Summary
Bacteria belonging to the genus Streptococcus are the first inhabitants of the oral cavity which can be acquired right after birth and thus play an important role in the assembly of the oral microbiota. In this chapter, we will discuss the different oral environments inhabited by streptococci and the species that occupy each niche. Special attention is given to the taxonomy of Streptococcus as this genus is now divided into 8 distinct groups where oral species are found in 6 of them. Oral streptococci produce an arsenal of adhesive molecules that allow them to efficiently colonize different tissues in the mouth. Also, they have a remarkable ability to metabolize carbohydrates via fermentation thereby generating acids as byproducts. Excessive acidification of the oral environment by aciduric species such as Streptococcus mutans is directly associated with the development of dental caries. However, less acid-tolerant species such as Streptococcus salivarius and Streptococcus gordonii produce large amounts of alkali displaying and important role in the acid-base physiology of the oral cavity. Another important characteristic of certain oral streptococci is their ability to generate hydrogen peroxide that can inhibit the growth of S. mutans. Thus, oral streptococci can also be beneficial to the host by producing molecules that are inhibitory to pathogenic species. Lastly, commensal and pathogenic streptococci residing in the oral cavity can eventually gain access to the bloodstream and cause systemic infections such as infective endocarditis.
The oral environment
Warm, moist, and rich in nutrients, the oral cavity provides an ideal environment for colonization by a community of bacteria, fungi, protozoa, archaea, and viruses, often in the form of a complex structure called biofilm, or plaque (1). In addition, several microbial niches exist within the oral cavity (e.g. cheek, gingiva, teeth, tongue, palate) that vary in nutrient content, pH, oxygen tension, and shear force due to salivary flow and mastication. These physico-chemical characteristics select for suitable microorganisms for each oral niche such that the microbial compositions can differ greatly from one site to another. Saliva is the biological fluid of the oral cavity and its microbial composition is a collection of bacteria that have shed from various oral niches. As an easily accessible body fluid, saliva can be used in assessment of general oral health, as the disappearance or emergence of certain taxa can indicate health or disease (2, 3). While biofilms from oral soft tissues such as tongue, cheeks and palate, and the supragingival biofilm (plaque on tooth surfaces above the gum line) are constantly bathed in saliva and subjected to an array of environmental fluctuations, the subgingival plaque (plaque on tooth surfaces below the gum line) comprises a unique niche with less abrupt environmental fluctuations. In particular, with the gum serving as a physical barrier, the oxygen tension and shear forces are reduced, and gingival crevicular fluid (serum) serves as a major nutrient source (4).
Taxonomy of oral streptococci
Streptococci are found in almost every location in the human body and are the dominant species in the human oral cavity and upper respiratory tract. Species within the genus Streptococcus were initially differentiated based on hemolysis patterns on blood agar plates with β-hemolysis (complete lysis), α-hemolysis (partial lysis), and γ-hemolysis (non-hemolytic) (5). Many β-hemolytic species can be further differentiated by carbohydrate “group” antigens (Lancefield groups) (6), such as the classic human pathogens S. pyogenes (Group A Streptococcus) and S. agalactiae (Group B Streptococcus) that are the subject of other chapters in this book. Historically, the oral streptococci were referred to as viridans streptococci (Streptococcus viridans) due to their high propensity to display partial hemolysis when cultured on blood agar plates, resulting in a green coloration surrounding the colonies. While the viridans streptococci or α-hemolytic designations are still used today to classify the oral streptococci, this term is not fully accurate as some isolates can display β-hemolysis or completely lack any hemolytic activity (γ-hemolysis). As more streptococcal species were identified, it became apparent that additional criteria were required to separate the growing number of species, prompting the development of biochemical and physiologic tests to differentiate streptococcal groups (7, 8). Initially, there were seven named groups of streptococci based on carbohydrate fermentation tests, chain length, and growth on gelatin: Streptococcus salivarius, Streptococcus mitis, Streptococcus anginosus, Streptococcus equinus, Streptococcus pyogenes, Streptococcus faecalis, and Streptococcus pneumoniae (pneumococcus), reviewed in (5). Eventually Streptococcus faecalis was reclassified, and is now a member of the genus Enterococcus (9, 10).
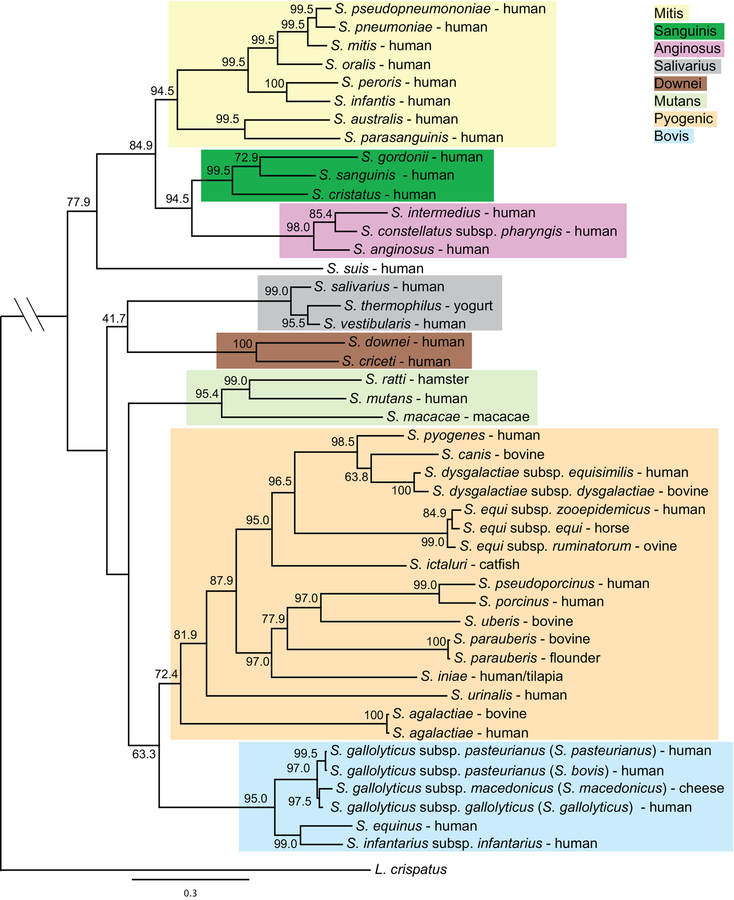
The advent of DNA sequencing introduced a new and more accurate approach to the identification of bacteria at the species level, and sequencing of the 16S rRNA region could be used to determine phylogenetic relationships (11). Initially, the oral streptococci were separated into 4 groups: anginosus, mitis, mutans, and salivarius, while the remaining streptococci isolated outside the oral cavity were classified into either the bovis or pyogenic groups (12). Recently, using a more robust phylogenetic approach, Richards et. al. separated the streptococci into 8 distinct groups: mitis, sanguinis, anginosus, salivarius, downei, mutans, pyogenic, and bovis (Fig. 1) (13). Presently, oral streptococci are found in all groups except the pyogenic and bovis groups. The mitis group is the largest of the groups found in the oral cavity, currently with 20 species. Species within the mitis group have been challenging to differentiate based on 16S RNA sequence alone, particularly S. oralis and S. mitis. Recent efforts to re-classify many of the species within this group indicate there are many strains that have been misclassified and new species descriptions may soon emerge (14).
Figure 1.
Phylogeny of the indicated streptococcal species derived from a core set of 136 concatenated genes. Numbers on branches show bootstrap support for each relationship. The color shading indicates the eight major groups.
Microbiome and metagenomics studies: The oral microbiome
While initial attempts to define the oral microbiome relied on culture-based techniques, 16S rRNA sequencing has identified more than 700 microbial species residing within the oral cavity. It is estimated that roughly one half of these species are non-culturable by current methods. The Human Microbiome Project (HMP) (http://commonfund.nih.gov/hmp/) launched in 2012 aimed to define the core microbiome of 18 different locations in the human body, including the oral cavity, and provided the first comprehensive analysis of bacterial diversity and abundance in humans (15, 16). Nearly all microbiome studies today use next-generation sequencing (NGS) technologies based on 16S rRNA sequence to determine the community composition (17), and large manually curated databases, such as www.homd.org, are now available to aid in the identification of bacterial species unique to the oral cavity. Based on these studies, we now can clearly appreciate that there are distinct communities of organisms living on the different surfaces of the mouth, which vary significantly depending on the site of sample collection, health status, and behavior of the host (18–20). Thus, as indicated in the beginning of the chapter, slight variations in the local environment within the mouth can have profound effects on the composition of the residing organisms (18, 19). However, regardless of the individual sampled, streptococci are universally present in all sites of the oral cavity, and are the dominant genus found in saliva and on soft tissues of the mouth (16, 18, 19). Critically, streptococci, especially members of the mitis group, are the first organisms detected in the mouth of newborn infants and are considered pioneer species (primary colonizer) that allow for the assembly of a complex microbiota.
Dental plaque is readily accessible on the surface of the teeth, and therefore is one of the most well characterized microbial communities in the human body (21, 22). Two of the most common human diseases, dental caries and periodontitis, are directly related to the presence and metabolic activities of dental plaque bacteria. A recent analysis that compared supragingival plaque microbiomes from carious lesions to healthy sites found substantial differences among children with different caries status (23). In particular, communities collected from dentin carious lesions contained notorious acidogenic (acid producing) and aciduric (acid tolerant) species, including S. mutans, Scardovia wiggsiae, Parascardovia denticolens, and Lactobacillus salivarius (23). In contrast, S. sanguinis, Neisseria species, and Leptotrichia species were found associated with samples collected from healthy sites. However, a decrease in species diversity was associated with dental caries progression, but an increased overall abundance of S. mutans has been demonstrated (24). Moreover, an association of S. mutans with other acidogenic species, such as Scardovia wiggsiae and bifidobacteria, in severe early childhood caries has shown (25). One study that focused on the microbiota that thrive at different pH ranges also found a decrease in microbial diversity in communities from acidic pH environments, but found Lactobacillus sp., Prevotella sp., Atopobium sp., Osenella sp., and Actinomyces sp. to be the most abundant taxa in dentinal caries (26). Microbiome studies of subgingival plaque identified high abundance of S. parasanguinis, S. sanguinis, S. mitis, S. intermedius and S. infantis associated with health (27–31). In diseased sites (gingivitis and periodontitis), a decrease in the proportions of health-associated Streptococcus (32) and an increase is anaerobic proteolytic bacteria such as Porphyromonas gingivalis is observed. However, S. constellatus and S. intermedius are often reported to be associated with periodontitis (33, 34).
Most microbiome studies are initially conducted with the intent of correlating the presence of disease with a particular signature microbiome. However, confounding factors such as genetic heterogeneity and phenotypic diversity within a species contribute to a more complex relationship between the presence of a particular bacterial species and the establishment of disease. Studies comparing the phenotypic diversity within streptococcal strains from the same species have identified major phenoptypic variations among isolates that are directly related to virulence or properties associated with healthy biofilms (35, 36). For this reason, the combination of 16S rRNA community profiling with other “-omics” technologies (i.e., transcriptomics, proteomics, and metabolomics) is essential for a global understanding of the specific contributions of oral bacteria to health and disease (17), providing a better understanding of how complex communities function and interact (37).
Sugar metabolism of oral streptococci
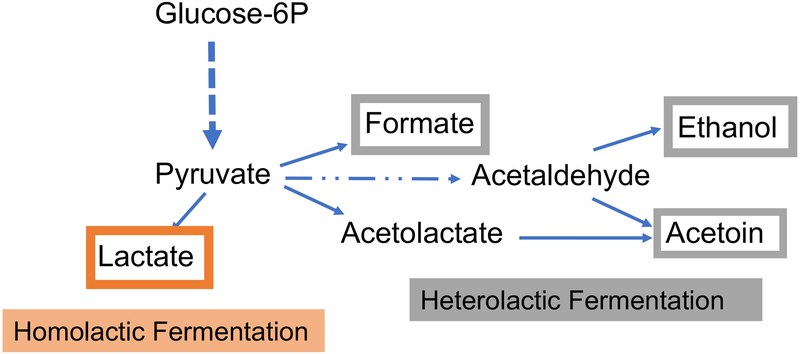
Streptococci are well known for their ability to assimilate a large array of carbohydrates by glycolysis, and for their enhanced tolerance to acidic pH. Employing the Embden-Meyerhof-Parnas (EMP) pathway, streptococci convert one molecule of glucose or other equivalent carbohydrates (e.g. fructose) into two molecules of pyruvate, with the concomitant generation of two molecules of ATP and two molecules of NADH. Under the conditions of excess carbohydrates and oxygen limitation, streptococci tend to perform homolactic fermentation, reducing pyruvate into lactic acid and regenerating NAD from NADH. The production of lactic acid results in rapid acidification of the environment, and allows streptococcal species to outcompete acid-sensitive microorganisms. When faced with carbohydrate limitation or increased oxygen tension, streptococci produce alternative fermentation products such as formate, acetate and ethanol (38) (Fig. 2). In fact, streptococci lack the capacity for oxidative phosphorylation and electron chain transport systems and depend exclusively on substrate-level phosphorylation for energy production. Therefore, most streptococci are considered facultative anaerobes and susceptible to growth inhibition by oxygen.
Figure 2.
Simplified schematic of homolactic fermentation and heterolactic fermentation. While homolactic fermentation generates almost exclusively lactate, heterolactic fermentation generates formate, acetoin, ethanol and reduced amounts of lactate.
Resulting from a reductive evolution as a consequence of adaptation to survival within human and animal hosts, streptococci possess an incomplete TCA cycle (tricarboxylic acid cycle, or Krebs cycle), with some enzymes of the pathway being preserved in certain species such as S. mutans and S. gordonii, and none present in species such as S. pyogenes or S. pneumoniae. As metabolites of the TCA cycle are often precursors required for amino acid biosynthesis, it follows that varying degrees of amino acid auxotrophy exist for different streptococcal species. Another outcome of this reductive evolution is the fact that streptococci did not maintain a functional Entner-Doudoroff pathway, an alternative pathway that utilizes glucose-6-Phosphate (glucose-6-P) to generate pyruvate through a series of enzymatic reactions that are distinct from glycolysis. Also, many species lack a complete pentose phosphate pathway (PPP) (38) which represents a parallel process to glycolysis as glucose-6-P is oxidized and then decarboxylated into a five-carbon product, ribulose-5-Phosphate (ribulose-5-P), yielding the important reducing equivalent NADPH. Further metabolism of ribulose-5-P by the non-oxidative phase creates precursors for biosynthesis of nucleic acids, histidine and aromatic amino acids. In bacteria lacking the oxidative phase of PPP (e.g. S. mutans), alternative reactions and enzymes (e.g., NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase or GapN) exist to compensate for the generation of both nucleotides and NADPH (39).
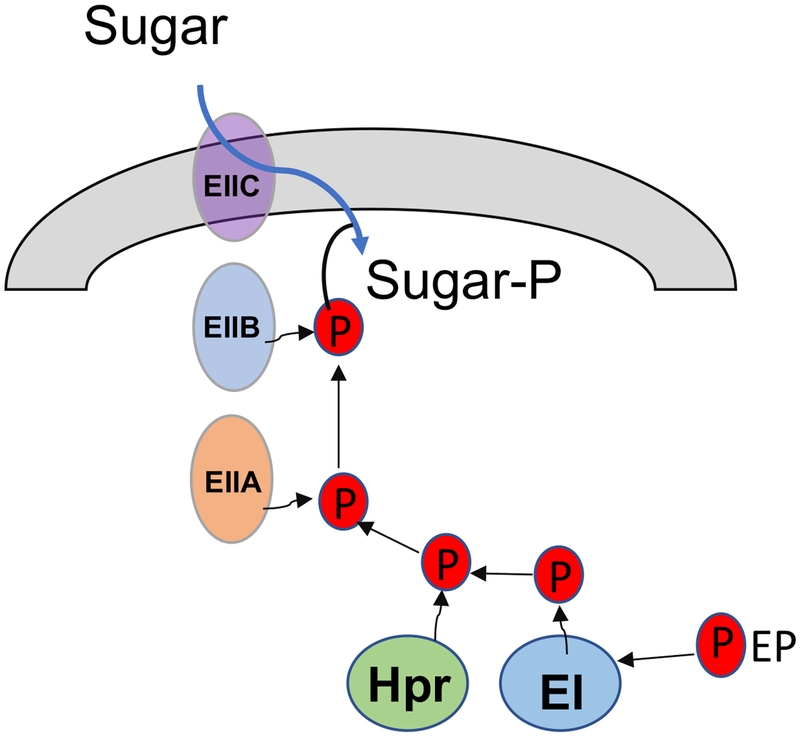
In contrast to these reductive evolutions, streptococci have maintained a significant arsenal of carbohydrate-uptake mechanisms, proteins and enzymes of the sugar:phosphotransferase system (PTS) (40, 41) and the ATP-binding cassette (ABC) transporter family (42). The PTS consists of two general enzymes, enzyme I (EI) and a phospho-carrier protein HPr, and a series of multi-domain, membrane-associated, substrate-specific sugar transporters called enzyme II (EII). Using phosphoenolpyruvate (PEP) as an energy source, EI and HPr initiate a cascade of phospho-relay reactions that funnels phosphoryl groups, via various EIIs, onto incoming carbohydrates (40) (Fig. 3). Recent sequencing projects of streptococci have uncovered large portions of these genomes as predicted carbohydrate transporters, many of which are apparent products of horizontal gene transfer and have origins in related bacterial species (43–45). Together, these transporters allow metabolic flexibility when glucose, the preferred energy source for most streptococci, is not available. A major coordinator of these processes is the global regulator CcpA (catabolite control protein A). CcpA oversees carbon catabolite repression (CCR), a regulatory mechanism by which enzymes associated with catabolism of non-preferred energy sources are repressed until glucose is no longer available (46, 47). When glucose is abundant, the cofactor HPr becomes phosphorylated at a serine residue. In association with this phosphorylated cofactor HPr(Ser-P), CcpA binds to the catabolite response element (cre) within the promoter region of a target gene to influence transcription (46). In addition to overseeing carbohydrate metabolism, CcpA has been associated with expression of virulence factors in streptococci (48–50). Notably, it has been demonstrated in some streptococci, e.g. S. mutans, S. pyogenes, and S. pneumoniae, that substrate-specific, CcpA-independent CCR plays profound roles in controlling carbohydrate prioritization and virulence expression [see next chapter and (51–55)]. Therefore, it appears that interactions among streptococci and with their host environment have shaped these bacteria toward a successful lifestyle that specializes in carbohydrate catabolism and environment acidification. As such, infectious diseases and virulence associated with streptococci are often heavily dependent on their metabolic activities, and consequently, their ecological relationships with commensal bacteria in each particular biofilm environment (45, 50, 56).
Figure 3.
Simplified schematic of the phosphoenolpyruvate sugar:phosphotransferase system (PTS).
Mechanisms of colonization
As important primary colonizers of the mouth, oral streptococci possess multiple high-affinity adhesins that mediate initial binding to tooth surfaces via interactions with substrates in the acquired salivary pellicle including albumin, proline-rich proteins, glycoproteins, mucins, and sialic acid. Bacterial extracellular polysaccharides as well as extracellular DNA are major constituents of the plaque biofilm matrices whose production is triggered by bacterial adherence (57, 58). These macromolecules further facilitate bacterial colonization and accumulation of multicellular clusters. Salivary amylase also appears to play a role in bacterial colonization of mammalian oral surfaces since amylase-binding bacteria (such as S. mitis, S. gordonii, S. salivarius and S. cristatus) were found to colonize only those hosts with salivary amylase activity (59). Late colonizers, both Gram-positive and Gram-negative species, build upon the initial streptococcal foundation through a series of co-aggregation and co-adhesion events until a mature plaque is achieved within any given individual (60). Once established, the plaque community is relatively stable, and under health conditions, the different species in the community exist in homeostasis.
Colonization as well as accumulation of oral streptococci on hard and soft tissue surfaces is guided by a series of adhesin-receptor interactions and regulated in response to environmental cues, including presence of other microbial species (61). The Antigen I/II (AgI/II) family of adhesins (SpaP, P1, PAc, SspA, SspB) is shared by most oral, as well as several of the pyogenic streptococci (62). These proteins mediate binding to multiple host proteins, with the salivary glycoprotein gp340 (DMBT1) being the most widely studied. Streptococcus gordonii possesses two paralogs of AgI/II, SspA and SspB, although the purpose for this duplication is not known (63). Notably, S. gordonii is a master of adherence and possesses numerous adhesive structures. Among them, GspB and Hsa are serine-rich repeat glycoproteins (Srr) that bind to sialylated glycans present on the soluble human salivary mucin MG2/MUC7, a component of the mucosal pellicle (64), and the platelet glycoprotein GPIb (65, 66). Srr proteins are important for biofilm formation and have been identified in many other oral streptococci, including Fap1 of S. parasanguinis (67) and S.oralis (68), SrpA of S. sanguinis (69) and S. cristatus (70), and SrpA, B and C of S. salivarius (71). Interestingly, Srr homologs are not present in the caries pathogen S. mutans. Genes encoding Srr family proteins are located within tightly coordinated genetic loci that also encode proteins that mediate their glycosylation, acetylation and export (72–74). Notably, instead of the general secretion machinery utilized for other surface proteins, an alternative secretion system including SecA2 and SecY2 is dedicated to the translocation of this family of serine-rich proteins (67, 75). In addition, CshA of S. gordonii and CshA-like proteins of S. oralis and S. sanguinis are multi-functional fibrillar adhesins that bind to multiple host proteins including fibronectin, as well as to other microorganisms(76, 77).
As detailed in the next chapter, production of extracellular glucans via glucosyltransferases GtfB, GtfC and GtfD is well known to facilitate adhesion of S. mutans to saliva-coated tooth surfaces (78). Secreted S. mutans Gtfs bind to the salivary pellicle and to other organisms, facilitating the recruitment of oral microorganisms to the dental biofilm. Although some other oral streptococci also express Gtfs, these enzymes have not been extensively studied like the Gtfs of S. mutans. A Gtf-negative mutant of S. gordonii showed diminished persistence on the teeth of sucrose fed rats, although initial colonization was not affected (79). In addition, deletion of the single gtf gene of S. gordonii, gtfG, impacted its ability to form mixed biofilms with Candida albicans (80). Streptococcus sanguinis also produces water soluble glucans from sucrose through a Gtf enzyme called GtfP. Its biofilm formation in monoculture in the presence of sucrose was significantly decreased upon inactivation of gtfP in S. sanguinis, although little effect on biofilm production was observed when the mutant strain was co-cultured with other oral streptococcal species (81). Additional studies are necessary to firmly establish the contribution of Gtfs from non-S. mutans oral streptococci to oral colonization and biofilm assembly in vivo.
Between good and evil – when things go wrong.
Dental caries, or simply cavities, is a multifactorial disease in which a carbohydrate rich diet and the oral microbial composition play key roles in the development and progression of the disease (see next chapter). The common main culprits associated with dental caries are two species from the mutans streptococcci group, Streptococcus mutans and Streptococcus sobrinus, but other organisms can also be involved in the process, as caries active subjects have been identified that do not harbor S. mutans or S. sobrinus. In healthy subjects, there is a balance between the numbers of cariogenic bacteria and non-cariogenic commensal species, with the latter keeping the cariogenic population in check through the production of alkali, hydrogen peroxide or other inhibitory substances. However, frequent exposure to dietary carbohydrates creates a dysbiotic environment, in which the generation of organic acids such as lactic acid as fermentation by-products allows S. mutans and other aciduric species to outcompete less acid-tolerant commensal bacteria present in the dental plaque. Eventually, the repetitive acidification of environmental pH below the critical point (pH 5.5) will lead to enamel demineralization and consequent cavitation of the tooth surface.
The distribution of oral streptococcal species differs within various ecological niches of the oral cavity (82). The mitis and sanguinis groups, including S. oralis, S. mitis, S. gordonii, and S. sanguinis, are the primary colonizers of the tooth surface and are commonly considered as commensals, although all have been implicated in cases of infective endocarditis. Oral bacteria can reach the bloodstream not only during invasive procedures such as dental extractions and oral surgery but also after simple daily activities such as mastication, brushing and flossing. Fortunately, our immune surveillance readily clears these bacteria from the bloodstream in a process that can take just a few seconds. However, in the rare occasions when these organisms successfully avoid the immune surveillance and cause systemic disease, interactions with platelets are especially important during the pathogenic process of infective endocarditis. Platelet binding by S. mitis was reported to be higher in strains that produce less exopolysaccharides, whereas high exopolysaccharide producers demonstrated less platelet aggregation (83). Another primary colonizer of the tooth surface that can cause transient bacteremia and endocarditis is S. parasanguinis. Its dual function lipoprotein FimA participates in both metal transport (84) and adherence to fibrin, critical for vegetation formation on damaged heart tissue (85).
The good Streptococci: commensal oral flora
In the human mouth, most microorganisms are embedded within a complex extracellular matrix known as biofilm or plaque (86). In a healthy individual, the oral biofilm is dominated by bacteria that are commensal in nature, engaging the host immunity in an intricate dialog that maintains a homeostasis integral to human health. Significantly, the commensal communities afford the host additional benefits by fending off pathogenic species. Dental health is often associated with a greater proportion of commensals with beneficial properties that include S. gordonii, S. sanguinis and S. parasanguinis, some of which can cause opportunistic diseases at different body sites as discussed above, but in the mouth, their presence is generally associated with lower proportions of cariogenic S. mutans and oral health. In the case of dental caries, repetitive exposures to acidic pH values, reduced salivary flow and a diet rich in fermentable carbohydrates are the known triggers of dysbiosis in dental plaque, tilting the balance towards acidogenic and aciduric populations (87). Here, we discuss a few mechanisms that are critical to understanding the antagonism between the commensal streptococci and dental pathogens: (i) production and neutralization of acids, (ii) generation of hydrogen peroxide (H2O2) that inhibits the persistence of S. mutans, and (iii) production and secretion of antimicrobial compounds.
Acid is the driving force behind dental caries, but pH in dental plaque is modulated in a complex way by the buffering capacities of salivary secretions, bacterial cells, certain foods and the metabolic products of the microbiota. As previously mentioned, commensal streptococci are early colonizers of the oral cavity and play pivotal roles in moderating biofilm acidification by, among other things, generating large amounts of alkali from salivary and dietary substrates. A primary route for alkali generation by oral streptococci is the arginine deiminase system (ADS), which consists minimally of three enzymes: arginine deiminase, a catabolic ornithine transcarbamylase (cOTC), and a catabolic carbamate kinase (cCK). The ADS acts on free arginine to generate one molecule of ornithine and CO2 plus two molecules of ammonia, with the concomitant production of ATP (88). Although free arginine is found in micromolar concentrations in ductal saliva, it is abundant in salivary proteins and peptides (89). The pH-modulating effects of ammonia production benefit the commensals and other species that are generally less aciduric, or overtly acid sensitive. Another well-characterized alkali-generating mechanism is the urease system identified in S. salivarius, a three-part enzyme complex and a set of accessory proteins that break down urea to release ammonia and CO2. The urease system could have considerable impact to oral bacterial ecology, given the millimolar concentration of urea present in saliva and gingival crevicular fluids. Of note, subjects with chronic renal failure who secrete 10-fold more urea in saliva than normal individuals, rarely developed dental caries despite being on a carbohydrate-rich diet (90, 91).
Importantly, there are multiple reasons to believe that the ADS and arginine both have a potent influence on the promotion of a microbiota that is less cariogenic and that prevents caries pathogens from emerging in substantial numbers. In a mixed-species biofilm model containing arginine in the growth medium that included S. mutans and S. gordonii, activities of ADS provided a two-fold beneficial impact: it promoted the dominance of commensals, and disruped the biofilm matrix of S. mutans by interfering with the exopolysaccharide (EPS) production (92). Multiple clinical studies involving both children and adults support that arginine and elevated ADS levels are correlated with dental health, whereas low ADS activity appears to be strongly associated with active disease (35, 93–95). As a result, arginine has been incorporated into oral health products around the globe to promote oral health.
A second strategy employed by oral commensal streptococci to achieve a competitive advantage over oral pathogens is the generation of hydrogen peroxide (H2O2), which inhibits the growth of S. mutans, the periodontal pathogen Porphyromonas gingivalis (96), and other oral pathogens. Among the known peroxigenic commensals, which include S. oralis, S. mitis, S. sanguinis, S. gordonii and S. oligofermentans, there are multiple enzymatic pathways for the generation of H2O2. A dominant mechanism in dental biofilms appears to involve the enzyme pyruvate oxidase (Pox, encoded by spxB), which is present in many abundant commensal streptococci. Pox catalyzes the conversion of pyruvate and inorganic phosphate to H2O2, CO2 and acetyl phosphate (AcP) in the presence of oxygen. Thus, not only does the commensal gain a competitive advantage over H2O2-sensitive pathogens, but also the AcP provides a bioenergetics benefit as it is further metabolized, via acetate kinase, to acetate with the production of ATP. Moreover, the production of CO2 contributes to some unknown degree the overall buffering capacity of the biofilm (97–99).
The H2O2-generating enzymes lactate oxidase (Lox) (100, 101), L-amino acid oxidase (LAAO) (102, 103) and NADH oxidase are also present in some commensal oral streptococci. A Lox enzyme has been identified in S. oligofermentans that is capable of utilizing the lactic acid generated by the glycolytic activities of lactic acid bacteria to yield pyruvate and H2O2, which has the added benefit of removing a strong acid (lactate) from the environment. S. oligofermentans also encodes a Pox enzyme, but there is evidence to suggest that these enzymes may not work synergistically because of the way the genes are regulated (100). Notably, in S. gordonii and S. sanguinis, expression of spxB is sensitive to carbon catabolite repression (CCR) by preferred carbohydrates like glucose (104). Thus, it remains to be determined to what degree H2O2 production by Pox and other enzyme systems is active under conditions that are conducive to the formation of caries, i.e. when the host is consuming large quantities of readily metabolizable sugars. Likewise, since mature dental biofilms tend to be depleted for oxygen and have a low redox potential, Pox and other systems may exert their benefit primarily in the early phases of dental plaque development and have a more potent influence on establishment of caries pathogens, whereas other pathways for competition become essential in more mature biofilms.
More recently, another mechanism whereby commensals may specifically affect fitness of S. mutans has come to light. Most isolates of S. mutans carry the ComDE two-component system, which is required for the activation of transcription of a number of different genes encoding lantibiotic and non-lantibiotic bacteriocins, small peptides that exhibit antimicrobial activity against other species. ComDE depends on a quorum sensing peptide CSP (competence stimulating peptide) for activity (105). S. gordonii DL-1 produces a protease, designated as challisin, which is able to degrade CSP and block the activation of bacteriocin gene expression by S. mutans (106). Interestingly, Streptococcus A12, a recently identified commensal with multiple strategies to interfere with S. mutans (105), also produces a challisin-like protease with similar activities against CSP and the ability to interfere with EPS production by S. mutans. Notably, A12 can also block a second peptide-dependent signaling pathway (XIP-ComRS) that is highly conserved in S. mutans (105).
Concluding remarks
As our understanding of oral infectious diseases evolves toward a multi-microbial, dysbiosis-based perspective, the field of oral microbiology has embraced a more holistic approach, exemplified by significant enhancements in our knowledge of molecular mechanisms underlying the physiological and ecological shifts that control the composition and activities of the microbiome in health and disease. Nevertheless, the metabolic plasticity, ability to colonize multiple oral surfaces, and the production of inhibitory substances make oral streptococci key players in oral homeostasis and disease.
Acknowledgement
Authors would like to thank Dr. Robert A. Burne for his careful reading and critical comments. The authors’ work in this area was supported by NIH-NIDCR grants DE019783, DE022559, DE008007, DE021789, DE024348, DE12236, DE19452, DE25348 and HHSN272200900007C.
REFERENCES
- 1.Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res 69:137–143. [DOI] [PubMed] [Google Scholar]
- 2.Giannobile WV, Wong DT. 2011. Salivary diagnostics: oral health and beyond! J Dent Res 90:1153–1154. [DOI] [PubMed] [Google Scholar]
- 3.Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DT. 2013. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev 26:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costalonga M, Herzberg MC. 2014. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett 162:22–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherman JM. 1937. The Streptococci. Bacteriol Rev 1:3–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lancefield RC. 1933. A Serological Differentiation of Human and Other Groups of Hemolytic Streptococci. J Exp Med 57:571–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beighton D, Hardie JM, Whiley RA. 1991. A scheme for the identification of viridans streptococci. J Med Microbiol 35:367–372. [DOI] [PubMed] [Google Scholar]
- 8.Facklam R 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15:613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig W, Seewaldt E, Kilpper-Balz R, Schleifer KH, Magrum L, Woese CR, Fox GE, Stackebrandt E. 1985. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol 131:543–551. [DOI] [PubMed] [Google Scholar]
- 10.Schleifer KH, Kilpper-Balz R. 1984. Transfer of Streptococcus faecalis and Streptococcus faecium to the Genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. International Journal of Systematic Bacteriology 34:31–34. [Google Scholar]
- 11.Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol 45:406–408. [DOI] [PubMed] [Google Scholar]
- 12.Whiley RA, Beighton D. 1998. Current classification of the oral streptococci. Oral Microbiol Immunol 13:195–216. [DOI] [PubMed] [Google Scholar]
- 13.Richards VP, Palmer SR, Pavinski Bitar PD, Qin X, Weinstock GM, Highlander SK, Town CD, Burne RA, Stanhope MJ. 2014. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol Evol 6:741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen A, Scholz CF, Kilian M. 2016. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus. Int J Syst Evol Microbiol 66:4803–4820. [DOI] [PubMed] [Google Scholar]
- 15.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methe BA, Huttenhower C. 2012. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol 10:e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huse SM, Ye Y, Zhou Y, Fodor AA. 2012. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nascimento MM, Zaura E, Mira A, Takahashi N, Ten Cate JM. 2017. Second Era of OMICS in Caries Research: Moving Past the Phase of Disillusionment. J Dent Res 96:733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. 2003. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol 30:644–654. [DOI] [PubMed] [Google Scholar]
- 20.Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ, Bretz W. 2013. The dental plaque microbiome in health and disease. PLoS One 8:e58487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakubovics NS. 2015. Intermicrobial Interactions as a Driver for Community Composition and Stratification of Oral Biofilms. J Mol Biol 427:3662–3675. [DOI] [PubMed] [Google Scholar]
- 22.Rosan B, Lamont RJ. 2000. Dental plaque formation. Microbes Infect 2:1599–1607. [DOI] [PubMed] [Google Scholar]
- 23.Richards VP, Alvarez AJ, Luce AR, Bedenbaugh M, Mitchell ML, Burne RA, Nascimento MM. 2017. Microbiomes of Site-Specific Dental Plaques from Children with Different Caries Status. Infect Immun 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner AC, Kent RL Jr., Holgerson PL, Hughes CV, Loo CY, Kanasi E, Chalmers NI, Johansson I. 2011. Microbiota of severe early childhood caries before and after therapy. J Dent Res 90:1298–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kianoush N, Adler CJ, Nguyen KA, Browne GV, Simonian M, Hunter N. 2014. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One 9:e92940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, Podar M, Leys EJ. 2012. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J 6:1176–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camelo-Castillo AJ, Mira A, Pico A, Nibali L, Henderson B, Donos N, Tomas I. 2015. Subgingival microbiota in health compared to periodontitis and the influence of smoking. Front Microbiol 6:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SM, Tian F, Huang QF, Zhao YF, Guo XK, Zhang FQ. 2011. Bacterial diversity of subgingival plaque in 6 healthy Chinese individuals. Exp Ther Med 2:1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng H, Xu L, Wang Z, Li L, Zhang J, Zhang Q, Chen T, Lin J, Chen F. 2015. Subgingival microbiome in patients with healthy and ailing dental implants. Sci Rep 5:10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaddox LM, Huang H, Lin T, Hou W, Harrison PL, Aukhil I, Walker CB, Klepac-Ceraj V, Paster BJ. 2012. Microbiological characterization in children with aggressive periodontitis. J Dent Res 91:927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7:1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colombo AP, Teles RP, Torres MC, Souto R, Rosalem WJ, Mendes MC, Uzeda M. 2002. Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J Periodontol 73:360–369. [DOI] [PubMed] [Google Scholar]
- 34.Rams TE, Degener JE, van Winkelhoff AJ. 2014. Antibiotic resistance in human chronic periodontitis microbiota. J Periodontol 85:160–169. [DOI] [PubMed] [Google Scholar]
- 35.Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol 24:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer SR, Miller JH, Abranches J, Zeng L, Lefebure T, Richards VP, Lemos JA, Stanhope MJ, Burne RA. 2013. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One 8:e61358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duran-Pinedo AE, Frias-Lopez J. 2015. Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect 17:505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willenborg J, Goethe R. 2016. Metabolic traits of pathogenic streptococci. FEBS Lett 590:3905–3919. [DOI] [PubMed] [Google Scholar]
- 39.Boyd DA, Cvitkovitch DG, Hamilton IR. 1995. Sequence, expression, and function of the gene for the nonphosphorylating, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase of Streptococcus mutans. J Bacteriol 177:2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vadeboncoeur C, Pelletier M. 1997. The phosphoenolpyruvate:sugar phosphotransferase system of oral streptococci and its role in the control of sugar metabolism. FEMS Microbiol Rev 19:187–207. [DOI] [PubMed] [Google Scholar]
- 42.Davidson AL, Chen J. 2004. ATP-binding cassette transporters in bacteria. Annu Rev Biochem 73:241–268. [DOI] [PubMed] [Google Scholar]
- 43.Buckwalter CM, King SJ. 2012. Pneumococcal carbohydrate transport: food for thought. Trends Microbiol 20:517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornejo OE, Lefebure T, Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, Stanhope MJ. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol Biol Evol 30:881–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shelburne SA, Davenport MT, Keith DB, Musser JM. 2008. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol 16:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deutscher J 2008. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11:87–93. [DOI] [PubMed] [Google Scholar]
- 47.Titgemeyer F, Hillen W. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek 82:59–71. [PubMed] [Google Scholar]
- 48.Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. 2008. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J Bacteriol 190:2340–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iyer R, Baliga NS, Camilli A. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 187:8340–8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shelburne SA 3rd, Keith D, Horstmann N, Sumby P, Davenport MT, Graviss EA, Brennan RG, Musser JM. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc Natl Acad Sci U S A 105:1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng L, Burne RA. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol Microbiol 75:1145–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol Microbiol 88:1176–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valdes KM, Sundar GS, Vega LA, Belew AT, Islam E, Binet R, El-Sayed NM, Le Breton Y, McIver KS. 2016. The fruRBA Operon Is Necessary for Group A Streptococcal Growth in Fructose and for Resistance to Neutrophil Killing during Growth in Whole Human Blood. Infect Immun 84:1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fleming E, Camilli A. 2016. ManLMN is a glucose transporter and central metabolic regulator in Streptococcus pneumoniae. Mol Microbiol 102:467–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fleming E, Lazinski DW, Camilli A. 2015. Carbon catabolite repression by seryl phosphorylated HPr is essential to Streptococcus pneumoniae in carbohydrate-rich environments. Mol Microbiol 97:360–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burne RA. 1998. Oral streptococci… products of their environment. J Dent Res 77:445–452. [DOI] [PubMed] [Google Scholar]
- 57.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45:69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kreth J, Merritt J, Qi F. 2009. Bacterial and host interactions of oral streptococci. DNA Cell Biol 28:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scannapieco FA, Solomon L, Wadenya RO. 1994. Emergence in human dental plaque and host distribution of amylase-binding streptococci. J Dent Res 73:1627–1635. [DOI] [PubMed] [Google Scholar]
- 60.Kolenbrander PE, Palmer RJ Jr., Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000. 42:47–79. [DOI] [PubMed] [Google Scholar]
- 61.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol Mol Biol Rev 73:407–450, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brady LJ, Maddocks SE, Larson MR, Forsgren N, Persson K, Deivanayagam CC, Jenkinson HF. 2010. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol 77:276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Demuth DR, Duan Y, Brooks W, Holmes AR, McNab R, Jenkinson HF. 1996. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol 20:403–413. [DOI] [PubMed] [Google Scholar]
- 64.Hannig C, Hannig M, Kensche A, Carpenter G. 2017. The mucosal pellicle - An underestimated factor in oral physiology. Arch Oral Biol 80:144–152. [DOI] [PubMed] [Google Scholar]
- 65.Bensing BA, Khedri Z, Deng L, Yu H, Prakobphol A, Fisher SJ, Chen X, Iverson TM, Varki A, Sullam PM. 2016. Novel aspects of sialoglycan recognition by the Siglec-like domains of streptococcal SRR glycoproteins. Glycobiology 26:1222–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bensing BA, Lopez JA, Sullam PM. 2004. The Streptococcus gordonii surface proteins GspB and Hsa mediate binding to sialylated carbohydrate epitopes on the platelet membrane glycoprotein Ibalpha. Infect Immun 72:6528–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. 2007. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol 189:1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Singh AK, Woodiga SA, Grau MA, King SJ. 2017. Streptococcus oralis Neuraminidase Modulates Adherence to Multiple Carbohydrates on Platelets. Infect Immun 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. 2014. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog 10:e1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Handley PS, Correia FF, Russell K, Rosan B, DiRienzo JM. 2005. Association of a novel high molecular weight, serine-rich protein (SrpA) with fibril-mediated adhesion of the oral biofilm bacterium Streptococcus cristatus. Oral Microbiol Immunol 20:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Couvigny B, Lapaque N, Rigottier-Gois L, Guillot A, Chat S, Meylheuc T, Kulakauskas S, Rohde M, Mistou MY, Renault P, Dore J, Briandet R, Serror P, Guedon E. 2017. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ Microbiol 19:3579–3594. [DOI] [PubMed] [Google Scholar]
- 72.Seepersaud R, Bensing BA, Yen YT, Sullam PM. 2012. The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J Bacteriol 194:5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seepersaud R, Sychantha D, Bensing BA, Clarke AJ, Sullam PM. 2017. O-acetylation of the serine-rich repeat glycoprotein GspB is coordinated with accessory Sec transport. PLoS Pathog 13:e1006558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou M, Wu H. 2009. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155:317–327. [DOI] [PubMed] [Google Scholar]
- 75.Bensing BA, Sullam PM. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol 44:1081–1094. [DOI] [PubMed] [Google Scholar]
- 76.McNab R, Holmes AR, Clarke JM, Tannock GW, Jenkinson HF. 1996. Cell surface polypeptide CshA mediates binding of Streptococcus gordonii to other oral bacteria and to immobilized fibronectin. Infect Immun 64:4204–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elliott D, Harrison E, Handley PS, Ford SK, Jaffray E, Mordan N, McNab R. 2003. Prevalence of Csh-like fibrillar surface proteins among mitis group oral streptococci. Oral Microbiol Immunol 18:114–120. [DOI] [PubMed] [Google Scholar]
- 78.Bowen WH. 2016. Dental caries - not just holes in teeth! A perspective. Mol Oral Microbiol 31:228–233. [DOI] [PubMed] [Google Scholar]
- 79.Tanzer JM, Thompson AM, Grant LP, Vickerman MM, Scannapieco FA. 2008. Streptococcus gordonii’s sequenced strain CH1 glucosyltransferase determines persistent but not initial colonization of teeth of rats. Arch Oral Biol 53:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ricker A, Vickerman M, Dongari-Bagtzoglou A. 2014. Streptococcus gordonii glucosyltransferase promotes biofilm interactions with Candida albicans. J Oral Microbiol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoshida Y, Konno H, Nagano K, Abiko Y, Nakamura Y, Tanaka Y, Yoshimura F. 2014. The influence of a glucosyltransferase, encoded by gtfP, on biofilm formation by Streptococcus sanguinis in a dual-species model. APMIS 122:951–960. [DOI] [PubMed] [Google Scholar]
- 82.Frandsen EV, Pedrazzoli V, Kilian M. 1991. Ecology of viridans streptococci in the oral cavity and pharynx. Oral Microbiol Immunol 6:129–133. [DOI] [PubMed] [Google Scholar]
- 83.Sullam PM, Costerton JW, Yamasaki R, Dazin PF, Mills J. 1993. Inhibition of platelet binding and aggregation by streptococcal exopolysaccharide. J Infect Dis 167:1123–1130. [DOI] [PubMed] [Google Scholar]
- 84.Oetjen J, Fives-Taylor P, Froeliger EH. 2002. The divergently transcribed Streptococcus parasanguis virulence-associated fimA operon encoding an Mn(2+)-responsive metal transporter and pepO encoding a zinc metallopeptidase are not coordinately regulated. Infect Immun 70:5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burnette-Curley D, Wells V, Viscount H, Munro CL, Fenno JC, Fives-Taylor P, Macrina FL. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect Immun 63:4669–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bowen WH, Burne RA, Wu H, Koo H. 2017. Oral Biofilms: Pathogens, Matrix, and Polymicrobial Interactions in Microenvironments. Trends Microbiol doi:10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res 8:263–271. [DOI] [PubMed] [Google Scholar]
- 88.Burne RA, Marquis RE. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193:1–6. [DOI] [PubMed] [Google Scholar]
- 89.Van Wuyckhuyse BC, Perinpanayagam HE, Bevacqua D, Raubertas RF, Billings RJ, Bowen WH, Tabak LA. 1995. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res 74:686–690. [DOI] [PubMed] [Google Scholar]
- 90.Peterson S, Woodhead J, Crall J. 1985. Caries resistance in children with chronic renal failure: plaque pH, salivary pH, and salivary composition. Pediatr Res 19:796–799. [DOI] [PubMed] [Google Scholar]
- 91.Epstein SR, Mandel I, Scopp IW. 1980. Salivary composition and calculus formation in patients undergoing hemodialysis. J Periodontol 51:336–338. [DOI] [PubMed] [Google Scholar]
- 92.He J, Hwang G, Liu Y, Gao L, Kilpatrick-Liverman L, Santarpia P, Zhou X, Koo H. 2016. l-Arginine Modifies the Exopolysaccharide Matrix and Thwarts Streptococcus mutans Outgrowth within Mixed-Species Oral Biofilms. J Bacteriol 198:2651–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Margolis HC, Duckworth JH, Moreno EC. 1988. Composition and buffer capacity of pooled starved plaque fluid from caries-free and caries-susceptible individuals. J Dent Res 67:1476–1482. [DOI] [PubMed] [Google Scholar]
- 94.Nascimento MM, Browngardt C, Xiaohui X, Klepac-Ceraj V, Paster BJ, Burne RA. 2014. The effect of arginine on oral biofilm communities. Mol Oral Microbiol 29:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Herrero ER, Slomka V, Bernaerts K, Boon N, Hernandez-Sanabria E, Passoni BB, Quirynen M, Teughels W. 2016. Antimicrobial effects of commensal oral species are regulated by environmental factors. J Dent 47:23–33. [DOI] [PubMed] [Google Scholar]
- 97.Garcia-Mendoza A, Liebana J, Castillo AM, de la Higuera A, Piedrola G. 1993. Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol 39:434–439. [DOI] [PubMed] [Google Scholar]
- 98.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu L, Tong H, Dong X. 2012. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol 78:2120–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol 63:872–880. [DOI] [PubMed] [Google Scholar]
- 102.Boggs JM, South AH, Hughes AL. 2012. Phylogenetic analysis supports horizontal gene transfer of L-amino acid oxidase gene in Streptococcus oligofermentans. Infect Genet Evol 12:1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tong H, Chen W, Shi W, Qi F, Dong X. 2008. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol 190:4716–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zheng LY, Itzek A, Chen ZY, Kreth J. 2011. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci 3:82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang X, Palmer SR, Ahn SJ, Richards VP, Williams ML, Nascimento MM, Burne RA. 2016. A highly arginolytic Streptococcus species that potently antagonizes Streptococcus mutans. Appl Environ Microbiol 82:2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang BY, Kuramitsu HK. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71:354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]