Summary
Differences in the growth and maturation of diverse forebrain tissues depend on region-specific transcriptional regulation. Individual transcription factors act simultaneously in multiple regions that develop very differently, raising questions about the extent to which their actions vary regionally. We found that the transcription factor Pax6 affects the transcriptomes and the balance between proliferation and differentiation in opposite directions in the diencephalon versus cerebral cortex. We tested several possible mechanisms to explain Pax6's tissue-specific actions and found that the presence of the transcription factor Foxg1 in the cortex but not in the diencephalon was most influential. We found that Foxg1 is responsible for many of the differences in cell cycle gene expression between the diencephalon and cortex and, in cortex lacking Foxg1, Pax6's action on the balance of proliferation versus differentiation becomes diencephalon like. Our findings reveal a mechanism for generating regional forebrain diversity in which one transcription factor completely reverses the actions of another.
Subject Areas: Neuroscience, Molecular Neuroscience, Developmental Neuroscience
Graphical Abstract
Highlights
-
•
Pax6 loss affects transcriptome profiles oppositely in cortex versus diencephalon
-
•
Pax6 promotes neuron differentiation in cortex, whereas proliferation in the diencephalon
-
•
Foxg1-null cortex develops a diencephalon-like profile of cell cycle gene expression
-
•
Foxg1 presence in cortex but not in diencephalon explains Pax6 tissue-specific actions
Neuroscience; Molecular Neuroscience; Developmental Neuroscience
Introduction
The mechanisms that create the brain's enormous inter-regional diversity of structure and function remain poorly understood. Early in embryogenesis, the anterior neural plate is patterned by the regional expression of transcription factors whose actions are essential for each part to acquire its correct size and cellular composition. These transcription factors are sometimes referred to as “master regulators” or “selectors” to reflect their powerful ability to control the hierarchies of gene expression that specify region-specific growth and identity (Allan and Thor, 2015). Although some transcription factors are expressed in discrete regions of the central nervous system, many of them do not instruct the development of unique regions but are expressed simultaneously in multiple regions whose morphologies and functions develop very differently. This raises questions about the degree to which the actions and mechanisms of action of individual transcription factors vary between different brain regions. Does a transcription factor regulate a particular process, such as cell proliferation, similarly in all regions? To what extent and how are its actions modified by the context in which it is expressed?
The transcription factor Pax6 is expressed simultaneously by large populations of progenitors in both major components of the forebrain, the telencephalon (rostrally) and the diencephalon (caudally) (Stoykova and Gruss, 1994). During this time, the telencephalon expands much more than the diencephalon, which it engulfs as it forms the cerebral cortex dorsally and the basal ganglia ventrally. The diencephalon forms the thalamus (Th) and prethalamus (PTh), which process and transmit signals to and from the overlying cortex. Pax6 regulates the proliferation of cortical cells by limiting the rate at which progenitors progress through the cell cycle. Pax6 deletion promotes a higher rate of proliferation of cortical radial glial progenitors (RGPs), whereas the opposite occurs if Pax6 is overexpressed within a physiological range (Georgala et al., 2011a, Georgala et al., 2011b, Manuel and Price, 2005, Mi et al., 2013). Previous studies have shown that Pax6 is required for normal thalamic and prethalamic development (Stoykova et al., 1996, Warren and Price, 1997), but whether it controls thalamic and prethalamic progenitors in the same way it regulates cortical RGPs is unknown.
We began by addressing this question. We compared the effects of acute Pax6 deletion on the transcriptomes of cells from embryonic cortex, Th, and PTh and found that the expression levels of genes associated with cell proliferation and differentiation were altered in opposite directions in the cortex and diencephalon. We went on to show that this corresponded with an opposite effect of Pax6 deletion on the balance between proliferation and differentiation in these two regions. We next explored the mechanisms that might account for these tissue-specific differences, including the possibility that the actions of Pax6 are affected by the presence or absence of another high-level transcription factor, Foxg1. Foxg1 regulates the cell cycles of cortical progenitors, is expressed by all telencephalic (cortical and basal ganglia) cells including RGPs, but is not expressed by diencephalic cells (Fasano et al., 2009, Kumamoto and Hanashima, 2017, Mariani et al., 2016, Martynoga et al., 2005, Xuan et al., 1995). We first showed that Foxg1 deletion from cortical cells makes their profile of cell cycle gene expression more diencephalon like. We went on to test how Foxg1-null cortical cells respond to Pax6 deletion. We found that this caused cell-autonomous changes that were opposite to those in Foxg1-expressing cortex and similar to those in diencephalic tissues, indicating that the region-specific actions of Pax6 on the balance of proliferation and differentiation in developing forebrain are Foxg1 dependent.
Results
Quality Control of RNA-Seq Data
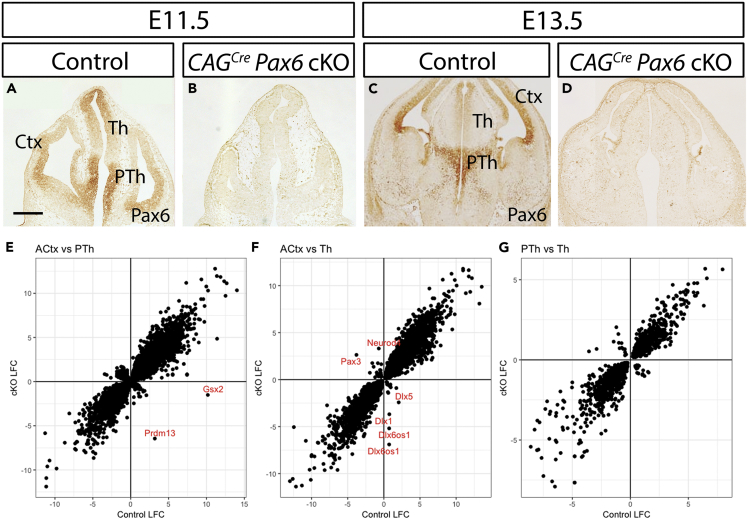
We started by using RNA sequencing (RNA-seq) to study the effects of tamoxifen-induced Pax6 deletion on gene expression in the cortex, Th, and PTh at embryonic day 13.5 (E13.5). We dissected only the anterior half of the cortex (ACtx), where cortical levels of Pax6 are higher. Administration of tamoxifen at E9.5 to Pax6fl/fl embryos ubiquitously expressing Cre recombinase from a CAGCreER allele caused Pax6 loss from E11.5 onward (Figures 1A–1D). These embryos are referred to here as CAGCreER Pax6 cKOs (conditional knockouts), and they are compared with CAGCreER Pax6fll+ littermate controls, which continue to express Pax6 in a normal pattern (Figures 1A and 1C). Heterozygosity for Pax6 does not detectably affect forebrain Pax6 protein levels or the proliferation of Pax6-expressing cells (Figure S1A; Mi et al., 2013). Accurate and consistent dissection of the Th, PTh, and the ACtx was guided by the DTy54 transgene, which expresses green fluorescent protein (GFP) under the control of all known Pax6 regulatory elements (Tyas et al., 2006a). This transgene faithfully reports the levels of Pax6 gene expression in cells in which the endogenous Pax6 locus can be either normal or null. PTh and ACtx are distinguished by high levels of Pax6/GFP expression; PTh has a sharp posterior boundary with Th, which expresses Pax6/GFP at lower levels (Figures S1B–S1E′).
Figure 1.
Effects of Pax6 Deletion on Transcriptomic Differences between Embryonic Anterior Cortex (ACtx), Thalamus (Th), and Prethalamus (PTh)
(A–D) Pax6 immunohistochemistry at (A and B) E11.5 and (C and D) E13.5 showing CAGCreER-induced loss of Pax6 throughout the forebrain following tamoxifen administration at E9.5. Ctx, cortex. Scale bar, 0.25 mm.
(E–G) Log2 fold change (LFC) of gene expression between (E) ACtx and PTh, (F) ACtx and Th, and (G) PTh and Th in controls and CAGCreERPax6 cKOs. Positive values indicate enrichment in PTh in (E) and in Th in (F and G). Negative values indicate enrichment in ACtx in (E and F) and in PTh in (G). Selected genes are labeled in red.
See also Figures S1 and S4 and Tables S1, S2, and S3.
Before RNA-seq, we quality controlled the accuracy and consistency of the separation of PTh and Th using quantitative real-time PCR (qRT-PCR) to measure the levels of expression of Dlx2, which is highly expressed in PTh, but not in Th, and Neurog2, which is highly expressed in Th and in only a small proportion of PTh cells (Figures S2A and S2B). None of the PTh samples included in the analysis contained mRNA for Wnt8b, confirming that they were not contaminated by adjacent eminentia thalami, which is Wnt8b-rich (Adutwum-Ofosu et al., 2016). At least three quality-controlled replicate samples representing each tissue and genotype from independent litters were analyzed by RNA-seq. After sequencing, we re-confirmed the accuracy of our diencephalic dissections by extracting the expression values of several reference genes with well-characterized differential expression between the PTh and Th. We found low levels of prethalamic markers (Dlx2, Gsx2 and Ascl1) in both control and cKO thalamic samples (red arrows, Figures S2C, S2E, and S2G) and low levels of thalamic markers (Neurog2, Gbx2, and Dbx1) in control and cKO prethalamic samples (red arrows, Figures S2D, S2F, and S2H). The RNA-seq data is at European Nucleotide Archive; www.ebi.ac.uk/ena; ENA: PRJEB9747. Principal component analyses (PCA) showed individual samples separating according to genotype along the axes of greatest variation in all three tissues (Figure S3).
Major Inter-regional Differences of Gene Expression in Control Embryos
We examined differences between the transcriptomes of ACtx, Th, and PTh in control embryos. Table S1 lists all genes showing significant (adjusted p < 0.05) enrichment in each control tissue over its level in each of the other two control tissues. There were about 3,000 significant differences between Th and PTh and about 10,000–11,000 between ACtx and Th or PTh. The expression patterns of genes showing the greatest inter-regional differential expression are shown in Figure S4. Many encode transcription factors or other molecules involved in regulating developmental processes. This analysis shows the great divergence in molecular identities and hence the context in which Pax6 operates in these three regions.
Major Inter-regional Differences of Gene Expression Remain after Pax6 Deletion
We then repeated this analysis on data from CAGCreER Pax6 cKOs to compare the differences between the transcriptomes of ACtx, Th, and PTh when Pax6 was deleted. The numbers of inter-regional differences increased to over 4,000 between Th and PTh and to over 12,000 between ACtx and either Th or PTh (Table S2). We then paired the regions—ACtx with Th, ACtx with PTh, Th with PTh—and calculated differential expression (in the form of log2 fold changes [LFCs]) between the members of each pair in controls and in cKOs. For each pair, we plotted control LFCs and cKO LFCs against each other, including all genes significantly differentially regulated by Pax6 loss in both members of each pair (Figures 1E–1G). The graphs showed that the vast majority of genes retained the direction of their inter-regional differential expression in cKOs (i.e., if they were higher in one region in controls, the same was true in cKOs). Exceptions included Gsx2, which is known from previous work to have its normally strong expression in PTh extinguished by Pax6 loss (Caballero et al., 2014), and Dlx family members, which are upregulated in cKO ACtx (Figures 1E and 1F); a full list of exceptions is provided in Table S3. We conclude that Pax6 is not required to maintain the vast majority of fundamental differences in molecular identity between these forebrain regions.
The Sets of Genes Regulated by Pax6 Vary between Forebrain Regions
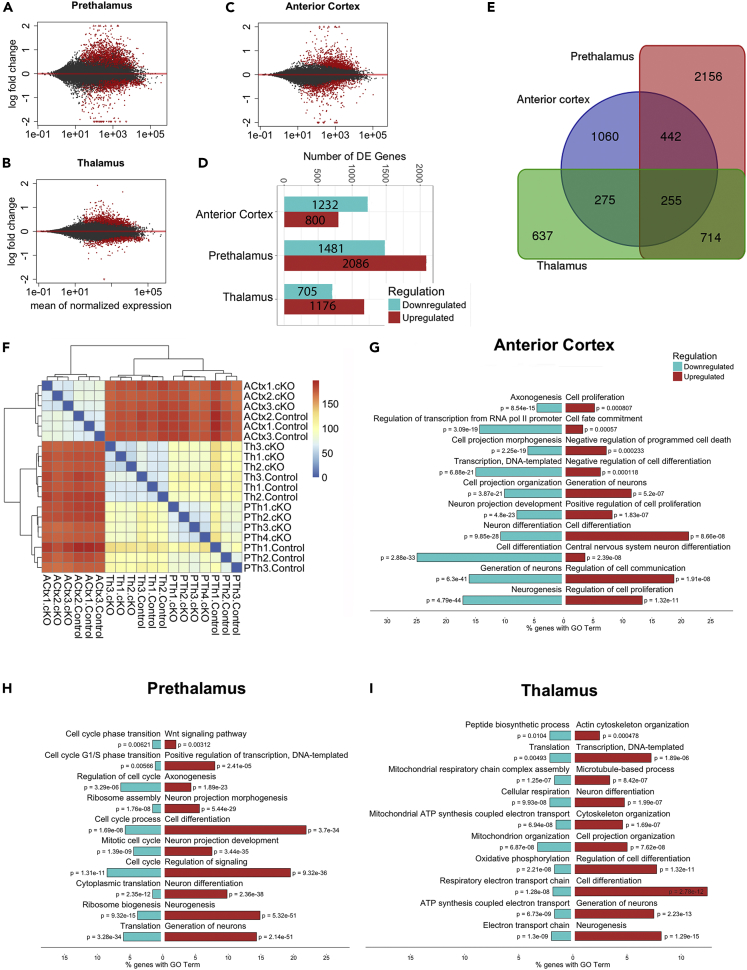
We next studied gene expression changes between controls and CAGCreER Pax6 cKOs within each individual tissue. Table S4 lists, for each tissue, all genes showing significant differential expression between controls and cKOs (adjusted p < 0.05). Figures 2A–2C plots, for each tissue, each gene's LFC in expression over its average expression (those with adjusted p < 0.05 in red), and Figure 2D summarizes the numbers of significantly up- and downregulated genes in cKOs. To explore the lists in Table S4 interactively, visit Differential Expression at https://pricegroup.sbms.mvm.ed.ac.uk/Pax6_diencephalon/. PTh showed the greatest number of differentially expressed genes, probably because Pax6 is expressed not only by all progenitors but also by many postmitotic neurons in PTh, whereas it is expressed only by progenitors in the cortex and Th. This likelihood is supported by the observation that genes showing the largest differential expression values in PTh include many encoding ion channels and receptors associated with postmitotic neurons, whereas these genes showed little or no significant differential expression in ACtx or Th (Table S4). Quite similar numbers of genes showed significant differential expression in ACtx and Th (Figure 2D). The numbers of significantly upregulated genes were ∼40%–60% higher than the numbers of downregulated genes in both Th and Pth, whereas this ratio was reversed in ACtx.
Figure 2.
The Effects of Pax6 Deletion on the Transcriptomes of Prethalamic, Thalamic, and Anterior Cortical Cells
(A–C) Scatter plots of log2 fold changes (on the y-axis) versus the mean of normalized counts (on the x-axis) in prethalamus (A), thalamus (B) and anterior cortex (C); red dots indicate statistically significant changes between genotypes (adjusted p values <0.05).
(D) An overview of the numbers of significantly differentially expressed (DE) genes resulting from Pax6 deletion in each region.
(E) Venn diagram showing the numbers of significantly DE genes in each region and common to multiple regions.
(F) Heatmap representing the results of hierarchical clustering of RNA-seq data from each sample from control and CAGCreERPax6 cKO.
(G–I) The 10 most highly enriched, non-redundant gene ontology (GO) terms with obvious relevance to developmental processes for up- and downregulated genes in each region.
See also Figures S2 and S3 and Tables S4 and S5.
A surprisingly small number of genes (255 of 5,539) showed significant differential expression across all three tissues (referred to here as “commonly deregulated genes”) (Figure 2E). Forty-five of these genes were upregulated, and only 12 were downregulated in all three tissues (listed in Table S5). Many of these 57 genes encode molecules implicated in intercellular interactions such as cell adhesion, cell-cell communication, and axon guidance. For example, proteins involved in Wnt signaling were strongly represented. Wnt3a, Wnt5a, Lrg4, Wif1, and Wls were commonly upregulated, and Dkk3 was commonly downregulated. Among the commonly downregulated genes were two related to retinoic acid signaling: Rlbp1 and Ripply3. To see the expression levels of any regulated gene in our dataset, visit Gene Expression Plots at https://pricegroup.sbms.mvm.ed.ac.uk/Pax6_diencephalon/. Interestingly, 8 of the 12 genes that are commonly downregulated have been reported to be directly regulated by Pax6 (Dkk3, Rlbp1) and/or show a peak in Pax6 chromatin immunoprecipitation sequencing (ChIP-seq) (Bmpr1b, Cldn12, Dkk3, Mlc1, Nr2e1, Rypply3, Rlbp1, and Sema3a; references in Table S5). The scarcity of commonly deregulated genes and the fact that the direction of change of 198 of 255 of them (i.e., whether they were up- or downregulated) varied between tissues suggests that Pax6 deletion affects aspects of cortical, thalamic, and prethalamic development in substantially different ways.
Pax6 Loss Affects Gene Expression Profiles Oppositely in Cortex and Diencephalon
To discover global similarities and differences in the inter-regional effects of Pax6 on gene expression, we carried out distance-based hierarchical clustering on our RNA-seq data. This identified two major clusters representing samples from the cortex and diencephalon (Figure 2F), suggesting that the reactions of the two diencephalic tissues, Th and PTh, to Pax6 loss might be more similar to each other than to that of the cortex.
To predict how Pax6 removal might affect biological processes in ACtx, Th, and PTh and to look for possible similarities and differences in how these regions respond to the deletion, we first carried out gene ontology (GO) term enrichment analysis on all genes showing significant differential expression in each tissue. Figures 2G–2I show the 10 most highly enriched, non-redundant GO terms with obvious relevance to developmental processes for up- and downregulated genes in each tissue. Regarding genes upregulated in the cortex, 3 of 10 terms contained “cell proliferation” with one being “positive regulation of cell proliferation” and 3 of 10 terms contained “differentiation” with one being “negative regulation of cell differentiation.” Regarding genes downregulated in the cortex, no terms related to proliferation but 2 of 10 terms contained “differentiation” and a further 4 of 10 terms related to processes that occur in differentiating cells, such as “axonogenesis” or “projection development.” By contrast, in diencephalic tissues, terms including “differentiation,” “axonogenesis,” or “projection development” were associated only with upregulated genes (5 of 10 in PTh and 4 of 10 in Th), whereas terms related to proliferation were associated only with downregulated genes (6 of 10 terms contained “cell cycle” in PTh). Overall, this analysis suggests that the effects of Pax6 loss on at least some developmental processes are likely to be opposite in the cortex and diencephalon. Pax6 loss might promote proliferation in the former but promote differentiation in the latter.
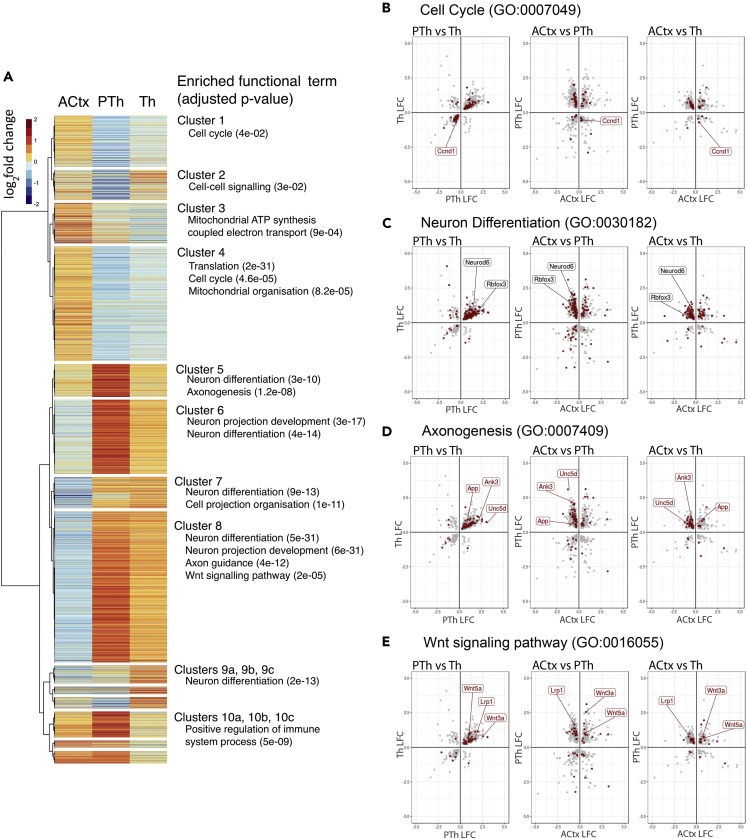
We tested this idea further by hierarchically clustering all genes showing significant differential expression in at least one tissue according to the direction and magnitude of their LFCs across all three tissues (Figure 3A). Most genes showed LFCs in the same direction in PTh and Th, but in an opposite direction in ACtx. The dendrogram was cut to produce 14 clusters suitable for GO term analysis. Figure 3A lists highly enriched, representative functional terms alongside each cluster (Table S6 contains a complete list of functional terms associated with each cluster). In Clusters 1 and 4, most of the genes showed positive LFCs in ACtx, but negative LFCs in Th and PTh. These clusters were strongly associated with the GO term “cell cycle.” Cluster 4 contains 21 cell-cycle-related terms with significant enrichment; cell cycle terms were exclusively enriched in these two clusters and not in others. Trends were opposite in clusters 6–8, with most genes showing negative LFCs in ACtx but positive LFCs in Th and PTh. These clusters were strongly associated with GO terms containing or related to “differentiation.” Many genes in clusters 5, 6, and 8, which are strongly associated with terms related to differentiation, showed the greatest LFCs in PTh, in agreement with the continued function for Pax6 in postmitotic neurons in this region. Interestingly, cluster 8 also showed enrichment for “Wnt signaling pathway,” indicating that this pathway might be upregulated specifically in the diencephalon.
Figure 3.
Pax6 Deletion Has Opposite Effects on the Transcriptomes of Cortical and Diencephalic (Thalamic and Prethalamic) Cells
(A) Hierarchical clustering of all genes that showed significant differential expression in at least one tissue according to the direction and magnitude of their log2 fold change (LFC) across all three tissues. The dendrogram is cut to generate 14 clusters, and enriched GO functional terms are listed against these clusters.
(B–E) LFCs of all genes showing significant differential expression (adjusted p < 0.05) in PTh and Th against each other, in ACtx and PTh against each other, and in ACtx and Th against each other. Genes associated with particular GO terms (“cell cycle” in A, “neuron differentiation” in B, “axonogenesis” in C, and “Wnt signaling pathway” in D) are marked on the plots (red).
See also Table S6.
Finally, we plotted the LFCs of all genes showing significant differential expression (adjusted p < 0.05) in (1) PTh and Th against each other, (2) ACtx and PTh against each other, and (3) ACtx and Th against each other (Figures 3B–3E). Genes associated with particular GO terms (“cell cycle,” “neuron differentiation,” “axonogenesis,” and “Wnt signaling pathway”) are marked on the graphs (in red). The LFCs of most genes associated with these and similar GO terms were directly related in Th and PTh (first column of graphs in Figures 3B–3E). However, the LFCs of large proportions of these genes were inversely related when Th or PTh were compared with ACtx (second and third columns of graphs in Figures 3B–3E). Selected genes associated with each function are highlighted in the graphs. For example, the cell cycle gene Ccnd1 (CyclinD1) was significantly downregulated in both Th and PTh but significantly upregulated in ACtx (Figure 3B). On the other hand, genes associated with differentiation, such as Neurod6 and Rbfox3 (also commonly known as NeuN), were significantly upregulated in Th and PTh but significantly downregulated in ACtx (Figure 3C). To identify in these graphs any individual gene or group of genes associated with any GO term, visit LFC-GO plots at https://pricegroup.sbms.mvm.ed.ac.uk/Pax6_diencephalon/.
These analyses showed that Pax6 deletion caused many genes to alter their expression levels in opposite directions in cortical versus diencephalic regions. They indicate that many of these oppositely regulated genes are associated with cell proliferation and differentiation. They suggest that Pax6 loss is likely to promote proliferation over differentiation in cortex but to have the opposite effect in both diencephalic regions.
Pax6 Has Opposite Effects on Neurogenesis in Cortex and Diencephalon
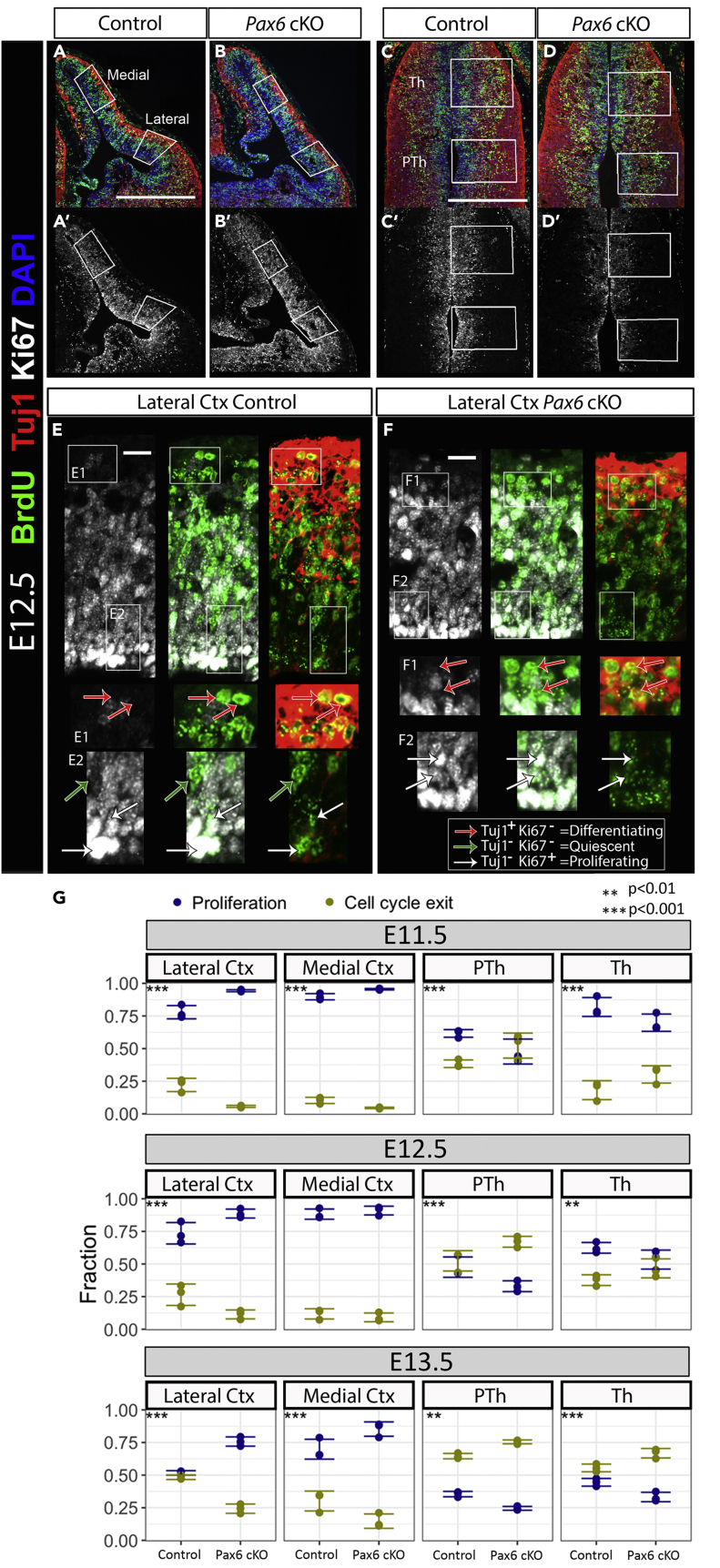
We then tested this prediction directly in embryonic brain. As before, we created CAGCreER Pax6 cKOs and littermate controls by giving tamoxifen at E9.5. We carried out the analysis at three different ages for all tissues (E11.5, E12.5, and E13.5), thereby allowing for the fact that diencephalic tissues develop slightly ahead of telencephalic tissues, with neurogenesis starting by E11.5 in the former (Suzuki-Hirano et al., 2011) and by E12.5 in the latter. We injected a single pulse of the S-phase marker bromodeoxyuridine (BrdU) 24 hr before collecting tissue for analysis using markers of proliferating cells (Ki67) and differentiating neurons (Tuj1). We counted from two regions of ACtx, one lateral and one medial, and one region each of Th and Pth (Figures 4A–4D′). We measured the proportions of BrdU+ cells that (1) remained proliferative (BrdU+, Ki67+, Tuj1−; white arrows in Figures 4E and 4F), (2) had started to differentiate (BrdU+, Ki67−, Tuj1+; red arrows in Figures 4E and 4F), and (3) were in an intermediate state (BrdU+, Ki67−, Tuj1−; green arrows in Figure 4E) in ACtx, Th, and PTh of control and Pax6 cKO embryos.
Figure 4.
The Effect of Pax6 Deletion on Proliferation and Differentiation in Cortical versus Diencephalic Neurons Is Opposite and Not Age Dependent
(A–D) Immunohistochemistry for BrdU and Tuj1 on E12.5 cortical (A and B) and diencephalic (C and D) tissues. Boxes show the areas of quantification. Scale bars, 0.25 mm.
(A′–D′) Same sections as A–D; channel showing Ki67 staining has been separated here for a clearer visualization.
(E and F) High-power details of E12.5 control (E) and CAGCreERPax6 cKO (F) lateral cortex showing separate channels. Zoomed boxed areas (E1, E2, F1, F2) show examples of cells attributed to each category in our quantifications (red arrows, differentiated cells, Brdu+/Tuj1+/Ki67-; green arrows, quiescent G0 state, Brdu+/Tuj1-/Ki67-; white arrows, proliferative cells, Brdu+/Tuj−/Ki67+). Scale bars, 10 μm.
(G) Analysis of the quantification of proliferation and cell cycle exit indexes in all ages (E11.5, E12.5, E13.5), tissues (lateral cortex, medial cortex, thalamus, and prethalamus), and genotypes (controls and CAGCreERPax6 cKO) showed that Pax6 loss produces an increase in proliferation and decrease in cell cycle exit in cortical tissues but the opposite is observed in diencephalic (thalamus and prethalamus) tissues. The data from all ages, regions, and genotypes were fitted to a generalized linear mixed model to test the effects of Pax6 inactivation depending on age and tissue. ANOVA was used with Tukey's method for multiple pairwise comparisons to obtain test statistics for contrasts in question. Ctx, cortex.
See also Figure S5.
Very few cells were BrdU+, Ki67− and Tuj1− in any condition, and their numbers changed in proportion to the fraction of differentiated cells (Figure S5). Most likely these cells had only recently entered G0 en route to undergoing differentiation. As there was no evidence that Pax6 deletion caused their numbers to increase, we decided to combine all BrdU+, Ki67− cells into one group, representing those that had exited the cell cycle to differentiate.
The data from all ages, regions, and genotypes were fitted to a generalized linear mixed model to test the effects of Pax6 inactivation depending on age and tissue (Figure 4G). In the cortex, the proportions of mitotic (BrdU+) cells that remained proliferative (Ki67+) after 24 hr were significantly higher in cKOs than in controls and the proportions that had exited the cell cycle were significantly lower in cKOs, with the sole exception of the medial cortex at E12.5, where we detected no significant effect. It appeared that Pax6 had less effect in the medial than the lateral cortex at all three ages, which might be explained by the normally lower levels of Pax6 in medial regions (Figures 1A and 1C). In both Th and PTh, effects of Pax6 loss were opposite to those in the cortex: the proportions of mitotic cells that remained proliferative after 24 hr were significantly lower in cKOs than in controls and the proportions that had exited the cell cycle were significantly higher.
These data also allowed us to compare the states of maturation of tissues of different ages in control animals. In E13.5 lateral cortex, for example, ∼45% of mitotic cells had exited the cell cycle during the last 24 hr. Similar rates of exit are found at E12.5 in Th and are likely to be reached between E11.5 and E12.5 in PTh. This validates our premise that the range of ages included in these experiments allows comparison of the effects of Pax6 at equivalent developmental stages in the three tissues. The effect observed is therefore tissue specific and not age dependent. The results agree with our prediction from the RNA-seq experiments that the effects of Pax6 on the balance between proliferation and differentiation are opposite in the cortex versus diencephalon.
Inter-regional Variation in Pax6 Levels and Exon Usage Do Not Correlate Closely with Its Function
We next considered possible mechanisms by which Pax6 might regulate the balance between proliferation and differentiation in opposite directions in cortical and diencephalic regions. The first possibility was that regional variation in attributes of the transcription factor itself causes it to operate differently in the cortex and diencephalon. Variation in Pax6's level and the ratio between its two major splice variants might modify its function (Haubst et al., 2004, Manuel et al., 2006).
Regional variation in levels of Pax6 protein in normal forebrain did not correlate with the effects of Pax6 on the balance of proliferation versus differentiation. Levels of Pax6 are the highest in PTh and ACtx, which show opposite changes in proliferation versus differentiation following Pax6 deletion, and are relatively low in Th, which shows changes in the same direction as PTh (Figures 1A–1D). Regarding alternative splicing, previous studies have suggested that the two major isoforms, Pax6 and Pax6(5a), the latter containing an additional 14 amino acids inserted into the paired domain, affect gene expression differently (Chauhan et al., 2004, Haubst et al., 2004, Pinson et al., 2005). We used our RNA-seq data to compare exon 5/5a usage in control ACtx, Th, and PTh (Figure S6). In each sample, we measured the average coverage across exons 5 and 5a and found that the only significant difference was an ∼2-fold difference in the 5/5a ratio between ACtx and PTh, with the ACtx having higher relative counts for exon 5a and therefore a lower 5/5a ratio. As with Pax6 levels, variation in relative levels of exon 5/5a usage did not correlate closely with the effects of Pax6 on the balance of proliferation versus differentiation. Pax6 is pro-proliferative in both Th and PTh and the opposite in ACtx, whereas the exon 5/5a ratio differed significantly between ACtx and only one of the diencephalic regions. Although regional differences in the exon 5/5a ratio might play a role, on their own they are unlikely to explain regional differences in Pax6 action on the balance of proliferation and differentiation. We went on to consider further possible mechanisms.
Pax6 Loss Causes Canonical Wnt Signaling Deregulation
We then considered the possibility that Pax6 might affect the balance between proliferation and differentiation in opposite directions in the cortex and diencephalon because of its differential effects on intercellular signaling. We examined Wnt signaling, because the aforementioned results identified it as a GO function that is regulated differentially between the cortical and diencephalic regions (Figure 3).
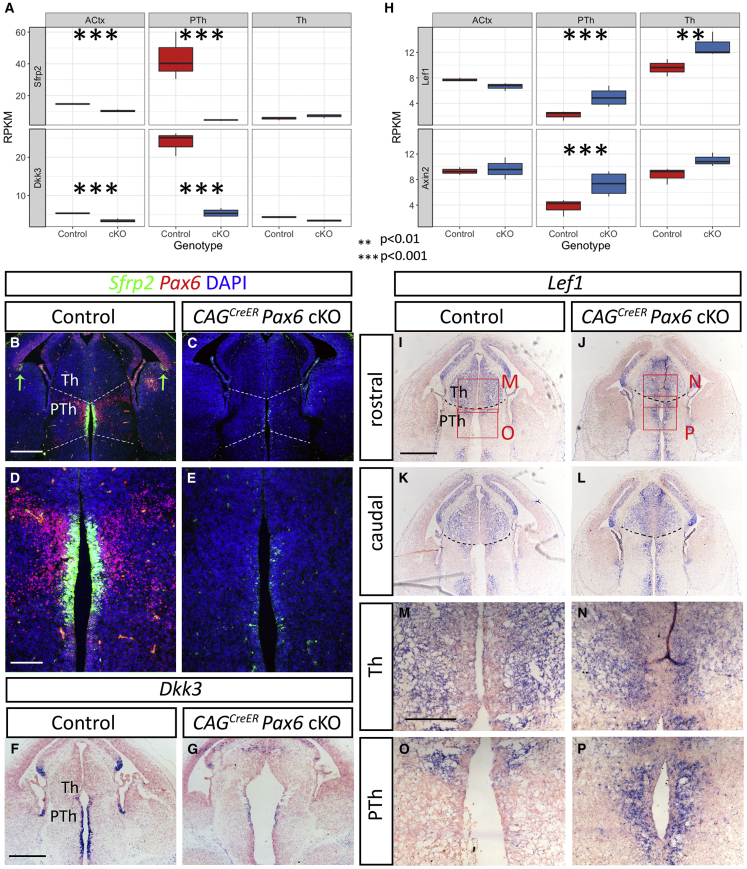
A number of genes involved in Wnt signaling were commonly deregulated across all three tissues, sometimes in different directions but sometimes in the same direction. Lrp1, which encodes low-density lipoprotein receptor-1, a potential negative regulator of canonical Wnt signaling (Willnow et al., 2007, Zilberberg et al., 2004), was upregulated in Th and PTh but downregulated in ACtx (Figure 3E). These changes correlated with regional differences in the effects of Pax6 on the balance of proliferation versus differentiation, whereas the others did not. Wnt3a and Wnt5a were upregulated across all three regions. The Wnt antagonists Sfrp2 and Dkk3 were strongly downregulated in prethalamic progenitors but were unaffected in Th, where their levels are normally very low (Figures 5A–5G). Sfrp2 and Dkk3 were significantly downregulated in ACtx, although their expression domains are limited mainly to extremely lateral cells around the pallial-subpallial boundary and medially at the cortical hem, respectively (Figures 5B, 5C, 5F, and 5G; Kim et al., 2001).
Figure 5.
Pax6-Loss-Induced Changes in Canonical Wnt Signaling in the Forebrain
(A and H) Box and whisker plots showing data from RNA-seq of reads per kilobase of transcript per million mapped reads (RPKM) for Sfrp2, Dkk3, Lef1, and Axin2 in ACtx, PTh, and Th from control and CAGCreERPax6 cKOs. Significance was tested by ANOVA followed by post-hoc Tukey comparison.
(B–E) In situ hybridization for Sfrp2 combined with immunohistochemistry for Pax6 in controls (B and D) and Pax6 cKOs (C and E) showing the drastic reduction of Sfrp2 in the prethalamic ventricular zone. Green arrows in B indicate Sfrp2+ cells at the pallial-subpallial boundary.
(F and G) In situ hybridization for Dkk3 showing a general downregulation in Pax6 cKOs (G) with respect to controls (F).
(I–P) In situ hybridization in E13.5 control (I, K, M, and O) and CAGCreER Pax6 cKOs (J, L, N, and P) detected an upregulation of the Wnt signalling readout Lef1, close to the ventricular zone of the prethalamus (compare O and P). Scale bars: 0.25 mm in (B, C, F, G, and I–L) and 0.1 mm in (D, E, and M–P).
To see if the changes in expression of individual Wnt pathway genes produced a net effect that correlated with regional differences in the effects of Pax6 on the balance of proliferation versus differentiation, we examined the expression of two bona fide readouts of the Wnt canonical pathway, Lef1 and Axin2 (Figures 5H–5P). Both genes were significantly upregulated in PTh and Lef1 was upregulated in Th, but neither gene was significantly altered in ACtx.
These analyses suggest that the effects of Pax6 loss on canonical Wnt signaling are largely restricted to the diencephalon, where the effects were greatest in PTh. These changes would likely affect the behaviors of diencephalic progenitors, whereas they do not provide a clear explanation for the effect of Pax6 loss on cortical progenitors. We went on, therefore, to explore other factors that might be important for explaining the nature of Pax6's cortical effect.
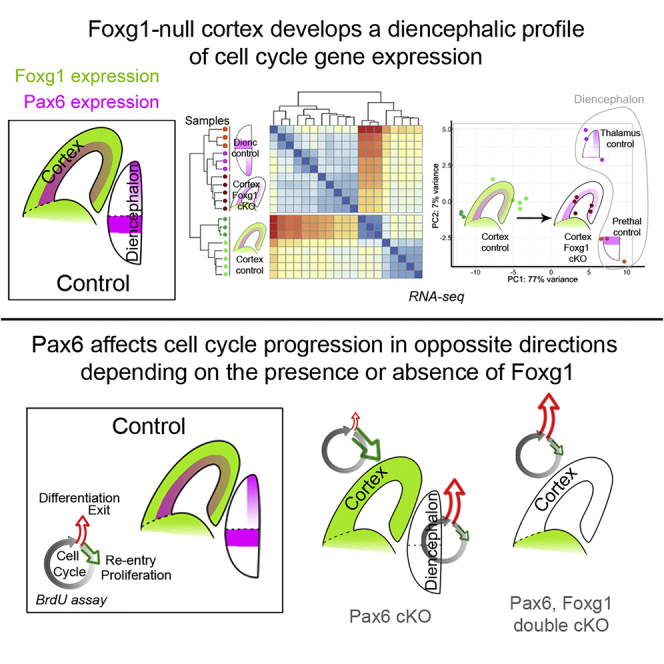
Foxg1-Null Cortex Develops a Diencephalon-like Profile of Cell Cycle Gene Expression
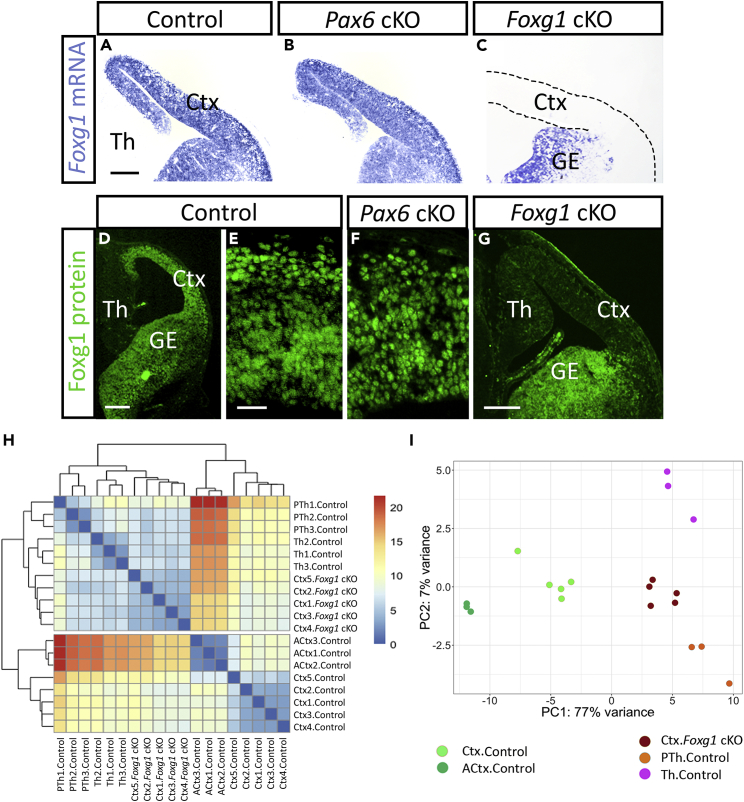
We next examined the possibility that Pax6 operates differently in the cortex because another transcription factor in this tissue moves the transcriptomes of cortical cells away from those of diencephalic cells, thereby altering the context within which Pax6 operates, thus modifying its actions. We postulated that such a factor should be present in the cortex and not the diencephalon and that it should not be regulated by Pax6. Transcripts for transcription factor Foxg1 showed the highest log2 fold enrichment between ACtx and Th (11.67) and the third highest between ACtx and PTh (9.57) (Table S1; Figure S4Y). Foxg1 is co-expressed with Pax6 by cortical progenitors and is an important positive regulator of their cell cycles (Martynoga et al., 2005, Vezzali et al., 2016, Xuan et al., 1995, Yip et al., 2012). Our RNA-seq data showed that its cortical expression is only marginally downregulated following Pax6 removal (LFC = −0.207; adjusted p = 0.0190), a change that was not visible in cortical sections (Figures 6A and 6B). Levels of Foxg1 protein and its nuclear distribution in cortical cells appeared unaltered by Pax6 loss (Figures 6D–6F). We hypothesized, therefore, that Foxg1 might modify the molecular context within which Pax6 operates and therefore its actions.
Figure 6.
Foxg1 Deletion in the Embryonic Cortex Makes Its Profile of Expression of Cell Cycle Genes Diencephalon Like
(A–C) In situ hybridizations showing Foxg1 telencephalic expression in (A) control, (B) Emx1CreERPax6 cKOs, and (C) Emx1CreERFoxg1 cKOs at E13.5. GE, ganglionic eminence. Scale bar, 0.2 mm.
(D–G) Immunohistochemistry showing Foxg1 protein expression in (D) control telencephalon, (E) control cortex, (F) Emx1CreERPax6 cKO cortex, and (G) Emx1CreERFoxg1 cKO telencephalon at E13.5. Scale bars: 0.2 mm in (D and G) and 0.05 mm in (E and F).
(H) Heatmap of hierarchical clustering of RNA-seq data on genes annotated by the “cell cycle” GO term (GO:0007049) that were significantly deregulated in the Foxg1 cKO. Data are from samples of control cortex, control ACtx, control PTh, control Th, and Emx1CreERFoxg1 cKO cortex.
(I) Principal component (PC) analysis on the same RNA-seq data as in (H).
See also Figures S7 and S8 and Tables S7, S8, and S9.
We first tested whether differences between the transcriptomes of cortical and diencephalic cells are created by Foxg1's presence in the cortex. We used RNA-seq to test whether Foxg1 deletion from the cortex moved its profile of gene expression toward that of diencephalic tissues. We gave tamoxifen at E9.5 to induce cortex-specific deletion of Foxg1 (Emx1CreER; Foxg1fl/fl, referred to here as Foxg1 cKO; the Foxg1fl allele was from Miyoshi and Fishell, 2012). This caused loss of Foxg1 from almost all cortical cells by E13.5 (Figures 6C and 6G). We carried out RNA-seq in E13.5 control (Emx1CreER; Foxg1fl/+) and Foxg1 cKO cortices (in these experiments the entire cortex was used). Five replicate samples for controls and cKOs from independent litters were sequenced. The raw data can be obtained from the European Nucleotide Archive (www.ebi.ac.uk/ena; ENA: PRJEB21349 ).
PCA showed that data from individual samples clustered by genotype along the axis of greatest variation (Figure S7A). Table S7 lists all genes with significant differential expression in Foxg1 cKOs (adjusted p < 0.05), and the numbers of up- and downregulated genes are shown in Figures S7B and S7C. The list of downregulated genes showed high enrichment for functional terms related to cell cycle and mitosis (Table S8), consistent with the fact that Foxg1 is a regulator of cortical proliferation (Martynoga et al., 2005, Xuan et al., 1995). This effect is unlikely to be mediated by an effect of Foxg1 on Pax6 levels or alternative splicing. Pax6 levels were only marginally decreased at the RNA level (LFC = −0.18; adjusted p = 0.007), Pax6 protein remained expressed in a normal pattern (Figure S7D), and there was no significant effect of Foxg1 deletion on the relative usage of Pax6's exons 5 and 5a (Figures S7E and S7F). We conclude that both Foxg1 and Pax6 operate largely independently with only very limited effects of either gene on the expression of the other.
To find out whether Foxg1 shifts the context within which Pax6 operates toward that in diencephalic tissues, we compared RNA-seq data from the cortex of Foxg1 cKOs with those from control cortex, Th, and PTh. Control cortical data were from the five samples obtained as controls for the Foxg1 cKOs and the three samples from ACtx obtained as controls for the CAGCreER Pax6 cKOs. Control thalamic and prethalamic data were from the samples obtained as controls for the CAGCreER Pax6 cKOs. We carried out distance-based hierarchical clustering including specifically those genes annotated by the “cell cycle” GO term that were significantly deregulated in the Foxg1 cKO. All eight control cortical samples clustered together and separated not only from control diencephalic samples but also from Foxg1-null cortical samples (Figure 6H; Table S9 lists the genes included in this analysis). Control diencephalic samples were clustered with Foxg1-null cortical samples. Only at the third level in the dendrogram did Foxg1 cKO cortical samples separate from Th control samples. PCA using the same set of genes revealed the same trend: Foxg1-null cortical samples, but not control cortical samples, clustered with samples from control Th and PTh along the axis of maximum variation, which represented 77% of the variance (Figure 6I). We repeated the same clustering experiments with sets of genes annotated by other GO terms, but none showed the same separation as observed for cell cycle genes (examples are shown in Figure S8).
These results suggest that the deletion of Foxg1 from the cortex shifts the profile of expression of genes whose actions are related to the cell cycle closer to that of normal diencephalic tissues. We then went on to test whether this shift alters the way in which cortical cells respond to Pax6 by asking whether Pax6 deletion from Foxg1−/− mutant cortex causes a response closer to that observed when Pax6 is lost from the wild-type diencephalon.
Foxg1 Alters Pax6's Effect on Expression of an Important Cell Cycle Regulator
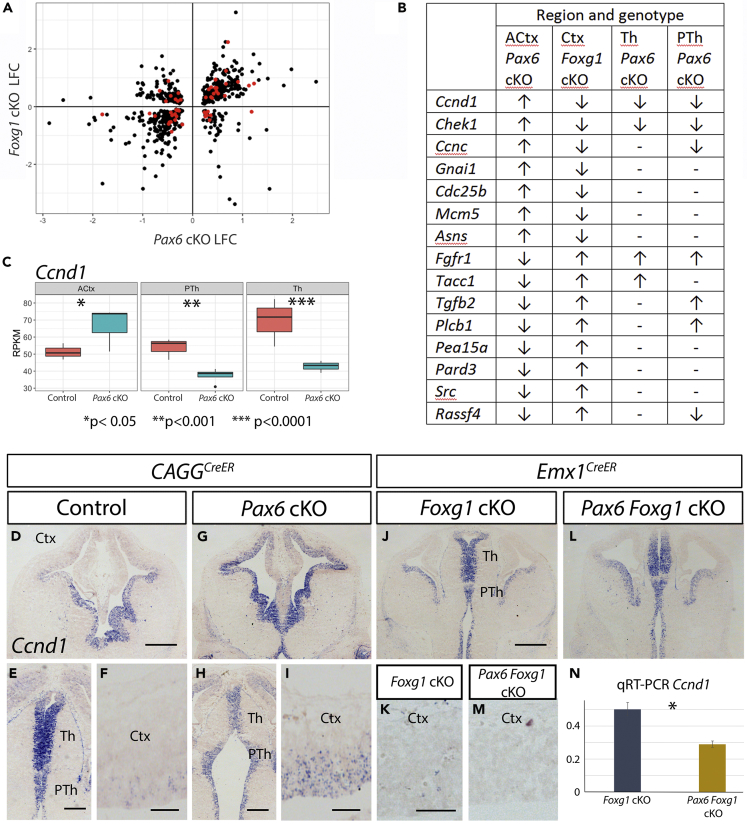
Before testing whether the presence of Foxg1 in the cortex modifies the effects of Pax6 on the balance between proliferation and differentiation, we sought molecular evidence for whether this was likely. We first focused on those genes that were regulated by both Pax6 and Foxg1 obtained from the intersection of our RNA-seq data from Pax6 cKOs and Foxg1 cKOs. We identified 678 genes whose cortical expression was regulated by both Pax6 and Foxg1. These genes are listed in Table S10, and their average LFCs in Pax6 cKO and Foxg1 cKO are plotted against each other in Figure 7A.
Figure 7.
Foxg1 Deletion in the Embryonic Cortex Reverses Effects of Pax6 Deletion
(A) Log2 fold changes (LFCs) of all genes showing significant regulation by both Pax6 (data from CAGCreERPax6 cKOs) and Foxg1 (data from Emx1CreERFoxg1 cKOs). Red dots mark cell-cycle-associated genes.
(B) Genes associated with the cell cycle regulated in opposite directions in CAGCreERPax6 cKOs versus Foxg1 cKOs; any significant changes following Pax6 removal from Th or PTh are indicated.
(C) Box and whisker plots of RPKM for Ccnd1 in ACtx, PTh, and Th from E13.5 control and CAGCreERPax6 cKOs. Significance was tested by ANOVA followed by post-hoc Tukey comparison.
(D–I) In situ hybridizations showing expression of Ccnd1 in the forebrains of E13.5 (D–F) controls and (G–I) CAGCreERPax6 cKOs. Scale bars: 0.25 mm in (D and G), 0.2 mm in (E and H), and 0.05 mm in (F and I).
(J–M) In situ hybridizations showing expression of Ccnd1 in the forebrains of E13.5 (J, K, and M) Emx1CreERFoxg1 cKOs and (L) Emx1CreERFoxg1 Pax6 cKOs. Scale bars: 0.25 mm in (J and L) and 0.05 mm in (K and M).
(N) Quantitative RT-PCR for Ccnd1 in E13.5 cortex of Emx1CreERFoxg1 cKOs and Emx1CreERFoxg1 Pax6 double cKOs. Means ± SEMs; n = 3 for each genotype; p < 0.05 Student's t test.
See also Table S10.
Pax6 and Foxg1 regulate many of these genes in the same direction. Included in this group are many downregulated genes with roles in cortical neurogenesis and differentiation, such as Neurog1, Neurod2, Neurod4, and Satb4. Pax6 and Foxg1 might act synergistically to promote the expression of these genes and drive neuronal precursors toward differentiation. Among the genes upregulated by either Pax6 or Foxg1 deletion, we found genes involved in signaling (Wls, Bmp2, Bmp5, Wnt3a, or Wnt8b) and several encoding transcription factors (Arx, Pax7, Zic3, or Sp5) (see Table S10). Pax6 and Foxg1 regulated others in opposite directions, and we focused on these genes for our subsequent analyses. We argued that this group would be the one most likely to include genes whose regulation by Pax6 is reversed by Foxg1.
Of the genes whose expression was deregulated in opposite directions by cortical deletion of Pax6 or Foxg1, 15 had GO annotations relating them with cell cycle control (red dots in Figure 7A). These genes are listed in Figure 7B along with the direction of their expression change when either Pax6 or Foxg1 was removed from each region. Seven of these genes showed directions of change in CAGCerER Pax6 cKOs that were opposite in ACtx versus Th and/or PTh. These gene expression changes, individually or collectively, might make a significant contribution to changing the balance between proliferation and differentiation in opposite directions in cortex versus the diencephalon after Pax6 deletion.
One of these genes was the cell cycle regulator Ccnd1, which we took as an exemplar of the principles outlined above. In our RNA-seq data, Ccnd1 showed significant Pax6-loss-induced differential expression in opposite directions in the cortex (upregulated) and diencephalon (downregulated in both Th and PTh) (Figure 7C). These changes can be seen with in situ hybridization in Figures 7D–7I. Ccnd1 is a G1 phase cyclin, which promotes entry into S-phase and hence cell cycle progression over cell cycle exit in many systems and cell types. Several previous studies have shown that altering its expression levels in the embryonic cortex alters the balance between proliferation and differentiation (Artegiani et al., 2011, Ferguson et al., 2000, Kollmann and Sexl, 2013, Lange et al., 2009, Matsushime et al., 1994, Morgan, 1997, Pilaz et al., 2009, Zerjatke et al., 2017). In the light of these findings, we studied changes in Ccnd1 expression further because they were likely to be good predictors of cellular responses to Pax6 removal in different contexts.
To test whether Foxg1 affects the actions of Pax6 on Ccnd1 expression, we tested the effect of deleting Pax6 from Foxg1-null cortex. We generated Emx1CreER;Foxg1fl/fl and Emx1CreER;Foxg1fl/fl;Pax6fl/fl embryos and induced deletion of Foxg1 or both Pax6 and Foxg1 by tamoxifen administration at E9.5 to produce embryos referred to here as Foxg1 cKOs or Pax6;Foxg1 dcKOs (double cKOs), respectively. Both Pax6 and Foxg1 protein were deleted from all but the most lateral cells of the cortex, where Emx1 is not expressed, by E13.5 (Figures 6C, 6G, 8I, and 8I′; see also Mi et al., 2013). Ccdn1 expression was significantly reduced in Foxg1 cKO cortex (Table S7), whereas the levels were even lower following Pax6 co-deletion (Figures 7J–7N). This Pax6-loss-induced change was opposite to the one that occurs following Pax6 removal from the cortex that expresses Foxg1. It was in the same direction as occurs in Th and PTh following Pax6 loss.
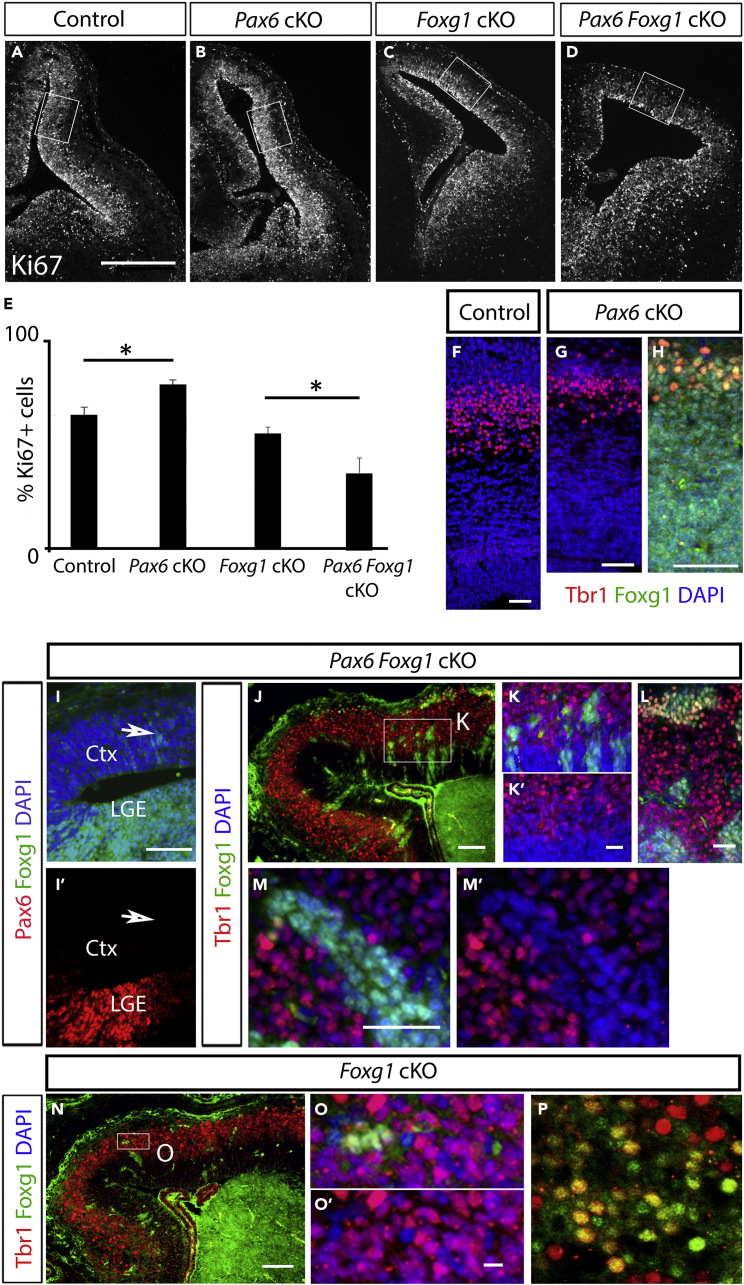
Figure 8.
Foxg1 Deletion in the Embryonic Cortex Reverses the Effects of Pax6 Deletion on the Balance of Proliferation versus Differentiation
(A–D) Immunohistochemistry of Ki67+ cells in the cortex of E13.5 (A) controls, (B) CAGCreERPax6 cKOs, (C) Emx1CreERFoxg1 cKOs, and (D) Emx1CreERFoxg1 Pax6 cKOs. Boxes show the areas selected for quantification. Scale bar, 0.25 mm.
(E) Quantifications of Ki67+ cells in the cortex of E13.5 controls, CAGCreERPax6 cKOs, Emx1CreERFoxg1 cKOs, and Emx1CreERFoxg1 Pax6 cKOs. Means ± SEMs; n = 3 for each genotype; p < 0.05 Student's t test.
(F–H) Immunohistochemistry for Tbr1 and Foxg1 in (F) control and (G and H) CAGCreERPax6 cKO at E15.5. Scale bars, 0.05 mm.
(I and I′) Immunohistochemistry for Pax6 and Foxg1 on the same section of E13.5 Emx1CreERFoxg1 Pax6 double cKO telencephalon. Arrows show that small numbers of cells in the cortex retained Foxg1 but not Pax6. Scale bars, 0.2 mm.
(J–M’) Immunohistochemistry for Foxg1 and Tbr1 on E15.5 Emx1CreER Foxg1 Pax6 double cKOs showing that most neurons expressing Foxg1 do not express Tbr1 and vice versa (K–M’). Box in J is shown in (K and K’). Scale bars: J, 0.05mm; (K–M)’, 0.025mm.
(N–P) Immunohistochemistry for Foxg1 and Tbr1 on E15.5 Emx1CreER Foxg1 single cKOs showing that most neurons expressing Foxg1 also express Tbr1 (O,O’, see double labelled cells in P). Box in N is shown in O,O’. Scale bars: N, 0.05mm; O–P, 0.01mm.
Altogether, these results increased the likelihood that Pax6's effects on the balance of proliferation versus differentiation in the cortex are Foxg1 dependent. We went on to test the hypothesis that in cortex lacking Foxg1, Pax6's effects would be more like those normally seen in the diencephalon.
Absence of Foxg1 Alters the Cellular Response of Cortex to Pax6 Removal
We first counted the proportions of cortical cells that were proliferative (Ki67+) in E13.5 control, Pax6 cKO, Foxg1 cKO, and Pax6;Foxg1 dcKO embryos (Figures 8A–8E). We found that, in contrast to the consequences of removing Pax6 from normal Foxg1-expressing cortex (Figures 4, 8A, and 8B), removing Pax6 from Foxg1−/− mutant cortex caused a significant reduction in the proportion of proliferating cells (Figure 8E; Student's t test, p = 0.02). This response was opposite to that of Foxg1-expressing cortex and in the same direction as that of Th and PTh (Figure 4).
We then tested the effect of Pax6 deletion from Foxg1-null cortex on the production of Tbr1-expressing cells. Tbr1 marks cortical cells as they transition from a proliferative progenitor state to a postmitotic neuronal state (Englund et al., 2005). In our RNA-seq data, Tbr1 levels were significantly reduced in cortical Pax6 cKOs (Table S2), fitting with the shift to proliferation. This was also clear with immunohistochemistry (Figures 8F–8H). Here we compared the effects of Pax6 removal from Foxg1-null versus Foxg1-expressing cortical cells on their production of Tbr1+ cells at E15.5.
We observed that in some E13.5 and E15.5 Foxg1 cKOs and Pax6;Foxg1 dcKOs, small clusters of cells continued to express Foxg1 protein. These clusters comprised only a few cells in the ventricular zone at E13.5 (Figure 8I) but were larger by E15.5 (Figures 8J–8P), as expected because their failure to delete Foxg1 would enhance their proliferation relative to Foxg1−/− cells. Foxg1+ clusters in Pax6;Foxg1 dcKOs did not express Pax6 (arrows in Figures 8I, and 8I′), suggesting that the floxed Pax6 allele deleted more efficiently than the floxed Foxg1 allele. This mosaicism allowed us to compare how the presence or absence of Foxg1 in neighboring cortical cells affected their response to Pax6 deletion.
In clusters that retained Foxg1 expression in E15.5 Pax6;Foxg1 dcKOs (i.e., single Pax6 cKO cells), only a small percentage of cells expressed Tbr1 (12% ± 3 SD). These had reached the outermost layer of the cortical plate (Figures 8J–8M), agreeing closely with the pattern and proportions of Tbr1-expressing cells (11% ± 2 SD) seen in E15.5 Pax6 single cKO embryos (Figures 8G and 8H). On the other hand, the majority (59% ± 7 SD) of cells that were null for both Pax6 and Foxg1 in dcKOs expressed Tbr1, indicating that neuronal differentiation was much more advanced in this population. To confirm that the differential outcome between Foxg1-positive and Foxg1-negative cells was a consequence of Pax6 removal, we compared clusters of Foxg1-positive and Foxg1-negative cells in Foxg1 single cKOs, in which Pax6 expression was normal. In these embryos, large proportions of both Foxg1-positive and Foxg1-negative cells throughout the cortical plate were Tbr1 positive (Figures 8N–8P).
These results indicate that although Pax6 normally promotes differentiation over proliferation in embryonic cortex, this effect is Foxg1 dependent. If Foxg1 is absent, Pax6 promotes cortical cell cycle progression, and Pax6 removal is anti-proliferative and pro-differentiative.
Discussion
Previous studies on non-neural cells from the lens of the eye, β-cells of the pancreas, and neural cells from the forebrain have reported that Pax6 has partially overlapping effects on the transcriptomes of these highly divergent Pax6-expressing cell types (Mitchell et al., 2017, Sun et al., 2015, Xie et al., 2013). How the molecular actions of Pax6 compare between subdivisions of the forebrain was not clear. In this study, we first explored the differences between the transcriptomes of embryonic cortical and diencephalic (thalamic and prethalamic) cells in control brains and in brains from which Pax6 had been deleted. In controls, there were many more differences between cortical and diencephalic cells (from either Th or PTh) than between thalamic and prethalamic cells. The gene expression changes that Pax6 deletion introduced within cortex were substantially different from those in the diencephalic tissues.
We found that whereas Pax6 regulates the balance between proliferation and differentiation in all three forebrain regions, it affects this process in opposite directions in cortex versus diencephalon. When we tested possible mechanisms to explain this, we found that the presence of Foxg1 in the cortex is a major factor. In vertebrates, the presence of Foxg1 protein is a hallmark of telencephalon (Kumamoto and Hanashima, 2017), the anteriormost part of the developing forebrain, which gives rise to the cerebral cortex dorsally and basal ganglia ventrally. Its expression is restricted to the telencephalic anlage in the anterior neural plate as this region folds, closes, and expands. It is present in the nuclei of telencephalic progenitors and affects their proliferation, maintaining a normal rate of cell cycle progression and preventing their exit from the cell cycle (Martynoga et al., 2005, Xuan et al., 1995). Its ability to bind DNA is required for it to keep progenitors in a proliferative state (Hanashima et al., 2002). This requirement is cell autonomous, as evidenced by our previous work on the cortex of Foxg1−/−;Foxg1+/+ chimeras showing that abnormalities of proliferation occur in Foxg1-null cells even if they are surrounded by wild-type cells (Manuel et al., 2011). Foxg1 is highly conserved not only in vertebrates but also in invertebrates that lack a telencephalon (Bredenkamp et al., 2007). In invertebrates, Foxg1 or its homologs are expressed in, and have an instructive role in the development of, cells situated anteriorly in the developing nervous system (Grossniklaus et al., 1994, Pani et al., 2012, Toresson et al., 1998). It is likely that the acquisition of Foxg1 by anterior neural tissue has been an important driver of this region's evolutionary expansion.
Like Foxg1, Pax6 is highly conserved across the animal kingdom and is essential for normal anterior neural development in both invertebrates and vertebrates (Manuel et al., 2015, Yuan et al., 2016). Unlike Foxg1, however, Pax6 promotes exit from the cell cycle in the mammalian cerebral cortex. On the face of it, this seems paradoxical because it would limit the production of progenitors and hence, ultimately, the numbers of neurons in a structure, one of whose major defining features is its rapid evolutionary expansion. It is conceivable that Pax6 actually evolved as a promoter of cell cycle re-entry, a feature that it demonstrates in the diencephalon, but that the superimposition of Foxg1 expression in the telencephalon has had the effect of reversing this activity. In a context where Foxg1 is generating a powerful drive to cell cycle re-entry, Pax6, although possessing the potential to promote proliferation, might have become a brake to Foxg1's drive, preventing the counterproductive retention of large numbers of cells in a progenitor state.
More broadly, the effects of the transcription factor Pax6 on the cell cycle are far from clear. For example, its role in different tumor cell types is contradictory. It has been shown to have both oncogenic and tumor suppressor effect depending on the tissue affected (Hegge et al., 2018, Muratovska et al., 2003, Zhang et al., 2015, Zhou et al., 2005). Here we show one mechanism by which this versatile transcription factor can control the cell cycle in opposite ways, depending on the presence or absence of another important transcription factor. This might be important for determining the nature of Pax6's role in different tumor types.
How the expression of Foxg1 leads to a reversal of Pax6's actions on proliferation is likely to be complex, with some or all of a number of possible mechanisms operating. Pax6 and Foxg1 might converge directly on the same downstream cell cycle genes to regulate their expression. For example, ChIP-seq studies on Pax6 and Foxg1 have shown that both transcription factors can bind genomic regions likely to regulate the expression of cell cycle regulators including Ccnd1 (Bulstrode et al., 2017, Sun et al., 2015). In the absence of Foxg1, Pax6 binding might activate expression of genes such as Ccnd1 that promote cell cycle re-entry. Pax6 binding might have an opposite effect if Foxg1 is present because Pax6 binding, although pro-proliferative, might interfere with a potentially more powerful pro-proliferative effect of Foxg1 binding, resulting in an inhibition of cell cycle re-entry. In other words, removal of Pax6 might allow Foxg1 freer rein to drive proliferation. This could occur through direct competition between Pax6 and Foxg1 for binding, a phenomenon observed in other systems (Hong and Wu, 2010, Ilsley et al., 2017, Ngondo-Mbongo et al., 2013, Norton et al., 2017, Wan et al., 2011, Zabet and Adryan, 2013). Another possibility is that, in common with other Fox transcription factors, Foxg1 might be a chromatin remodeling (or pioneer) transcription factor that opens the chromatin to allow proteins such as Pax6 to access sites that they would not otherwise be able to bind, thereby changing its function (Golson and Kaestner, 2016, Iwafuchi-Doi and Zaret, 2016, Magnani et al., 2011). Another possibility is that Foxg1 modifies Pax6's function by binding to it rather than to DNA.
Although Ccnd1 emerges as a clear example of a gene that may explain the context-dependent role of Pax6, it is likely to be just one of a cohort of genes that act as effectors of Pax6 and Foxg1. Genes showing opposite regulation in cortex versus diencephalon and opposite regulation in cortex after Foxg1 versus Pax6 loss, such as Chek1 or Ccnc (Figure 7B), could represent good candidates. Fgfr1 emerges as a powerful contender to be directly co-regulated by both transcription factors because ChIP-seq data show it has overlapping peaks for Pax6 and Foxg1 binding in an intronic region (Bulstrode et al., 2017, Sun et al., 2015).
Pax6 and Foxg1 might not converge directly on the same downstream cell cycle genes, rather they might act on progenitors independently through separate routes. Our data indicate that there are many more differences than similarities in the genes regulated by Pax6 and Foxg1, with both regulating many other transcription factors that have actions on progenitor proliferation. It is possible that these Pax6-and Foxg1-regulated transcription factors then control the activity of more direct cell cycle regulators. Indeed, given the breadth of Pax6's and Foxg1's actions, it seems likely that multiple mechanisms will need to be invoked to explain how Pax6 affects the balance of proliferation and differentiation in opposite directions in cortex and diencephalon. In the future, it would be interesting to assess the interplay between Pax6 and Foxg1 by co-misexpression experiments, for example, in neural stem cell culture or by in utero electroporation.
One possibility we considered was that regional variation in an attribute of Pax6 itself, such as its expression level or its exon usage, might cause it to operate differently in the cortex and diencephalon. Variation in Pax6's level and the ratio between its two major splice variants, Pax6 and Pax6(5a), can modify its function (Chauhan et al., 2004, Haubst et al., 2004, Manuel et al., 2006). We found, however, that regional variation in levels of Pax6 protein and in the ratio between exon 5/5a usage did not correlate in a straightforward way with the effects of Pax6 on the balance of proliferation versus differentiation. Previous work has shown that both Pax6 and its rarer variant Pax6(5a) can suppress proliferation and that the level of suppression is dose dependent across the cortex (Haubst et al., 2004, Manuel et al., 2006, Mi et al., 2013). Levels of Pax6 are, however, very high in PTh, where our new evidence indicates that Pax6 promotes proliferation, an effect that is shared with neighboring Th, where Pax6 levels are relatively low. It is unclear how the ∼2-fold difference in exon 5/5a usage between ACtx and PTh, but not between ACtx and Th, might contribute to the opposite effects of Pax6 on proliferation in ACtx versus both PTh and Th. It is conceivable that the actions of the two forms might oppose each other in the context of prethalamic cells and that higher levels of Pax6 relative to Pax6(5a) in PTh might promote proliferation in this region, but at present there is no evidence for or against this possibility.
We also considered the possibility that a differential effect on canonical Wnt signaling might contribute. In eye development, Pax6 directly and positively regulates the expression of Wnt inhibitors, such as Sfrp1, Sfrp2, and Dkk1, thereby suppressing canonical Wnt signaling, with absence of Pax6 leading to aberrant canonical Wnt activity (Machon et al., 2010). In the forebrain, our results indicated that Pax6 loss caused significant upregulation of canonical Wnt signaling (indicated by increased Lef1 and Axin2 expression) in diencephalic structures but not in the cortex. It appears that Pax6's effects on canonical Wnt signaling are largely restricted to the diencephalon, where many are striking, such as the almost complete loss of the Wnt antagonist Sfrp2 from PTh progenitors. Although these changes would likely affect the behaviors of diencephalic progenitors, previous studies would predict increased canonical Wnt signaling to promote proliferation (Stolz and Bastians, 2015), whereas we observed the opposite following Pax6 loss from Th and PTh. It seems very likely that multiple synergistic and antagonistic factors combine to generate the tissue specificity of Pax6's net cellular effects, but we need a much better understanding of how contributing factors act and interact in the different contexts presented by different brain regions.
In conclusion, we have discovered that Pax6 has opposite molecular and cellular effects on the balance of proliferation and differentiation in two major forebrain regions, the cortex and diencephalon. We provide evidence that a major reason for these differences is the presence of Foxg1 in cortical cells, which reverses the cortical actions of Pax6 so that they occur in the same direction as in the diencephalon.
Limitations of the Study
Our cell cycle parameter quantifications were performed at the population level, assuming that all cell progenitors are homogeneous and divide at equivalent rates. We know the cortex, and probably the Th, contain at least two main classes of progenitors (apical and basal) with different functions and dynamics. With our tools, we could not discern between these two types of progenitor pools. In the future it would be very interesting to use in vivo cell imaging or single-cell transcriptomics to tackle this matter.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
An MRC UK Research Grant (N012291), a BBSRC UK Research Grant (N006542), a Royal Society UK Fellowship (NF151396), and a Marie Curie Fellowship EC (624441) funded the work; the Foxg1fl mice were a gift from Dr. Goichi Miyoshi (Tokyo Women's Medical University) and Dr. Gordon Fishell (Harvard Medical School). RNA-seq was performed at Edinburgh Genomics (University of Edinburgh). We thank Chrysoula Giasafaki for her help in the Wnt study, Dr. Crispin Jordan for help with the statistical analysis, Dr. J Pinson for western blots, and Dr. U Borrello for the Ccnd1 probe.
Author Contributions
I.Q.-U. and D.J.P. were involved in overall conceptualization, data interpretation, and manuscript preparation. I.Q.-U., S.M., T.T., and M.M. carried out wet-laboratory experiments; I.Q.-U. and Z.K. did the bioinformatics and statistics. J.O.M., D.J.P., and I.Q.-U. acquired funding, and J.O.M. and D.J.P. supervised the work.
Declaration of Interests
The authors declare no competing interests.
Published: December 21, 2018
Footnotes
Supplemental Information includes Transparent Methods, 8 figures, and 10 tables and can be found with this article online at https://doi.org/10.1016/j.isci.2018.11.031.
Data and Software Availability
RNA-seq raw data of the Pax6 deletion experiment can be obtained from the European Nucleotide Archive (www.ebi.ac.uk/ena; Project PRJEB9747). RNA-seq raw data of the Foxg1 deletion experiment can be obtained from the European Nucleotide Archive (www.ebi.ac.uk/ena; PRJEB21349).
Supplemental Information
Adjusted p < 0.05.
Adjusted p < 0.05.
References
- Adutwum-Ofosu K.K., Magnani D., Theil T., Price D.J., Fotaki V. The molecular and cellular signatures of the mouse eminentia thalami support its role as a signalling centre in the developing forebrain. Brain Struct. Funct. 2016;221:3709–3727. doi: 10.1007/s00429-015-1127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan D.W., Thor S. Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip. Rev. Dev. Biol. 2015;4:505–528. doi: 10.1002/wdev.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artegiani B., Lindemann D., Calegari F. Overexpression of cdk4 and cyclinD1 triggers greater expansion of neural stem cells in the adult mouse brain. J. Exp. Med. 2011;208:937–948. doi: 10.1084/jem.20102167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenkamp N., Seoighe C., Illing N. Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev. Genes Evol. 2007;217:227–233. doi: 10.1007/s00427-006-0128-x. [DOI] [PubMed] [Google Scholar]
- Bulstrode H., Johnstone E., Marques-Torrejon M.A., Ferguson K.M., Bressan R.B., Blin C., Grant V., Gogolok S., Gangoso E., Gagrica S. Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev. 2017;31:757–773. doi: 10.1101/gad.293027.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I.M., Manuel M.N., Molinek M., Quintana-Urzainqui I., Mi D., Shimogori T., Price D.J. Cell-autonomous repression of Shh by transcription factor Pax6 regulates diencephalic patterning by controlling the central diencephalic organizer. Cell Rep. 2014;8:1405–1418. doi: 10.1016/j.celrep.2014.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan B.K., Yang Y., Cveklová K., Cvekl A. Functional interactions between alternatively spliced forms of Pax6 in crystallin gene regulation and in haploinsufficiency. Nucleic Acids Res. 2004;32:1696–1709. doi: 10.1093/nar/gkh334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C., Fink A., Charmaine L., Pham D., Ray D.A.M., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano C.A., Phoenix T.N., Kokovay E., Lowry N., Elkabetz Y., Dimos J.T., Lemischka I.R., Studer L., Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson K.L., Callaghan S.M., O’Hare M.J., Park D.S., Slack R.S. The Rb-CDK4/6 signaling pathway is critical in neural precursor cell cycle regulation. J. Biol. Chem. 2000;275:33593–33600. doi: 10.1074/jbc.M004879200. [DOI] [PubMed] [Google Scholar]
- Georgala P.A., Carr C.B., Price D.J. The role of Pax6 in forebrain development. Dev. Neurobiol. 2011;71:690–709. doi: 10.1002/dneu.20895. [DOI] [PubMed] [Google Scholar]
- Georgala P.A., Manuel M., Price D.J. The generation of superficial cortical layers is regulated by levels of the transcription factor Pax6. Cereb. Cortex. 2011;21:81–94. doi: 10.1093/cercor/bhq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golson M.L., Kaestner K.H. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Cadigan K.M., Gehring W.J. Three maternal coordinate systems cooperate in the patterning of the Drosophila head. Development. 1994;120:3155–3171. doi: 10.1242/dev.120.11.3155. [DOI] [PubMed] [Google Scholar]
- Hanashima C., Shen L., Li S.C., Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 2002;22:6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubst N., Berger J., Radjendirane V., Graw J., Favor J., Saunders G.F., Stoykova A., Gotz M. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development. 2004;131:6131–6140. doi: 10.1242/dev.01524. [DOI] [PubMed] [Google Scholar]
- Hegge B., Sjøttem E., Mikkola I. Generation of a PAX6 knockout glioblastoma cell line with changes in cell cycle distribution and sensitivity to oxidative stress. BMC Cancer. 2018;18:1–19. doi: 10.1186/s12885-018-4394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.W., Wu L.C. ZAS3 represses NFκB-dependent transcription by direct competition for DNA binding. BMB Rep. 2010;43:807–812. doi: 10.5483/BMBRep.2010.43.12.807. [DOI] [PubMed] [Google Scholar]
- Ilsley M.D., Gillinder K.R., Magor G.W., Huang S., Bailey T.L., Crossley M., Perkins A.C. Krüppel-like factors compete for promoters and enhancers to fine-tune transcription. Nucleic Acids Res. 2017;45:6572–6588. doi: 10.1093/nar/gkx441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M., Zaret K.S. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.S., Anderson S.A., Rubenstein J.L., Lowenstein D.H., Pleasure S.J. Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J. Neurosci. 2001;21:RC132. doi: 10.1523/JNEUROSCI.21-05-j0002.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann K., Sexl V. CDK6 and p16INK4A in lymphoid malignancies. Oncotarget. 2013;4:1858–1859. doi: 10.18632/oncotarget.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto T., Hanashima C. Evolutionary conservation and conversion of Foxg1 function in brain development. Dev. Growth Differ. 2017;59:258–269. doi: 10.1111/dgd.12367. [DOI] [PubMed] [Google Scholar]
- Lange C., Huttner W.B., Calegari F. Cdk4/CyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Machon O., Kreslova J., Ruzickova J., Vacik T., Klimova L., Fujimura N., Lachova J., Kozmik Z. Lens morphogenesis is dependent on Pax6-mediated inhibition of the canonical Wnt/Beta-catenin signaling in the lens surface ectoderm. Genesis. 2010;48:86–95. doi: 10.1002/dvg.20583. [DOI] [PubMed] [Google Scholar]
- Magnani L., Eeckhoute J., Lupien M. Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends Genet. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Manuel M., Price D.J. Role of Pax6 in forebrain regionalization. Brain Res. Bull. 2005;66:387–393. doi: 10.1016/j.brainresbull.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Manuel M., Georgala P.A., Carr C.B., Chanas S., Kleinjan D.A., Martynoga B., Mason J.O., Molinek M., Pinson J., Pratt T. Controlled overexpression of Pax6 in vivo negatively autoregulates the Pax6 locus, causing cell-autonomous defects of late cortical progenitor proliferation with little effect on cortical arealization. Development. 2006;134:545–555. doi: 10.1242/dev.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M.N., Martynoga B., Molinek M.D., Quinn J.C., Kroemmer C., Mason J.O., Price D.J. The transcription factor Foxg1 regulates telencephalic progenitor proliferation cell autonomously, in part by controlling Pax6 expression levels. Neural Dev. 2011;6:1–12. doi: 10.1186/1749-8104-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel M.N., Mi D., Mason J.O., Price D.J. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 2015;9:1–21. doi: 10.3389/fncel.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J., Coppola G., Zhang P., Abyzov A., Tomasini L., Amenduni M., Szekely A., Palejev D., Wilson M., Gerstein M. Foxg1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell. 2016;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B., Morrison H., Price D.J., Mason J.O. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 2005;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Matsushime H., Quelle D.E., Shurtleff S.A., Shibuya M., Sherr C.J., Kato J.Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi D., Carr C.B., Georgala P.A., Huang Y.T., Manuel M.N., Jeanes E., Niisato E., Sansom S.N., Livesey F.J., Theil T. Pax6 Exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and Hypophosphorylation of pRb. Neuron. 2013;78:269–284. doi: 10.1016/j.neuron.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R.K., Nguyen-Tu M.-S., Chabosseau P., Callingham R.M., Pullen T.J., Cheung R., Leclerc I., Hodson D.J., Rutter G.A. The transcription factor Pax6 is required for pancreatic β cell identity, glucose-regulated ATP synthesis, and Ca 2+ dynamics in adult mice. J. Biol. Chem. 2017;292:8892–8906. doi: 10.1074/jbc.M117.784629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G., Fishell G. Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron. 2012;74:1045–1058. doi: 10.1016/j.neuron.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D.O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Muratovska A., Zhou C., He S., Goodyer P., Eccles M.R. Paired-Box genes are frequently expressed in cancer and often required for cancer cell survival. Oncogene. 2003;22:7989–7997. doi: 10.1038/sj.onc.1206766. [DOI] [PubMed] [Google Scholar]
- Ngondo-Mbongo R.P., Myslinski E., Aster J.C., Carbon P. Modulation of gene expression via overlapping binding sites exerted by ZNF143, Notch1 and THAP11. Nucleic Acids Res. 2013;41:4000–4014. doi: 10.1093/nar/gkt088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton L.J., Hallal S., Stout E.S., Funnell A.P.W., Pearson R.C.M., Crossley M., Quinlan K.G.R. Direct competition between DNA binding factors highlights the role of Krüppel-like Factor 1 in the erythroid/megakaryocyte switch. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-03289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani A.M., Mullarkey E.E., Aronowicz J., Assimacopoulos S., Grove E.A., Lowe C.J. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483:289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L.-J., Patti D., Marcy G., Ollier E., Pfister S., Douglas R.J., Betizeau M., Gautier E., Cortay V., Doerflinger N. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc. Natl. Acad. Sci. USA. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinson J., Mason J.O., Simpson T.I., Price D.J. Regulation of the Pax6: Pax6(5a) mRNA ratio in the developing mammalian brain. BMC Dev. Biol. 2005;5:4–7. doi: 10.1186/1471-213X-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Bastians H. Fresh WNT into the regulation of mitosis. Cell Cycle. 2015;14:2566–2570. doi: 10.1080/15384101.2015.1064569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Gruss P. Roles of Pax-genes in developing and adult brain as suggested by expression patterns. J. Neurosci. 1994;14:1395–1412. doi: 10.1523/JNEUROSCI.14-03-01395.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoykova A., Fritsch R., Walther C., Gruss P. Forebrain patterning defects in Small eye mutant mice. Development. 1996;122:3453–3465. doi: 10.1242/dev.122.11.3453. [DOI] [PubMed] [Google Scholar]
- Sun J., Rockowitz S., Xie Q., Ashery-Padan R., Zheng D., Cvekl A. Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015;43:6827–6846. doi: 10.1093/nar/gkv589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Hirano A., Ogawa M., Kataoka A., Yoshida A.C., Itoh D., Ueno M., Blackshaw S., Shimogori T. Dynamic spatiotemporal gene expression in embryonic mouse thalamus. J. Comp. Neurol. 2011;519:528–543. doi: 10.1002/cne.22531. [DOI] [PubMed] [Google Scholar]
- Toresson H., Martinez-Barbera J.P., Bardsley A., Caubit X., Krauss S. Conservation of BF-1 expression in amphioxus and zebrafish suggests evolutionary ancestry of anterior cell types that contribute to the vertebrate telencephalon. Dev. Genes Evol. 1998;208:431–439. doi: 10.1007/s004270050200. [DOI] [PubMed] [Google Scholar]
- Tyas D.A., Ian Simpson T., Carr C.B., Kleinjan D.A., van Heyningen V., Mason J.O., Price D.J., Mason -JohnMason J.O. BMC Developmental Biology Functional conservation of Pax6 regulatory elements in humans and mice demonstrated with a novel transgenic reporter mouse. BMC Dev. Biol. 2006;6:21. doi: 10.1186/1471-213X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzali R., Weise S.C., Hellbach N., Machado V., Heidrich S., Vogel T. The FOXG1/FOXO/SMAD network balances proliferation and differentiation of cortical progenitors and activates Kcnh3 expression in mature neurons. Oncotarget. 2016;7:37436–37455. doi: 10.18632/oncotarget.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan P., Hu Y., He L. Regulation of melanocyte pivotal transcription factor MITF by some other transcription factors. Mol. Cell. Biochem. 2011;354:241–246. doi: 10.1007/s11010-011-0823-4. [DOI] [PubMed] [Google Scholar]
- Warren N., Price D.J. Roles of Pax-6 in murine diencephalic development. Development. 1997;124:1573–1582. doi: 10.1242/dev.124.8.1573. [DOI] [PubMed] [Google Scholar]
- Willnow T.E., Hammes A., Eaton S. Lipoproteins and their receptors in embryonic development: more than cholesterol clearance. Development. 2007;134:3239–3249. doi: 10.1242/dev.004408. [DOI] [PubMed] [Google Scholar]
- Xie Q., Yang Y., Huang J., Ninkovic J., Walcher T., Wolf L., Vitenzon A., Zheng D., Götz M., Beebe D.C. Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS One. 2013;8:e54507. doi: 10.1371/journal.pone.0054507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan S., Baptista C.A., Balas G., Tao W., Soares V.C., Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yip D.J., Corcoran C.P., Alvarez-Saavedra M., DeMaria A., Rennick S., Mears A.J., Rudnicki M.A., Messier C., Picketts D.J. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev. Cell. 2012;22:871–878. doi: 10.1016/j.devcel.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Wang W., Hu B., Pan C., Chen M., Ke L., Yang L., Chen J. Cloning and functional analysis of Pax6 from the hydrothermal vent tubeworm Ridgeia piscesae. PLoS One. 2016;11:1–17. doi: 10.1371/journal.pone.0168579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabet N.R., Adryan B. The effects of transcription factor competition on gene regulation. Front. Genet. 2013;4:1–10. doi: 10.3389/fgene.2013.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjatke T., Gak I.A., Kirova D., Fuhrmann M., Daniel K., Gonciarz M., Müller D., Glauche I., Mansfeld J. Quantitative cell cycle analysis based on an endogenous all-in-one reporter for cell tracking and classification. Cell Rep. 2017;19:1953–1966. doi: 10.1016/j.celrep.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang X., Wang J., Liang T., Gu Y., Yang D. Down-regulation of PAX6 by promoter methylation is associated with poor prognosis in non small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015;8:11452–11457. [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-H., Wu X., Tan F., Shi Y.-X., Glass T., Liu T.J., Wathen K., Hess K.R., Gumin J., Lang F. PAX6 suppresses growth of human glioblastoma cells. J. Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- Zilberberg A., Yaniv A., Gazit A. The low density lipoprotein receptor-1, LRP1, interacts with the human frizzled-1 (HFz1) and down-regulates the canonical Wnt signaling pathway. J. Biol. Chem. 2004;279:17535–17542. doi: 10.1074/jbc.M311292200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Adjusted p < 0.05.
Adjusted p < 0.05.