Abstract
Background
Glioblastoma (GBM) is the most common and the most malignant glioma subtype. Among numerous genetic alterations, miRNAs contribute to pathogenesis of GBM and it is suggested that also to GBM recurrence and resistance to therapy. Based on publications, we have selected 11 miRNAs and analyzed their expression in GBM. We hypothesized that selected miRNAs are differentially expressed and involved in primary as well as in recurrent GBM, that show significant expressional differences when different treatment options are in question, and that are related to certain patients and tumor characteristics.
Patients and methods
Paraffin embedded tissues, obtained from primary and corresponding recurrent tumor from 83 patients with primary GBM were used. Eleven miRNAs (miR-7, miR-9, miR-15b, miR-21, miR-26b, miR-124a, miR-199a, let-7a, let-7b, let-7d, and let-7f) were selected for qPCR expression analysis. For patients who received temozolamide (TMZ) as chemotherapeutic drug, O6-methylguanine-DNA methyltransferase (MGMT) methylation status was defined using the methyl-specific PCR.
Results
There was a significant change in expression of miR-7, miR-9, miR-21, miR-26b, mirR-124a, miR-199a and let-7f in recurrent tumor compared to the primary. In recurrent tumor, miR-15b, let-7d and let-7f significantly changed comparing both treatment options. We also observed difference in progression free survival between patients that received radiotherapy and patients that received radiotherapy and chemotherapy, and longer survival for patients who received chemotherapy after second surgery compared to not treated patients. miR-26b showed correlation to progression free survival and let-7f to overall survival. We did not find any expression difference between the tumors with and without methylated MGMT.
Conclusions
Our data suggest that analyzed miRNAs may not only contribute to pathogenesis of primary GBM, but also to tumor progression and its recurrence. Moreover, expression of certain miRNAs appears to be therapy-dependent and as such they might serve as additional biomarker for recurrence prediction and potentially predict a therapy-resistance.
Key words: glioblastoma, recurrent, radiotherapy, chemotherapy, miRNA, expression
Introduction
Gliomas are the most common primary brain tumors and are classified on the basis of histopathological characteristics. Glioblastoma (GBM) is the most common and malignant glioma subtype characterized by rapid growth and poor prognosis.1 Like cancer in general, GBM develop as a consequence of genetic alterations that accumulate with tumor progression. However, pathogenesis of GBM recurrence is still poorly understood. Certain molecular pathways and novel biomarkers have been established as diagnostic, prognostic and predictive markers, such as is epidermal growth factor receptor (EGFR) amplification, O6-methylguanine-DNA methyltransferase (MGMT) methylation and others.1, 2, 3, Among novel biomarkers are non-coding RNAs, of which the most studied are microRNAs (miRNAs). The miRNAs are small RNAs that act as endogenous regulators of gene expressions and function in different physiological processes, contributing to pathogenesis of different types of cancer, including gliomas. miRNA expression might be a potential biomarker for GBM used as diagnostic support or for prognostic and therapeutic application.4, 5, 6, However, differential expression of certain miRNAs might be also involved in pathophysiological mechanisms of GBM recurrence, but there are limited data regarding miRNA expression as GBM progresses (at its recurrence).7, 8, Comprehensive knowledge and understanding of the molecular pathways underlying disease progression, tumor recurrence and response to therapy might be of great importance in future development of more efficient therapies in GBM.
Local infiltration and progressive growth of GBM tumor cells into the adjacent brain tissue invalidate any possibility for a radical surgical tumor resection.9 Consequently, recurrence is observed in almost all tumors.7 Additionally, the difficulty in treatment of this lethal disease is in tumor acquisition of several mechanisms of radio and chemoresistance. In addition to methylation of MGMT10, a change in miRNA’s expression is also a cause for drug resistance in GBM.11 Several miRNAs have been related to radio- and chemo-sensitivity as well as radio- and chemo-resistance of glioma cells, and recognized as crucial regulators during glioma pathogenesis and progression.12 However, a certain degree of this aberrant regulation may also be the consequence of treatment.
In aim to characterize at least some miRNAs that might be involved in GBM formation/progression and recurrence, and may also contribute to therapy resistance, we selected 83 consecutive patients with operable recurrent GBM, where samples from first and from second surgery were available. We selected ten miRNAs, which expression was up-regulated (miR-9, miR-15b, miR-21, miR-26b, let-7a, let-7b, let-7d and let-7f) or down-regulated (miR-7 and miR-124a) in previous studies of GBM, and were differentially expressed from other glioma subtypes.13 We additionally included miR-199a, which is involved in tumorigenesis of various types of cancer, including GBM.14 The majority of these miRNAs have already been shown to be involved in different tumorigenic processes in GBM or glioma cell lines (Table 1).4,15 We hypothesized that: (i) selected miRNAs are deregulated in primary as well as in recurrent GBM; (ii) some miRNAs are also differently expressed between primary and recurrent tumor; (iii) expression of selected miRNAs in recurrent GBM is therapy dependent; (iv) progression free survival and overall survival is different between distinct therapies; (v) expression of some miRNAs is related to progression free survival and overall survival; and (vi) methylation status of MGMT is in correlation to certain miRNAs.
TABLE 1.
List of the analysed miRNAs with known functions in glioma pathogenesis
| MicroRNAs | Invasion | Proliferation | Tu growth | Migration | Cell cycle | cell Survival/death | Other functions |
|---|---|---|---|---|---|---|---|
| miR-7 | + | + | + | nd | + | + | Differentiation, Vasculogenic mimicry |
| miR-9 | nd | + | nd | + | nd | nd | Self-renewal, Vasculogenic mimicry |
| miR-15b | + | + | + | nd | + | + | Angiogenesis |
| miR-21 | + | + | + | + | + | + | / |
| miR-26b | + | + | + | + | nd | + | Vasculogenic mimicry |
| miR-124 | + | + | + | + | + | + | Differentiation, Angiogenesis |
| miR-199a | nd | + | nd | nd | nd | nd | / |
| let-7a | nd | + | + | + | nd | nd | / |
| let-7b | + | nd | nd | + | nd | nd | / |
| let-7d | nd | nd | nd | nd | nd | nd | / |
| let-7f | + | + | nd | + | nd | nd | / |
Patients and methods
Patients and tissue samples
Our retrospective study included eighty-three patients with recurrent GBM. The data were obtained from the patients charts from hospital registry of the Institute of Oncology and from the Cancer Registry of Slovenia. Each patient underwent the first surgery and the second one after the first recurrence of GBM between January 1997 and November 2011 at the University Clinical Centre Ljubljana, Slovenia. The selected patients were consecutive; of the 1117 that underwent the first surgery for GBM, 83 underwent also the second surgeryat tumor recurrence since these patients were capable of re-operation according to clinical standards (Karnofsky performance status over 80). Patients received radiotherapy (RT) after the surgery, and RT with concomitant and adjuvant chemotherapy (ChT) after 2005.
Tissue samples were fixed in 10% buffered formalin and embedded in paraffin (FFPE) for routine diagnostics. The grading criteria based on WHO classification was used.14 Paired tissue samples were retrieved from the archive of the Institute of Pathology, Faculty of Medicine, University of Ljubljana. Samples were classified in two groups, i.e. primary GBM (sample tissue collected at first surgery) and corresponding recurrent GBM (sample tissue collected at second surgery). As control sample RNA FirstChoice Human Brain Reference Total RNA (Cat. no. 6050, Ambion; Invitrogen, USA) was used.
The patients’ data, including age, gender and therapy are summarized in Table 2.
TABLE 2.
Demographic and clinicopathological characteristics of patients included in the study
| Histopathological diagnosis | GBM, grade WHO IV | |
|---|---|---|
| Number of cases | 83 | |
| Mean age (Year) | 50 (min ± 12.8 8, max 71) | |
| Male | 50 (60%) | |
| Gender | ||
| Female | 33 (40%) | |
| Karnofsky performance status ≥ 80 | 83 | |
| radiotherapy after first surgery | 83/83 (100%) | |
| chemotherapy after first surgery | 54/83 (65%) | |
| Adjuvant treatment | ||
| radiotherapy after second surgery | 15/83 (18%) | |
| chemotherapy after second surgery | 47/83 (56%) | |
| MGMT (tested methylated in a subset of patients treated with TMZ) | Primary GBM | 38/47 (81%) |
| Recurrent GBM | 46/47 (98%) | |
| Time to recurrence (months) | 0.84–67.30 | |
| Survival after second surgery (months) | 0.03–25.75 | |
| Overall survival (months) | 0.84–70.48 | |
MGMT = methyl guanine methyl transferase; TMZ = temozolamide
RNA isolation from FFPE tissue samples
Tissue samples were cut at 10 μm from FFPE tissue blocks and for the isolation procedure six to eight 10 μm sections were used. Total RNA isolation was performed using miRNeasy FFPE Kit (Qiagen, Germany) according to the manufacturer’s protocol. The RNA was eluted in 30 μl of nuclease-free water. The yield was measured spectrophotometrically using the NanoDrop-1000 (Thermo Scientific, USA) and the quality was evaluated on the Bioanalyzer 2100 (Agilent Technologies, USA).
Quantitative real-time PCR (qPCR)
All the reagents were from Qiagen, except where otherwise indicated. Quantitative PCR (qPCR) was carried out using the Rotor Gene-Q Real-Time PCR System and all the qPCR reactions were performed in duplicates or triplicates. Prior to qPCR analysis, two pools of RNA samples were created from FFPE tissue samples (primary, recurrence) and, along with reference RNA, tested for qPCR efficiency. Reverse transcription using miScript reverse transcription kit was performed in a 10 μl reaction master mix with 50 ng of total RNA according to manufacturer instruction. The resulting cDNA was diluted 100-fold and qPCR reaction was carried out in 10 μl PCR master mix, according to manufacturer instruction. As the reference genes, RNU6B and SNORD25 were used based on efficiency results. Tested miRNAs were: miR-7, miR-9, miR-15b, miR-21, miR-26b, miR-124a, miR-199a, let-7a, let-7b, let-7d, and let-7f.
DNA isolation
In a subset of patients that were treated with temozolamide (TMZ) (n = 47), MGMT methylation status was determined. Tissue samples from the first surgery and the surgery at first recurrence were cut at 10 μm from FFPE tissue blocks and for the isolation procedure, six to eight 10 μm sections were used. Total DNA isolation was performed using QIAamp DNA FFPE Tissue Kit (Qiagen) according to the manufacturer’s protocol. The DNA was eluted in 60 μl of nuclease-free water. The yield was measured fluorescently using the Quant-It (Life Technologies) according to manufacturer instruction and Rotor Gene Q (Qiagen).
MGMT methylation status analysis
For determining the MGMT methylation status, methyl-specific polymerase chain reaction (MSP) was used in a two-step approach with primers previously described.16 Briefly, prior to MSP, 500 ng of DNA was used for bisulfite conversion using innuCONVERT Bisulfite Basic Kit according to manufacturer instruction (Analytik Jena) and stored at -20°C for subsequent MSP. For MSP, 15 ng of bisulfite converted DNA was used with 0.2 μM of each primer for methylated form and 0.3 μM of each primer for unmethylated form, 2 mM of dNTP and 0.25 U of Hot Master Polymerase (5 Prime), all in 10 μl reaction. Amplification was performed according to manufacturer instruction using 59°C for primer annealing. In each run, fully methylated (EpiTect Control DNA, methylated, Qiagen) as well as fully unmethylated controls (EpiTect Control DNA, unmethlyated, Qiagen) were used as assay controls. Results were analyzed using 2% agarose gel. The investigator who analyzed the GBM samples was blinded to all clinical information.
Statistical analysis
To present a relative gene expression the 2-ΔΔCt method was used.17 For paired samples (primary, recurrent), the calculated ΔCt was tested for statistical significance using Wilcoxon Signed Rank test. For independent group of samples, ΔΔCt was calculated relatively to Human brain Reference RNA and used for Mann-Whitney test (treatment, MGMT status, etc.). Kaplan-Meier curve was used to analyze difference in progression free survival (time to recurrence) and overall survival (different treatment options, miRNA expression). Correlations between scale variables (miRNA expression, survival) were calculated using Pearson’s and Spearman’s correlation coefficient. For all statistical tests, the SPSS analytical software (ver.24 SPSS Inc. IL) was used (cut-off point at p < 0.05). The workflow of comparisons is summarized in Figure 1.
FIGURE 1.
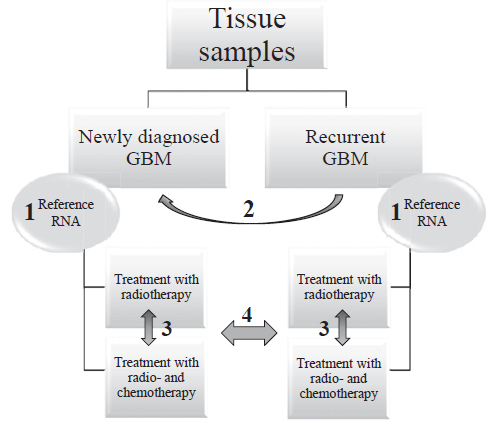
Schematic representation of the workflow and statistical comparisons.
GBM = glioblastoma; Reference RNA = Human Brain Reference RNA; 1 = Willcoxon Signed Rank Test between primary glioblastoma and corresponding recurrent glioblastoma using ΔCt; 2 = normalization to Human Brain reference RNA resulting in ΔΔCt; 3 and 4 = Mann-Whitney test between independent groups of samples using ΔΔCt (normalization of glioblastoma samples to Human Brain Reference RNA)
Compliance with ethical standards
Study has been approved by Slovene National Medical Ethics Committee (Number 113/05/13).
Results
Expression of miRNAs is changed in primary and recurrent GBM
Comparing expression of analyzed miRNAs to Human Brain Reference RNA (ΔΔCt), we found altered expression for all 11 analyzed miRNAs both in primary as well as in recurrent GBM. Whereas miR-9 and miR-21 were mainly up-regulated and miR-15b was mainly down-regulated in primary and recurrent GBM, all other miRNAs showed more heterogeneous expression when compared to the Human Brain Reference RNA. Results are summarized in Figure 2.
FIGURE 2.
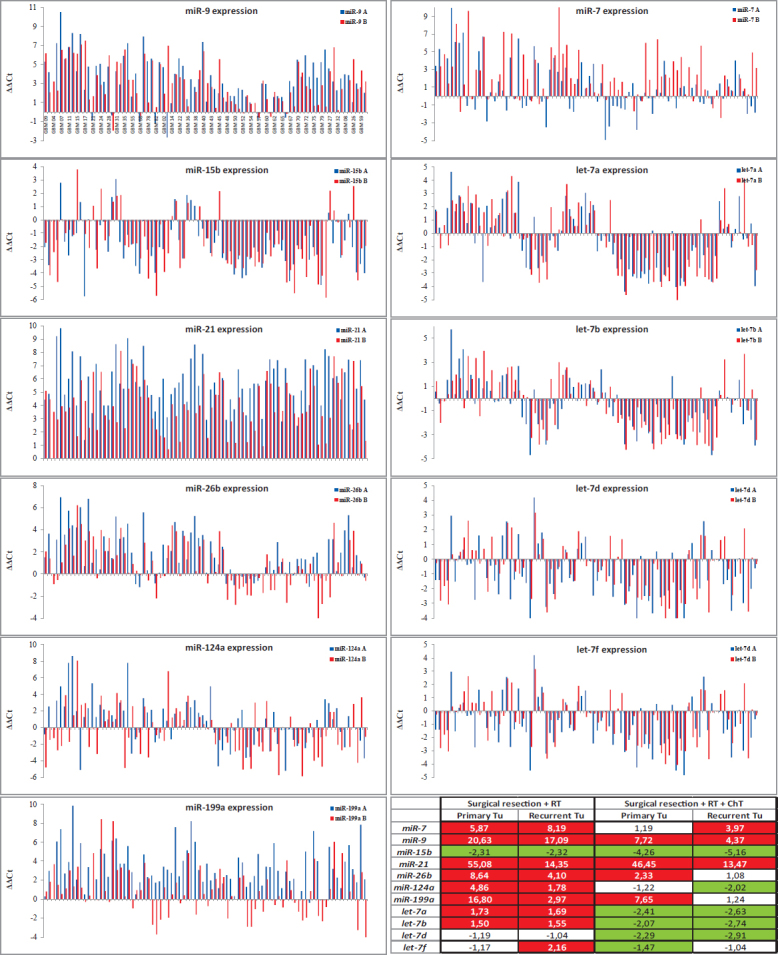
Expression of miRNAs in primary and recurrent GBM (compared to the Human Brain Reference RNA, ΔΔCt) with Table presenting Average Fold Changes.
ChT = chemotherapy; RT = radiotherapy; A = primary GBM; B = recurrent GBM
miRNAs in recurrent GBM are differentially expressed compared to primary GBM
Using calculated ΔCt and Wilcoxon Signed Rank test, we have observed significant alteration in expression for 7 miRNAs between primary and recurrent GBM. Five miRNAs, miR-9 (p = 0.016), miR-21 (p < 0.001), miR-26b (p < 0.001), miR-124 (p = 0.029) and miR-199a (p < 0.001), were down-regulated, and miR-7 (p = 0.001) and let-7f (p < 0.001) were up-regulated in recurrent tumors compared to first time surgery. However, all of these miRNAs were differentially expressed also in a subgroup treated with RT and ChT, whereas only four of these showed statistically significant differential expression in a subgroup treated with RT alone. Results are summarized in Figure 3 and Figure 2 (Average Fold Change Table).
FIGURE 3.
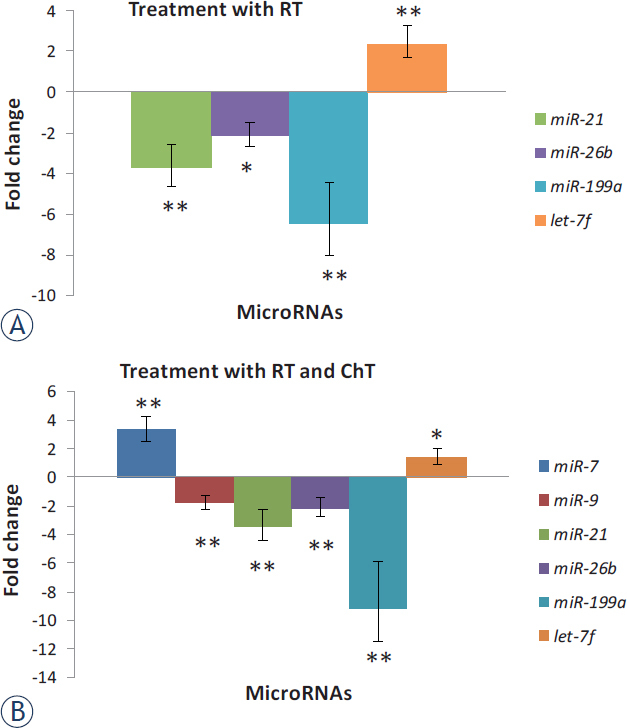
Expression of miRNAs in recurrent glioblastoma (GBM) after treatment compared to expression in primary GBM. Results are represented separately for patients treated with RT and those treated with RT and ChT.
ChT = chemotherapy; RT = radiotherapy; * = significant differences in expression of miRNAs, p < 0.05; ** = p < 0.01
Distinctive expression of miRNAs after different treatment options in recurrent GBM
Using Mann-Whitney test for comparing subgroup of patients treated with RT alone to patients treated with RT and ChT (analyzed miRNAs normalized to Human Brain Reference RNA, ΔΔCt), we observed that expression of certain miRNAs was different before treatment (at first surgery) as well as after treatment (at second surgery). Therefore, only those miRNAs that showed significantly differential expression only after treatment (at second surgery) were considered truly as a consequence of treatment. Both miR-15b and miR-7d were down-regulated compared to Human Brain Reference RNA after treatment, and there was significant change in expression between patients that received RT alone and those that received RT and ChT (p = 0.013 for miR-15b and p = 0.008 for let-7d). In patients treated with RT alone, let-7f was up-regulated compared to Human Brain Reference RNA at second surgery, whereas it was unchanged in patients that were treated with RT and ChT. There was significant change in expression of let-7f between both groups (p = 0.007). Results are summarized in Figure 4.
FIGURE 4.
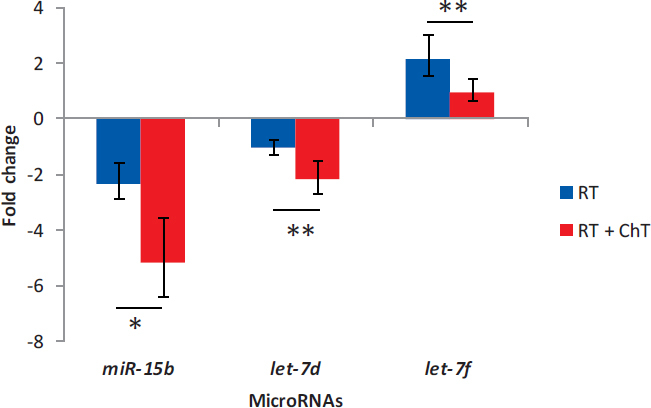
Distinctive expression of miRNAs after different treatment options in recurrent glioblastoma
ChT = chemotherapy; RT = radiotherapy; * = p < 0.05; ** = p < 0.01
Correlation between progression free survival, survival after second surgery, overall survival, treatment and miRNA expression
There was weak, but statistically significant change in progression free survival between patients who received RT alone and those who received both, RT and ChT after first surgery (7 months and 11 months, respectively; p = 0.045). Results are summarized in Figure 5A
FIGURE 5.
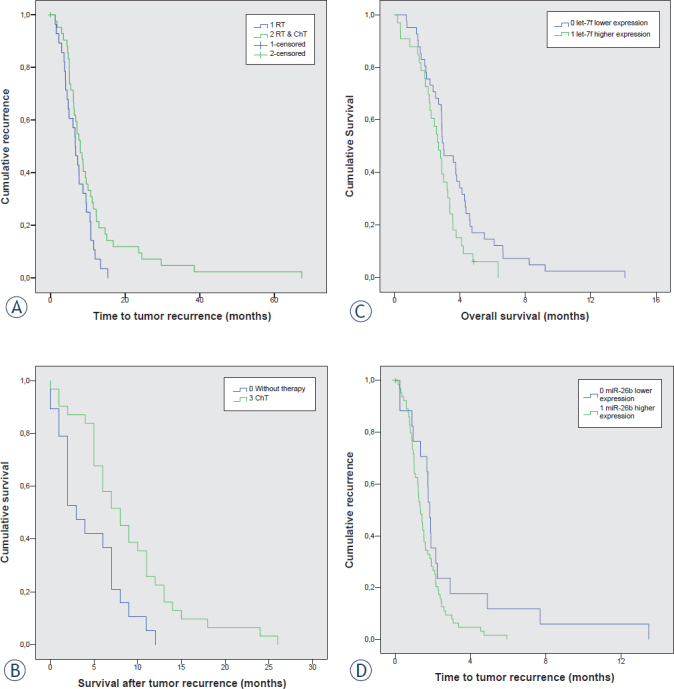
Survival curve analyses; (A) Progression free survival between patients that received RT alone or both, RT and ChT; (B) Survival time after second surgery for patients who received no therapy versus patients who received ChT; (C) Progression free survival between patients with up- and downregulated miR-26b; (D) Progression free survival between patients with up- and downregulated let-7f.
ChT = chemotherapy; RT = radiotherapy
We have also calculated the survival time from second surgery. The number of patients who received RT alone or RT and ChT was too small for reliable analysis of survival time. However, there was statistically significant longer overall survival for patients who received ChT compared to those that were not treated after second surgery (8 months and 5 months, respectively; p = 0.004). Results are summarized in Figure 5B.
We further calculate Spearman’s and Pearson’s correlation coefficient between miRNA expression (analyzed miRNAs normalized to Human Brain Reference RNA, ΔΔCt) in primary GBM and progression free survival and overall survival, and found weak inverse correlation for let-7b and progression free survival (r = -0.220, p = 0.049) and weak inverse correlation between let-7f and overall survival (r = -0.226, p = 0.050). We further analysed difference in Kaplan-Meier curve for patients with up- and down-regulated miRNAs and found, that there is a difference in progression free between patients with up- and down-regulated miR-26b (p = 0.05, Figure 5C), and difference in overall survival for up- and down regulated let-7f (p = 0.021).
miRNAs and MGMT methylation status
Among patients treated with ChT, there were 47 treated with TMZ. Among them, there were 9 patients with unmethylated and 34 with methylated MGMT. All, except one, became methylated in recurrent tumor. Comparing miRNA expression normalized to Human Brain Reference RNA between MGMT methylated and MGMT unmethylated tumors revealed no correlation of investigated miRNA expression to MGMT methylation status.
Discussion
In our 83 consecutive patients operated for primary GBM and re-operated at the first recurrence of GBM, we have analyzed the expression of 11 selected miRNAs (miR-7, miR-9, miR-15b, miR-21, miR-26b, miR-124a miR-199a, let-7a, let-7b, let-7d, and let-7f). However, to the best of our knowledge, this is the first report of these miRNAs expression in recurrent GBM.
Expression of analyzed miRNAs in primary and recurrent GBM
We observed differential expression of the majority of analyzed miRNAs in both samples. In primary as well as in recurrent GBM we have observed overall miR-7 over-expression, although was expression heterogeneous among tumors. In spite of numerous studies on miR-7 expression, its expression in recurrent GBM has not been analyzed yet. Increased expression of miR-7 was found to suppress GBM cell proliferation, induce apoptosis, inhibit cell migration in vitro, and reduce tumorigenicity in vivo.4 However, in contrast to our study, other research group showed miR-7 downregulation in patients with GBM.18 Possible explanation for different outcomes might be in different normalization strategies used for qPCR (i.e. different reference genes used). miR-9 over-expression, observed in our primary and recurrent GBM samples, might contribute to tumor progression since it has been identified that up-regulation of miR-9 predicts an unfavorable prognosis and correlates with tumor progression.19 Similarly to other cancer research20, we observed increased expression of oncogenic miR-21 in primary and recurrent GBM, although its’ up-regulation in recurrent tumors is significantly lower compared to primary tumors. miR-26b was also found to be mainly over-expressed, although there are samples with its downregulation, but in contrast to our results, it has been shown that miR-26b was decreased in glioma cell lines and inversely correlated with the grade of glioma in humans.21 The discrepancy could be due to different normalization strategies. We normalized expression of miRNAs in GBM samples to Human Brain Reference RNA, in others the normal brain tissue was used for normalization.21 Expression of miR-124a in our research showed up-regulation in certain samples of primary tumors. Published data showed decreased miR-124a expression in majority of GBM samples due to hyper-methylation of promoter region of miR-124a.22 Speculatively, different methylation status of miR-124a might be observed in our samples. We also observed mainly higher expression level of miR-199a in primary GBM samples and near to normal level in recurrent GBM. miR-199a up-regulation in gliomas has been related to hypo-methylation of one of two copies of miR-199a within genome consequently leading to its up-regulation.14 miR-199a is believed to have diverse biological functions in different tissues. It behaves as a tumor-suppressor in certain cancer types14; however, its exact role in GBM is yet to be defined. miRNA of let-7 family (let-7a/b/d/f) showed the most heterogeneous expression across both, primary and recurrent tumor samples.15,23
Expression of miRNA in recurrent and primary GBM and its correlation to the choice of therapy
Our data also showed statistically significant alteration of expression of seven miRNAs (miR-7, miR-9, miR-21, miR-26b, miR-124a, miR-199a and let-7f) between primary and recurrent tumors. After receiving RT and ChT, we observed higher levels of miR-7 in recurrent compared to the primary GBM, whereas in patients treated solely using RT there was no significant difference in expression. Based on glioma cell line models, up-regulation of miR-7 increases radio-sensitivity.24 Several target genes that might be involved in sensitivity of glioma cells to therapy has been shown for miR-7, including EGFR, IRS1 and IRS2 regulating differentiation, invasion and proliferation4, as well as IGF-1R/Akt signalling pathway, regulating cellular growth and glucose metabolism in gliomas.25,26 We did observe a decreased expression of miR-9 in recurrent compared to primary GBM, however, miR-9 is still over-expressed in recurrent GBM. miR-9 has a role in chemotherapeutic resistance by regulating the p38 mitogen activated protein kinase signaling network, involved in regulation of cell stress and cell differentiation; it influences proliferation and self-renewal of glioma cells by targeting SOX2; and through miRNA regulation of PTCH1, receptor in Sonic Hedgehog signaling.26 miR-21 has been shown to be up-regulated in more radio-resistant cell lines 12 mediating the resistance of GBM cells against RT27 and TMZ.20 miR-21 is substantially implicated in pathogenesis of GBM by targeting PTEN, RECK, PDCDC4, TIMP3, BCL2, SPRY2, MTAP, SOX5, JMY, TGFRBR2, TGFBR3, TP73L, APAFI, BMPR2, TOPORS, DAXX, TP53BP2, and PPIF. All this target genes are involved either in pathways of apoptosis, invasion, proliferation and tumor growth either in chemo-resistance.4 Therefore, all these results suggest that lower expression of miR-21 after treatment might implicate less resistant glioma cells to therapy; however, as well as miR-9, it is still up-regulated in recurrent GBM, possibly contributing to tumor progression.
Decrease expression of miR-26b in our recurrent samples compared to primary GBM, could be a consequence of tumor progression. Accordingly, it was found that ectopic expression of miR-26b inhibits proliferation, migration and invasion of human glioma cells by repressing the endogenous level of EphA2 protein.21 We further observed decrease in miR-124a expression in recurrent tumors compared to primary, which might be a consequence of tumor progression. It has been shown that miR-124a radio-sensitizes human glioma cells by targeting CDK4.28 Several other targets have been also demonstrated for miR-124a, as are CDK6, SCP1, PTBP1, ITGB1, LAMCI, IQGAPI, RB and STAT3, regulating cell cycle, differentiation, invasion and proliferation of glioma cells and T cell-mediated immune clearance of glioma.4,29 All these processes may be involved in sensitivity and survival of glioma cells.
In recurrent GBM, but not in primary GBM, we found significant change in expression between samples of patients who were treated with RT and ChT and those that were treated with RT alone for only miR-15b, let-7d and let-7f. Let-7f was over-expressed in recurrent compared to the primary GBM; however, it was up-regulated only in patients treated with RT after first surgery. It has been shown that members of let-7 miRNA family (including let-7f) were mostly up-regulated in more radio-resistant cell lines12 suggesting radio-resistant function in glioma cells, since its expression after receiving RT and ChT was back to normal. There was observed normal expression of let-7d in GBM patients after receiving RT, however, after treating patients with RT and ChT, its expression was decreased and similar to that in primary GBM. There are limited data about the role of let-7d in the cell response to radiation and chemotherapeutic drugs in glioma, since depends on dose, time after irradiation and source of oxidative stress.30 There are indication that let-7d down-regulation in GBM might be involved in modulation of chemo-resistance to TMZ.11 One of the target genes of let-7d is oncogene HMGA231 and studies suggest that overexpression of HMGA2 may participate in regulating tumor cell invasion in high grade gliomas.32 However, whether HMGA2 is up-regulated in GBM due to let-7d down-regulation, is yet to be defined. Another miRNA, miR-15b, was found in our study to be significantly under-expressed in patients treated with RT and ChT compared to those treated with RT alone. Similarly, to let-7d, decrease of miR-15b did not change in recurrent GBM compared to primary GBM. miR-15b was demonstrated to suppress proliferation and induce apoptosis in glioma cells by targeting CCNE1.33 miR-15b expression decreases with the ascending histopathological grade and its low expression is associated with poor overall survival of patients with high grade gliomas.34 Already known functions about radio-and chemoresistance/sensitivity and analyzed miRNAs are summarized in Figure 6.
FIGURE 6.
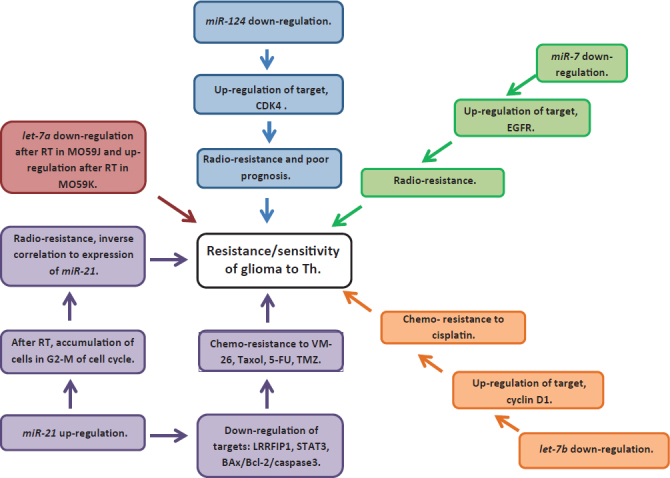
Summary of known functions in radio- and chemoresistance and sensitivity of miRNAs analyzed in our study.
Time to recurrence, survival and overall survival in correlation to therapy and miRNA expression
Finally, in our patients we have observed longer disease free survival in patients treated with RT and ChT after first surgery compared to those treated with RT alone. The standard care treatment of GBM altered importantly after year 2005 when RT alone was replaced by RT associated with chemotherapeutic TMZ in great majority of our patients. Similarly to our results, it has been shown that for patients treated with surgery and RT alone median survival was shorter (12–12.1 months) compared to patients treated additionally with TMZ (14.2–14.6 months).35
Investigating miRNAs expression and survival in patients with GBM revealed that, both, miR-26b and let-7f represent better prognosis when downregulated. Lower expression of miR-26b is in correlation to progression free survival, whereas lower expression of let-7f is in correlation to overall survival. To the best of our knowledge, this is the first report describing expression of these two miRNAs to the function of survival in the GBM.
MGMT and miRNAs
Although we did not found any differential expression of investigated miRNAs between methylated and unmethylated MGMT, we did observed change in MGMT methylation status from primary to recurrent GBM. Eight of nine unmethylated samples in primary GBM were methylated in recurrent GBM. This change in methylation status of MGMT is not surprising, since there are several publications describing similar percentage of change; however all changes were from more to less favorable state for tumor progress ion.36, 37, 38, 39
Conclusions
In conclusion, our results suggest that in recurrent GBM patients, miRNAs might play important role not only in pathogenesis of GBM, but also in tumor progression and its recurrence, as well as in response to therapy, in overall survival and progression free survival.
Acknowledgements
This work is a part of the PhD thesis of candidate Boštjan Matos, MD. This work was supported by the Slovenian Research Agency (ARRS) (Program P3-0054 ).
Disclosure
No potential conflicts of interest were disclosed.
References
- 1.Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134:505–12. doi: 10.1007/s11060-017-2379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegal T. Clinical impact of molecular biomarkers in gliomas. J Clin Neurosci. 2015;22:437–44. doi: 10.1016/j.jocn.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Karsy M, Arslan E, Moy F. Current progress on understanding micro-RNAs in glioblastoma multiforme. Genes Cancer. 2012;3:3–15. doi: 10.1177/1947601912448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silber J, James CD, Hodgson JG. microRNAs in gliomas: small regulators of a big problem. Neuromolecular Med. 2009;11:208–22. doi: 10.1007/s12017-009-8087-9. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci (Landmark Ed) 2012;17:700–12. doi: 10.2741/3952. PMID: 22201769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilhan-Mutlu A, Wöhrer A, Berghoff AS, Widhalm G, Marosi C, Wagner L. Comparison of microRNA expression levels between initial and recurrent glioblastoma specimens. J Neurooncol. 2013;112:347–54. doi: 10.1007/s11060-013-1078-6. et al. [DOI] [PubMed] [Google Scholar]
- 8.Yan D, Hao C, Xiao-Feng L, Yu-Chen L, Yu-Bin F, Lei Z. Molecular mechanism of Notch signaling with special emphasis on microRNAs: implications for glioma. J Cell Physiol. 2018 doi: 10.1002/jcp.26775. [DOI] [PubMed] [Google Scholar]
- 9.Bucci MK, Maity A, Janss AJ, Belasco JB, Fisher MJ, Tochner ZA. Near complete surgical resection predicts a favorable outcome in pediatric patients with nonbrainstem, malignant gliomas: results from a single center in the magnetic resonance imaging era. Cancer. 2004;101:817–24. doi: 10.1002/cncr.20422. et al. [DOI] [PubMed] [Google Scholar]
- 10.Cabrini G, Fabbri E, Lo Nigro C, Dechecchi MC, Gambari R.. Regulation of expression of O6-methylguanine-DNA methyltransferase and the treatment of glioblastoma (Review) Int J Oncol. 2015;47:417–28. doi: 10.3892/ijo.2015.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tezcan G, Tunca B, Bekar A, Preusser M, Berghoff AS, Egeli U. micro-RNA expression pattern modulates temozolomide response in GBM tumors with cancer stem cells. Cell Mol Neurobiol. 2014;34:679–92. doi: 10.1007/s10571-014-0050-0. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhry MA, Sachdeva H, Omaruddin RA. Radiation-induced micro-RNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010;29:553–61. doi: 10.1089/dna.2009.0978. [DOI] [PubMed] [Google Scholar]
- 13.Lages E, Guttin A, El Atifi M, Ramus C, Ipas H, Dupré I. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One. 2011;6:e20600. doi: 10.1371/journal.pone.0020600. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu S, Cheung HH, Lee TL, Lu G, Poon WS, Chan WY. Molecular mechanisms of regulation and action of microRNA-199a in testicular germ cell tumor and glioblastomas. PLoS One. 2013;8:e83980. doi: 10.1371/journal. pone.0083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Yan K, Fang J, Qu Q, Zhou M, Chen F. Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J Exp Clin Cancer Res. 2013;32:41. doi: 10.1186/1756-9966-32-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cankovic M, Mikkelsen T, Rosenblum ML, Zarbo RJ. A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin-fixed paraffin-embedded tissue. Lab Invest. 2007;87:392–7. doi: 10.1038/labinvest.3700520. [DOI] [PubMed] [Google Scholar]
- 17.Latham GJ. Normalization of microRNA quantitative RT-PCR data in reduced scale experimental designs. Methods Mol Biol. 2010;667:19–31. doi: 10.1007/978-1-60761-811-9_2. [DOI] [PubMed] [Google Scholar]
- 18.Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan W. Overexpressed let-7a inhibits glioma cell malignancy by directly targeting K-ras, independently of PTEN. Neuro Oncol. 2013;15:1491–501. doi: 10.1093/neuonc/not107. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Z, Wang L, Li G, Liu H, Fan F, Li Z. Increased expression of microRNA-9 predicts an unfavorable prognosis in human glioma. Mol Cell Biochem. 2013;384:263–8. doi: 10.1007/s11010-013-1805-5. et al. [DOI] [PubMed] [Google Scholar]
- 20.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu N, Zhao X, Liu M, Liu H, Yao W, Zhang Y. Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One. 2011;6:e16264. doi: 10.1371/journal.pone.0016264. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tivnan A, Zhao J, Johns TG, Day BW, Stringer BW, Boyd AW. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumour Biol. 2014;35:1459–65. doi: 10.1007/s13277-013-1200-6. et al. [DOI] [PubMed] [Google Scholar]
- 23.Yan S, Han X, Xue H, Zhang P, Guo X, Li T. Let-7f inhibits glioma cell proliferation, migration, and invasion by targeting periostin. J Cell Biochem. 2015;116:1680–92. doi: 10.1002/jcb.25128. et al. [DOI] [PubMed] [Google Scholar]
- 24.Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol. 2011;101:171–6. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Sun F, Dong N, Sun Z, Diao Y, Zheng C. MicroRNA-7 directly targets insulin-like growth factor 1 receptor to inhibit cellular growth and glucose metabolism in gliomas. Diagn Pathol. 2014;9:211. doi: 10.1186/s13000-014-0211-y. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Hamo R, Efroni S.. Gene expression and network-based analysis reveals a novel role for hsa-miR-9 and drug control over the p38 network in glioblastoma multiforme progression. Genome Med. 2011;3:77. doi: 10.1186/gm293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chao TF, Xiong HH, Liu W, Chen Y, Zhang JX. MiR-21 mediates the radiation resistance of glioblastoma cells by regulating PDCD4 and hMSH2. J Huazhong Univ Sci Technolog Med Sci. 2013;33:525–9. doi: 10.1007/s11596-013-1153-4. [DOI] [PubMed] [Google Scholar]
- 28.Deng X, Ma L, Wu M, Zhang G, Jin C, Guo Y. miR-124 radiosensitizes human glioma cells by targeting CDK4. J Neurooncol. 2013;114:263–74. doi: 10.1007/s11060-013-1179-2. et al. [DOI] [PubMed] [Google Scholar]
- 29.Wei J, Wang F, Kong LY, Xu S, Doucette T, Ferguson SD. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73:3913–26. doi: 10.1158/0008-5472.CAN-12-4318. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizoguchi M, Guan Y, Yoshimoto K, Hata N, Amano T, Nakamizo A. Clinical implications of microRNAs in human glioblastoma. Front Oncol. 2013;3:19. doi: 10.3389/fonc.2013.00019. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolenda T, Przybyła W, Teresiak A, Mackiewicz A, Lamperska KM. The mystery of let-7d - a small RNA with great power. Contemp Oncol (Pozn) 2014;18:293–301. doi: 10.5114/wo.2014.44467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolenda T, Przybyła W, Teresiak A, Mackiewicz A, Lamperska KM. Overexpression of RKIP inhibits cell invasion in glioma cell lines through upregulation of miR-98. Biomed Res Int. 2013;2013:695179. doi: 10.1155/2013/695179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia H, Qi Y, Ng SS, Chen X, Chen S, Fang M. MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem Biophys Res Commun. 2009;380:205–10. doi: 10.1016/j.bbrc.2008.12.169. et al. [DOI] [PubMed] [Google Scholar]
- 34.Sun G, Yan S, Shi L, Wan Z, Jiang N, Li M. Decreased expression of miR-15b in human gliomasi is associated with poor prognosis. Cancer Biother Radiopharm. 2015;30:169–73. doi: 10.1089/cbr.2014.1757. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. et al. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal S, Suri V, Sharma MC, Sarkar C. Therapy and progression--induced O6-methylguanine-DNA methyltransferase and mismatch repair alterations in recurrent glioblastoma multiforme. Indian J Cancer. 2015;52:568–73. doi: 10.4103/0019-509x.178403. [DOI] [PubMed] [Google Scholar]
- 37.Smrdel U, Popovic M, Zwitter M, Bostjancic E, Zupan A, Kovac V. Longterm survival in glioblastoma: methyl guanine methyl transferase (MGMT) promoter methylation as independent favourable prognostic factor. Radiol Oncol. 2016;50:394–401. doi: 10.1515/raon-2015-0041. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pala A, Schmitz AL, Knoll A, Schneider M, Hlavac M, König R. Is MGMT promoter methylation to be considered in the decision making for recurrent surgery in glioblastoma patients? Clin Neurol Neurosurg. 2018;167:6–10. doi: 10.1016/j.clineuro.2018.02.003. et al. [DOI] [PubMed] [Google Scholar]
- 39.Wick W, Osswald M, Wick A, Winkler F. Treatment of glioblastoma in adults. Ther Adv Neurol Disord. 2018;11:1756286418790452. doi: 10.1177/1756286418790452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visani M, de Biase D, Marucci G, Cerasoli S, Nigrisoli E, Bacchi Reggiani ML. Expression of 19 microRNAs in glioblastoma and comparison with other brain neoplasia of grades I-III. Mol Oncol. 2014;8:417–30. doi: 10.1016/j.molonc.2013.12.010. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]


