH, Mannheim, Germany) was used.
2.5. CART analysis
Classification And Regression Tree (CART) analysis is a nonparametric, supervised statistical learning technique that combines variable values, here CSF Aβ42, tau, and Ptau, such that these best discriminate between classes, in our case AD dementia and controls. The optimal combination of variables and possible cutoff values used for classification is determined through an exhaustive search of all possibilities by the CART algorithm [18]. The results are presented in a decision tree with several splits based on the selected variables and cutoffs, ending with the class labels. The Gini criterion was applied to minimize node impurity after splitting, and cross-validation was performed to prune the tree based on the minimum deviance, that is, the minimum (proportion) deviance improvement for proceeding with a new split [19]. We split our data set (controls n = 442; AD n = 1004) into a randomly selected training and validation set. The training data set included 50% of control subjects (n = 221) and was balanced with an equal amount of AD subjects (n = 221). The remaining subjects constituted the validation data set (controls n = 221; AD n = 783).
2.6. Validation
The decision tree was validated three-fold. First, internal validation applying cross-validation was used to build the tree. Second, the resulting tree was externally validated using an unseen validation data set. For the validation data set, accuracy (ACC), sensitivity (SE), and specificity (SP) were compared to the “typical” AD-like CSF biomarker profile consisting of decreased Aβ42 (<813 pg/ml) and increased tau concentrations (>375 pg/ml), for which cutoffs were previously defined [2], [17], [20] (see Table 2 for overview). Third, we further validated the performance of the tree to discriminate between MCI subjects who showed progression to AD dementia and MCI subjects who remained stable or had progressed to a non-AD–type dementia. We compared the ACC, SE, and SP with the previously defined cutoffs. CART analysis was performed using the R package “tree” in R (version 3.3.1, 2016-05-03). The predictor variables entered into the CART analysis included Aβ42, tau, and Ptau levels; apolipoprotein E (APOE) genotype; sex; and age.
Table 2.
Comparison of test characteristics
| Aβ42 (pg/ml) | tau (pg/ml) | Accuracy,% | SE, % | SP, % | |
|---|---|---|---|---|---|
| AD vs controls | |||||
| Previous cutoff∗ | 813 | 375 | 80 [78-83] | 78 [75-81] | 89 [84-93] |
| Tree training | 801 and 647 | 374 | 90 [87-93] | 93 [89-96] | 88 [83-92] |
| Tree validation | 801 and 647 | 374 | 86 [84-88] | 86 [83-88] | 87 [82-91] |
| MCI stable vs progression to AD | |||||
| Previous cutoff∗ | 813 | 375 | 76 [71-80] | 77 [69-84] | 75 [69-81] |
| Tree | 801 and 647 | 374 | 76 [71-80] | 84 [77-90] | 70 [63-76] |
Comparison of decision tree with typical AD biomarker–like profile using previously defined cutoff values for Aβ42 (813 pg/ml) and tau (375 pg/ml).
The decision tree has two cutoffs for Aβ42. Accuracy, SP, and SE calculations with 95% CI.
Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; SE, sensitivity; SP, specificity; CI, confidence interval; Aβ42, amyloid β1-42.
Cutoff for Aβ42 is based on drift corrected data.
2.7. Statistical analysis
Test characteristics consisting of ACC, SE, and SP, together with 95% confidence intervals were calculated with the epi.tests function, part of the “epiR” package in R. Statistical significance between test characteristics was obtained by comparing 95% confidence intervals. For the calculations of subject characteristics, and the subgroup comparisons, independent t-test, Kruskal-Wallis test, and chi-square test were applied. A P-value < .05 was considered significant.
3. Results
3.1. Subject characteristics
Subject characteristics according to diagnostic group are shown in Table 1. In brief, the training and validation cohorts used to build and validate the decision tree did not differ in subject characteristics. In both training and validation data sets, the controls were somewhat younger than AD patients, more often female, and had a higher level of education. As expected, Mini-Mental State Examination scores, and CSF Aβ42, tau, and Ptau levels were abnormal in AD patients compared to controls in both data sets. Furthermore, AD patients more often carried an APOE e4 allele compared to controls in both data sets.
Table 1.
Demographics AD cohort and controls
| Total |
Training |
Validation |
||||
|---|---|---|---|---|---|---|
| Controls | AD | Controls | AD | Controls | AD | |
| N (%) | 442 (31) | 1004 (69) | 221 (50) | 221 (50) | 221 (22) | 783 (78) |
| Age, years (SD) | 64.4 (6.3) | 67.3 (7.2)* | 64.4 (6.5) | 67.3 (7.0)* | 64.4 (6.0) | 67.2 (7.2)* |
| Females, n (%) | 273 (62) | 490 (49)* | 132 (60) | 110 (50)* | 141 (64) | 380 (49)* |
| Level of education (SD)† | 5.4 (1.3) | 4.8 (1.3)* | 5.4 (1.3) | 4.9 (1.4)* | 5.4 (1.2) | 4.8 (1.3)* |
| MMSE score (SD) | 28 (1.7) | 21 (4.9)* | 28 (1.7) | 20 (5.1)* | 28 (1.7) | 21 (4.9)* |
| APOE ε4 allele carriers, n (%) | 157 (36%) | 598 (60%) | 73 (33%) | 131 (59%)* | 84 (38%) | 467 (60%)* |
| CSF Aβ1-42 (SD), pg/ml | 1055 (259) | 656 (168)* | 1049 (249) | 672 (179)* | 1062 (269) | 651 (165)* |
| CSF Tau (SD), pg/ml | 318 (206) | 706 (406)* | 320 (204) | 686 (408)* | 317 (208) | 711 (406)* |
| CSF Ptau (SD), pg/ml | 51 (24) | 87 (39)* | 51 (25) | 86 (38)* | 51 (23) | 87 (38)* |
Data are mean (SD), unless otherwise specified. Independent T-test, Kruskal-Wallis test, or chi square test was used when applicable. *P < .05: AD differs from SCD.
Abbreviations: SCD, Subjective Cognitive Decline, served as controls; AD, Alzheimer's disease; MMSE, Mini-Mental State Examination; APOE, Apolipoprotein E; Aβ42, amyloid β 1-42; Ptau, tau phosphorylated at threonine 181; SD, standard deviation.
According to the Verhage system.
3.2. CART analysis
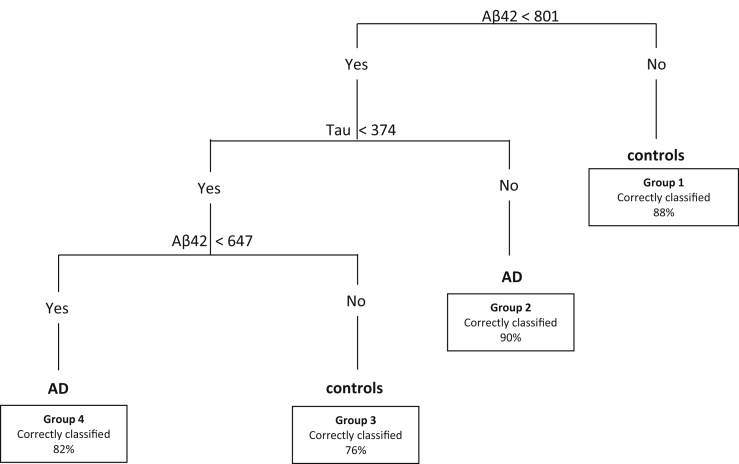
Fig. 1 shows the decision tree generated by the CART analysis on the training set. The decision tree identified two cutoff values for Aβ42 and one cutoff for tau. The first variable selected for decision-making was CSF Aβ42: subjects with Aβ42 higher than 801 pg/ml are classified as controls (group 1; Aβ42 ≥ 801). When Aβ42 levels were lower than 801 pg/ml, tau provided the most significant split at a cutoff level of 374 pg/ml. If tau concentrations were equal or higher than 374 pg/ml, no further splits were made and subjects were assigned the AD label (group 2; Aβ42 < 801 and tau ≥374). When tau concentrations were lower than 374 pg/ml, a second Aβ42 cutoff at 647 pg/ml provided the best split. This split separated a second group of controls with Aβ42 concentrations between 647 and 801 pg/ml (group 3; 647 < Aβ42 < 801 and tau <374) and a second group of AD patients with Aβ42 concentrations lower than 647 pg/ml (group 4; Aβ42 < 647 and tau <374). Group 1 contained most observed controls (n = 182; 82%) and group 2 contained most observed AD patients (n = 167; 76%). Additional variables, such as APOE, Ptau, sex, and age, did not add to the discriminative power of the decision tree and were not selected by the algorithm.
Fig. 1.
Decision tree. Decision tree calculated by Classification And Regression Tree (CART) analysis. The tree was built in the training cohort and is validated in the validation cohort. Numbers in the boxes indicate the percentage of correctly classified subjects out of the total number of subjects in that group for the training cohort in which the tree was built. Groups are numbered from 1 to 4. Aβ42 and tau concentrations are shown in pg/ml. Abbreviations: SCD, subjective cognitive decline; AD, Alzheimer's disease; Aβ42, amyloid β 1-42; tau, total tau.
3.3. Validation: diagnostic performance AD vs controls
As shown in Table 2, the decision tree had an overall SE of 93% [89-96] and SP of 88% [83-92] to detect AD dementia in the training cohort, and a SE of 86% [83-88] and SP of 87% [82-91] to detect AD dementia in the validation cohort. The overall diagnostic ACC was: 90% [87-93] and 86% [84-88] in the training and validation cohorts, respectively.
The number of correctly labeled subjects per group differed from the training cohort in which the tree was built compared to the validation cohort (see Supplementary Table 1). Especially, the control group with lower Aβ42 concentrations and normal tau concentrations (group 3; 647 < Aβ42 < 801 and tau <374) contained only 29% (n = 12/41) correctly classified subjects in the validation cohort, indicating that this control group mostly consisted of clinical AD subjects. In contrast, in the training cohort, 76% (n = 16/21) were correctly classified as controls.
We next compared the SE and SP levels obtained by the decision tree to those obtained with the previous binary cutoffs in the validation cohort. The decision tree with multiple cutoffs for Aβ42 had a significantly higher diagnostic ACC (86% [84-88]) in discriminating controls from AD compared to the previously defined single cutoff's for Aβ42 and tau together (80% [78-83]). Moreover, the SE of the decision tree was also higher than the previously defined cutoffs (86% [83-88] and 78% [75-81], respectively).
3.4. Validation: predictive performance for progression to AD
Our next step was to evaluate the predictive performance of the decision tree. Therefore, we used a cohort of MCI subjects with the follow-up available (average follow-up time: 2.5 ± 1.5 years). Subject characteristics are shown in Table 3. In brief, MCI subjects who progressed to AD were slightly older and were less often female than MCI subjects who did not progress to AD dementia. They also had lower CSF Aβ42 levels and higher tau and Ptau levels compared to the MCI subjects who did not progress to AD dementia.
Table 3.
Subject characteristics MCI cohort
| MCI | Stable | Progression to AD |
|---|---|---|
| n, (%) | 220 (61%) | 143 (39%) |
| Age, years (SD) | 67.3 (6.5) | 69.3 (7.4)* |
| Females, n (%) | 144 (65) | 73 (51)* |
| Level of education, mean (SD)† | 5.1 (1.4) | 5.0 (1.4) |
| APOE ε4 allele carriers, n (%) | 93 (42%) | 87 (61%) |
| MMSE score, mean (SD) | 27 (2.3) | 26 (2.8) |
| CSF Aβ1-42 (SD), pg/ml | 934 (299) | 679 (122)* |
| CSF tau (SD), pg/ml | 375 (227) | 668 (350)* |
| CSF Ptau (SD), pg/ml | 58 (29) | 70 (34)* |
| Follow-up duration, years (SD) | 2.5 (1.6) | 2.4 (1.3) |
Abbreviations: MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; APOE, apolipoprotein E; Aβ42, amyloid β 1-42; Ptau, tau phosphorylated at threonine 181; SD, standard deviation; CSF, cerebrospinal fluid.
Data are mean (SD), unless otherwise specified. Independent T-test, Kruskal-Wallis test, or chi square test was used when applicable. *P < .05.
According to the Verhage system.
The tree correctly identified 70% (n = 154 of 220) of MCI subjects who did not develop AD and predicted progression correctly in 84% (n = 120 of 143) of MCI subjects who clinically progressed to AD. Most incorrectly classified subjects (n = 50 of 89, 56%) had clinically not progressed to AD dementia but were classified as progressors by the decision tree, and vice versa, the remaining 44% had clinically progressed to AD but were classified as nonprogressors by the tree. The overall predictive ACC was comparable to that of the previously defined cutoffs (see Table 2) [17], [20].
3.5. Subtype comparisons
We further compared the four subgroups with different CSF profiles that were identified by the CART analysis. Groups 1 and 3 were labeled as control groups and had either high Aβ42 levels irrespective of tau (group 1: Aβ42 ≥ 801) or intermediate Aβ42 and low tau levels (group 3: 647 < Aβ42 < 801 and tau <374). Groups 2 and 4 were labeled as AD, and had low Aβ42 and high tau levels (group 2:Aβ42 < 801 and tau ≥374), or strongly decreased Aβ42 and low tau levels (group 4: Aβ42 < 647 and tau<374).
Across the total cohort (Table 4), Mini-Mental State Examination scores were highest in control group 1, followed by control group 3, AD group 2, and AD group 4. Control group 3, showed a similar proportion of APOE e4 carriers compared to AD groups 2 and 4. Groups 2 and 4 showed more atrophy for all magnetic resonance imaging parameters than control group 1. Control group 3 showed similar temporal lobe atrophy as AD groups 2 and 4, and parietal cortical atrophy and global cortical atrophy appeared to be in-between control group 1 and AD group 2.
Table 4.
Subgroup comparison within total cohort
| Predicted label | Group 1 controls |
Group 3 |
Group 2 AD |
Group 4 |
|---|---|---|---|---|
| n = 374 | n = 62 | n = 805 | n = 105 | |
| Demographics | ||||
| Observed label: controls/AD | 363/111 | 28/34 | 42/763 | 9/96 |
| Age, years (SD) | 65.2 (6.9)b,d | 65.3 (6.2) | 67.2 (7.2)a | 67.0 (6.5)a |
| Females, n (%) | 275 (58) | 37 (60) | 386 (48) | 65 (62) |
| Level of education (SD) | 5.2 (1.4)b,d | 5.0 (1.3) | 4.9 (1.3)a | 4.8 (1.2)a |
| Global cognition | ||||
| MMSE score (SD) | 27 (3.9)b,c,d | 24 (5.7)a,b,d | 21 (5.2)a,c | 21 (4.9)a,c |
| APOE genotype | ||||
| APOE ε4 carriers, n (%) | 152 (34)b,c,d | 37 (65)a | 509 (70)a | 74 (78)a |
| CSF biomarkers | ||||
| CSF Aβ1-42 (SD), pg/ml | 1110.8 (205.1)b,c,d | 715.3 (42.5)a,b,d | 617.9 (95.1)a,c,d | 542.2 (69.3)a,b,c |
| CSF tau (SD), pg/ml | 334.3 (222.1)b | 267.3 (81.5)b | 800.6 (391.5)a,c,d | 283.9 (63.8)b |
| CSF Ptau (SD), pg/ml | 51.5 (22.1)b | 45.1 (13.9)b | 96.9 (36.4)a,c,d | 45.5 (11.7)b |
| MRI biomarkers | ||||
| MTA, median (IQR) | 0.5 (0-1)b,c,d | 1.0 (0.5-2)a | 1.5 (1-2)a | 1.5 (1-2)a |
| PCA, median (IQR) | 1 (0-1)b,d | 1.0 (0-1)b | 1.0 (1-2)a,c | 1.0 (1-1.5)a |
| GCA, median (IQR) | 0.5 (0-1)b,d | 1.0 (0-1)b | 1.0 (1-2)a,c | 1 (1-1)a |
Comparison between different CSF profiles as identified by the CART analysis. Data are mean (SD), unless otherwise specified. Kruskal-Wallis test or chi square test was used when applicable. a: P < .05 different from group 1; b: P < .05 different from group 2; c: P < .05 different from group 3; d: P < .05 different from group 4.
Abbreviations: APOE, apolipoprotein E; AD, Alzheimer's disease; CSF, cerebrospinal fluid; MMSE, Mini-Mental State Examination; Aβ42, amyloid β 1-42; tau, total tau; Ptau, tau phosphorylated at threonine 181; MRI, magnetic resonance imaging; MTA, medial temporal lobe atrophy; PCA, Posterior Cortical Atrophy; GCA, Global Cortical atrophy; SD, standard deviation; IQR, interquartile range.
Within the MCI cohort (Table 5), group 2 labeled as progressors contained more females than group 1 (nonprogressors). Moreover, control group 1, labeled as nonprogressors, contained the least APOE e4 carriers, whereas group 3 contained nearly as much as APOE e4 carriers as groups 2 and 4 who were labeled as progressors.
Table 5.
Subgroup comparison within MCI population
| Predicted label | Group 1 nonprogressors |
Group 3 |
Group 2 progressors |
Group 4 |
|---|---|---|---|---|
| n = 196 | n = 35 | n = 214 | n = 29 | |
| Demographics | ||||
| Observed label: nonprogressors/progressors | 186/17 | 31/6 | 95/120 | 18/11 |
| Age, years (SD) | 66.8 (6.8)b | 68.5 (7.8) | 70.0 (6.9)a | 68.1 (6.1) |
| Females, n (%) | 52 (28)b | 12 (32) | 105 (49)a,d | 8 (28)b |
| Level of education (SD) | 5.0 (1.3) | 5.1 (1.4) | 5.1 (1.4) | 5.6 (1.5) |
| Time to progression (SD), yrs | 2.1 (0.8) | 2.7 (2.0) | 2.3 (1.4) | 2.2 (1.2) |
| Global cognition | ||||
| MMSE score (SD) | 26.8 (2.5)b | 27.2 (2.1) | 26.1 (2.5)a | 27.1 (1.9) |
| APOE genotype | ||||
| APOE ε4 carriers, n (%) | 57 (31)b,c | 20 (61)a | 146 (77)a | 15 (58)a |
| CSF biomarkers | ||||
| CSF Aβ1-42 (SD), pg/ml | 1100.1 (11.1)b,c,d | 722.4 (26.0)a,b,d | 636.8 (10.8)a,c | 551.5 (29.3)a,c |
| CSF tau (SD), pg/ml | 327.3 (17.1)b | 250.0 (40.0)b | 707.6 (16.6)a,c,d | 288.7 (45.2)b |
| CSF Ptau (SD), pg/ml | 52.4 (26.4)b | 43.2 (14.8)b | 92.9 (31.0)a,c,d | 48.5 (14.0)b |
| MRI biomarkers | ||||
| MTA, median (IQR) | 0.5 (0-1) | 1.0 (0-1.5) | 1.0 (0-1.5) | 0.5 (0.5-1.9) |
| PCA, median (IQR) | 1.0 (0-1) | 1.0 (0-1) | 1.0 (0-1) | 1.0 (0-1) |
| GCA, median (IQR) | 1.0 (0-1) | 1.0 (0-1) | 1.0 (1-1) | 1.0 (0-1) |
Comparison between different CSF profiles as identified by the CART analysis. Data are mean (SD), unless otherwise specified. Kruskal-Wallis test or chi square test was used when applicable. a: P < .05 different from group 1; b: P < .05 different from group 2; c: P < .05 different from group 3; d: P < .05 different from group 4.
Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; MMSE, Mini-Mental State Examination; Aβ42, amyloid β 1-42; tau, total tau; Ptau, tau phosphorylated at threonine 181; MRI, magnetic resonance imaging; MTA, medial temporal lobe atrophy; PCA, Posterior Cortical Atrophy; GCA, global cortical atrophy; SD, standard deviation; IQR, interquartile range.
4. Discussion
The main finding of this study is that a decision tree, consisting of two Aβ42 cutoff values at 801 pg/ml and 647 pg/ml and one tau cutoff at 374 pg/ml, best distinguished between controls and AD dementia. As a result, two AD and two control subgroups were identified who showed distinct CSF biomarker profiles. Age, Ptau, sex, and APOE status did not contribute to classification. Compared to the classical AD-like biomarker profile with previously defined cutoffs (813 pg/ml for Aβ42 and 375 pg/ml for Tau [17], [20]), the decision tree performed better in terms of diagnostic characteristics (diagnostic ACC 86% [84-88]; SE 86% [83-88]; SP 87% [82-91]). The predictive ACC of the decision tree in MCI subjects was similar to that of the previously defined cutoffs (76% [71-80] and 76% [71-80], respectively). Using CART analysis, we derived a robust decision tree that is readily applicable in the clinic.
Previous attempts to make a decision tree to distinguish AD from controls were made by Galasko et al. (1998) using Classification tree analysis [21]. Like in our present study, they also identified two cutoff values for CSF Aβ42. However, their cohort was relatively small. Here, we reproduce and further extend these findings showing strong external validation supporting the robustness of the approach, which is a prerequisite for implementation [21].
Others have suggested the use of regression formulas or the tau/Aβ42 ratio to combine multiple biomarkers for AD classification [20], [22], [23], [24]. Although such approaches can also provide a cut point, a benefit of the decision tree approach is that it allows clinicians to combine information from multiple biomarkers, providing intuitive interpretation of all markers involved, in contrast to, for example, a ratio when an abnormal value may be caused by either the numerator and/or denominator of the ratio. This could lead to false positive results, for example, an increase in tau can also occur in patients with other neurological conditions, such as minor stroke or Creutzfeldt-Jakob Disease, resulting in a high, abnormal ratio value, despite normal Aβ42 concentrations [14].
A decision tree does not suffer from those issues and is in better alignment with clinical decision paths, logically clustering signs and symptoms [25]. Furthermore, logistic regression models can be used to test whether a variable can separate a case from a control, provided that these labels are present a priori. A benefit of the CART analysis is that it allows for the existence of subgroups, even when these are not known a priori. In our case, we discovered subgroups 3 and 4, which would not have been identified in by a logistic regression model [18].
Ptau, age, sex and APOE status were not selected as part of the decision tree, suggesting that these variables did not add to the discriminative value of the tree. Previous studies showed that CSF levels of Aβ42 are lower in APOE carriers, and thus, we expected that APOE allele status would become part of the model [26]. A possible explanation for APOE not being included may be that the influence of APOE is mediated via Aβ42 levels, such that subgrouping based on APOE is of no further value [27]. A recent study of our group in which a data-driven cutoff is defined with Gaussian mixed modeling similarly found the cutoff to be independent of APOE allele status [16]. Nevertheless, this same study found a higher of Aβ42 cutoff when age increases [16], whereas in our study, age was not selected by the CART algorithm. A possible explanation could be that our cohort was considerably young and did not reflect a range of relatively young to old ages. The absence of Ptau in the model is in agreement with previous studies and is probably due to its high correlation with tau [14].
An additional finding of this study was the identification of two AD subgroups that showed distinct CSF biomarker profiles. The first AD subtype had a typical AD-like biomarker profile (low Aβ42 and high tau or Ptau; Aβ42 < 801 and tau ≥374) (group 2). The second AD subtype contained subjects with an “atypical” CSF profile with strongly decreased Aβ42 concentrations and normal CSF tau levels (group 4; Aβ42 < 647 and tau <374). Despite the different CSF biomarkers profiles, the subgroup analysis showed that both AD groups did not differ in clinical characteristics or magnetic resonance imaging measures. In view of the pathological heterogeneity often seen in AD patients, it might be that group 4, with the atypical CSF profile, is a subgroup of AD with a different underlying pathology leading to the disease [28]. For example, in a previous study, a subgroup similar to our atypical AD subtype (group 4) was identified, with strongly decreased CSF Aβ42 concentrations (513 pg/ml) and close to normal CSF tau concentrations (392 pg/ml) [5]. Interestingly, 15% of this AD subgroup had AD pathology with Lewy body inclusions at postmortem pathological examinations. Whether this explains our atypical group can only be defined after postmortem analysis.
Within controls, we also detected two subgroups: group 1 (n = 182) with normal Aβ42 levels (≥801 pg/ml) irrespective of tau levels, and group 3, with slightly reduced Aβ42 concentrations (801- 640pg/ml), and normal tau concentrations (<374 pg/ml). Group 3 contained a similar proportion of APOE e4 carriers as both subgroups with CSF AD profiles (groups 2 and 4). According to the recent NIA-AA criteria, the controls in group 3 with already lowered Aβ42, but normal tau concentrations have Alzheimer's pathologic change, and are at the beginning of the Alzheimer's continuum, meaning that these controls could have preclinical AD and might develop tau pathology over time [2], [29], [30]. Previous research has shown that Aβ42 can be present twenty years before the onset of clinical AD [31], [32]. So far, insufficient amount of follow-up information was available to determine whether this group was at risk to show clinical progression to MCI or dementia. Moreover, group 3 mainly consisted of observed controls in the training cohort, whereas it mostly consisted of observed AD patients in the validation cohort. With this group (group 3), the decision tree might have identified a “gray” area for Aβ42 values that fall within the two cutoffs, where the discriminatory performance of the biomarker is not sufficient enough to state whether the disease is present or absent.
We studied the prognostic ACC of the decision tree in MCI patients, which was comparable with the prognostic ACC of the previously defined cutoffs. However, the decision tree had a lower SP (70%), mostly due to misidentifying MCI subjects who remained stable. These MCI subjects had abnormal CSF Aβ42 concentrations according to the decision tree and did not progress to AD dementia within the time they were followed. Thus, it cannot be excluded that if they were followed for a longer period, they may have still showed progression.
Limitations of our study are the use of clinical diagnosis as a gold standard to derive the decision tree. Because clinical diagnoses are not always correct, even in an expert center, this can result in a bias of the diagnostic performance and suboptimal cutoff values of the decision tree [33]. The use of SCD subjects over healthy subjects as controls could be seen as a limitation. However, we believe SCD subjects fit our study design better than healthy controls because they represent the population that present themselves to the doctor with memory complaints more so than healthy controls. A strength of our study is the availability of a large data set that enabled us to validate the decision tree in two independent cohorts.
In conclusion, the CART analysis identified several CSF biomarker profiles to classify AD based on two Aβ42 cutoff values and one tau cutoff. This led to an improved diagnostic ACC when compared to the regularly used AD-like biomarker profile based on single cutoffs. The results incorporated into a decision tree facilitate interpretation of CSF biomarker results in clinical practice.
Research in Context.
-
1.
Systematic review: Traditional sources (e.g., PubMed) and information gained from abstracts and presentations were used for this systematic review. Previous literature has demonstrated that cerebrospinal fluid (CSF) biomarkers can aid in the diagnosis of Alzheimer's disease (AD). However, several studies and daily clinical practice indicate that not all AD patients have a clear abnormal pattern of all three biomarkers. The diversity of possible CSF biomarker profiles that accompany AD can be confusing for clinicians, thus complicating the interpretation of CSF biomarker results in daily clinical practice.
-
2.
Interpretation: Our decision tree analysis identified two CSF AD biomarker profiles in a large clinical cohort, which were incorporated in a readily applicable decision tree for daily clinical practice. Moreover, the decision tree improved diagnostic accuracy compared to current methods.
-
3.
Future directions: The article proposes further understanding of the underlying pathological mechanism of the AD subjects with different CSF biomarker profiles.
Acknowledgments
Research of the VUmc Alzheimer center is part of the neurodegeneration research program of the Neuroscience Amsterdam. The VUmc Alzheimer Center is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. The clinical database structure was developed with funding from Stichting Dioraphte.
Authors' contributions: Statistical analysis was performed by BMT and RBM. WMVDF, CET, BMT, NSMS, and RBM interpreted the data. RBM wrote the article. PS, WMVDF, CET, BMT, NSMS, and PJV revised the article.
Disclosures: RBM and NSMS had no disclosures to report. BMT received grant funding from the ZonMW Memorabel grant program #73305056 and #733050824. PS has acquired grant support (for the institution) from Piramal. In the past 2 years, he has received consultancy/speaker fees (paid to the institution) from Biogen and Roche (diagnostics). He is PI of studies with Probiodrug and EIP Pharma. PJV serves as an advisory board member of Eli-Lilly, is consultant for Janssen, and has received grants from GE healthcare and Biogen. Research programs of WMVDF have been funded by ZonMW, NWO, EU-FP7, Alzheimer Nederland, CardioVasculair Onderzoek Nederland, Stichting Dioraphte, Gieskes-Strijbis fonds, Biogen MA Inc, Boehringer Ingelheim, Piramal Neuroimaging, Roche BV, Janssen Stellar, Combinostics. WMVDF has performed contract research for Biogen MA Inc and Boehringer Ingelheim. WMVDF has been an invited speaker at Boehringer Ingelheim and Biogen MA Inc. All funding is paid to her institution. CET received grant funding from the European Commission, the Dutch Research Council (ZonMW), Association of Frontotemporal Dementia/Alzheimer's Drug Discovery Foundation, Alzheimer Netherlands. CET has functioned in advisory boards of Fujirebio and Roche received nonfinancial support in the form of research consumables from ADx Neurosciences and Euroimmun, performed contract research or received grants from Probiodrug, Janssen prevention center, Boehringer, Brains online, Axon Neurosciences, EIP pharma, Roche. These funding sources had no influence on study analyses or article writing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dadm.2018.10.004.
Supplementary Data
References
- 1.Scheltens P., Blennow K., Breteler M.M.B., de Strooper B., Frisoni G.B., Salloway S. Alzheimer's disease. Lancet. 2016;388:505–517. doi: 10.1016/S0140-6736(15)01124-1. [DOI] [PubMed] [Google Scholar]
- 2.Jack C.R., Bennet D.A., Blennow K., Carillo M.C., Dunn B., Haeberlein S.B. NIA-AA Research Framework: Toward a biological definition of Alzheimer's disease. Alzheimer's Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois B., Feldman H.H., Jacova C., Hampel H., Molinuevo J.L., Blennow K. Advancing research diagnostic criteria for Alzheimer’s disease: The IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- 4.Skillback T., Farahmand B.Y., Rosen C., Mattsson N., Nagga K., Kilander L. Cerebrospinal fluid tau and amyloid-beta1-42 in patients with dementia. Brain. 2015;138:2716–2731. doi: 10.1093/brain/awv181. [DOI] [PubMed] [Google Scholar]
- 5.Iqbal K., Flory M., Khatoon S., Soininen H., Pirttila T., Alafuzoff I. Subgroups of Alzheimer's disease based on cerebrospinal fluid molecular markers. Ann Neurol. 2005;58:748–757. doi: 10.1002/ana.20639. [DOI] [PubMed] [Google Scholar]
- 6.van der Vlies A.E., Verwey N.A., Bouwman F.H., Blankenstein M.A., Klein M., Scheltens P. CSF biomarkers in relationship to cognitive profiles in Alzheimer disease. Neurology. 2009;72:1056–1061. doi: 10.1212/01.wnl.0000345014.48839.71. [DOI] [PubMed] [Google Scholar]
- 7.van der Flier W.M., Scheltens P. Amsterdam Dementia Cohort: Performing Research to Optimize Care. J Alzheimer's Dis. 2018;62:1091–1111. doi: 10.3233/JAD-170850. Perry G, Avila J, Zhu X, editors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Flier W.M., Pijnenburg Y.A., Prins N., Lemstra A.W., Bouwman F.H., Teunissen C.E. Optimizing patient care and research: The Amsterdam dementia cohort. J Alzheimer's Dis. 2014;41:313–327. doi: 10.3233/JAD-132306. [DOI] [PubMed] [Google Scholar]
- 9.McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Kawas C.H. The diagnosis of dementia due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen R.C., Smith G.E., Waring S.C., Ivnik R.J., Tangalos E.G., Kokmen E. Mild cognitive impairment: clinical characterization and outcome [published erratum appears in Arch Neurol 1999 Jun;56(6):760] Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 11.Albert M.S., Dekosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C. The diagnosis of mild cognitive impairment due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verhage F. [Intelligence and Age: study with Dutch people aged 12 to 77]. Assen Van Gorcum; Assen, The Netherlands: 1964. Intelligentie en leeftijd: onderzoek bij Nederlanders van twaalf tot zevenzeventig jaar. [Google Scholar]
- 13.del Campo M., Mollenhauer B., Bertolotto A., Engelborghs S., Hampel H., Simonsen A.H. Recommendations to standardize preanalytical confounding factors in Alzheimer's and Parkinson's disease cerebrospinal fluid biomarkers: an update. Biomark Med. 2012;6:419–430. doi: 10.2217/bmm.12.46. [DOI] [PubMed] [Google Scholar]
- 14.Duits F.H., Teunissen C.E., Bouwman F.H., Visser P.J., Zetterberg H., Blennow K. The cerebrospinal fluid “alzheimer profile”: Easily said, but what does it mean? Alzheimer's Dement. 2014;10:713–723. doi: 10.1016/j.jalz.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Willemse E.A.J., van Uffelen K.W.J., van der Flier W.M., Teunissen C.E. Effect of long-term storage in biobanks on cerebrospinal fluid biomarker Aβ1-42, T-tau, and P-tau values. Alzheimer's Dement Diagnosis. Assess Dis Monit. 2017;8:45–50. doi: 10.1016/j.dadm.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertens D., Tijms B.M., Scheltens P., Teunissen C.E., Visser P.J. Unbiased estimates of cerebrospinal fluid β-amyloid 1–42 cutoffs in a large memory clinic population. Alzheimers Res Ther. 2017;9:8. doi: 10.1186/s13195-016-0233-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tijms B.M., Willemse E.A.J., Zwan M.D., Mulder S.D., Visser P.J., van Berckel B.N.M. Unbiased Approach to Counteract Upward Drift in Cerebrospinal Fluid Amyloid-β 1–42 Analysis Results. Clin Chem. 2018;64:576–585. doi: 10.1373/clinchem.2017.281055. [DOI] [PubMed] [Google Scholar]
- 18.Lemon S.C., Roy J., Clark M.A., Friedmann P.D., Rakowski W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann Behav Med. 2003;26:172–181. doi: 10.1207/S15324796ABM2603_02. [DOI] [PubMed] [Google Scholar]
- 19.Breiman L., Friedman J.H., Olshen R.A., Stone C.J. Wadsworth Stat. Ser; 1984. Classification and Regression Trees. [Google Scholar]
- 20.Mulder C., Verwey N.A., van der Flier W.M., Bouwman F.H., Kok A., van Elk E.J. Amyloid-beta(1-42), total tau, and phosphorylated tau as cerebrospinal fluid biomarkers for the diagnosis of Alzheimer disease. Clin Chem. 2010;56:248–253. doi: 10.1373/clinchem.2009.130518. [DOI] [PubMed] [Google Scholar]
- 21.Galasko D., Chang L., Motter R., Clark C.M., Kaye J., Knopman D. High cerebrospinal fluid tau and low amyloid beta42 levels in the clinical diagnosis of Alzheimer disease and relation to apolipoprotein E genotype. Arch Neurol. 1998;55:937–945. doi: 10.1001/archneur.55.7.937. [DOI] [PubMed] [Google Scholar]
- 22.Hulstaert F., Blennow K., Ivanoiu A., Schoonderwaldt H.C., Riemenschneider M., De Deyn P.P. Improved discrimination of AD patients using beta-amyloid(1-42) and tau levels in CSF. Neurol. 1999;52:1555–1562. doi: 10.1212/wnl.52.8.1555. [DOI] [PubMed] [Google Scholar]
- 23.Mattsson N., Zetterberg H., Hansson O., Andreasen N., Parnetti L., Jonsson M. CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA. 2009;302:385–393. doi: 10.1001/jama.2009.1064. [DOI] [PubMed] [Google Scholar]
- 24.Schoonenboom N.S.M., Reesink F.E., Verwey N.A., Kester M.I., Teunissen C.E., van de Ven P.M. Cerebrospinal fluid markers for differential dementia diagnosis in a large memory clinic cohort. Neurology. 2012;78:47–54. doi: 10.1212/WNL.0b013e31823ed0f0. [DOI] [PubMed] [Google Scholar]
- 25.Marshall R.J. The use of classification and regression trees in clinical epidemiology. J Clin Epidemiol. 2001;54:603–609. doi: 10.1016/s0895-4356(00)00344-9. [DOI] [PubMed] [Google Scholar]
- 26.Verghese P.B., Castellano J.M., Garai K., Wang Y., Jiang H., Shah A. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci U S A. 2013;110:E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapiola T., Pirttilä T., Mehta P.D., Alafuzoff I., Lehtovirta M., Soininen H. Relationship between apoE genotype and CSF β-amyloid (1-42) and tau in patients with probable and definite Alzheimer's disease. Neurobiol Aging. 2000;21:735–740. doi: 10.1016/s0197-4580(00)00164-0. [DOI] [PubMed] [Google Scholar]
- 28.Dong A., Toledo J.B., Honnorat N., Doshi J., Varol E., Sotiras A. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer's disease: links to cognition, progression and biomarkers. Brain. 2017;140:735–747. doi: 10.1093/brain/aww319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Harten A.C., Visser P.J., Pijnenburg Y.A., Teunissen C.E., Blankenstein M.A., Scheltens P. Cerebrospinal fluid Aβ42 is the best predictor of clinical progression in patients with subjective complaints. Alzheimers Dement. 2013;9:481–487. doi: 10.1016/j.jalz.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Jack C.R., Knopman D.S., Jagust W.J., Shaw L.M., Aisen P.S., Weiner M.W. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jack C.R., Holtzman D.M. Biomarker modeling of alzheimer's disease. Neuron. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fagan A.M., Xiong C., Jasielec M.S., Bateman R.J., Goate A.M., Benzinger T.L. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer's disease. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3007901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coart E., Barrado L.G., Duits F.H., Scheltens P., van der Flier W.M., Teunissen C.E. Correcting for the Absence of a Gold Standard Improves Diagnostic Accuracy of Biomarkers in Alzheimer's Disease. J Alzheimers Dis. 2015;46:889–899. doi: 10.3233/JAD-142886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.